Abstract
Glioblastoma (GBM) is the most malignant human brain tumour, characterized by rapid progression, invasion, intense angiogenesis, high genomic instability, and resistance to therapies. Despite countless experimental researches for new therapeutic strategies and promising clinical trials, the prognosis remains extremely poor, with a mean survival of less than 14 months. GBM aggressive behaviour is due to a subpopulation of tumourigenic stem-like cells, GBM stem cells (GSCs), which hierarchically drive onset, proliferation, and tumour recurrence. The morbidity and mortality of this disease strongly encourage exploring genetic characteristics of GSCs. Here, using array-CGH platform, we investigated genetic and genomic aberration profiles of GBM parent tumour (n = 10) and their primarily derived GSCs. Statistical analysis was performed by using R software and complex heatmap and corrplot packages. Pearson correlation and K-means algorithm were exploited to compare genetic alterations and to group similar genetic profiles in matched pairs of GBM and derived GSCs. We identified, in both GBM and matched GSCs, recurrent copy number alterations, as chromosome 7 polysomy, chromosome 10 monosomy, and chromosome 9p21deletions, which are typical features of primary GBM, essential for gliomagenesis. These observations suggest a condition of strong genomic instability both in GBM as GSCs. Our findings showed the robust similarity between GBM mass and GSCs (Pearson corr.≥0.65) but also highlighted a marked variability among different patients. Indeed, the heatmap reporting Gain/Loss State for 21022 coding/noncoding genes demonstrated high interpatient divergence. Furthermore, K-means algorithm identified an impairment of pathways related to the development and progression of cancer, such as angiogenesis, as well as pathways related to the immune system regulation, such as T cell activation. Our data confirmed the preservation of the genomic landscape from tumour tissue to GSCs, supporting the relevance of this cellular model to test in vitro new target therapies for GBM.
1. Introduction
Glioblastoma (GBM) is the most common malignant primary tumour of the central nervous system (CNS). The standard first-line management for this cancer consists in maximum surgical resection, but its ability to deeply infiltrate makes complete resection quite impossible. GBM is characterized by rapid progression and invasion, cell infiltration, intense angiogenesis, resistance to radio- and chemotherapies, and high frequency of relapse. This aggressive behaviour is responsible for the poor prognosis, with a median survival of about 14 months and a 5-year survival rate of 5.1% [1]. GBM resistance to therapies and the high frequency of recurrence are mainly due to a subpopulation of tumourigenic stem-like cells, known as GBM stem cells (GSCs), able to hierarchically initiate, maintain, and spread the neoplasm. GSCs are self-renewing, pluripotent, highly proliferative, and genetically unstable. Unlike normal stem cells, which acquire a quiescence after DNA damage, GSCs express a plethora of proteins that promote cell survival [2].
Despite the advances in the GBM biology knowledge, comprising behaviour, molecular features, and the heterogeneous genetic landscape, only few targeted therapies have been developed and applied in clinics [3–5]. However, an increasing number of studies that exploit genomic approaches have identified genetic markers typical of glioma and useful for diagnosis. These evidence led the World Health Organization to introduce new diagnostic guidelines based upon molecular diagnosis in 2016 (WHO) [6]. Therefore, nowadays, GBMs are distinguished in IDH-wildtype or IDH-mutated, depending on the presence of recurrent hotspot mutations in isocitrate dehydrogenases IDH1 and IDH2. Notably, large sample studies reported that IDH mutation is found in the majority of secondary GBMs (70-80%) and only rarely in primary GBMs [7]. GBMs are well characterized also from an epigenetic point of view and the DNA-hypermethylated phenotype, called CpG island methylator phenotype (CIMP), has been correlated to a good prognosis [8]. In particular, the most established improvement in predicting GBM patients' outcome is the methylation of the O6-Methylguanine-DNA methyltransferase (MGMT) promoter, which has been associated to a positive response to temozolomide [9, 10]. However, the constant improvement in genetic characterization of GBMs is still failing to be translated to clinical practice, suggesting that other discovery paradigms should be considered. At DNA level, GBMs are usually characterized by high levels of genomic instability. The GBM genome shows many copy number alterations (CNAs) that have been catalogued by computational methodologies [11–13]. These studies collectively have identified frequently amplified genes such as EGFR, MET, PDGFRA, MDM2, PIK3CA, CDK4, and CDK6 and deleted genes including CDKN2A/B, PTEN, and RB1. During the last years, array comparative genomic hybridization (array-CGH) has successfully contributed to improve the detection rate of genomic unbalances and alterations in cancer and to correlate recurrent CNAs to cancer pathomechanisms and prognosis [14, 15]. Considering the importance of CNA data, we focused on the comparison between genetic and genomic aberration profiles of GBM tumour masses and their primarily derived GSCs, investigating if GSC population harbours typical alterations different from the tumour bulk. Since GSCs are widely exploited in in vitro disease studies, uncovering dissimilarities with the corresponding tumour masses could be useful to set more reliable GBM models and to test new potential targeted therapies that could be effective also on the cell populations that are therapy resistant. In order to address these questions, we analysed recurrent hotspot mutations and performed array-CGH to compare CNAs between GBM and their derived GSCs at whole genome level.
2. Materials and Methods
2.1. Study Population
Ten GSCs were derived from matched primary (n = 9) and recurrent (n = 1) GBMs provided by the Neurosurgery Unit of the Fondazione IRCCs Ca' Granda Ospedale Maggiore Policlinico (Milan), according to the ethical requirements of the institutional committee on human experimentation.
Patients recruited between January 2014 and May 2016 were randomly included in the study, after informed consent was signed. The histological analyses were based on the 2016 WHO criteria for CNS tumours [3]. Demographic and clinical data are reported in Table 1.
Table 1.
Clinicopathological characteristics of the patients enrolled in the study.
| Patient ID | Sex | Age (yrs) | Dx | OS | KPS | Ki67 |
|---|---|---|---|---|---|---|
| Pt 2 | M | 60 | GBM | 4 | 60% | 15% |
| Pt 3 | F | 50 | GBM(R) | 16 | 70% | 40% |
| Pt 9 | F | 82 | GBM | 5 | 60% | 15% |
| Pt 10 | F | 63 | GBM | 5 | 90% | 35% |
| Pt 15 | M | 36 | GBM | 54 | 80% | 60% |
| Pt 33 | F | 65 | GBM | 11 | 90% | 70% |
| Pt 56 | M | 75 | GBM | 2 | 60% | 40% |
| Pt 60 | M | 49 | GBM | 16 | 80% | 70% |
| Pt 85 | M | 59 | GBM | 12 | 80% | 30% |
| Pt 90 | F | 58 | GBM | 17 | 90% | 30% |
Pt: patient; M: male; F: female; yrs: years; Dx: diagnosis; R: relapse; OS: overall survival expressed in months; KPS: Karnofsky Performance Score.
2.2. GBM Sample Collection
GBM sample processing was performed as previously reported [16]. Briefly, after each surgical intervention, the tumour specimen was collected and maintained in Dulbecco's modified Eagle's medium: Nutrient Mixture F-12 (DMEM/F12, Thermo Fisher, Waltham, Massachusetts) containing 1% penicillin/streptomycin (Thermo Fisher) at 4°C, for up to 24 h. An aliquot of tissue was fixed in a 4% paraformaldehyde (PFA) solution (Sigma-Aldrich, Basel, Switzerland) in Dulbecco's phosphate-buffered saline (D-PBS) and subsequently embedded in paraffin (Sigma-Aldrich). Another aliquot was firstly mechanically minced with a surgical scalpel and then enzymatically digested with 0.625Wu/mL Liberase Blendzyme 2 (Roche, Mannheim, Germany) for 1 h at 37°C [17]. A cell suspension was obtained after passing dissociated through a 0.70 μm pore size filter (Thermo Fisher).
2.3. Primary GSC Isolation
GSCs were isolated as previously described [17]. Briefly, GBM cells were cultured in the appropriate medium for isolation of GSCs, consisting of DMEM/F12 supplemented with 20 ng/mL epidermal growth factor (EGF) (R&D Systems, Minneapolis, USA), 10 ng/mL fibroblast growth factor-2 (bFGF) (PeproTech Inc., Rocky Hill, NY, USA), and 1% penicillin/streptomycin. Cells were cultured in a humidified incubator with 5% CO2, 5% O2 at 37°C, and split once a week. At each passage (P), GSC neurospheres were dissociated with Tryple Select (Gibco, Grand Island, NY, USA) and Trypan Blue dye exclusion assay (Gibco) was used to assess cell viability. Cells were routinely observed with an inverted phase-contrast microscope (Nikon Eclipse TE300; Nikon, Shinjuku, Tokyo, Japan), and images were acquired with a digital camera (Zeiss Axiovision, Zeiss, Oberkochen, Germany).
2.4. Immunophenotypic Analyses
Flow cytometric analyses were performed to assess the immunophenotypic profile of GSCs. The assay was conducted as previously described [17]. Briefly, each GSC line (5 × 104 cell/tube) was incubated with phycoerythrin- (PE-) or fluorescein isothiocyanate- (FITC-) conjugated antibody to rate the expression of the following stemness/progenitor cell markers: CD15, CD31, CD45, CD133 (Miltenyi Biotec, Bisley, Surrey, UK), and CD90 (Millipore Temecula, CA, USA). Notably, 7-aminoactinomycin D (7-AAD, BD) was added to each tube to exclude dead cells from the analysis. After an incubation of 30 min at room temperature, washing and fixation with 4% PFA, immunomarked GSCs were scanned on a fluorescence-activated cell sorting (FACS) flow cytometer and analysed by CellQuest software (BD Biosciences, San Jose, CA). FlowJo software (Tree Star Inc., Ashland) was used to perform postprocessing analyses.
2.5. DNA Isolation
DNA from the FFPE tumour samples was obtained using a Biostic FFPE tissue DNA isolation kit (MO BIO Laboratories, Carlsbad, CA, USA), following manufacturer's instructions. DNA extraction was performed from FFPE tissue sections with at least 70% of tumour content, assessed by hematoxylin/eosin staining. DNA was extracted from GSCs, at a passage ranging from P4 and P10, using the QIAamp DNA Micro Kit (Qiagen, Hilden, Germany), according to manufacturer's protocol.
2.6. IDH1, IDH2, and TERT Mutation Analysis
The assessment of hotspot mutations in IDH1, IDH2, and TERT promoter was performed as previously described [18]. In brief, DNA was amplified by multiplexed primer mix targeting codon 132 and 172 of IDH1 and IDH2, respectively, and position c.-124 and c.-146 of TERT promoter. PCR, SAP, and IPLEX reactions were conducted as described in manufacturer's protocol (Agena Bioscience, San Diego, CA, USA).
Samples were transferred to a SpectroCHIP (Agena Bioscience, San Diego, CA, USA) and analysed by mass spectrometry. The spectral profiles generated by MALDI-TOF mass spectrometry were evaluated using Typer v.4.0 software (Agena Bioscience, San Diego, CA, USA).
2.7. MGMT Promoter Methylation Evaluation
MGMT methylation was assessed both in FFPE tumour tissue and GSCs. DNA was modified with sodium bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research Corp., Irvine, CA). The methylation analysis was implemented as previously described [10, 19]. PCR was performed on at least 20 ng of bisulfite-treated DNA and about 10 pmol primers. Quantitative DNA methylation analysis was carried out on the Pyro Mark ID instrument using Pyro Gold Reagents (Qiagen) and 1 pmol of sequencing primer. Methylation data were analysed by the Q-CpG software v1.9 (Qiagen) and the levels of methylation of each sample are represented by the mean of the methylation percentages at each CpG site of the investigated region.
2.8. Array-CGH
Array-CGH analysis was performed as previously described [19], using 60-mer oligonucleotide probe technology (SurePrint G3 Human CGH 8 × 60 K, Agilent Technologies, Santa Clara, CA, USA), according to manufacturer's instructions. Agilent Feature Extraction was exploited to generate raw data, which were further analysed using Cytogenomics 2.7 with the ADAM-2 algorithm (Agilent Technologies, Santa Clara, CA, USA). In order to increase the accuracy of the results, the Diploid Peak Centralization algorithm was applied. A minimum of three consecutive probes/region was considered as filter. The threshold for genomic deletion is x = −1; the threshold for genomic gain is x = +0.58. Notably, in a mosaic scenario, the threshold is between -1 and 0 for deletions and between 0 and +0.58 for duplications [20]. Amplifications and homozygous deletions are considered with threshold >+2 and <-1, respectively. Variants reported as population variants in the public databases were not listed. Genomic coordinates are according to the build 37 assembly (March 2009) of the Human Genome Reference consortium (GRch37/hg19).
2.9. Statistical Analysis
Statistical analysis has been performed by using R software and complex heatmap [21] and corrplot packages (https://github.com/taiyun/corrplot) were exploited to figure out the data. Pearson correlation was used to compare genetic alteration profiles in matched pairs of GBM and derived GSCs. In order to group similar genetic profiles among samples, K-means algorithm was performed. Moreover, we used the chi-square test to compare MGMT methylation levels of GBM with the matched GSC samples. GO-Term enrichment analysis was developed using WebGestalt (http://www.webgestalt.org/option.php).
3. Results
3.1. Clinical Features of Patients Affected by GBM
Our GBM cohort has an even proportion of male and female patients with a mean age at diagnosis of 59 years (ranging from 36 to 82 years old). All tumours had been examined and classified according to the WHO guidelines and all GBMs were IDH-wildtype; GBM of patient (Pt) 3 is the first relapse. The observed overall survival (OS), in terms of time between the date of surgery and the date of death, varies between 2 and 54 months, with a mean value of 14.2 and a median value of 11.5 months. The Karnofsky Performance Score (KPS) was performed during hospitalization, before surgery, and assessed between 60% and 90%. As a cellular marker for proliferation, the Ki-67 protein was evaluated by immunohistochemistry, in a range between 15% and 70%.
3.2. GSCs Differ in Stemness Marker Expression
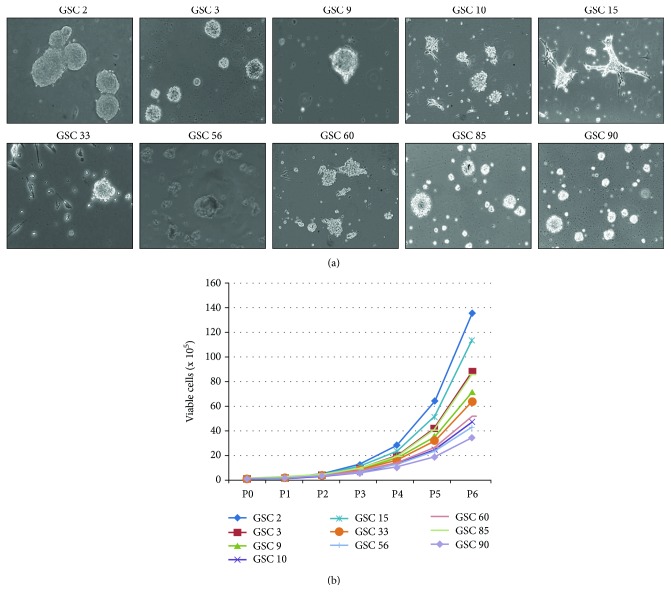
About 30% of GSC lines experienced both an adherent and a floating phase (GSC 15, GSC 33, and GSC 56), still maintaining stemness properties (Figure 1(a)). Notably, estimating the number of viable cells from P0 to P6 (30 days), we observed different growth rates, thus each GSC line shows a specific proliferation curve, anyhow growing with an exponential rate (Figure 1(b)). Analysing GSCs, at a passage ranging from P4 to P10, by flow cytometry, we determined the expression of CD133, CD90, CD15, CD31, and CD45 as stem/progenitor cell markers, reporting different expression patterns among patients. As shown in Table 2, all the investigated stemness markers were expressed in our GSCs, with CD90 as the highest one. Notably, performing flow cytometry at higher passages, we observed that marker expression remains rather stable in 90% of GSCs, with a variability of about 5-10%.
Figure 1.
GSC isolation and propagation. (a) Representative images of GSCs of each patient, captured at passage 5. Magnification 10x with an inverted phase-contrast microscope. (b) Proliferation curves of GSCs. Each passage was done every 5 days.
Table 2.
Stemness marker analyses in GSCs revealed different expression patterns among GBM patients.
| Sample | Passage | CD15 | CD31 | CD34 | CD45 | CD133 | CD90 |
|---|---|---|---|---|---|---|---|
| GSC 2 | 10 | + | +++ | + | + | +++ | +++ |
| GSC 3 | 4 | + | +++ | + | + | +++ | + |
| GSC 9 | 4 | + | +++ | ++ | + | + | + |
| GSC 10 | 4 | + | + | + | ++ | + | +++ |
| GSC 15 | 4 | + | ++ | ++ | + | ++ | +++ |
| GSC 33 | 4 | ++ | + | ++ | ++ | +++ | +++ |
| GSC 56 | 4 | + | + | + | + | ++ | +++ |
| GSC 60 | 5 | + | + | ++ | + | + | +++ |
| GSC 85 | 4 | + | ++ | ++ | + | ++ | ++ |
| GSC 90 | 6 | + | + | + | +++ | +++ | +++ |
+: 0-33%; ++: 34-66%; +++: 67-100%.
3.3. Recurrent Hotspot Alterations
We investigated the most frequently reported genetic alterations for GBM IDH-wildtype in all GBM-GSC-matched couples at a passage ranging from P4 to P10, as in correspondence of the flow cytometric analysis. The presence of the hotspot mutations c.-124C>T and c.-146C>T in the promoter region of TERT was evaluated. As shown in Table 3, 8 out of 10 tumours were mutated in TERT, 6 harboured the c.-124C>T mutation (Pt 2, Pt 10, Pt 15, Pt 56, Pt 85, and Pt 90), while 2 had the c.-146C>T mutation (Pt 9 and Pt 60). For all samples, the same alteration found in the GBM was present also in the GSCs, confirming the driver function of these mutations. Moreover, the high prevalence of mutated cases in our small cohort of patients is concordant with the one reported by previous large studies on GBM IDH-wildtype [22, 23], further corroborating the relevance of such marker in this type of glioma.
Table 3.
Genetic profile of the main markers of IDH-wildtype GBM in our cohort of GBMs and GSCs.
| Gene/ID | Pt 2 | Pt 3 | Pt 9 | Pt 10 | Pt 15 | Pt 33 | Pt 56 | Pt 60 | Pt 85 | Pt 90 | % tumours altered∗ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TERT promoter mutation | GBM | c.-124C>T | WT | c.-146C>T | c.-124C>T | c.-124C>T | WT | c.-124C>T | c.-146C>T | c.-124C>T | c.-124C>T | 72-90% |
| GSC | c.-124C>T | WT | c.-146C>T | c.-124C>T | c.-124C>T | WT | c.-124C>T | c.-146C>T | c.-124C>T | c.-124C>T | ||
|
| ||||||||||||
| MGMT methylation | GBM | 5% | 4% | 4% | 12% | 31% | 15% | 3% | 2% | 2% | 58% | 40-50% |
| GSC | 4% | 2% | 3% | 9% | 14% | 20% | 3% | 2% | 1% | 86% | ||
|
| ||||||||||||
| MGMT CN | GBM | NA | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | / |
| GSC | NA | NA | LOSS | LOSS | NA | LOSS | LOSS | LOSS | LOSS | LOSS | ||
|
| ||||||||||||
| EGFR CN | GBM | GAIN (4) | AMP | GAIN | GAIN (4) | AMP | AMP | AMP | AMP | GAIN | GAIN | 35-45% amplified |
| GSC | GAIN | GAIN | GAIN | GAIN | AMP | GAIN | GAIN | AMP | GAIN | GAIN | ||
|
| ||||||||||||
| PTEN/10q CN | GBM | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | LOSS | 75-95% deleted |
| GSC | LOSS | NA | LOSS | LOSS | NA | LOSS | LOSS | LOSS | LOSS | LOSS | ||
|
| ||||||||||||
| CDKN2A CN | GBM | LOSS | DEL | DEL | DEL | DEL | DEL | NA | LOSS | DEL | DEL | 35-50% deleted |
| GSC | DEL | GAIN | DEL | DEL | LOSS | DEL | DEL | DEL | DEL | DEL | ||
|
| ||||||||||||
| NFKB1 CN | GBM | NA | NA | LOSS | NA | NA | NA | NA | NA | NA | NA | 25% deleted |
| GSC | NA | NA | LOSS | NA | NA | NA | NA | NA | NA | GAIN | ||
|
| ||||||||||||
| PDGFRA CN | GBM | NA | NA | AMP | NA | NA | NA | NA | NA | NA | NA | 13% amplified |
| GSC | NA | NA | AMP | NA | NA | NA | NA | NA | NA | GAIN | ||
|
| ||||||||||||
| MDM4 CN | GBM | NA | NA | NA | NA | NA | AMP | NA | NA | NA | NA | 7% amplified |
| GSC | NA | GAIN | NA | NA | NA | AMP | NA | NA | NA | NA | ||
|
| ||||||||||||
| MET CN | GBM | GAIN | GAIN | GAIN | GAIN | NA | GAIN | GAIN | GAIN | GAIN | GAIN | 4% amplified |
| GSC | GAIN | GAIN | GAIN | GAIN | NA | GAIN | GAIN | GAIN | GAIN | GAIN | ||
∗Percentage of mutated tumours as defined by the reported references. CN: copy number; NA: not altered; DEL: log2 values <-1; LOSS: log2 values <0 and >-1; GAIN: log2 values >0 and <2; AMP: log2 values >2.
We analysed the MGMT promoter methylation, which is the principal prognostic factor in primary GBM [24]. One out of 10 GBMs (Pt 90) was substantially hypermethylated at this promoter region, while Pt 10, Pt 15, and Pt 33 were only partially methylated; the remaining GBMs were nonmethylated (Table 3). Analogously to TERT mutations, the methylation levels of MGMT promoter were fairly concordant between GBM and GSCs of each patient. Particularly, nonmethylated GBMs showed nonmethylated GSCs. Regarding the methylated cases, Pt 10 and Pt 33 showed almost equal methylation percentages in the two components, considering the 5% of sensitivity limitation of the pyrosequencing technique [25], while Pt 15 and Pt 90 displayed more different values (p value = 1.12% for Pt 15 and 1.96% for Pt 90). In particular, Pt 90 showed an increment in methylation in GSCs (86% vs. 58% in the tumour, Table 3), which could be explained considering that GBM is more heterogeneous and nonneoplastic cells could partially alter the methylation level. Conversely, GSC 15 methylation is lower than GBM 15 (14% vs. 31% in the tumour, Table 3). In order to deepen the differences in methylation levels in Pt 15 and Pt 90, we verified the presence of deletions encompassing the MGMT locus (on chromosome 10q26.3) (Table 3) and both GBM 90 and GSC 90 exhibited a monosomy of chromosome 10 with a mosaic distribution. It means that not all cells harbour a deletion of one MGMT allele and that the heterogeneity is more evident in tumour bulk. However, Pt 15 displays deletions of MGMT with a mosaic pattern only in the GBM component, while the GSCs have no genomic aberration at this locus, which could partially explain the lower methylation levels in GSCs (Table 3).
3.4. Array-CGH Profiles
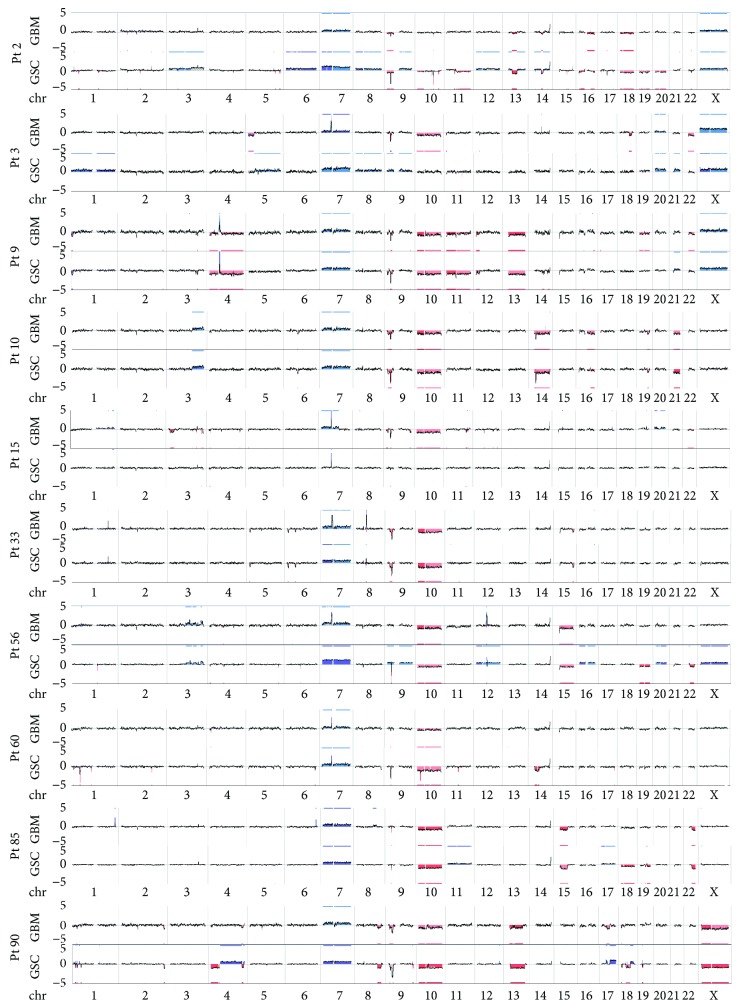
Array-CGH experiments confirmed the robust similarity between GBM mass and GSCs of each case (Figure 2) but also highlighted a marked variability among different cases. Common alterations, present in almost all tumours, were polysomy of chromosome 7, monosomy of chromosome 10, and homozygous 9p21 deletion.
Figure 2.
Genome view of GBM and GSCs of each patient displaying high intrapatient similarity and interpatient discordance. Gains or losses of chromosomal regions are computed by comparing signal intensity in GBM and GSCs with the background signal of the reference and then applying a segmenting and smoothing algorithm. Line plots show chromosomal changes seen in GBMs; the y-axis indicates the log2 value ranging from -5 to 5; blue indicates gain/amplifications, red losses/deletions.
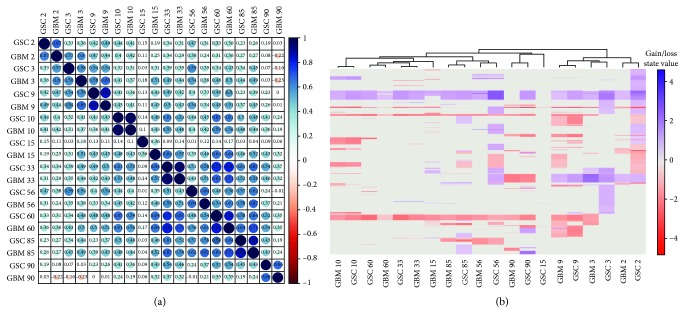
To measure the genomic relationship between GBM and derived GSCs, we performed Pearson correlation metric (Figure 3(a)). The mean Pearson correlation is 0.43, range -0.23/+1. By setting the correlation threshold to 0.65 (calculated from our data considering the mean correlation added to the standard deviation), we found that all patients, except for Pt 3, Pt 15, and Pt 56, showed a good correlation (Pearson correlation ≥ 0.65), denoting the concordance between GBM and matched GSC.
Figure 3.
Hierarchical clustering results in our cohort of 10 patients highlighted the intrapatient similarity. (a) Correlation map reporting Pearson correlation values for each comparison. The bar on the left of the map indicates the color legend of the Pearson corr. values calculated for each couple of samples in the matrix. (b) Heatmap reporting Gain/Loss State for 21022 coding/noncoding genes (y-axis) in all samples (x-axis). Red and blue colors represent, respectively, losses and gains.
Notably, Pt 3 and Pt 56 had, respectively, Pearson corr. values of 0.58 and 0.64, thus below the threshold, but fairly close to it, whereas Pt 15 Pearson correlation was 0.36, corroborating the mentioned increase in dissimilarity between GBM and GSCs for this patient, which is already appreciable from the genome view reported in Figures 2 and 3(a).
Moreover, we obtained the p values of our correlation metric using cor.mtest function in R that performs the significance test for each pair of correlations; p values < 0.05 indicate a significant correlation between samples. Notably, all p values resulted <0.05, showing a significant correlation between GBM and GSC of each patient, also for Pt 15.
Noteworthy, Pt 15 has the lowest number of genomic aberrations among our cohort. This could be related to his long-term survival (OS 54 months), significantly higher than the mean GBM OS (14.2 months) and to his young age (36 yrs, with respect to the GBM patient median age of 59 yrs). Unfortunately, the fact that Pt 15 is the only patient with these characteristics, due to the great rarity of such high OS cases, prevented further considerations about his genomic pattern.
To further assess the intrapatient and interpatient correlation, we investigated the distribution of CNAs (gains and losses) in GBM and GSCs along genes (coding and noncoding). As reported in Figure 3(b), except Pt 15, all GBM-GSC-matched pairs are clustered together, highlighting the strong correspondence of GBM and GSCs in each patient. This analysis also quite demonstrated the divergence between GBM and GSCs of Pt 15, further corroborating the abovementioned considerations.
Taken together, the data show an increment of alterations in GSCs than in GBM, i.e., GSC 90 at chromosomes 4, 17, and 18 compared to the matched GBM 90 (Figure 1).
Therefore, to deepen the comparison of GBM and GSCs, we decided to focus on copy number patterns of well-characterized genetic markers in primary GBMs that could be also exploited for the development of new treatments: EGFR, PDGFRA, MDM4, and MET amplification and CDKN2A, PTEN, and NFKB1 deletions (Table 3). We highlighted that our cohort of patients almost completely recapitulates the information reported in literature, ensuring that not selection biases were made.
All of our samples, except again Pt 15, displayed a complete polysomy of chromosome 7, encompassing both EGFR and MET genes. The chromosome 7 polysomy is considered a fundamental event driving GBM tumourigenesis. Indeed, it was present in almost all GBMs and derived GSCs together with the monosomy of chromosome 10, encompassing the PTEN locus and the focal deletion of chromosome 9p21 surrounding CDKN2A [26–29]. Regarding EGFR, it was further substantially amplified in 5 out of 10 samples (Pt 3, Pt 15, Pt 33, Pt 56, and Pt 60), comprehending also Pt 15 that had only the focal amplification of EGFR locus in both GBM and GSCs. It is noteworthy that 3 out of 5 EGFR-amplified GBM lost this marker in the derived GSCs (Pt 3, Pt 33, and Pt 56). Moreover, none of the samples had MET amplification, further supporting the rarity of this condition [4]. As for the polysomy of chromosome 7, almost all GBMs and GSCs harboured monosomy of chromosome 10, thus at least one allele of PTEN is deleted in all GBMs and matched GSCs, confirming the driver function of this alteration in gliomagenesis [26–29].
Another marker present in almost all samples was the deletion of chromosome 9p21 involving CDKN2A. It was present at homozygous status in the majority of patients, with some discordance between GBM and GSC, probably due to the heterogeneity of the samples, such as for GBM 56. Remarkably, Pt 3, the relapse sample, exhibited two opposite patterns of alteration at this locus in his GBM and GSCs, del and gain, respectively, (Table 3).
Finally, the other markers were present each in only one sample, confirming the low frequency of these alterations, particularly, Pt 9 (GBM and GSCs) lost NFKB1 in a mosaic pattern and displayed the amplification of PDGFRA. Pt 33 (GBM and GSCs) instead was the only sample with MDM4 amplification.
3.5. Clustering of Similar Genetic CNA Profiles in GBM and GSC Samples
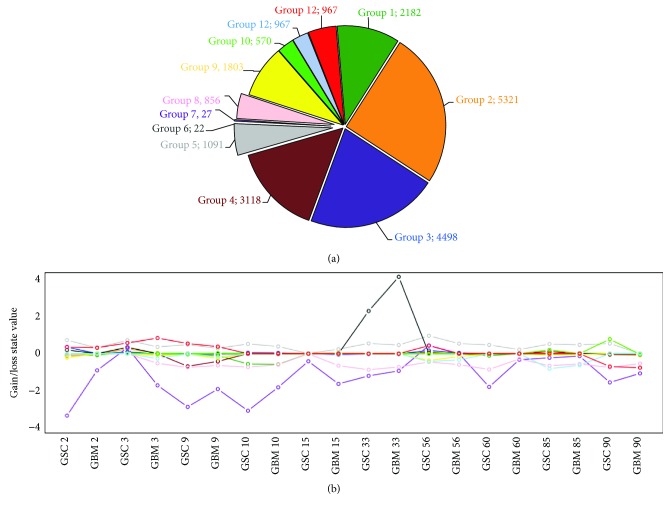
In order to further evaluate the genetic patterns, we used K-means algorithm to find genes with similar profiles among samples. We chose k = 12 because it maximizes the variance interclusters and minimizes the variance intracluster (Supplementary Material (available here)). Results are shown in Figure 4, reporting the average genetic alteration profile line for each cluster. All clusters seemed to profile randomly among samples, except clusters 5, 7, and 8. Cluster 5 (grey area and grey line in Figures 4(a) and 4(b), respectively) contains 1091 genes (including EGFR), which were amplified on average in all samples, except GBM 90. Clusters 7 (purple area and line in Figures 4(a) and 4(b)) and 8 (pink area and line in Figures 4(a) and 4(b)) contain, respectively, 27 and 856 genes (encompassing CDKN2A and PTEN, respectively), which were deleted in about 90% of samples.
Figure 4.
K-means results to cluster similar genetic CNA profiles in our GBM and GSC samples in 12 groups. (a) Pie chart reporting the number of genes gathered in each cluster according to K-means algorithm. (b) Centroid profiles for each cluster. Each line depicts the intracluster CNA mean values among samples (x-axis). Grey line represents the cluster 5 profile, containing genes amplified in all patients except GBM 90. Purple and pink lines are referred to clusters 7 and 8, containing genes deleted in all samples except for GSC 3 and GSC 56.
We performed GO-Term enrichment analysis using WebGestalt tool to investigate the pathways of genes involved in these aberrations and found that there is an impairment of pathways related to cancer development and progression (Supplementary Material). In particular, we identified clear distinctive pathways included in the three clusters analysed. In cluster 5, we identified genes involved in the new vessel formation and in nitrogen and oxygen response. In cluster 7, the main identified pathways are related to the immune response as “regulation of cytokine-mediated signaling pathway,” whereas in cluster 8, we identified genes associated at two distinctive pathways, metabolic process pathway and interestingly brain development.
4. Discussion
GBM is highly heterogeneous and resistant to conventional chemo- and radiotherapies and several recent studies suggested that it is driven and maintained by GSCs. The development of more personalized therapies is still urgent and in vitro models able to reliably recapitulate the original tumour are yet under discussion. Here, we focused on the characterization of the genomic patterns of GBM masses and respective isolated GSCs in 10 primary GBM patients to additionally establish the reliability of GSCs as in vitro model and to search for genetic features typical of GSCs and able to explain the resistance and aggressiveness of these cells.
Remarkably, the characterization of primary GSCs in the analysed cases revealed patient-specific profiles, both in terms of growth rate and surface stemness marker expression. Particularly, we observed a high interpatient variability and a peculiar immunophenotypic profile in each GSC population, in accordance with the well-known heterogeneity of GBMs [30]. Interestingly, our cytofluorimetric analysis revealed that all GSCs were all positive, but at different levels, for the markers of stemness and endothelial progenitors: CD15, CD133, CD90, CD31, and CD45.
Although some authors have demonstrated that in vitro growth of GSCs and their stemness marker expression represents a significant predictor of clinical outcome [31–33], the real association between the biological features of GSCs and GBM profile has yet to be established.
From a genomic perspective, the typical genetic features of primary GBMs were markedly concordant between GBM/GSC couples, confirming the high similarity between the tumour mass and its respective GSC population. Regarding TERT mutations, 8 out of 10 GBMs and matched GSC cases were mutated in the promoter region. Of note, TERT mutations have a prognostic role in gliomas, being a negative prognostic factor in primary GBM without MGMT methylation [34]. In our cohort, two cases were methylated at the MGMT promoter (Pt 15 and Pt 90), but unfortunately, the number of samples included prevented the possibility to perform statistically significant analyses about the link between TERT promoter mutations, MGMT methylation, and patients' survival. Nevertheless, this was beyond the purposes of this study. The almost similar levels of MGMT promoter methylation between GBM and GSCs suggested that also this epigenetic marker is conserved in the GSC population, which is usually responsible for the high frequency of recurrence in this type of tumour. This evidence is in accordance with the fact that generally the level of methylation between primary GBMs and respective relapses is fairly comparable [35]. Moreover, the increased methylation levels of GSC 90 could be explained by the fact that it is a more homogeneous sample than the whole GBM mass [3]; in addition, a sample heterogeneity cannot be excluded, as already suggested [3, 36]. Conversely, the decreased methylation level in GSC 15 compared to GBM 15 could be due to the absence of deletions at this locus. Nevertheless, the methylation levels in both GSC 15 and GSC 90, along with the presence of mosaic deletions of MGMT in GSC 90, suggest that also the GSC population is heterogeneous and not all cells are MGMT methylated.
Almost all GBM and GSC couples had polysomy of chromosome 7, monosomy of chromosome 10 (PTEN), and deletions at chromosome 9p21 (CDKN2A) that are typical features of primary GBM, essential for gliomagenesis and usually conserved in the whole GBM mass [13, 26, 28, 29, 37]. The only two patients showing different patterns of these markers between GBM and GSCs were Pt 3 and Pt 15. Pt 3 showed opposite molecular features at the 9p21 region between GBM (9p21 loss) and GSCs (9p21 gain). This GBM is the first tumour recurrence, meaning that the patient had been already treated and this could have affected the genomic configuration of the GBM and/or GSCs, although the Pearson corr. is quite close to the threshold of 0.65. Another possible mechanism to explain the peculiar molecular profile of Pt 3 could be the “parallel evolution” model proposed by Baysan et al. [26]. By this theory, different clones could evolve separately from a common ancestor, giving rise independently to fairly equal genomic patterns between primary and recurrent tumours.
Pt 15 is the only young long-term survival patient included in the study and he has substantially less genomic aberration both in GBM and in GSCs. In details, Pt 15 showed the amplification of EGFR without the polysomy of the chromosome 7 in both GBM and GSCs, the monosomy of chromosome 10 only in GSCs, and the deletion of CDKN2A in a mosaic fashion in GSCs. The asset of these GBM peculiar markers in long-term GBM survivors is still discussed; therefore, it is quite complex to define their association with overall survival [1, 38–40].
Regarding EGFR amplification, 5 out of 10 analysed cases exhibit this alteration in GBM, but only 2 of them retained it in GSC population. This could be due to the negative selective pressure induced by EGF addition to the medium, as described by [41, 42]. However, this phenomenon is still controversial; indeed, not all GSCs derived from EGFR-amplified GBM lack the amplification of this marker, as reported in several studies [43–45].
The polysomy of chromosome 7 involves also the MET locus, which is never amplified in our cohort, neither in the cases without EGFR amplification. Indeed, MET is usually overexpressed in mesenchymal/proneural subgroup of GBMs EGFR-wildtype and PTEN-lost [46], thus configuring as an alternate way to deregulate the tyrosine kinase signaling pathway. Other alternative mechanisms to impair the signaling pathway mediated by tyrosine kinases are the PDGFRA amplification and the NFKB1A deletion [4, 47].
Pt 9 displayed a normal pattern of EGFR and harbours both PDGFRA amplification and NFKB1A deletion in GBM and GSCs. Finally, only GBM and GSCs of Pt 33 harboured MDM4 amplification that is an alternative and quite infrequent route to inactivate p53 [4].
Therefore, almost all samples show concordance between GBM and GSCs, as demonstrated by the Pearson correlation analysis (Pearson corr.≥0.65), corroborating the fact that GSCs resemble their parental tumour at a genomic level [45, 48]. Besides this, the heatmap highlights the interpatient variability between GBM and GSC, further supporting the high heterogeneous behaviour of this type of tumour.
The slight intrapatient genomic differences between GBM and GSCs, and particularly the partial enrichment of alterations in GSCs, could be related to the heterogeneity of GBM and that some low frequent alterations could be masked by the presence of nonneoplastic cells in the analysed specimens. Nevertheless, the presence of these dissimilarities could also be due to the in vitro growing conditions of GSCs. Indeed, even if different GSCs did not acquire in vitro identical genomic alterations under the same selective pressure, it is anyway possible that in vitro culture could select small subpopulations of cells in the parental GBM mass harbouring alterations not detectable in the tumour bulk. Moreover, it was recently demonstrated that discordant inheritance of chromosomal and extrachromosomal DNA elements between primary tumour mass, neurosphere cultures, and xenografts exists and is strictly related to the dynamic GBM evolution [44]. Therefore, on the one side, elevated concordance between GSCs and the primary tumour masses corroborates the use of these as reliable in vitro and in vivo models of GBM; on the other side, the dissimilarities could be useful to understand clonal evolution of the tumour and to test new targeted treatments. Indeed, the K-means algorithm identified three clusters of genes equally altered among the GBM and GSC samples. These three clusters involved the already described genes EGFR, CDKN2A, and PTEN. However, besides these loci, GO-term enrichment analysis pointed out defects in other genes, with similar CNA profiles among GBM/GSC couples, encompassing pathways essential for the development and progression of cancer, such as pathway related to angiogenesis, as well as pathways related to the immune system regulation. This last point is particularly interesting, considering the promising advances in immunotherapies.
Despite the limitation of our work, related to the small number of patients analysed, our findings contribute to add a little piece to the complicated puzzle of GBM. The detailed analyses of genomic aberrations in GSCs further testify that they represent a good model since they maintain the main genetic markers of GBM. Indeed, as GSCs are patient-specific and radio- and chemoresistant, they represent a challenging target for therapies. In the era of precision medicine, indeed, a better understanding of the GSC molecular landscape is pivotal to design effective target therapies for GBM.
Acknowledgments
We would like to show our gratitude to the Fondazione Floriani which, with great competence and humanity, has been a professional guide and a valid support for our patients in all the complex phases from diagnosis to therapy. We are also immensely grateful to A-Tono: The World in Your Hand and to Dr. Giuseppe Caspani for the precious help in communication strategy with dissemination of our research activity. The research was funded by the Italian Ministry of Health Regione Lombardia Ricerca Finalizzata 2011-2012, RF-2011-02347106 (PI Monica Miozzo), and by the Italian Ministry of Health, RC2015/2016 (PI Giovanni Marfia). The study was also carried out by the support of Fondazione Floriani and A-Tono: The World in Your Hand.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Chiara Pesenti and Stefania Elena Navone contributed equally to this work.
Supplementary Materials
Supplementary Material: an Excel File in which there are reported lists of genes included in n = 12 clusters, grouped by K-means algorithm, with the relative location, values, and GO-Term enrichment analysis performed by WebGestalt. Moreover, all the output files achieved by WebGestalt are included.
References
- 1.Wipfler K., Cornish A. S., Guda C. Comparative molecular characterization of typical and exceptional responders in glioblastoma. Oncotarget. 2018;9(47):28421–28433. doi: 10.18632/oncotarget.25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou-Antoun T. J., Hale J. S., Lathia J. D., Dombrowski S. M. Brain cancer stem cells in adults and children: cell biology and therapeutic implications. Neurotherapeutics. 2017;14(2):372–384. doi: 10.1007/s13311-017-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sottoriva A., Spiteri I., Piccirillo S. G. M., et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wick W., Dettmer S., Berberich A., et al. N2M2 (NOA-20) phase I/II trial of molecularly matched targeted therapies plus radiotherapy in patients with newly diagnosed non-MGMT hypermethylated glioblastoma. Neuro-Oncology. 2018;21(1):95–105. doi: 10.1093/neuonc/noy161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis D. N., Perry A., Reifenberger G., et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Yan H., Parsons D. W., Jin G., et al. IDH1 and IDH2 mutations in gliomas. The New England Journal of Medicine. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes J. A., Paulsson A. K., Page B. R., et al. Genomic predictors of patterns of progression in glioblastoma and possible influences on radiation field design. Journal of Neuro-Oncology. 2015;124(3):447–453. doi: 10.1007/s11060-015-1858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M., Garcia-Foncillas J., Andion E., et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. The New England Journal of Medicine. 2000;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 10.Fontana L., Tabano S., Bonaparte E., et al. MGMT-methylated alleles are distributed heterogeneously within glioma samples irrespective of IDH status and chromosome 10q deletion. Journal of Neuropathology & Experimental Neurology. 2016;75(8):791–800. doi: 10.1093/jnen/nlw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mermel C. H., Schumacher S. E., Hill B., Meyerson M. L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology. 2011;12(4, article R41) doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beroukhim R., Getz G., Nghiemphu L., et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedemeyer R., Brennan C., Heffernan T. P., et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13(4):355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rausch V., Krieg A., Camps J., et al. Array comparative genomic hybridization of 18 pancreatic ductal adenocarcinomas and their autologous metastases. BMC Research Notes. 2017;10:p. 560. doi: 10.1186/s13104-017-2886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patmore H. S., Ashman J. N. E., Stafford N. D., et al. Genetic analysis of head and neck squamous cell carcinoma using comparative genomic hybridisation identifies specific aberrations associated with laryngeal origin. Cancer Letters. 2007;258(1):55–62. doi: 10.1016/j.canlet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Guarnaccia L., Navone S. E., Trombetta E., et al. Angiogenesis in human brain tumors: screening of drug response through a patient-specific cell platform for personalized therapy. Scientific Reports. 2018;8, article 8748 doi: 10.1038/s41598-018-27116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marfia G., Campanella R., Navone S. E., et al. Autocrine/paracrine sphingosine-1-phosphate fuels proliferative and stemness qualities of glioblastoma stem cells. Glia. 2014;62(12):1968–1981. doi: 10.1002/glia.22718. [DOI] [PubMed] [Google Scholar]
- 18.Pesenti C., Paganini L., Fontana L., et al. Mass spectrometry-based assay for the molecular diagnosis of glioma: concomitant detection of chromosome 1p/19q codeletion, and IDH1, IDH2, and TERT mutation status. Oncotarget. 2017;8(34):57134–57148. doi: 10.18632/oncotarget.19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronchetti D., Todoerti K., Tuana G., et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer Journal. 2012;2(11):e96–e98. doi: 10.1038/bcj.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valli R., Marletta C., Pressato B., et al. Comparative genomic hybridization on microarray (a-CGH) in constitutional and acquired mosaicism may detect as low as 8% abnormal cells. Molecular Cytogenetics. 2011;4(1):p. 13. doi: 10.1186/1755-8166-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 22.Killela P. J., Reitman Z. J., Jiao Y., et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killela P. J., Pirozzi C. J., Healy P., et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–1525. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masui K., Mischel P. S., Reifenberger G. Molecular classification of gliomas. Handbook of Clinical Neurology, Vol. 134. 1st. 2016. pp. 97–120. [DOI] [PubMed]
- 25.Tsiatis A. C., Norris-Kirby A., Rich R. G., et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. The Journal of Molecular Diagnostics. 2010;12(4):425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baysan M., Woolard K., Cam M. C., et al. Detailed longitudinal sampling of glioma stem cells in situ reveals Chr7 gain and Chr10 loss as repeated events in primary tumor formation and recurrence. International Journal of Cancer. 2017;141(10):2002–2013. doi: 10.1002/ijc.30887. [DOI] [PubMed] [Google Scholar]
- 27.Cimino P. J., Kim Y., Wu H. J., et al. Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma. Genes & Development. 2018;32(7-8):512–523. doi: 10.1101/gad.312157.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crespo I., Vital A. L., Nieto A. B., et al. Detailed characterization of alterations of chromosomes 7, 9, and 10 in glioblastomas as assessed by single-nucleotide polymorphism arrays. The Journal of Molecular Diagnostics. 2011;13(6):634–647. doi: 10.1016/j.jmoldx.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tunici P., Bissola L., Lualdi E., et al. Genetic alterations and in vivo tumorigenicity of neurospheres derived from an adult glioblastoma. Molecular Cancer. 2004;3(1):p. 25. doi: 10.1186/1476-4598-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayin N. S., Modrek A. S., Placantonakis D. G. Glioblastoma stem cells: molecular characteristics and therapeutic implications. World Journal of Stem Cells. 2014;6(2):230–238. doi: 10.4252/wjsc.v6.i2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallini R., Ricci-Vitiani L., Montano N., et al. Expression of the stem cell marker CD133 in recurrent glioblastoma and its value for prognosis. Cancer. 2011;117(1):162–174. doi: 10.1002/cncr.25581. [DOI] [PubMed] [Google Scholar]
- 32.Zeppernick F., Ahmadi R., Campos B., et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clinical Cancer Research. 2008;14(1):123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 33.Kong B. H., Moon J. H., Huh Y. M., et al. Prognostic value of glioma cancer stem cell isolation in survival of primary glioblastoma patients. Stem Cells International. 2014;2014:6. doi: 10.1155/2014/838950.838950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arita H., Yamasaki K., Matsushita Y., et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathologica Communications. 2016;4(1):79–93. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandes A. A., Franceschi E., Paccapelo A., et al. Role of MGMT methylation status at time of diagnosis and recurrence for patients with glioblastoma: clinical implications. The Oncologist. 2017;22(4):432–437. doi: 10.1634/theoncologist.2016-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sciuscio D., Diserens A. C., van Dommelen K., et al. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clinical Cancer Research. 2011;17(2):255–266. doi: 10.1158/1078-0432.CCR-10-1931. [DOI] [PubMed] [Google Scholar]
- 37.Verhaak R. G. W., Hoadley K. A., Purdom E., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1 . Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H., Han F., Hu R., et al. PTEN gene mutations correlate to poor prognosis in glioma patients: a meta-analysis. OncoTargets and Therapy. 2016;9:3485–3492. doi: 10.2147/ott.s99942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraus J. A., Glesmann N., Beck M., et al. Molecular analysis of the PTEN, TP53 and CDKN2A tumor suppressor genes in long-term survivors of glioblastoma multiforme. Journal of Neuro-Oncology. 2000;48(2):89–94. doi: 10.1023/A:1006402614838. [DOI] [PubMed] [Google Scholar]
- 40.Felsberg J., Hentschel B., Kaulich K., et al. Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: prognostic role and comparison between primary and recurrent tumors. Clinical Cancer Research. 2017;23(22) doi: 10.1158/1078-0432.CCR-17-0890. [DOI] [PubMed] [Google Scholar]
- 41.Liffers K., Lamszus K., Schulte A. EGFR amplification and glioblastoma stem-like cells. Stem Cells International. 2015;2015:11. doi: 10.1155/2015/427518.427518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.William D., Mokri P., Lamp N., et al. Amplification of the EGFR gene can be maintained and modulated by variation of EGF concentrations in in vitro models of glioblastoma multiforme. PLoS One. 2017;12(9, article e0185208) doi: 10.1371/journal.pone.0185208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stockhausen M. T., Broholm H., Villingshøj M., et al. Maintenance of EGFR and EGFRvIII expressions in an in vivo and in vitro model of human glioblastoma multiforme. Experimental Cell Research. 2011;317(11):1513–1526. doi: 10.1016/j.yexcr.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 44.deCarvalho A. C., Kim H., Poisson L. M., et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nature Genetics. 2018;50(5):708–717. doi: 10.1038/s41588-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis B., Shen Y., Poon C. C., et al. Comparative genomic and genetic analysis of glioblastoma-derived brain tumor-initiating cells and their parent tumors. Neuro-Oncology. 2016;18(3):350–360. doi: 10.1093/neuonc/nov143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boccaccio C., Comoglio P. M. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Research. 2013;73(11):3193–3199. doi: 10.1158/0008-5472.CAN-12-4039. [DOI] [PubMed] [Google Scholar]
- 47.Bredel M., Scholtens D. M., Yadav A. K., et al. NFKBIA deletion in glioblastomas. The New England Journal of Medicine. 2011;364(7):627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J., Kotliarova S., Kotliarov Y., et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material: an Excel File in which there are reported lists of genes included in n = 12 clusters, grouped by K-means algorithm, with the relative location, values, and GO-Term enrichment analysis performed by WebGestalt. Moreover, all the output files achieved by WebGestalt are included.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.