Abstract
Background
The prion protein (PrP) is known to bind certain soluble aggregates of the amyloid β-protein (Aβ), and two regions of PrP, one centered around residues 19–33, and the other around 87–112, are thought to be particularly important for this interaction. When either of these sequences are grafted into a human IgG the resulting antibodies react with disease-associated PrP conformers, whereas the parental b12 IgG does not.
Methods
Human antibodies containing grafts of PrP 19–33 or 87–112 were prepared as before (Solforosi et al. 2007) and tested for their ability to recognize synthetic and Alzheimer’s disease (AD) brain-derived Aβ. Since aqueous extracts of AD brain contain a complex mixture of active and inactive Aβ species, we also assessed whether PrP-grafted antibodies could protect against neuritotoxicity mediated by AD brain-derived Aβ. For these experiments, human iPSC-derived neurons were grown in 96-well plates at 5000 cells per well and on post-induction day 21, AD brain extracts were added +/− test antibodies. Neurons were imaged for 3 days using an IncuCyte live-cell imaging system, and neurite number and density quantified.
Results
Grafted antibodies bound a significant portion of aggregated Aβ in aqueous AD extracts, but when these antibodies were co-incubated with neurons treated with brain extracts they did not reduce toxicity. By contrast, PrP and the PrP fragment N1 did protect against Aβ.
Conclusions
These results further demonstrate that not all Aβ oligomers are toxic and suggest that PrP derivatives may allow development of agents that differentially recognize toxic and innocuous Aβ aggregates.
Keywords: Alzheimer`s Disease, prion protein, neuritotoxicity
1. Introduction
Neurodegenerative diseases are an enormous public health problem, which affect tens of millions of sufferers. A hallmark of disorders such as Alzheimer’s disease (AD), Parkinson’s disease and transmissible spongiform encephalopathies (TSEs) is the presence of aggregates formed from neuronal proteins of unrelated sequences (Selkoe, 2003; Silveira et al., 2005; Soto, 2003; Walsh and Selkoe, 2016). Consequently, there is considerable interest in the therapeutic targeting of these aggregates and on their detection as diagnostic indicators. Two of the most widely studied polypeptides involved in neurodegeneration are the cellular prion protein (PrPC) and the amyloid β-protein (Aβ).
PrP is critical for the neurotoxicity that underlies the degenerative neurological disorders known as prion diseases or TSEs. Prions are infectious and propagate by converting α-helix-rich PrPC into β-sheet-rich PrPSc. Persuasive evidence indicates that oligomerization and fibrillization of Aβ are key events in AD (Karran and De Strooper, 2016; Selkoe and Hardy, 2016), and over the past decade, data have emerged which suggest that PrPC may serve as a receptor that mediates certain aspects of Aβ toxicity (Gunther and Strittmatter, 2010; Smith and Strittmatter, 2017). In addition to binding soluble aggregates of Aβ (aka Aβ oligomers), the N-terminal domain of PrPC is known to bind aggregated forms of PrP and PrPSc, conformers of a yeast prion protein and certain designed β-sheet-rich peptides, leading some to speculate that PrP may be capable of binding a variety of aggregated, amyloid-prone proteins (Beland and Roucou, 2012; Resenberger et al., 2011). Special attention has focused on the interaction between PrP and Aβ oligomers. Notably, while the pathological significance of Aβ binding to PrP has been contentious, there is complete agreement that PrP binds Aβ oligomers with high affinity and that PrP is a sub-stoichiometric inhibitor of Aβ aggregation (Chen et al., 2010; Fluharty et al., 2013; Nieznanska et al., 2018; Nieznanski et al., 2012; Younan et al., 2013). There is also general agreement that two sites within PrP (one around residues 23–33 and the other around 87–112) are important for binding Aβ oligomers (Figure 1A) (Chen et al., 2010; Fluharty et al., 2013; Nieznanska et al., 2018; Younan et al., 2013). N1 (23–111) is a naturally occurring soluble form of PrP (Biasini et al., 2012; Mange et al., 2004) which retains all of the potential residues involved in Site I and most (if not all) of the residues implicated in Site II. Importantly, several groups have shown that N1 or similar fragments which retain the Aβ binding sites exhibit binding and aggregation inhibition activity comparable to that of full-length PrP (Fluharty et al., 2013; Nieznanska et al., 2018; Nieznanski et al., 2012; Um et al., 2012). Thus, it may be possible to exploit the PrP-Aβ oligomer binding sequences to develop tools to allow the measurement and/or targeting of toxic Aβ. However, since full length PrP is itself a potential neurotoxin (Chesebro et al., 2005) other derivatives of PrP would be more desirable (Biasini et al., 2012). In prior studies, short overlapping sequences covering the entire PrP molecule were used to replace the extended heavy chain complementary-determining region 3 (CDR3) of a human IgG, and their ability to bind PrPSc was investigated (Moroncini et al., 2004; Solforosi et al., 2007). Analysis of 20 different grafted monoclonal antibodies (mAbs) revealed three high affinity PrPSc recognition motifs, centered around 19–33, 89–112, and 136–158 (Solforosi et al., 2007). Given that 2 of 3 PrPSc binding sites have been implicated in Aβ binding we investigated whether such antibodies could also bind Aβ.
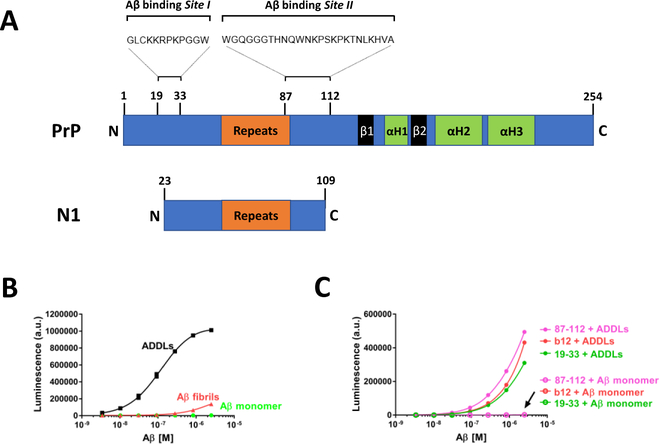
Figure 1. Recombinant murine PrP, but not PrP-motif grafted antibodies bind ADDLs with high affinity.
(A) Schematic of PrP and N1. The precise amino acid sequences constituting oligomer binding Site I and Site II are not yet known but appear to be centered around residues 19–33 and 87–112, respectively. The octapeptide repeat domain, β-sheets (β)1 and 2, and α-helices (H)1–3 are indicated. PrP can undergo endoproteolytic cleavage yielding N-terminal secreted products referred to as N1 (23–111 or 23–112). We used a PrP fragment comprising the amino acid residues 23–109 for our study. (B) Direct binding of ADDLs, Aβ fibrils, and Aβ monomers was assessed using an ELISA-like microtiter plate assay. Preparations of Aβ were incubated for 1 hour on PrP-coated plates and bound Aβ was detected with biotinylated anti-Aβ mAb 4G8. ADDLs bind PrP in a dose-dependent manner, whereas Aβ monomers and fibrils show little affinity for PrP. Each data point is the average ± SEM of three independent experiments. (C) Antibodies grafted with and without PrP residues 19–33 and 87–112 were coated onto microtiter plate wells and incubated with ADDLs (filled circles) or Aβ monomers (open circles) for 1 hour, and then detected with the mAb 4G8. Red circles denote b12 mAb. Antibodies grafted with PrP motifs 19–33 and 87–112 are colored in light green and pink, respectively. Each data point is the average ± SEM of two technical replicates. Where the error bars are not visible, the SEM is smaller than the size of the symbol. Results are representative of at least 2 individual experiments.
Here, we show that while recombinant PrP can bind synthetic Aβ oligomers with high affinity, grafted PrP mAbs do not. Yet PrP-grafted antibodies can immunoprecipitate certain aggregates of Aβ from the soluble phase of AD brain. These results and their implications for understanding and targeting toxic and innocuous forms of Aβ are discussed.
2. Results
2.1. Recombinant PrP, but not PrP-motif grafted antibodies, bind oligomers of synthetic Aβ
The cellular prion protein (PrP) is known to specifically and tightly bind Aβ oligomers and this interaction is believed to be mediated by 2 distinct binding sites within the N-terminus of PrP (Site I and Site II, Figure 1A). These two binding sites have also been implicated in the interaction between PrPC and PrPSc (Biasini et al., 2008; Moroncini et al., 2004; Solforosi et al., 2007), and it has been shown that mAbs containing grafts of Site I and Site II can bind PrPSc (Solforosi et al., 2007). Here, we investigated whether PrP-grafted mAbs known to bind PrPSc could also bind Aβ oligomers. Three distinct preparations of Aβ (see Supplementary Figure 1 for detailed characterization) were tested for their ability to bind to plate-immobilized recombinant PrP (Figure 1B). As in prior studies Aβ-derived diffusible ligands (ADDLs), a recipe-based preparation of Aβ1–42 which contains monomers, protofibrils, and globular oligomers, bound tightly to PrP with a mean EC50 of 113 nM (95% CI 96 – 133 nM), based on 3 independent experiments. In contrast, Aβ monomer (EC50 not determinable) and fibrils (EC50 not determinable) showed no or little affinity for PrP (Figure 1B). Next, we examined whether plate-immobilized PrP-grafted mAbs could bind Aβ. None of the mAbs bound Aβ monomers, but grafted mAbs 19–33 and 87–112 and the parental mAb, b12, each bound ADDLs (Figure 1C). However, the finding that b12 and the grafted mAbs bind ADDLs to similar extents indicates that binding is not PrP graft-specific, but rather a result of a generic interaction between the b12 backbone and some component of ADDLs.
The lack of specific binding by PrP-grafted mAbs could have resulted due to inactivation of the mAbs which can occur during purification, or as a consequence of steric obstruction preventing 4G8 from detecting oligomers bound to grafted mAbs. We addressed these possibilities first by testing if grafted mAbs could bind PrPSc (Supplementary Figure 2 A) and second by using an alternative anti-Aβ mAb, 6E10, for detection (Supplementary Figure 2 B and C). Grafted mAbs 19–33 and 87–112 specifically recognized PrPSc, but not PrPC. The parent mAb b12 did not immunoprecipitate (IP) either PrPC or PrPSc (Supplementary Figure 2 A). These results confirm that the grafted PrP mAbs used are active. Furthermore, when the N-terminal anti-Aβ mAb, 6E10, was used in place of 4G8, PrP-grafted and b12 parent mAbs produced similar binding curves (compare Figure 1B and Supplementary Figure 2 B). Again, monomers showed no binding to PrP. However, unlike 4G8 use of 6E10 allowed appreciable detection of fibrils. Nonetheless, even with 6E10, the EC50 for ADDLs (234 nM, 95% CI 218 – 251 nM) was an order of magnitude tighter than for Aβ fibrils (1731 nM, 95% CI 1601 – 1870 nM). Thus, irrespectively of the anti-Aβ mAb used, PrP preferentially bound ADDLs. Hence, it is apparent that the inability of PrP-grafted mAbs to bind ADDLs does not result due to trivial experimental artifacts.
2.2. PrP-grafted antibodies immunoprecipitate aggregates of Aβ from AD brain extracts
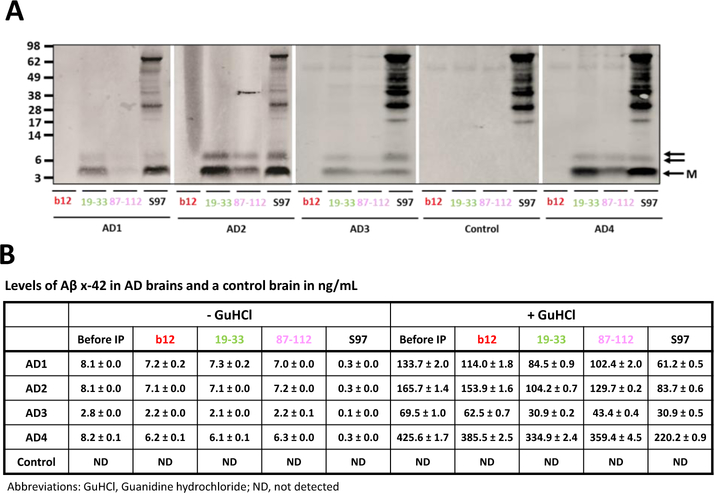
Given that PrP-grafted mAbs were capable of immunoprecipitating PrPSc from infected mouse brains and that it is uncertain if synthetic Aβ assemblies are relevant to Aβ structures that occur in vivo, we asked whether PrP-grafted antibodies could IP aggregated Aβ from human brain. Extensive data indicate that aqueous extracts of AD brain contain an array of Aβ species of which only a portion has disease-relevant activity (Hong et al., 2018). Here, we examined whether PrP-grafted antibodies 19–33 and 87–112 could IP Aβ from aqueous extracts of 4 end-stage AD brains. Grafted mAbs each IP`d a portion of Aβ from all AD brain extracts examined, whereas the parent b12 did not bind any Aβ. In all cases 19–33 bound more Aβ than 87–112 (Figure 2A), whereas no Aβ was detected in IPs of control brain extract. IP/WB detected 2 distinct Aβ-immunoreactive bands, one consistent with Aβ monomer (~4 kDa) and the other with Aβ dimer (~7 kDa). Previous work has shown that the ~7 kDa species most likely represents a mixture of covalently cross-linked Aβ dimers (Brinkmalm et al., 2018; Mc Donald et al., 2015; Vazquez de la Torre et al., 2018) and some aberrantly migrating monomer (Watt et al., 2013). Samples before and after IP were also analyzed using an Aβx-42 MSD-based immunoassay. Treatment of brain extracts with the chaotropic agent GuHCl dissociates Aβ-aggregates to monomers, and enables their detection (Mc Donald et al., 2015). Thus, by measuring Aβx-42 in extracts +/− GuHCl treatment, it is possible to quantify the amount of native Aβ42 monomer (- GuHCl), and Aβ42 aggregates that can be broken down to monomer by incubation with GuHCl (+ GuHCl). Relative to the starting extract (Before IP), immunoprecipitation with S97, a polyclonal pan-Aβ antibody, depleted Aβ monomer from AD brain extracts by 90 – 96%, and effectively reduced aggregated Aβ by 48 – 56% in AD1 – AD4 (Figure 2B). Treatment with PrP-motif grafted mAb 19–33 reduced aggregated Aβ in AD1, AD2, and AD3 brains by 36 – 56%, and in AD4 by 21%. It is noteworthy that AD4 contained a much higher starting level of aggregated Aβ (425.6 ng/mL) than the other three AD brain extracts (69.5 – 165.7 ng/mL). PrP-motif grafted antibody 87–112 reduced aggregated Aβ by 16 – 38%. The finding that grafted antibody 87–112 is less effective at reducing the level of soluble aggregates than grafted antibody 19–33 is in keeping with IP/WB results which indicate that 19–33 IP`d approximately twice as much Aβ as 87–112. In AD brain extracts Aβ monomer levels were modestly decreased relative to starting extracts, but unchanged versus parent b12 treatment. Thus, grafted mAbs 19–33 and 87–112 can IP a pool of aggregates, but do not IP Aβ monomer.
Figure 2. PrP-motif grafted antibodies immunoprecipitate aggregated Aβ from AD, but not control brain.
(A) Water-soluble extract from four AD brains (AD1 to AD4) and one control brain were incubated with PrP-motif grafted antibodies. The b12 antibody was used as negative control and the polyclonal antiserum, S97, as a positive control. Anti-human IgG coupled to paramagnetic beads was used to capture b12 and b12-PrP grafted mAbs. S97 was captured with Protein A agarose beads. Precipitated Aβ was detected via Western blot using anti-Aβ mAbs, HJ2 and 21F12. Molecular weight markers are indicated on left, and the antibodies used for IP are indicated at the bottom of the gel. M (single arrow) denotes Aβ monomer and double arrow refers to SDS-stable ~7kDa Aβ. (B) Starting brain extracts and IP supernates were analyzed using an Aβx-42 MSD immunoassay with (right panel) and without (left panel) preincubation with 5 M GuHCl. ND denotes non-detected.
2.3. PrP and PrP N1, but not PrP-grafted antibodies protect against Aβ-induced neuritotoxicity
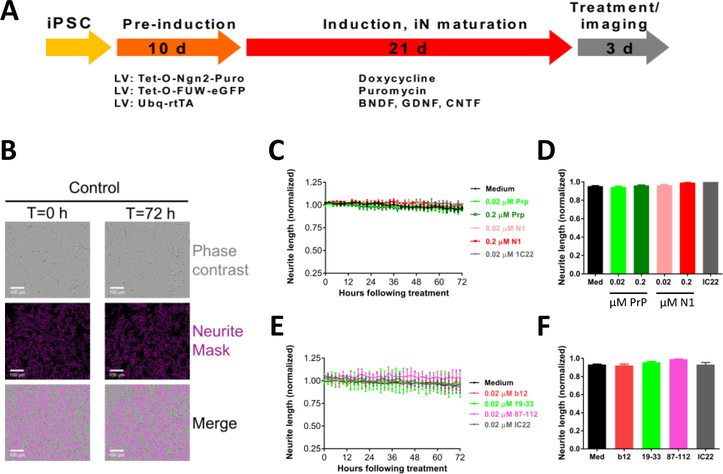
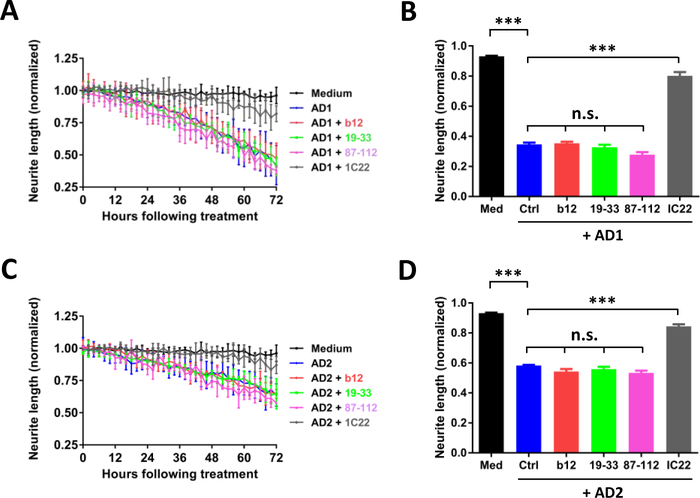
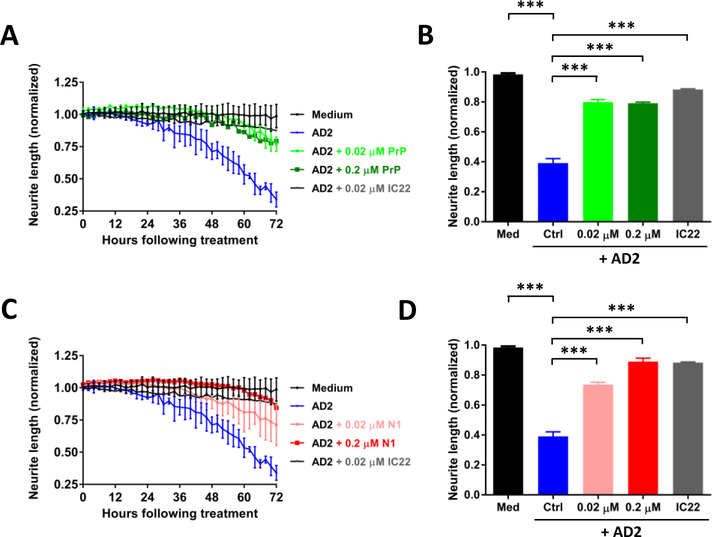
Having established that PrP-motif grafted antibodies can bind aggregated Aβ in AD brain extracts, we next assessed whether these mAbs recognize bioactive Aβ species. Recently, we described a live-cell imaging paradigm to measure the effect of AD brain extracts on human neurons (Hong et al., 2018; Jin et al., 2018). Differentiated iPSC-derived human neurons (iNs) (see Figure 3A and 3B for a details) were treated with AD brain extracts and imaged every two hours for 72 hours. Exposure to AD1 brain extract caused a time-dependent decrease in neurite length (Figure 4A and B) (Medium, black; AD1, blue; p < 0.001, medium vs. AD1; one-way ANOVA). Co-administration of the aggregate specific anti-Aβ mAb, IC22 (Jin et al., 2018; Mably et al., 2015), was used as a positive control to rescue Aβ-dependent neuritotoxicity. Co-administration of IC22 conferred near complete protection against neuritotoxicity induced by AD1 (AD1, blue; AD1 + IC22, grey; p<0.001, AD1 vs. AD1 + IC22; one-way ANOVA). However, none of the PrP-motif grafted antibodies were protective (AD1, blue; b12, red; 19–33, light green; 87–112, pink; p > 0.05, AD1 vs. b12, AD1 vs. 19–33, AD1 vs. 87–112; one-way ANOVA). To test the generalizability of these results, we used a second AD brain extract. As with AD1, AD2 caused a time-dependent loss of neurite length, which was rescued with IC22 but not with PrP-grafted antibodies (Figure 4C and D). To test whether PrP can reduce neuritotoxicity induced by AD brain extract, we treated cells with AD2 brain extract in the presence and absence of PrP or the PrP fragment N1. At concentrations equal to or greater than used for IC22 (Figure 4, 0.02 μM), PrP and the N1 fragment provided almost complete protection against AD2 induced neuritotoxicity (AD2, blue; AD1 + 0.02 μM PrP, light green; p<0.001, AD2 vs. AD2 + 0.02 μM PrP; one-way ANOVA) (Figure 5 A and B); (AD2, blue; AD1 + 0.02 μM N1, light red; p<0.001, AD2 vs. AD2 + 0.02 μM N1; one-way ANOVA) (Figure 5 C and D). Importantly, when tested alone, none of the mAbs, PrP, and N1 did not affect neurite integrity (Figure 3 C–F).
Figure 3. Live-cell imaging and analysis of neurite length in iPSC-derived human neurons.
(A) Schematic shows the timeline to generate, mature, and image iPSC-derived human neurons (iNs). (B) Upper panels: phase contrast images of iNs at the start (T=0 hours) and end of the imaging period (T=72 hours). Middle panels: images were analyzed using IncuCyte NeuroTrack algorithm for the identification of neurites (pink). Lower panels: NeuroTrack identified neurites are superimposed on the phase contrast image. The scale bar is 100 μm. To test whether mAbs, PrP, and N1 protein are compatible with our assay, iNs were treated with mAbs, PrP, or N1 in the indicated concentrations, and cells imaged for 72 hours. (C + D) Time-course plots are similar for iN cells treated +/− mAbs, PrP or N1 protein. Each data point is the average of 3 wells ± SD. (D + E) Neuritic length was averaged over the last 6 h of imaging, and values normalized to pre-treatment measurements.
Figure 4. PrP-grafted antibodies do not protect against Aβ-induced neuritotoxicity.
Live-cell imaging was used to monitor the effects of Aβ-containing AD brain extracts on iPSC-derived neurons (iNs) and whether or not PrP-grafted mAbs could protect neurons. On post-induction date 21, iNs were treated with medium alone (black circles), AD extract (blue), or AD extract plus b12 (red), 19–33 (green), 87–112 (pink), or IC22 (gray). Each antibody was used at a final concentration of 0.02 μM. (A) Time-course plot shows that AD1 extract caused neuritotoxicity and this could be attenuated by IC22, but not PrP-grafted mAbs. Each data point is the average of 3 wells ± SD. (C) Similar results were obtained using extracts from AD2. (B and D) Neuritic length was averaged over the last 6 h of imaging, and values normalized to pre-treatment measurements. The results are representative of two independent experiments. Differences in means were assessed with ANOVA followed by Bonferoni`s post-hoc test. n.s., non-significant; p>0.05; *** p<0.001.
Figure 5. PrP and N1 protect against Aβ-induced neuritotoxicity.
The same paradigm as in Figure 4 was employed to monitor the effects of the Aβ-containing AD 2 brain extract on iNs and whether or not PrP and N1 protein could protect neurons. On post-induction date 21, iNs were treated with medium alone (black circles), AD 2 extract (blue), or AD 2 extract plus 0.02 μM PrP (light green), 0.2 μM PrP (dark green), or IC22 (gray). (A) Time-course plot shows that AD2 extract caused neuritotoxicity and this could be attenuated by treatment with either PrP or IC22. Each data point is the average of 3 wells ± SD. (C) Similar results could be obtained by treatment with N1 protein. Coincubation of iNs with AD 2 extract plus 0.02 μM N1 protein (light red) and 0.2 μM N1 protein (dark red) reduced AD brain extract induced neuritotoxicity. (B and D) Neuritic length was averaged over the last 6 h of imaging, and values normalized to pre-treatment measurements. Differences in means were assessed with ANOVA followed by Bonferoni`s post-hoc test. The results shown are from a single experiment. *** p<0.001.
Collectively, our results indicate that while PrP-grafted mAbs show little affinity for synthetic Aβ aggregates (ADDLs) they do bind certain Aβ aggregates in extracts of AD brain, but they do not immuno-neutralize the neurotoxic forms therein. In contrast, forms of recombinant PrP and the N1 fragment of PrP, which contains Aβ binding Sites I and II, do attenuate Aβ neuritotoxicity.
3. Discussion
Compelling evidence indicates that PrP can bind certain soluble aggregates of Aβ and that this interaction is mediated by two distinct sites within the flexible N-terminus of PrP. Intriguingly, the sites believed to be important for Aβ binding have also been implicated in the interaction between PrPC and PrPSc (Lau et al., 2007; Moroncini et al., 2004; Solforosi et al., 2007; Turnbaugh et al., 2012) and it is known that antibodies into which these sites have been grafted can bind PrPSc (Biasini et al., 2008; Moroncini et al., 2004; Novitskaya et al., 2006; Solforosi et al., 2007). These observations prompted us to investigate whether PrP-grafted mAbs can bind the most disease relevant form of Aβ, protein extracted from AD brain.
Initial experiments involved synthetic Aβ and recombinant PrP. As in numerous prior studies PrP tightly bound soluble Aβ aggregates (ADDLs), but showed little affinity for Aβ monomers (Chen et al., 2010; Fluharty et al., 2013; Freir et al., 2011; Lauren et al., 2009; Nicoll et al., 2013; Nieznanski et al., 2012; Resenberger et al., 2011; Um et al., 2012). In parallel experiments, PrP-grafted mAbs bearing Aβ binding Sites I (19–33) and II (87–112) only very weakly bound ADDLs, and this binding was indistinguishable from that produced by the parental b12 mAb that lacked any PrP sequence. Since it is not clear whether structures such as ADDLs are ever present in the human cerebrum, we went on to test if PrP-grafted mAbs could bind disease relevant forms of Aβ extracted from AD brain. Prior studies used an IP/WB paradigm to demonstrate that Site I and II-containing grafted mAbs do not bind PrP monomer (i.e. PrPC) PrPSc, but bind both infectious and (Moroncini et al., 2004; Solforosi et al., 2007), non-infectious pathological aggregates of PrP (Biasini et al., 2008). We adapted a similar approach using aqueous extracts of human brain alongside extracts of scrapie-infected mouse brain. As in earlier reports, b12 19–33 and b12 87–112 specifically IP’d PrPSc from infected brain but did not IP PrPC. In two separate experiments b12 87–112 IP’d noticeably more PrPSc than b12 19–33. Both grafted mAbs also IP’d a portion of soluble Aβ from the bioactive extracts of two AD brains, but in these samples b12 19–33 IP’d more Aβ than did b12 87–112. Differences in the relatively affinity of b12 19–33 and b12 87–112 for PrP and Aβ aggregates while interesting should be interpreted with caution due to the small number of brain samples tested and limited quantitative precision of IP/WB paradigms. On the other hand, the finding that neither of the PrP-grafted mAbs IP’d native Aβ monomer in 4 different AD brain extracts is clear cut, and demonstrates that like recognition of PrPSc, binding of Aβ, is aggregation specific. Interestingly, mAb 15B3 which selectively recognizes aggregates of PrP can also bind aggregated but not monomeric forms of Aβ (Stravalaci et al., 2016). These and our results indicate that certain Aβ and PrP aggregates have a common structure. Future studies should explore whether the common recognition of Aβ and PrP aggregates extends to other disease-related proteins (Corbett et al., 2018).
While Aβ aggregation is widely acknowledged to be central to AD pathogenesis, an increasing body of evidence indicates that only a sub-population of Aβ aggregates are bioactive (Brinkmalm et al., 2018; Hong et al., 2018; Lesne, 2013). As with PrP and prion disease (Chiesa et al., 2008; Sandberg et al., 2011; Sandberg et al., 2014), the form or forms of Aβ which mediate toxicity are not yet defined. Hence, rather than relying on simple binding assays to understand and target the small portion of Aβ that is toxic, it is essential to utilize bioactivity assays. To address whether PrP-grafted mAbs recognize toxic Aβ we utilized a very recently developed medium-throughput video-microscopy paradigm to monitor disruption of neurite integrity by AD brain derived Aβ (Hong et al., 2018; Jin et al., 2018). In prior studies we demonstrated that certain anti-Aβ mAbs could attenuate AD brain Aβ mediated neuritotoxicity (Jin et al., 2018). Here, we show that even though a single round of IP with b12 19–33 and b12 87–112 can pull down an appreciable portion of Aβ in bioactive AD brain extracts (~36% and ~23% of respectively), neither grafted mAb altered the time-course nor the extent of neuritotoxicity. By contrast, when used at the same concentration as PrP-grafted mAbs (0.02 μM), our in-house anti-Aβ aggregate-preferring mAb, 1C22 (Mably et al., 2015), nearly fully protected against AD brain Aβ. Thus, it appears that b12 19–33 and b12 87–112 cannot effectively immunoneutralize neuritotoxic Aβ.
PrP and a PrP-fragment (N1) both of which contain Aβ binding Sites I and II conferred protection against AD brain Aβ. This is in line with prior studies which reported that N1 attenuated Aβ-mediated toxicity (Fluharty et al., 2013; Nieznanski et al., 2012). Why PrP-grafted mAbs should be able to bind at least some soluble aggregates of Aβ, but not impact Aβ toxicity is not yet clear, but it is tempting to speculate that the lack of reactivity with bioactive Aβ is because the grafted mAbs used are monospecific. That is, b12 19–33 contains two 19–33 grafts and no 87–112, and b12 87–112 contains two 87–112 grafts and no 19–33. Binding of bioactive Aβ by PrP, and by extension PrP-grafted mAbs, may require both binding Sites I and II, and it is plausible that the presence of two of the same binding sites on the same grafted mAb may restrict the forms and sizes of Aβ assemblies that can be bound. In future studies it will be interesting to generate and test whether bispecific mAbs containing both Sites I and II can protect against AD brain Aβ, and to employ our all-human video-microscopy paradigm to compare the protection afforded by full length PrP, N1, and PrP-grafted bispecific mAbs.
4. Material and Methods
4.1. Reagents and antibodies
Size exclusion standards were obtained from Bio-Rad (Hercules, CA, USA). All other chemicals and reagents were from Millipore-Sigma (Burlington, MA) unless otherwise noted. Antibodies are described in Table 1.
Table 1.
Primary antibodies and their antigens, dilutions, and source
| Antibody | Antigen/Epitope | Dilution | Source | Reference |
|---|---|---|---|---|
| 6E10 | Aβ 6–10 | 1 μg/ml (ELISA) | Biolegend | Kim et al., 1988 |
| 4G8 | Aβ 18–22 | 1 μg/ml (ELISA) | Biolegend | Chang et al., 2007 |
| 266 | Aβ 13–28 | 3 μg/ml (MSD) | Janssen | Johnson-Wood et al., 1997 |
| HJ2 | Ap terminating at Val40 | 1 μg/ml (WB) | D. Holtzman | Pankiewicz et al., 2014 |
| 21F12 | Aβ terminating at Ile42 | 1 μg/ml (WB) 1/8,000 (MSD) | Janssen | Johnson-Wood et al., 1997 |
| IC22 | Aβ aggregates | 3 μg/ml (0.02 μM) (Neurotoxicity assay) | Walsh lab | Mably et al., 2015 |
| MI0131 | PrP 23–51 | 1 μg/ml (WB) | Medimmune | Ondrejcak et al., 2018 |
| 6D11 | PrP 93–109 | 1 μg/ml (WB) | Biolegend | Pankiewicz et al., 2006 |
| ICSM35 | PrP 95–105 | 5 μg/ml (IP) | J. Collinge | White et al., 2003 |
4.2. Preparation of Recombinant Prion Protein 23–231 and 23–109
Escherichia coli strain BL21(DE3)pLysS (New England BioLabs, Ipswich, MA) was transformed with the pTrcHis B vector expressing murine PrP23–231 (Jackson et al., 1999; Nicoll et al., 2013; Zahn et al., 1997), with minor modifications. Ampicillin resistant colonies were selected, expanded and expression was induced by the addition of 1 mM IPTG for 16 hours at 37 °C and 225 RPM. Cultures were harvested by centrifugation at 6,000 × g for 15 minutes, washed in PBS and lysed by sonication (2 × 120 seconds bursts at 30% output) in extraction buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 0.1% Tween-20, 50 U/mL Benzonase, 10 μg/mL lysozyme). Suspensions were sedimented at 10,000 × g for 30 min and the inclusion body-enriched pellets were extracted by sonication (2 × 120 seconds bursts at 30% output) in solubilization buffer (6 M GuHCl / 50 mM Tris-HCl pH 8.0 / 0.8% β-mercaptoethanol). Suspensions were sedimented at 21,000 × g for 45 minutes and the PrP-enriched supernatant was clarified with 5 μm and 0.45 μm syringe filters prior to on-column oxidative refolding. Filtered supernatants were loaded onto 5 mL HisTrap HP columns (GE Life Sciences, Marlborough, MA) at 1 mL/minute using a BioLogic DuoFlow FPLC system (Bio-Rad, Hercules, CA) and washed with 10 column volumes (CV) of Buffer A (6 M GuHCl, 10mM Tris-HCl, 100 mM Na2HPO4, 10mM Glutathione pH 8.0) at 1 mL/minute. Bound PrP was refolded in a linear gradient of Buffer A to Buffer B (10mM Tris-HCl, 100 mM Na2HPO4, pH 8.0) for 30 CV at 0.213 mL/minute (11 ¾ hours). The following day, the column was eluted in a linear gradient of Buffer B to Buffer C (10mM Tris-HCl, 100 mM Na2HPO4, 1 M imidazole, pH 5.8) for 3 CV at 0.5 mL/minute and the fractions containing PrP were buffer exchanged with 2 kDa dialysis cassettes (ThermoFisher, Waltham, MA) overnight at 4 °C against 1000 volumes of Bis-Tris HCl, pH 6.5. The ploy-histidine tag was cleaved from PrP using 50 U restriction grade thrombin (Novagen, Madison, WI) overnight with agitation, and cleaved PrP was separated from the free histidine tag using a 5 mL HisTrap HP column. The fractions containing purified PrP were dialyzed overnight at 4 °C against 1000 volumes of Bis-Tris HCl, pH 6.5. Protein purity was determined by SDS-PAGE/sliver staining and mass spectrometry, and secondary structure was analysed by circular dichroism (Jasco J-815, Jasco, Easton, MD). Concentration was determined by measuring absorbance at 280 nm and using the predicted extinction coefficient (63370, ε280 = M−1cm−1).
For production of PrP23–109 (N1), Escherichia coli strain BL21 Star was transformed with the pET101 vector expressing murine PrP23–109. This vector was expressed and purified as described previously (McDonald et al., 2014), with minor modifications. Cells were lysed, then purified with an ÄKTA purification system (GE Healthcare, Marlborough, MA, USA) using a Ni2+-immobilized metal ion affinity column. Protein was eluted with 5 M guanidine HCl, 0.1 M Tris acetate, 0.1 M potassium phosphate (pH 4.5). A280 positive fractions spanning the elution peak were combined, and the pH was raised to 8 by addition of potassium acetate. The neutralized solution was desalted into 20 mM potassium acetate, pH 5.5 using a HiPrep 26/10 desalting column (GE Healthcare, Marlborough, MA, USA). This material was then purified using a C4 reverse-phase HPLC column (Grace/Vydac, Columbia, MD, USA). Fractions containing the purified protein were pooled, lyophilized, and stored at −80 °C. Protein stocks were reconstituted in 0.22 μm filtered water. Protein purity was determined by SDS-PAGE/Coomassie staining. N1 was further characterized by Western Blot using anti-PrP mAbs directed against Aβ binding Sites I and II (Supplementary Figure 3 A and B). N1 and PrP bind ADDLs with similar affinity, and show little affinity for fibrils and monomers in an ELISA like assay (Supplementary Figure 3 C).
4.3. Preparation of Aβ conformers
Aβ1–40 monomers and Aβ1–42 fibrils were prepared as previously described (O’Malley et al., 2016). To remove pre-existing aggregates peptides were incubated in 50 mM Tris-HCl, pH 8.5, containing 5 mM EDTA and 7 M guanidine HCl for at least 16 hours. Aβ1–40 monomers were isolated by chromatographing disaggregated peptide on a Superdex 75 10/300 SEC column (GE Life Sciences, Marlborough, MA, USA) eluted with 50 mM ammonium bicarbonate, pH 8.5 (Supplementary Figure 1A). Peak fractions were collected and concentration determined by measuring UV absorbance at 275 nm (O’Malley et al., 2014). Aliquots were flash-frozen in liquid nitrogen and stored at −80 °C. To generate fibrils Aβ1–42 monomer was isolated using the same column as for Aβ1–40, but this time eluted in in 20 mM sodium phosphate, pH 8.0. Peak fractions were pooled, the concentration measured, and the solution diluted to 20 μM in 20 mM sodium phosphate, pH 8.0. A portion of the sample was mixed with Thioflavin T (20 μM) to monitor formation of fibrils (Supplementary Figure 1B). The remainder was incubated without Thioflavin T. When fibril formation had reached its maximum (Supplementary Figure 1B) fibrils were harvested by centrifugation at 100,000 × g for 60 minutes. Fibrils were resuspended in an equal volume of phosphate-buffered saline, pH 7.4, and washed 2 times. Aβ fibrils were flash-frozen in liquid nitrogen and stored in small aliquots at −80 °C. Aβ-derived diffusible ligands (ADDLs) were prepared as described (Freir et al., 2011). In brief, 1 mg Aβ1–42 was dissolved in HFIP, and incubated at 37 °C for 1 hour in a 2 mL lobind protein Eppendorf tube (Eppendorf, Hamburg, Germany). The HFIP was evaporated under a gentle stream of Helium to produce a clear film, and then stored over desiccant at −20 °C. The peptide film was dissolved in anhydrous DMSO and diluted to 100 μM in phenol red-free DMEM/F12 media (Thermo Scientific, Waltham, MA, USA). Samples were incubated at 4 °C for 48 hours, centrifuged at 14,000 × g for 10 minutes, and the supernatant flash-frozen in liquid nitrogen and stored at −80 °C. ADDLs contained a mixture of Aβ monomer and soluble aggregates which appeared on EM as imperfect spheres of ~5 nm diameter and protofibrils up to 200 nm in length (Supplementary Figure 1C).
4.4. Electron microscopy
Sample or buffer (10 μL) was loaded onto 200 mesh formvar/carbon-coated copper grids and left for 60 seconds, and fixed with 0.5% glutaraldehyde for 60 seconds. Grids were wicked dry and washed with 0.22 μm filtered ultra-pure water. Finally, samples were stained with 2% uranyl acetate for 120 seconds, wicked dry and then allowed to air dry.
4.5. Expression and purification of PrP-grafted antibodies
PDR12 vector containing the b12 light chain and the b12 human γ1 heavy chain gene for the parental b12 antibody, and the b12 antibody with PrP 19–33 and 87–112 grafts were described previously (Biasini et al., 2008; Moroncini et al., 2004; Solforosi et al., 2007). The cysteine residue 22 in the PrP sequence 19–33 was mutated to alanine to avoid unwanted dimerization. Expi293 cells were transfected at a ratio of 90 μg of plasmid to 1.75 × 108 cells using ExpiFectamin 293 reagent (Gibco, Grand Island, NY, USA) and 15 μg pAdVantage vector (Promega, Madison, WI, USA). Cells were grown in vented 500 mL Erlenmeyer flasks with shaking (122 rpm) at 37 °C and 5% CO2 for 96 hours. Cell medium was collected and clarified by sequential centrifugation at 150 × g for 6 minutes and 3,500 × g for 10 minutes, followed by filtration through a 0.45 μm filter. Antibodies were purified using a Hitrap Protein A column (GE, Boston, MA, USA) and eluted with 0.1 M sodium citrate buffer, pH 3, into 1 M Tris, pH 9.0. Immediately after purification, antibodies were buffer exchanged into phosphate-buffered saline, pH 7.4 using a 40 k cut-off Zeba column (Thermo Scientific, Waltham, MA, USA), and then passed through a 0.22 μm filter. Purity was assessed by SDS-PAGE/Coomassie staining.
4.6. Mice and preparation of brain lysates
Intracerebral inoculations of C57BL/6 were performed at Boston University with the Rocky Mountain Laboratory strain of scrapie (RML) as described previously (Fang et al., 2016). Mice were inoculated intracerebrally with 1% brain homogenate (30 μL) from the brain of a terminally ill RML infected mouse. The inoculated mice were monitored until the appearance of clinical signs (~170 days after the inoculation). At that time animals were euthanized. Brains were collected, and stored at −80 °C. All animal procedures were performed in accordance with the National Institutes of Health Policy on the Use of Animals in Research and were approved by the Boston University medical centre institutional animal care and use committee. Ten percentage (w/v) homogenates of mouse hemibrains were prepared in phosphate buffer saline, pH 7.2, containing 1% Triton X-100 and EDTA-free Complete protease inhibitors (Roche, Indianapolis, IN, USA) using 1 mm glass beads (Benchmark Scientific, Edison, NJ, USA) and a Beadbug microtube homogenizer (Benchmark Scientific). Brain homogenates were passed once through a 0.5 cc insulin syringe with a 28-gauge needle (Becton Dickinson, Franklin Lakes, NJ, USA) and cleared by sequential centrifugation at 1,000 × g for 30 seconds and 16,000 × g for 5 minutes at 4 °C. Protein concentration was measured with a BCA protein kit (Pierce, Rockford, IL, USA) and adjusted to 1 mg/mL.
4.7. Preparation of human brain extracts
Human specimens were obtained from the Massachusetts ADRC Neuropathology core at the Massachusetts General Hospital in accordance with the Partners Institutional Review Board (Walsh BWH2011). Frozen temporal cortical tissue was obtained from 5 cases, 4 end-stage AD cases, and 1 subject who died free of AD (Table 2). All cases met current post-mortem and clinical diagnosis criteria. The post-mortem interval was 36 hours or less. Approximately 4 g of cortical tissue was dissected from each case. Extracts were prepared by homogenizing 1 g of tissue in 5 volumes of ice-cold artificial cerebrospinal fluid (aCSF) (124 mM NaCl, 2.8 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, pH 7.4) containing 5 mM EDTA, 1 mM EGTA, 5 μg/mL leupeptin, 5 μg/mL aprotinin, 2 μg/mL pepstatin, 120 μg/mL Pefabloc and 5 mM NaF with 25 strokes of a motorized Teflon-glass Dounce homogenizer (Fisher, Ottawa, Canada) (Jin et al., 2018; Wang et al., 2017). Homogenates were centrifuged at 200,000 × g and 4 °C for 110 minutes. The upper 80% of the supernatant was removed and dialyzed against a 100-fold excess of aCSF using a 2000 MWCO Slide-A-Lyzer G2 dialysis cassettes (Fisher Scientific, Waltham, MA, USA) at 4 °C. Samples were dialyzed for at least 72 hours with three buffer changes.
Table 2.
Demographic details of the cases used in this study
| Case | Age | Gender | PMI (h) | Clinical diagnosis | Neuropathology diagnosis | B&B, CERAD |
|---|---|---|---|---|---|---|
| AD1* | 68 | F | 36 | AD | AD | VI, C |
| AD2* | 84 | F | 15 | AD | AD | VI, C |
| AD3 | 75 | F | 24 | AD | AD | NA, C |
| AD4 | 83 | F | NA | AD | AD | NA, C |
| NC | 58 | F | 18 | Not demented | Control | II, NA |
Abbreviations: AD, Alzheimer`s disease; B&B, Braak stage; CERAD, consortium to establish a registry for AD score; F, female; PMI, post-mortem interval; NA, information not available
Denotes samples used in bioactivity assays
4.8. Recombinant PrP and PrP-grafted mAbs immunoassay
Black half-area high binding 96-well plates (Greiner Bio-One, Frickenhausen, Germany) were coated with PrP (0.5 μM) or mAbs (13.33 nM) in 50 mM carbonate coating buffer, pH 9.6 at 30 μL/well and incubated at 37°C for 1 hour on an orbital shaker (Woodbridge, NJ, USA) at 300 rpm. Wells were washed 4 times with PBS containing 0.05% Tween 20 (TBS-T) and then blocked with SuperBlock (Thermo Scientific, Waltham, MA, USA) for 2 hours at 37°C at 300 rpm. All remaining steps were conducted at room temperature. Wells were washed 4 times with PBS-T. Samples were analyzed in duplicate (30 μL/well). Standards and samples were diluted with PBS-T containing 0.1% BSA (w/v). All samples were incubated for 50 minutes at 37 °C on an orbital shaker at 300 rpm. Thereafter, wells were washed 4 times with PBS-T and 30 μL of biotinylated 4G8 (b4G8) or biotinylated 6E10 (b6E10) in PBS-T containing 0.5% (w/v) BSA was added to each well and incubated for 1 hour with shaking on an orbital shaker. Wells were washed four times and 30 μL of HRP-conjugated streptavidin (R&D, Minneapolis, MN, USA) in PBS-T containing 0.5% (w/v) BSA was added and incubated on an orbital shaker for 20 minutes. Subsequently, wells were washed 4 times with PBS-T and incubated with 50 uL of Pico Super Signal Chemiluminescence substrate (Thermo Scientific, City, USA). Chemiluminescence was measured using a Synergy H1 plate reader (Biotek, Winooski, VT, USA). Standard curves were fitted using non-linear regression using least squares (ordinary) fit with Graph Pad Prism (LaJolla, CA, USA).
4.9. MSD Aβx-42 immunoassay
This assay was performed as described before (Mably et al., 2015). All incubation steps were conducted at room temperature using an orbital shaker (Woodbridge, NJ, USA) at 300 rpm. MULTI-ARRAY 96 well black microplates (MSD, Rockville, MD, USA) were coated with 3 μg/mL of the monoclonal anti-Aβ antibody 266 in PBS, pH7.4 (40 μl per well). Thereafter, wells were blocked with 150 μL of 5% (w/v) blocker A (MSD, Rockville, MD, USA) in TBS for 1 hour. Plates were washed 3 times with TBS containing 0.05% Tween 20 (TBS-T). Incubation with guanidine HCl dissociates soluble Aβ aggregates allowing detection of Aβx-42 monomer (Mc Donald et al., 2015). Therefore, samples were analyzed with and without pre-treatment with 5 M GuHCl. In brief, 20 μL of sample was incubated with 50 μL of 7 M guanidine HCl overnight at 4 °C. Samples containing 5 M guanidine HCl were diluted 20-fold in TBS-T containing 1% (w/v) blocker A on the day of analysis. Consequently, blanks and Aβ1–42 standards were also prepared in TBS-T containing 1% (w/v) blocker A and 0.25 M guanidine HCl (5 M divided by the dilution factor of 20). After capture, wells were washed 3 times in TBS-T and incubated with biotinylated 21F12 antibody in TBS-T containing 1% (w/v) blocker A. Simultaneously, 1 μg/mL of the reporter reagent (SULFO-TAG labeled streptavidin) (MSD, Rockville, MD, USA) was added and incubated for 2 hours. Finally, wells were washed for 3 times with TBS-T and 150 μL 2 × MSD reading buffer was added to allow for electrochemiluminescence detection with a SECTOR imager (MSD, Rockville, MD, USA). The lower limit of quantification (LLoQ) was defined as the lowest standard with a signal higher than the average signal for the blank plus 9 SDs and allows a percent recovery ≥ 100 ± 20% and a coefficient of variance (CV) ≤20%. In this study, LLoQ of the assay was either 9.8 or 19.5 pg/mL.
4.10. Immunoprecipitation/Western Blot analysis
PrP-grafted antibodies were used to immunoprecipitate (IP) PrPSc from mouse brain extracts as described previously (Moroncini et al., 2004). Polyclonal goat anti-human or goat anti-mouse IgG (Fab`)2 (Thermo Scientific, Waltham, MA, USA) was conjugated to M280 Tosyl-activated paramagnetic Dynabeads (Thermo Scientific, Waltham, MA, USA) according to the manufacturer`s instructions. Beads (5 mg) were washed and resuspended in 250 μL 0.1 M sodium phosphate, pH 7.4 containing 1.2 M ammonium sulphate and 100 μg IgG. Beads were then incubated with IgG for 16 hours at 37 °C on a rotating mixer. For all IPs, half ml aliquots of brain extract were diluted 1:1 in IP buffer (phosphate buffered saline, pH 7.2 containing 1% (v/v) Triton X-100 and 1 × Complete Protease inhibitor). Each aliquot was incubated with 5 μg of PrP-grafted antibody or ICSM35 for 2 hours at 25 °C on a rotating wheel. Then, 10 μL of IgG-conjugated Dynabeads (anti-human or anti-mouse, as appropriate) was added, and incubated for another 2 hours at 25 °C. Beads were collected and washed 10 times with 1 mL of IP buffer for 10 minutes. Finally, pellets were resuspended in 20 μL of non-reducing 2× Laemmli loading buffer (150 mM Tris, pH 6.8 containing 6% SDS (w/v), 0.3% bromophenol blue (w/v), 30% glycerol (v/v)), and heated to 96 °C for 5 minutes. Samples were centrifuged for 15 seconds at 16,000 × g and electrophoresed on 4–12% pre-cast NuPAGE MES Bis-Tris gels (Thermo Scientific, Waltham, MA, USA). Proteins were transferred onto 0.2 μm PVDF (100 V for 30 minutes). PrP was detected with 6D11 (1 μg/mL) and bands visualized employing ECL.
IPs of 0.5 mL aliquots of human brain lysates were performed as described above with minor modifications. Human brain extracts were incubated with 5 μg PrP-grafted antibodies or 20 μL S97 antiserum for 2 hours at 4 °C on a rotating wheel. Then, 10 μL of IgG-conjugated paramagnetic beads (for PrP-grafted antibodies) or 30 μL of Protein A agarose beads (Merck Millipore, Burlington, MA, USA) (for S97) was added, and incubated overnight at 4 °C on a rotating wheel. All subsequent steps were carried out as described above, but proteins were transferred to 0.2 μm nitrocellulose membranes at 400 mA and 4 °C for 2 hours. Blots were microwaved in PBS, Aβ was detected using the anti-Aβ40 and anti-Aβ42 antibodies, HJ2 (1 μg/mL) and 21F12 (1 μg/mL) and visualized using the Li-COR Odyssey infrared imaging system (LiCor, Lincoln, NE, USA).
4.11. Western Blot analysis of recombinant prion protein
Proteins were electrophoresed on pre-cast 4–12% NuPAGE MES Bis-Tris gels (Thermo Scientific, Waltham, MA, USA) and transferred onto 0.2 μm nitrocellulose membranes at 400 mA for 2 hours. Blots were incubated in Odyssey blocking buffer (Li-COR, Lincoln, NE, USA) for 1 hour at room temperature, and then incubated with primary antibody in Odyssey blocking buffer (1 μg/mL MI0131 or 1 μg/mL ICSM35) overnight at 4 °C. Next day, blots were washed with phosphate buffered saline with 0.05% Tween-20 (PBS-T) 4 × 10 minutes. Membranes were then incubated with infra-red labeled anti-human or anti-mouse secondary antibodies (Li-COR, Lincoln, NE, USA) for 1 hour at room-temperature, followed by 4 × 10 minutes washing with PBS-T and a final washing step in PBS. Immunoreactive bands were visualized using the Li-COR Odyssey infrared imaging system (LiCor, Lincoln, NE, USA).
4.12. Production of induced neurons (iNs) from human induced pluripotent stem cells (iPSCs)
Neurogenin 2 (Ngn2)-induced human neurons (Zhang et al., 2013) were prepared as described in Figure 1A (Hong et al., 2018; Jin et al., 2018). In brief, YZ1 iPSCs were maintained in medium containing DMEM/F12, Knockout Serum Replacement, penicillin/streptomycin, L-glutamine, MEM-NEAA, and β-mercaptoethanol (all from Invitrogen, Carlsbad, CA, USA) with addition of 10 μg/mL basic fibroblast growth factor (bFGF; Millipore-Sigma, Burlington, MA, USA). iPSCs were plated at a density of 95,000 cells/cm2 for viral infection. Ultrapure lentiviral titers were obtained from Alstem (Richmond, CA, USA) and used at the following concentrations: Tet-O-Ngn2-puro: 0.1 μL/50,000 cells; Tet-O-FUW-EGFP: 0.05 μL/50,000 cells; FUdeltaGW-rtTA: 0.11 μL/50,000 cells. To induce Ngn-2 expression, doxycycline was added on iN day 1 at a concentration of 2 μg/mL. On iN day 2, puromycin was added at 20 μg/mL and was maintained in medium thereafter. On iN day 4, cells were plated on Matrigel (Corning, NY)-coated 96-well plates (5,000 cells/well) and maintained in Neurobasal media (Gibco, Carlsbad, CA, USA) containing Glutamax, 20% dextrose, MEM-NEAA, B27 and 10 ng/mL BDNF, CNTF, GDNF (PeproTech, Rocky Hill, NY, USA). On iN day 21, cells were used to investigate effects of AD brain extracts +/− mAbs.
4.13. Treatment of iNs with AD brain extracts and mAbs and live-cell imaging
Treatment of iNs and live-cell imaging was performed as previously described (Hong et al., 2018; Jin et al., 2018). Aliquots (2, 0.5 mL) of AD brain extracts were thawed on ice for 30–60 minutes, centrifuged at 16,000 × g for 2 minutes, and buffer-exchanged into iN medium using a 5 ml HiTrap desalting column (GE, Boston, MA, USA). Immediately prior to treatment, cells were placed in an IncuCyte Zoom live-cell imaging instrument (Essen Bioscience, Ann Arbor, MI, USA), and imaged every 2 hours for a total of 6 hours. After baseline imaging 100 μL of medium was removed from each well of iN cells leaving a total volume of ~ 100 μL. Cells were then treated with 50 μL of medium ± AD extract and a further 50 μL of medium ± test mAbs. Antibodies were tested at 0.02 μM (3 μg/mL). PrP and N1 were each tested at 0.2 and 0.02 μM, i.e., 4.6 and 0.46 μg/mL for PrP, and 1.82 and 0.18 μg/mL for N1. Cells were then returned to the IncuCyte instrument and imaged every 2 hours for a total of 72 hours. Images (Figure 3B) were analyzed using the Neural Track algorithm of IncuCyte Zoom 2016A software (Essen Bioscience, Ann Arbor, MI, USA. Neurite length were normalized to the average value measured in the 6 hours prior to addition of samples.
4.14. Statistical analysis
Statistical analyses were carried out using GraphPad Prism, version 5 (LaJolla, CA, USA). One-way ANOVA followed by Bonferroni’s post-hoc test was performed to compare means. The significance threshold was set to a two-sided p ≤ 0.05.
Supplementary Material
Highlights.
Recombinant PrP binds synthetic Aβ in an aggregation-dependent manner
IgGs into which PrP sequences 19–33 and 87–112 are grafted do not bind synthetic Aβ
PrP-grafted antibodies bind a fraction of Aβ in aqueous extracts of AD brain
PrP-grafted antibodies do not prevent Aβ-induced neuritotoxicity
PrP derivatives may be exploited to differentiate between active and inactive Aβ
Acknowledgments
This work was made possible by support to DMW from National Institutes of Health (AG047505) and Medimmune Plc. DM received support through a Research Fellowship from the Fritz Thyssen Foundation. GTC is recipient of a T32 fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None of the authors have biomedical financial interests or potential conflicts of interest.
References
- Beland M, Roucou X, 2012. The prion protein unstructured N-terminal region is a broad-spectrum molecular sensor with diverse and contrasting potential functions. J Neurochem. 120, 853–68. [DOI] [PubMed] [Google Scholar]
- Biasini E, Seegulam ME, Patti BN, Solforosi L, Medrano AZ, Christensen HM, Senatore A, Chiesa R, Williamson RA, Harris DA, 2008. Non-infectious aggregates of the prion protein react with several PrPSc-directed antibodies. J Neurochem. 105, 2190–204. [DOI] [PubMed] [Google Scholar]
- Biasini E, Turnbaugh JA, Unterberger U, Harris DA, 2012. Prion protein at the crossroads of physiology and disease. Trends Neurosci. 35, 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmalm G, Hong W, Wang Z, Liu W, O`Malley T, Sun X, Frosch MP, Selkoe DJ, Portelius E, Zetterberg H, Blennow K, Walsh DM, 2018. Identification of neurotoxic cross-linked Aβ dimers in Alzheimer brain. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yadav SP, Surewicz WK, 2010. Interaction between human prion protein and amyloid-b (Ab) oligomers: role of N-terminal residues. Journal of Biological Chemistry. 285, 26377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, Oldstone M, 2005. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 308, 1435–9. [DOI] [PubMed] [Google Scholar]
- Chiesa R, Piccardo P, Biasini E, Ghetti B, Harris DA, 2008. Aggregated, wild-type prion protein causes neurological dysfunction and synaptic abnormalities. J Neurosci. 28, 13258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett GT, Asfaw A, Hall TC, Wang Z, Liu W, Collinge J, Perkinton M, Billinton A, Walsh DM, 2018. A recipe for neurotoxicity: the prion protein, promiscuous binding, and soluble protein aggregates? Society for Neuroscience. Abstract #: 713.02, San Diego. [Google Scholar]
- Fang C, Imberdis T, Garza MC, Wille H, Harris DA, 2016. A Neuronal Culture System to Detect Prion Synaptotoxicity. PLoS Pathog. 12, e1005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluharty BR, Biasini E, Stravalaci M, Sclip A, Diomede L, Balducci C, La Vitola P, Messa M, Colombo L, Forloni G, Borsello T, Gobbi M, Harris DA, 2013. An N-terminal fragment of the prion protein binds to amyloid-b oligomers and inhibits their neurotoxicity in vivo. Journal of Biological Chemistry. 288, 7857–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freir D, Nicoll AJ, Klyubin I, Panico S, Mc Donald JM, Risse E, Asante EA, Farrow MA, Sessions RB, Saibil HR, Clarke AR, Rowan MJ, Walsh DM, Collinge J, 2011. Interaction between prion protein and toxic amyloid β assemblies can be therapeutically targeted at multiple sites. Nature Communications. 2, 1341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther EC, Strittmatter SM, 2010. Beta-amyloid oligomers and cellular prion protein in Alzheimer’s disease. J Mol Med (Berl). 88, 331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Wang Z, Liu W, O’Malley TT, Jin M, Willem M, Haass C, Frosch MP, Walsh DM, 2018. Diffusible, highly bioactive oligomers represent a critical minority of soluble Abeta in Alzheimer’s disease brain. Acta Neuropathol. 136, 19–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson GS, Hill AF, Joseph C, Hosszu L, Power A, Waltho JP, Clarke AR, Collinge J, 1999. Multiple folding pathways for heterologously expressed human prion protein. Biochim Biophys Acta. 1431, 1–13. [DOI] [PubMed] [Google Scholar]
- Jin M, O’Nuallain B, Hong W, Boyd J, Lagomarsino VN, O’Malley TT, Liu W, Vanderburg CR, Frosch MP, Young-Pearse T, Selkoe DJ, Walsh DM, 2018. An in vitro paradigm to assess potential anti-Abeta antibodies for Alzheimer’s disease. Nat Commun. 9, 2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran E, De Strooper B, 2016. The amyloid cascade hypothesis: are we poised for success or failure? J Neurochem. 139 Suppl 2, 237–252. [DOI] [PubMed] [Google Scholar]
- Lau AL, Yam AY, Michelitsch MM, Wang X, Gao C, Goodson RJ, Shimizu R, Timoteo G, Hall J, Medina-Selby A, Coit D, McCoin C, Phelps B, Wu P, Hu C, Chien D, Peretz D, 2007. Characterization of prion protein (PrP)-derived peptides that discriminate full-length PrPSc from PrPC. Proc Natl Acad Sci U S A. 104, 11551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauren J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM, 2009. Cellular prion protein mediates impairment of synaptic plasticity by amyloid b oligomers. Nature. 457, 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesne SE, 2013. Breaking the Code of Amyloid-beta Oligomers. Int J Cell Biol. 2013, 950783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mably AJ, Liu W, Mc Donald JM, Dodart JC, Bard F, Lemere CA, O’Nuallain B, Walsh DM, 2015. Anti-Abeta antibodies incapable of reducing cerebral Abeta oligomers fail to attenuate spatial reference memory deficits in J20 mice. Neurobiol Dis. 82, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mange A, Beranger F, Peoc’h K, Onodera T, Frobert Y, Lehmann S, 2004. Alpha- and beta-cleavages of the amino-terminus of the cellular prion protein. Biol Cell. 96, 125–32. [DOI] [PubMed] [Google Scholar]
- Mc Donald JM, O’Malley TT, Liu W, Mably AJ, Brinkmalm G, Portelius E, Wittbold WM 3rd, Frosch MP, Walsh DM, 2015. The aqueous phase of Alzheimer’s disease brain contains assemblies built from approximately 4 and approximately 7 kDa Abeta species. Alzheimers Dement. 11, 1286–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald AJ, Dibble JP, Evans EG, Millhauser GL, 2014. A new paradigm for enzymatic control of alpha-cleavage and beta-cleavage of the prion protein. J Biol Chem. 289, 803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroncini G, Kanu N, Solforosi L, Abalos G, Telling GC, Head M, Ironside J, Brockes JP, Burton DR, Williamson RA, 2004. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci U S A. 101, 10404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll AJ, Panico S, Freir DB, Wright D, Terry C, Risse E, Herron CE, O’Malley T, Wadsworth JD, Farrow MA, Walsh DM, Saibil HR, Collinge J, 2013. Amyloid-beta nanotubes are associated with prion protein-dependent synaptotoxicity. Nature Communications. 4, 2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieznanska H, Bandyszewska M, Surewicz K, Zajkowski T, Surewicz WK, Nieznanski K, 2018. Identification of prion protein-derived peptides of potential use in Alzheimer’s disease therapy. Biochim Biophys Acta. 1864, 2143–2153. [DOI] [PubMed] [Google Scholar]
- Nieznanski K, Choi JK, Chen S, Surewicz K, Surewicz WK, 2012. Soluble prion protein inhibits amyloid-b (Ab) fibrillization and toxicity. Journal of Biological Chemistry. 287, 33104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitskaya V, Makarava N, Bellon A, Bocharova OV, Bronstein IB, Williamson RA, Baskakov IV, 2006. Probing the conformation of the prion protein within a single amyloid fibril using a novel immunoconformational assay. J Biol Chem. 281, 15536–45. [DOI] [PubMed] [Google Scholar]
- O’Malley TT, Oktaviani NA, Zhang D, Lomakin A, O’Nuallain B, Linse S, Benedek GB, Rowan MJ, Mulder FA, Walsh DM, 2014. Abeta dimers differ from monomers in structural propensity, aggregation paths and population of synaptotoxic assemblies. Biochem J. 461, 413–26. [DOI] [PubMed] [Google Scholar]
- O’Malley TT, Witbold WM 3rd, Linse S, Walsh DM, 2016. The Aggregation Paths and Products of Abeta42 Dimers Are Distinct from Those of the Abeta42 Monomer. Biochemistry. 55, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resenberger U, Harmeier A, Woerner A, Goodman J, Muller V, Krishnan R, RM V, Kretzschmar H, Lindquist S, Hartl F, Multhaup G, Winklhofer Ka.J. T, 2011. The cellular prion protein mediates neurotoxic signalling of b-sheet-rich conformers independent of prion replication. EMBO Journal. 30, 2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J, 2011. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 470, 540–2. [DOI] [PubMed] [Google Scholar]
- Sandberg MK, Al-Doujaily H, Sharps B, De Oliveira MW, Schmidt C, Richard-Londt A, Lyall S, Linehan JM, Brandner S, Wadsworth JD, Clarke AR, Collinge J, 2014. Prion neuropathology follows the accumulation of alternate prion protein isoforms after infective titre has peaked. Nat Commun. 5, 4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, 2003. Folding proteins in fatal ways. Nature. 426, 900–4. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ, Hardy J, 2016. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 8, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B, 2005. The most infectious prion protein particles. Nature. 437, 257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Strittmatter SM, 2017. Binding Sites for Amyloid-beta Oligomers and Synaptic Toxicity. Cold Spring Harb Perspect Med. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solforosi L, Bellon A, Schaller M, Cruite JT, Abalos GC, Williamson RA, 2007. Toward molecular dissection of PrPC-PrPSc interactions. J Biol Chem. 282, 7465–71. [DOI] [PubMed] [Google Scholar]
- Soto C, 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 4, 49–60. [DOI] [PubMed] [Google Scholar]
- Stravalaci M, Tapella L, Beeg M, Rossi A, Joshi P, Pizzi E, Mazzanti M, Balducci C, Forloni G, Biasini E, Salmona M, Diomede L, Chiesa R, Gobbi M, 2016. The Anti-Prion Antibody 15B3 Detects Toxic Amyloid-beta Oligomers. J Alzheimers Dis. 53, 1485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh JA, Unterberger U, Saa P, Massignan T, Fluharty BR, Bowman FP, Miller MB, Supattapone S, Biasini E, Harris DA, 2012. The N-terminal, polybasic region of PrP(C) dictates the efficiency of prion propagation by binding to PrP(Sc). J Neurosci. 32, 8817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Nygaard HB, Heiss JK, Kostylev MA, Stagi M, Vortmeyer A, Wisniewski T, Gunther EC, Strittmatter SM, 2012. Alzheimer amyloid-beta oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat Neurosci. 15, 1227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez de la Torre A, Gay M, Vilaprinyo-Pascual S, Mazzucato R, Serra-Batiste M, Vilaseca M, Carulla N, 2018. Direct Evidence of the Presence of Cross-Linked Abeta Dimers in the Brains of Alzheimer’s Disease Patients. Anal Chem. 90, 4552–4560. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Selkoe DJ, 2016. A critical appraisal of the pathogenic protein spread hypothesis of neurodegeneration. Nat Rev Neurosci. 17, 251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jackson RJ, Hong W, Taylor WM, Corbett GT, Moreno A, Liu W, Li S, Frosch MP, Slutsky I, Young-Pearse TL, Spires-Jones TL, Walsh DM, 2017. Human Brain-Derived Abeta Oligomers Bind to Synapses and Disrupt Synaptic Activity in a Manner That Requires APP. J Neurosci. 37, 11947–11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AD, Perez KA, Rembach A, Sherrat NA, Hung LW, Johanssen T, McLean CA, Kok WM, Hutton CA, Fodero-Tavoletti M, Masters CL, Villemagne VL, Barnham KJ, 2013. Oligomers, fact or artefact? SDS-PAGE induces dimerization of beta-amyloid in human brain samples. Acta Neuropathol. 125, 549–64. [DOI] [PubMed] [Google Scholar]
- Younan ND, Sarell CJ, Davies P, Brown DR, Viles JH, 2013. The cellular prion protein traps Alzheimer’s Ab in an oligomeric form and disassembles amyloid fibers. Federation of American Societies for Experimental Biology Journal. 27, 1847–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R, von Schroetter C, Wuthrich K, 1997. Human prion proteins expressed in Escherichia coli and purified by high-affinity column refolding.FEBS Lett. 417, 400–4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Sudhof TC, 2013. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 78, 785–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.