Abstract
The c-Myb gene encodes a transcription factor that regulates cell proliferation, differentiation, and apoptosis through protein-protein interaction and transcriptional regulation of signaling pathways. The protein is frequently overexpressed in human leukemias, breast cancers, and other solid tumors suggesting that it is a bona fide oncogene. c-MYB is often overexpressed by translocation in human tumors with t(6;7)(q23;q34) resulting in c-MYB-TCRβ in T cell ALL, t(X;6)(p11;q23) with c-MYB-GATA1 in acute basophilic leukemia, and t(6;9)(q22–23;p23–24) with c-MYB-NF1B in adenoid cystic carcinoma. Antisense oligonucleotides to c-MYB were developed to purge bone marrow cells to eliminate tumor cells in leukemias. Recently small molecules that inhibit c-MYB activity have been developed to disrupt its interaction with p300. The Dmp1 (cyclin D binding myb-like protein 1; Dmtf1) gene was isolated through its virtue for binding to cyclin D2. It is a transcription factor that has a Myb-like repeat for DNA binding. The Dmtf1 protein directly binds to the Arf promoter for transactivation and physically interacts with p53 to activate the p53 pathway. The gene is hemizygously deleted in 35–42% of human cancers, and is associated with longer survival. The significances of aberrant expression of c-MYB and DMTF1 proteins in human cancers and their clinical significances are discussed.
Keywords: c-MYB, DMP1 (DMTF1), expression, leukemia, solid tumor, breast cancer, therapy, anti-sense oliodeoxynucleotides, super enhancer, small molecule
Introduction
The c-Myb proto-oncogene encodes the transcription factor that was originally identified as the cellular proto-oncogene for the v-Myb found in two different chicken leukemia viruses: Avian Myeloblastosis Virus (AMV) and E26 (1–3; Fig. 1A). The v-Myb retroviruses are highly oncogenic and are capable of transforming immature hematopoietic cells in culture and induce acute leukemias in animals. The c-Myb gene is expressed in hematopoietic cell precursors, colonic crypts, intestines, kidneys, and brain (4, 5). In humans, amplification or rearrangement of c-MYB, caused by chromosomal abnormalities in the region 6q22–24, were originally linked to acute myelogenous leukemia (6). More recently, deregulated or overexpressed c-MYB has been detected in a wide range of human cancers and is associated with poorly differentiated tumors in many cell types, including hematopoietic malignancies (7–14), breast cancers (BCs, 15–20), colon cancers (21–25), pancreatic cancers (26, 27), glioblastomas (28–30), melanomas (31–33), head and neck cancers (34) and esophageal cancers (35), vulvar cancers (36), and lacrimal gland cancers (37). The widespread association of c-Myb with many types of tumors in a diverse set of tissues suggests that it plays a critical role in tumorigenesis (38 for a review). The mechanisms of aberrant expression of c-MYB in human cancers compatible with its oncogenic role include gene amplification, chromosomal translocation, increased transcription, mRNA stabilization due to loss of microRNA-binding sites (34, 39).
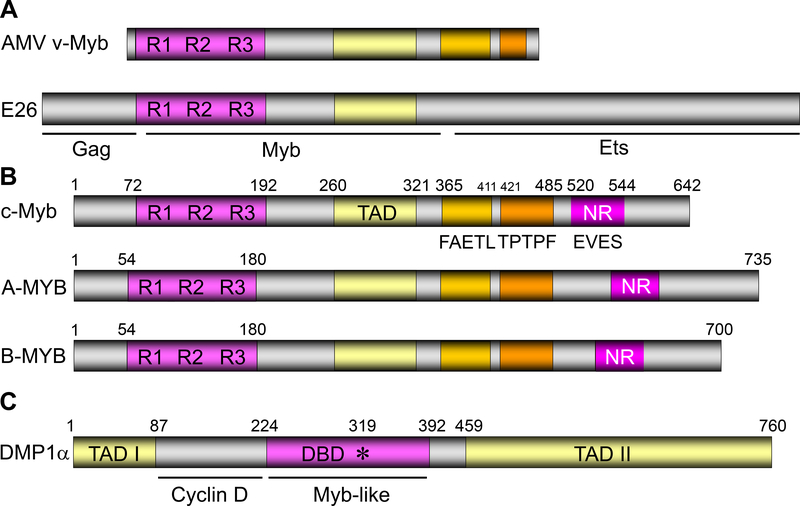
Figure 1. The domain structures of Myb-like proteins.
A. The structure of AMV and E26 proteins that encode v-Myb. R1–3 represents the three repeats within the Myb protein responsible for DNA binding. See refs. 1–3, 105–111.
B. The domain structures of A-Myb, B-Myb, and c-Myb (53, 170). TAD: transactivation domain; NR: negative regulatory domain. The “FAETL” domain (171) is required for oncogenic activity, the “TPTPF” domain conserved in the other Myb proteins, and the “EVES” domain (172) that is involved in intra‑molecular interactions and negative regulation.
C. The domain structure of the human DMTF1α protein (58). DBD: DNA-binding domain. This protein has three tandem MYB-like repeats and two transactivation domains.
Recently, three types of evidence have provided strength to the argument that c-MYB is a bona fide oncogene. The first was the finding that the c-MYB gene is frequently involved in genomic duplications in two subsets of pediatric T-cell acute lymphocytic leukemias (T-ALL) (7–9, 14). In patients harboring the reciprocal translocation t(6;7)(q23;q34), the c-MYB gene became juxtaposed to the TCRβ gene leading to c-MYB overexpression (8, 40; Fig. 2A). The second evidence comes with acute basophilic leukemia (ABL) where the c-MYB gene is overexpressed as a result of t(X;6)(p11;q23) translocation resulting in c-MYB-GATA1 expression (41, 42; Fig. 2B). The third type of evidence was the discovery of a recurrent fusion between the c-Myb and Nuclear Factor 1B (NF1B) genes in translocations t(6;9)(q22–23;p23–24), which occurs in adenoid cystic carcinomas of the breast, salivary glands of the head and neck, and other organs (17, 34, 36, 37, 38, 43; Fig. 3A). The translocation results in the expression of chimeric transcripts in which the normal 3’-UTR of the c-MYB transcript is replaced by a portion of the NF1B mRNA. The chimeric transcript lacks a number of binding sites for microRNAs that would normally down-regulate the expression of c-MYB protein, resulting in overexpressed c-MYB in human tumors (17, 34, 43). Recently, small molecules to c-MYB have been developed to disrupt its interaction with p300 promising their therapeutic effects in human cancer overexpressing c-MYB (44, 45 for review). The general consensus is that super enhancers (46, 47) to c-MYB as a consequence of chromosomal translocation cause overexpression of c-MYB resulting in carcinogenesis. These discoveries that the human c-MYB gene is involved in recurrent translocations in tumors provide evidence that c-MYB is a bona fide human proto-oncogene.
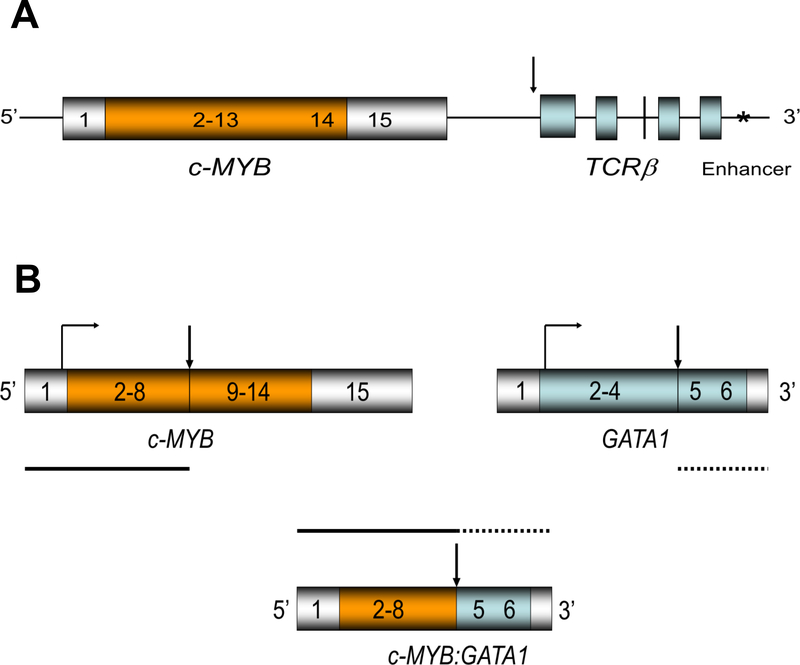
Figure 2. The genomic structures of translocations involving c-MYB.
A: The genomic structure for the chimeric c-MYB-TCRβ locus (8, 38, 40). The transcription of the entire c-MYB locus is enhanced by the telomeric enhancer for TCRβ.
B: The genomic structure for the c-MYB, GATA1, and c-MYB:GATA1 chimeric loci (39, 40) implicated in human acute basophilic leukemia. The vertical arrows represent the breakpoint of translocation.
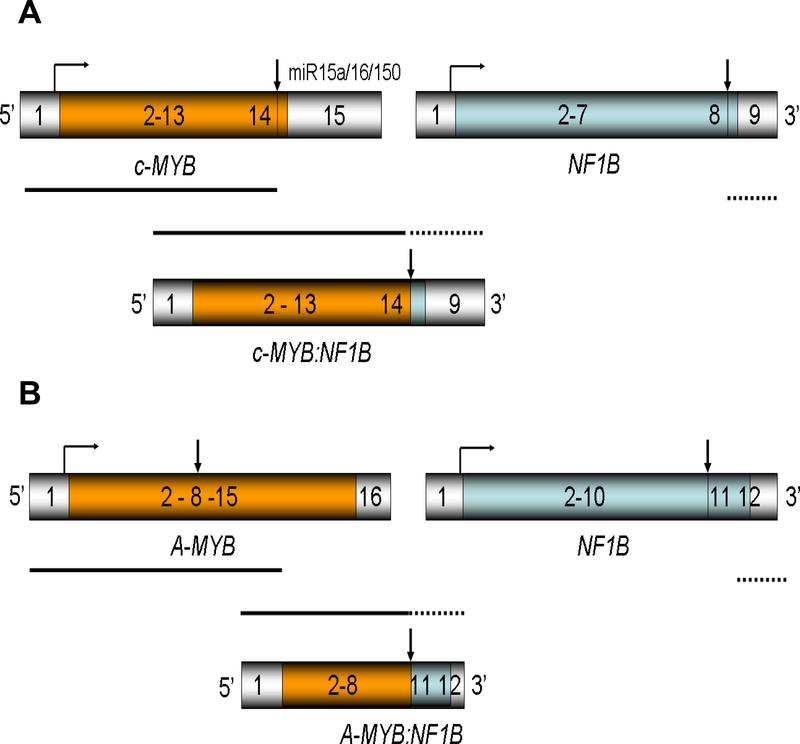
Figure 3. The genomic structures and protein products of translocations involving c-MYB and A-MYB in adenoid cystic carcinomas.
A. The genomic structure for the c-MYB, NF1B, and chimeric c-MYB:NF1B locus with t(6:9) (17, 41). The vertical arrows represent the breakpoint of translocation. This translocation has been found in adenoid cystic carcinomas in the breast, head and neck, vulva, lacrimal gland, and other organs regardless of the site of origin.
B. The genomic structure for the A-MYB, NF1B, and chimeric A-MYB:NF1B locus without t(6:9) (146).
Scientists have been engaged in the development of antisense oligonucleotides that are more effective for eliminating leukemic cells from the bone marrow (48). They have also been used to treat chronic myelogenous leukemia (CML) by systemic infusion with some success (49). Genomic abnormalities that decrease c-MYB expression have been reported in human melanomas suggesting its tumor-suppressive role in this neoplasm (50, 51).
There are two other Myb family members in mammals: A-Myb (MYBL1), B-Myb (MYBL2), in addition to c-Myb (52–56; Fig. 1B). These two genes had been isolated during screening of human cDNA libraries at low stringency with the v-Myb probe (51). Orthologues of all these three are present in both mice and humans while two vertebrates, Drosophila melanogaster and sea urchins have a single homolog of Myb (56).
The Myb-like transcription factor DMP1 (cyclin D binding Myb-like Protein 1; DMTF1; Fig. 1C) governs the activity of the ARF-p53 tumor suppressor pathway by binding to the ARF promoter in response to oncogenic stresses driven by Myc/Ras/cyclin D1 (57–73) and through physical interaction with p53 in response to DNA damage (74, 75; 76–83 for reviews of DMP1; 84–86 for reviews of ARF). DMP1 also regulates p16INK4a transcription involved in the RB tumor suppressor pathway (87). It encodes transcriptional activator of amphiregulin, thrombospondin-1, JunB, and Egr1 indicating that it is a mediator of a variety of signal transduction pathways (69). One unique feature of the DMTF1 locus is the fact that the locus generates at least three splice variants: DMTF1α, β, and γ with antagonizing activities (88, 89; 80, 90 for review). The hDMTF1α gene corresponds to mouse Dmtf1α with tumor suppressive activity (89). The hDMTF1-ARF-MDM2-p53 pathway provides cell autonomous tumor surveillance that detects and force early stage cancer cells to undergo senescence and/or apoptosis to prevent the development of cancer (71). The hDMTF1 expression is suppressed by WT1 in leukemic cells via direct binding to an EGR/SP1 site (91). Since the WT1 gene is aberrantly overexpressed in human leukemic cells, it can be used as a molecular marker of the minimal residual disease (92–99). These studies delineated a new oncogenic WT1-mediated mechanism of control of cell proliferation in the hematopoietic cells (99).
We have shown that Eμ-Myc, K-RasLA, HER2/neu, or cyclin D1-driven tumor development was significantly accelerated in both Dmtf1+/− and Dmtf1−/− mice with no significant differences in the survival between the two cohorts, suggesting that Dmtf1 is haplo-insufficient tumor suppressor (83 for review). In Eμ-Myc lymphomas, the combined frequencies of p53 mutation and Arf deletion in mice of Dmtf1+/− or Dmtf1−/− background were significantly lower than that in Dmtf1+/+ littermates, indicating that Dmtf1 is a physiological regulator of the Arf-p53 pathway in vivo (63). Consistently, Kobayashi and Srour reported that Dmtf1 regulates hematopoietic stem cell function under both steady-state and stress conditions through regulation of Arf and p21Cip1 (70).
Loss of heterozygosity (LOH) of the hDMTF1 locus was found in 42 % of human breast cancer (71) and 35 % of non-small cell lung cancer (67, 100) in mutually exclusive fashion with that of INK4a/ARF or p53 (67, 71). Recent studies suggest the critical roles of oncogenic splice variants from human genomic loci in carcinogenesis (reviewed in 80, 101–104). We found overexpression of the splice variant DMTF1β in human breast cancer primary samples and conducted clinic-pathological and transgenic mouse studies focusing on DMTF1 (88).
In this review, we will summarize the finding on aberrant c-MYB protein expressions in cancer (both overexpression and underexpression) that had been reported in the literatures focusing on c-MYB and DMTF1. We also make comments on overexpression of other MYB proteins in cancer. Understanding the mechanisms for altered expression of c-MYB and DMTF1 proteins is important not only for the understanding of pathophysiology of cancer, but also play significant roles in the development of novel therapies targeting these molecules.
Aberrant expression of c-Myb in cancer
c-Myb: cloning and expression
In 1979, Beug et al. (1) reported that cells transformed by AMV and E26 viruses resemble myeloblasts in that they weakly express Fc receptors, phagocytic capacity, and macrophage cell surface antigen, but strongly express myeloblast cell surface antigen and ATPase activity. AMV-infected cells contain two viral mRNAs: 7.5 kilobase (kb) genomic mRNA and 2.5 kb subgenomic mRNA, which contains the AMV-specific sequences, v-Myb (1). In vitro translation of AMV virion RNA size-fractionated by sucrose density gradient centrifugation yielded 76-, 56-, 48.5-, 47 -, and 32- kilo dalton (kDa) products. Three of these might represent the product of the AMV Myb gene (2). In 1982, the oncogene (v-Myb) of AMV and its cellular homolog (c-Myb: chicken) have been molecularly cloned and sequenced (105). Comparisons between the sequences of v-Myb and c-Myb indicated that transduction of c-Myb to form v-Myb possibly resulted from an initial DNA rearrangement and the subsequent use of a spliced RNA (105). The data indicate that the c-Myb gene contains at least eight exons which span a total of about 16 kbps (106).
Nucleotide sequencing has shown that mouse clones have both 5’ and 3’ to the sequences homologous to the v-Myb oncogenes of AMV and avian leukemia virus E26 (1, 2) with an open reading frame of 1,944 nucleotides encoding a protein highly homologous to the chicken c-Myb protein (106). Examination of the predicted amino acid sequence of the murine c-Myb protein revealed the presence of a 3 tandem repeat of 52 amino acid residues near the N-terminus of the protein (i.e. Myb repeats responsible for DNA-binding; Fig. 1A) having a high α-helix content, a basic region toward the N-terminus of the protein, and an overall globular configuration (Fig. 1A). c-Myb and v-Myb comparison suggested that the v-Myb protein lacks the N-terminal region of c-Myb (106–109; Fig 1A, B).
Global and conditional knockout mouse models for c-Myb
Loss of c-Myb function in mice results in embryonic lethality due to failure of fetal hepatic hematopoiesis (110). Although embryonic erythropoiesis was not impaired by the c-Myb alteration, adult-type erythropoiesis, which first takes place in the fetal liver, was greatly diminished in c-Myb mutants (110). Additional hematopoietic lineages were similarly affected. These results are consistent with a role for c-Myb in maintaining the proliferative potential of hematopoietic progenitor cells.
Since the global knockout of c-Myb was embryonic lethal, Emambokus et al. (111) generated a knockdown allele of c-Myb (c-MybLoxP/LoxP), expressing low levels of the protein, which enabled them to investigate further the involvement of c-Myb in hemopoiesis (111). Low levels of c-Myb were sufficient to allow progenitor expansion but, importantly, the progression of progenitors towards terminal differentiation was significantly impaired (111). Decreased levels of c-Myb expression allowed differentiation of macrophage and megakaryocytes, while higher levels seem to be important in the control of erythro- and lymphopoiesis. The transition from the CFU-E to erythroblasts was critically dependent on c-Myb levels (111). During T cell development, c-Myb regulated immature cell numbers and differentiation prior to expression of CD4 and CD8. Overall, their results pointed to a complex involvement of c-Myb in the regulation of proliferation and commitment within the hematopoietic hierarchy (111).
Liu and Reddy disrupted the c-Myb proto-oncogene specifically in adult bone marrow (BM; i.e. conditional knockout) to demonstrate that this transcription factor was a regulator of proliferation and differentiation of adult hematopoietic stem cells (HSCs, ref. 112). Targeted disruption of the c-Myb gene resulted in depletion of the HSC pool. In addition, BM hematopoiesis in adult mice was impaired in c-Myb-deficient cells, resulting in profound reductions of nearly all hematopoietic lineages (112). Serial BM transplantation into lethally irradiated recipient mice indicated an essential role for c-Myb in self-renewal process. In conclusion, their data indicated a critical role for c-Myb self-renewal in adult BM hematopoiesis and multi-lineage differentiation of adult HSCs (112).
The same group later showed that conditional disruption of the c-Myb proto-oncogene resulted in dramatic reductions in common myeloid progenitors (CMP; CD34+CD16/CD32−), granulocyte macrophage progenitors (GMP; CD34+CD16/CD32+), and megakaryocyte erythrocyte progenitors (MEP; CD34−CD16/CD32−) in adult mice, leading to a reduction of neutrophils, basophils, monocytes and platelets in peripheral blood (113). In short, c-Myb plays an essential role in the regulation of multiple stages in adult myeloid hemopoiesis (113).
The colonic crypt is a functional unit of the colon mucosa with a central role in ion and water reabsorption (114). The distal colonic crypt has a single stem cell at its base that gives rise to highly proliferative progenitor cells differentiating into columnar, goblet, and endocrine cells. Malaterre et al. (114) studied three genetically distinct hypomorphic c-Myb-mutant mouse strains (i.e. c-mybPlt3/plt3, c-mybPlt4/Plt4, and c-mybM303V/M303V); all of which show reduced colonic crypt size. In vivo proliferation and cell cycle marker studies suggested that these mice had a progenitor cell proliferation defect mediated in part by reduced cyclin E1 expression. To evaluate the extent to which c-Myb was required for colonic crypt homeostasis, they created a tissue-specific, mouse knockout model for c-Myb deletion, where they showed that c-Myb was required for crypt integrity, differentiation, and proliferation (114).
Mouse models for aberrant overexpression of c-Myb in thrombopoiesis
Mice null for the thrombopoietin (TPO) receptor Mpl are profoundly thrombocytopenic, as are humans with mutations in the MPL gene (115). Levin et al. (116) studied whether 5-FU produced thrombocytosis in c-Mpl−/− mice (116). They found that mice can produce a normal level of platelets after administration of 5-FU by producing large numbers of megakaryocytic, myeloid, and erythroid progenitors through a TPO - independent mechanism (116). To identify mutations for causing thrombocytopenia and to define the molecular pathways regulating platelet production, Carpinelli et al. (117) performed a suppressor screen in Mpl-null mice using N-ethyl-N-nitrosourea (ENU). They showed that mutations in the c-Myb gene caused a myeloproliferative syndrome and expansion of megakaryocytes and platelets in the absence of TPO signaling, demonstrating the utility of large scale ENU mutagenesis suppressor screens for the discovery and validation of therapeutic targets in mice (117).
Sandberg et al. (118) created knock-in mice with a homozygous Met303 to Val (M303V) mutation in the c-Myb gene where the interaction between c-Myb and the transcriptional coactivator p300 was disrupted (used in study 114 as well). The biologic consequences of the mutation included thrombocytosis, megakaryocytosis, anemia, lymphopenia, and absence of eosinophils. Detailed analysis of hematopoiesis in mutant mice revealed distinct blocks in T-cell, B-cell, and red blood cell development, as well as a 10-fold increase in the number of hematopoietic stem cells (118). Cell cycle analysis showed that twice as many hematopoietic stem cells were actively cycling in mutant mice compared with wild type mice. In conclusion, c-MYB, through its interaction with p300, controls the proliferation and differentiation of hematopoietic stem and progenitor cells (118).
Overexpression of c-MYB in human cancer
c-MYB: human chromosomal mapping and cytogenetics
In 1982, Dalla-Favera et al. (119) assigned the c-MYB proto-oncogene to chromosome 6 through study of somatic cell hybrids. Harper et al. (120) then conducted detailed mapping of the c-MYB locus to chromosome 6q22–6q24 by in situ hybridization, which was confirmed by Winqvist et al. (121) and Janssen et al. (122). These studies indicated that the c-MYB gene was localized at the breakpoints of translocations frequently involved in T-cell acute lymphatic leukemia (T-ALL), ovarian cancers, and melanomas.
c-MYB expression in human hematopoietic malignancies – role of translocations
It has been reported that Alu repeat clusters act as mediators of recurrent chromosomal alterations in tumors (123). The human c-MYB locus was found to be flanked by 257-bp Alu repeats and the duplication was mediated somatically by homologous recombination between the flanking Alu elements on sister chromatids (124). This Alu - mediated c-MYB tandem duplication could be one of the mechanisms for genomic duplication for c-MYB in human T-ALL (8, 14), MYST3-linked acute myeloid leukemia (AML; ref. 11), and BRCA1-mutated BC (15).
Overexpression of the c-MYB gene has been reported most frequently in human hematopoietic malignancies. Barletta et al. and others (125, 126) found that deletions of the long arm of chromosome 6 (6q-), was frequently found in acute lymphoblastic leukemias (ALLs), non-Hodgkin lymphomas, and myeloid leukemias with high levels of c-MYB expression although the c-MYB locus was not allelic. The c-MYB gene was retained on 6q22, which was bordered by chromosomal breakpoints in both interstitial and terminal 6q- deletions.
In human T-ALL, recurrent chromosomal translocations t(6;7)(q23;q34) involving the TCRβ and c-MYB loci have been reported (8, 38; Fig. 2A). This translocation led to the juxtaposition of the c-MYB proto-oncogene near the TCRβ regulatory sequence suggesting aberrant expression of the protein (Fig. 2A). Moreover, Lahortiga et al. (9) reported that a duplication of the c-MYB gene was found in 8.4 % of patients with T-ALL, associated with a 3-fold increase in c-MYB expression. Consistently, knockdown of c-MYB initiated T cell differentiation indicating that c-MYB depletion is a therapeutic target for T-ALL.
Acute basophilic leukemia (ABL) is a rare subtype of acute leukemia with clinical features related to hyperhistaminemia because of excessive growth of basophils. Cases of t(X;6)(p11;q23) translocation have been reported in this disease (41). Quelen et al. (42) reported the four cases of ABL with a t(X;6)(p11;q23) translocation occurring in infants. The in situ hybridization and rapid amplification of cDNA ends revealed that the translocation generated a MYB-GATA1 fusion gene (42; Fig. 2B). Expression of MYB-GATA1 in mouse lineage(−) cells committed them to the granulocyte lineage and blocked them at an early stage of differentiation. These results establish a link between chromosomal translocation and the development of ABL with t(X;6)(p11;q23) translocation (42).
Although human cancers have complex genotypes and genomically unstable, they are often dependent on the continued presence of oncogenic mutation(s) that caused the disease - a phenomenon called “oncogene addiction” (127). Such dependencies have also been demonstrated in mouse models, where conditional expression of oncogenes to initiate cancer is required for tumor maintenance and progression; thus they can be used as pathways for therapeutic targets (e.g. dox-neuNT; refs. 128, 129). Zuber et al. (127) performed an integrative approach that combines genetically modified mouse models, transcriptional profilings, and inducible RNAis to characterize cellular programs that underlie addiction to the MLL-AF9 fusion protein found in aggressive AML. They showed that MLL-AF9 strengthened c-Myb-governed programs for aberrant self-renewal associates with leukemic stem cell activity and poor prognosis in AML (127). c-Myb suppression precisely mimicked MLL-AF9 withdrawal and eradicated aggressive AML in vivo without affecting normal myelopoiesis, indicating that strategies to inhibit Myb-dependent self-renewal programs are promising in cancer therapeutics (127).
Self-renewal is a characteristic of both hematopoietic stem cells (HSCs) and leukemic stem cells (LSCs); therefore, the identification of mechanisms required their functions could provide therapeutic opportunities that are more effective and less toxic than current ones. Zhu et al. (130) performed an in vivo short hairpin RNA (shRNA) screen and identified jumonji domain - containing protein JMJD1C as an important driver of MLL-AF9 leukemia. Using a conditional mouse model, they showed that loss of Jmjd1c substantially decreased LSC frequency and caused differentiation of MLL-AF9- and homeobox A9 (HOXA9)- driven leukemias (130). They found that JMJD1C directly interacted with HOXA9 and modulated a HOXA9 - controlled gene expression program. In contrast, loss of Jmjd1c led to only minor defects in blood homeostasis and modest effects on HSC self-renewal (130). Together, these data establish JMJD1C as an important mediator of MLL-AF9- and HOXA9- driven LSC function that is largely dispensable for normal HSC function.
Sroczynska et al. (131) used a mouse model of human AML induced by the MLL-AF9 fusion oncogene; they also performed screening with a shRNA library to find novel drug targets. One of the best candidate drug targets identified in these screens was again Jmjd1c. Depletion of JMJD1C impaired the growth and colony formation of mouse MLL-AF9 cells in vitro (131). Depletion of JMJD1C impaired expansion and colony formation of human leukemic cell lines, with the strongest effect observed in the MLL-rearranged ALL cell line. The growth defect upon JMJD1C depletion was caused by increased apoptosis indicating that JMJD1C as a potential therapeutic target for human leukemias (131). Overexpression of c-Myb or c-Myc significantly provided a growth advantage over Jmjd1c-depleted cells, indicating the functional relationship between c-Myb/c-Myc and Jmjd1c in colony formation of leukemic cells (131).
Antisense oligonucleotides to c-MYB in cancer therapy
For the past decades, scientists have been engaged in the development of antisense oligodeoxynucleotide (ODN) drugs that might be more effective for the treatment of leukemias and other human malignancies. c-MYB, c-MYC, and BCR/ABL genes have been chosen since they are often overexpressed in human malignancies and have short half-life mRNAs/proteins.
Since the c-Myb gene encodes proteins that are critical for hematopoietic cell proliferation and development, Gewirtz et al. (132) developed a model for testing the in vivo efficacy of phosphorothioate antisense ODNs for disrupting c-MYB. Their model was a human leukemia - SCID mouse chimera with K562 cells derived from a patient with chronic myelogenous leukemia (CML). The tumor cells carried the Philadelphia chromosome: Ph1 and the BCR/ABL hybrid gene (133, 134) that could be used to track human cells in the mouse host. Animals treated with antisense c-MYB survived at least 3.5 times longer than controls. Moreover, animals receiving antisense c-MYB DNA had significantly less disease at sites most frequently involved by leukemic cell infiltration (CNS and ovary), promising the future direction of AS-ODN therapy (134).
ODNs to c-MYB had been developed to purge marrow autografts administered to allograft BMT-ineligible CML patients. Luger et al. (135) purged CD34(+) marrow cells with ODN to c-MYB for either 24 or 72 hours. After treatment, c-MYB mRNA levels declined significantly in ~50 % of patients. Analysis of BCR/ABL expression in long-term culture-initiating cells suggested that purging had been successful in more than 50 % of patients. Overall 6 of 14 patients (43 %) had achieved a major cytogenetic response (135). These results lead to the speculation that enhanced delivery of ODNs targeted to critical proteins of short half-life lead to the development of effective therapeutic agents in the future.
In the Phase I systemic infusion study, Myb AS-ODN was delivered by continuous infusion at dose levels ranging between 0.3 to 2 mg/kg/day for 2 months (132). One BC patient survived ~14 months with transient restoration of chronic phase disease. These studies indicate that ODN may be administered safely to leukemic patients through systemic infusion, which may eventually demonstrate therapeutic utility in the treatment of human leukemias (132, 135). The Gewirtz lab also reported the increase efficiency with phosphorothioated 2’-deoxy-2’-fluoro-beta-D-arabinonucleic acid modified antisense c-MYB ODNs in leukemic cell killing in comparison to unmodified ODNs (136). The modifications and synergisms with other antisense therapy should be pursued further to increase the therapy of antisense c-MYB in leukemia therapy. Although antisense MYB therapy have been studied mostly in CML due to the presence of universal breakpoint marker BCR/ABL, the possibility for purging should be pursued in human AMLs using other breakpoint markers such as PML/RARA (137), AML1/ETO (138), or by using WT1 as an universal marker for human acute leukemias (92, 94, 96–98), the expression of which increases on leukemic relapse (95).
Small molecules that antagonize c-MYB activity
Although these antisense approaches to c-MYB looked very promising in leukemia therapy, it was not pursued later clinically to remove leukemic cells in patients. Instead, small molecules to inhibit c-MYB activity, e.g. by disrupting interactions between c-MYB and its co-regulator p300, has been was attempted to destabilizing the protein itself (139–142; 143 for review). Uttakar et al. (139) identified the triterpenoid Celastrol as a potent low-molecular-weight inhibitor of the interaction of Myb with its cooperation partner p300. They reported that Celastrol inhibited the proliferative of acute myeloid leukemia (AML) cells while not affecting normal hematopoietic progenitor cells. Moreover, Celastrol prolonged the survival of mice in a model of an aggressive AML. Their work demonstrates the therapeutic potential of a small molecule inhibitor Celastrol that disrupt the Myb-p300 interaction for the treatment of AML through a combination of molecular biological, chemical biological, and in vivo experiments with primary samples from AML patients (139). The same group later proved the effectiveness of nathoquinone blumbagin using the same method (140). These outstanding studies provide a starting point for the further development of Myb-inhibitory compounds for the treatment of leukemia and possibly other tumors driven by deregulated Myb.
Coulibaly et al. (141) showed that helenalin acetate inhibited C/EBPβ by binding to the N-terminal part of C/EBPβ, thereby disrupting the cooperation of C/EBPβ with the co-activator p300. They reported that helenalin acetate is the first small-molecule C/EBPβ inhibitor by direct binding (141). The same group extended the study to clarify the mechanism of effectiveness of the c-Myb inhibitor Celastrol in cancer therapy. They became aware that the reporter system used for c-Myb inhibitor screening also responded to inhibition of C/EBPβ, a transcription factor known to cooperate with c-Myb in myeloid cells (142, 143). They found that Celastrol strongly inhibited the activity of C/EBPβ by disrupting its interaction with the Taz2 domain of p300. Helanalin Acetate independently targets c-Myb and C/EBPβ by disrupting the interaction of both transcription factors with p300 (142). c-Myb, C/EBPβ, and p300 cooperate in myeloid-specific gene expression and are associated with ‘super-enhancers’ (46, 47) in AML cells that have been implicated in the maintenance of the leukemia (144, 145). They posit that the ability of Celastrol to disrupt the activity of a transcriptional Myb-C/EBPβ-p300 module might explain its promising anti-leukemic activity (142).
MYB-NF1B translocation and c-MYB overexpression in adenoid cystic carcinomas (ACCs)
Chromosomal translocations involving the c-MYB gene have been reported in other types of human tumors than leukemias. The t(6;9)(q22–23;p23–24) translocation was found in adenoid cystic carcinomas (ACCs) of the breast, head and neck, vulva, lacrimal gland and other tissues (17, 36, 37), which consistently result in MYB-NF1B fusion transcripts consisting of c-MYB exon 14 linked to the last coding exon of nuclear factor 1B (NF1B). This leads to loss of c-MYB exon 15, which encodes the 3’-UTR, where several highly conserved target sites for microRNAs (miR15a/16 and miR-150) are located (Fig. 3A). Thus, the translocation appears to deregulate c-MYB by removing the microRNA binding sites, and the product is different from that of wild type c-Myb.
Mitani et al. (34) did an extensive analysis with 123 primary tumors of the salivary gland, including primary and metastatic ACCs, and non-ACC salivary carcinomas. The c-MYB-NF1B fusion genes were identified by reverse transcriptase-PCR. The MYB-NF1B fusion was detected in 28% primary and 35% metastatic ACCs, but not in any of the non-ACC salivary carcinomas (34). Different exons in both the c-MYB [13, 8b, 11, 15, 9b, 8a, 16 (the order of frequency)] and NF1B [12, 11, 9, 9’] genes were involved in the fusions, resulting in expression of multiple chimeric variants (34). c-MYB was overexpressed in the vast majority of the ACCs although c-MYB expression was significantly higher in tumors carrying the c-MYB-NF1B fusion. They concluded that the c-MYB-NFIB fusion characterizes a subset of ACCs contributing to c-MYB overexpression (34).
In 2016, Mitani et al. (146) reported the genetic alterations in ACC lacking the classical translocation and fusion transcript for t(6;9)(q22–23;p23–24) and identified new abnormalities in translocation (+) tumors (reviewed in 147, 148) (Fig. 3B). They identified a novel MYBL1-NF1B (i.e. A-MYB-NF1B) gene fusion as a result of t(8;9) translocation and multiple rearrangements in the MYBL1 (A-MYB) gene in 35 % of the t(6;9)(−) ACC (146; Fig. 3B). All MYBL1 alterations involved deletion of the C-terminal negative regulatory domain showed high MYBL1 expression (146). Reciprocal relationship between c-MYB and MYBL1 (A-MYB) expression was consistently found in ACC. The breakpoints in the four tumors with MYBL1-NF1B fusions were located in introns 8 and 14 of the MYBL1 gene and in intron 10 of the NF1B gene. Accordingly, only the last two exons of NF1B were part of the gene fusions (146; Fig. 3B). Three of the t(6:9)(+)/MYB-NF1B (−) tumors had the 5’ end of the NF1B gene fused to different gene partners (non-MYB/MYBL1) including XRCC4, NKAIN2, PTPRD and AIG1, suggesting a biologic importance of the C-terminal part of these fusions (146). The role of MYBL1 in translocation in ACC was also reported by a different group (149). Brayer et al. (149) analyzed ACC tumors with t(8;9) and t(8;14) translocations and found that the MYBL1 (A-MYB) gene was fused to the NFIB and RAD51B genes, respectively. Interestingly, tumors with c-MYB and MYBL1 translocations showed similar gene expression profiles in RNA-seq, correlated with clinical outcomes, suggesting that the related MYB proteins are interchangeable oncogenic drivers in ACC (149).
Driver of c-MYB expression in human cancer
Accumulating studies show that c-MYB overexpression itself caused by translocation rather than creation of chimeric protein by translocation is essential for the pathogenesis of ACC (150, 151). Drier et al. (151) identified the juxtaposition of super-enhancer regions to the c-MYB locus as the unifying feature of ACC translocations. Detailed genomic and epigenomic analyses of ACCs revealed alternate rearrangements that translocated super-enhancers in the NFIB and TGFBR3 loci either upstream or downstream of the c-MYB gene. c-MYB protein bound these super-enhancers, which loop to the c-MYB promoter, thereby establishing a positive feedback loop that sustains expression of this master regulator (151). They also showed that c-MYB bound to a larger repertoire of enhancers genome-wide, which appeared to support alternate ACC expression signatures in the myoepithelial and luminal epithelial compartments of ACC. Consistently, BET bromodomain inhibitors, which disrupt enhancer functions, slowed tumor growth in ACC primagraft models in vivo (151). In conclusion, the major driver of c-MYB overexpression in cancer is the translocation of enhancer(s), with the protein fusion event playing a minor part. The situation is similar to WT1 overexpression in human leukemias where the gene overexpression itself rather than translocation(s) play a major role in leukemogenesis (92–99).
Expression of c-MYB and MYB-like protein DMTF1 in breast cancer
Breast cancer (BC) is important for c-MYB because 1) ACC is found in BC, 2) c-MYB expression correlates strongly with ER positivity (19, 20). This observation was confirmed by analysis of data from studies of a large number of tumors by using microarray expression profiling (16). In many cell types, c-MYB expression appears to be regulated by transcriptional repression in the first intron (152), which involves a region potentially capable of forming a stem-loop structure in the transcript and an adjacent poly(dT) region (153, 154). Intriguingly, this motif is frequently mutated in colon carcinomas, but not in BC (153), suggesting that another mechanism is responsible for overcoming attenuation in BC cells. Elevated levels of c-MYB mRNA in human cancers can also be caused by genomic amplification in hereditary BRCA1(+) breast cancer (15, 155).
To determine the frequency and patterns of inactivation of the hDMTF1-ARF-Hdm2-p53 pathway in human BCs, we extracted DNA from 110 pairs of clinical samples and conducted LOH analyses for hDMTF1, INK4a/ARF, p53, and gene copy number assay for Hdm2 (71). LOH for as found in 27 samples with the 5’ probe (25 %), 30 cases (27 %) with the 3’ probe, and 46 of 110 cases (42 %) with either the 5’ or 3’ probes. None of the 61 samples we studied showed methylation of the hDMTF1 promoter (71) suggesting that complete inactivation of hDMTF1 does not happen in tumor cells. Detailed mapping of the genomic fragment deleted in BC showed that gene deletion was limited to the hDMTF1 locus in 30 of 32 cases of LOH (94 %) (71), suggesting that the gene deletion was selective to the hDMTF1 locus (71). With INK4a/ARF probes, LOH or homozygous deletion was detectable in 19 cases with the 5’ probe (17 %), 10 cases (9 %) with the 3’ probe, and 22 of 110 (20 %) with either of these. Likewise, LOH for the TP53 locus was detectable in 22 cases (20 %) with the 5’ probe, 30 with the 3’ probe (27 %), and 37 of 110 (34 %) with either of these (71). Overexpression of the p53 protein by immunohistochemistry was found in 6 of 13 p53 LOH(+) cases (46 %), but not in any of the p53 LOH(−) BCs, consistent with the previous report that showed association of p53 mutations with loss of the p53 allele in BC (156). LOH for hDMTF1 and INK4a/ARF was found to be mutually exclusive in 62 of 65 cases (95 %, p = 0.0027, χ2 = 9.0) (71). Likewise, LOH for hDMTF1 and p53 was also mutually exclusive in 63 of 73 cases (86 %, p = 0.025, χ2 = 5.0). The Hdm2 gene amplification was found in 14 of 110 samples (13 %), which occurred independently of the LOH for hDMTF1. Thus, our data demonstrate that 1) LOH for hDMTF1 is frequently found in BCs with wild-type INK4a/ARF and p53 genomic loci, and 2) LOH for hDMTF1 and Hdm2 amplification occur at random. We also found significant correlation between cases with hDMTF1 LOH (+) BC with low Ki67 expression (p = 0.027, χ2 = 4.9) and diploid content of genomic DNA (p = 0.046, χ2 = 4.0). Conversely, BCs with LOH for p53 were associated with high Ki67 (p = 0.015, χ2 = 5.9) and aneuploidy (p = 0.014, χ2 = 6.0) suggesting their association with specific BC subtype(s) with poor prognosis. Indeed we found that hDMTF1 LOH (+) BCs were significantly associated with luminal A group of BCs (p = 0.0085; χ2 = 6.9) while p53 LOH(+) BCs were associated with non-luminal A subtype (p = 0.023; χ2= 5.1) (71).
Consistent with these findings, BCs with LOH for hDMTF1 had longer relapse-free survival than those without LOH (p = 0.0092, χ2 = 6.8) (71). Conversely, LOH for p53 had negative impact on patients’ disease-free survival (p = 0.021, χ2 = 5.4) (71) consistent with our observation that ~50 % of p53 LOH cases showed mutation of the remaining p53 allele (71). LOH for INK4a/ARF had no impact on patients’ survival while BC with Hdm2 amplification showed significantly shorter survival than those without gene amplification (p = 0.022, χ2 = 5.3) (71). Together, our data indicate that the more downstream the molecule is localized in DMTF1-ARF-Hdm2-p53 signaling, the more negative impact the marker shows on BC patients’ survival.
Correlation of DMTF1 protein expression with hDMTF1 LOH and HER2 status in human BC
BC samples without LOH for hDMTF1 showed more intense nuclear staining for hDMTF1 (mostly grades 2–3) while tumors with LOH showed weaker staining (mostly grades 0–1) (p = 0.0006). Normal breast epithelial cells also showed weak (1+) hDMTF1 staining. We found a significant increase in hDMTF1 staining in BCs that showed HER2 overexpression (2+ or 3+) (p = 0.0038), regardless of the genomic status for hDMTF1 (68, 81). Together, our data show that: 1) hDMTF1 protein is downregulated in clinical samples that showed LOH for hDMTF1 and 2) HER2 and hDMTF1 protein expression levels are positively correlated.
The impact of hemizygous loss of DMTF1 on breast cancer or mammary tumors
Our study shows that LOH for hDMTF1 is associated with low Ki67 index and increased frequency of diploid DNA, both of which are indicators for favorable prognoses of BCs (157–159). Nevertheless, in wild type MMTV-neu tumors, loss of Dmtf1 was associated with higher histological grades with increased local invasion (68), and thus with more aggressive disease than wild type tumors without Dmtf1 involvement. Likewise, K-RasLA or cyclin D1-driven mouse models of cancer exhibit more aggressive tumors with invasion/metastasis in Dmtf1-deficient mice (67, 72) when p53 is wild type. Moreover our GeneChip Microarray (69) showed that i) Dmtf1α upregulates Thbs1, an inhibitor of angiogenesis, ii) it increases the transcription of Egr1 that inhibits cancer metastasis through regulation of TGFβ1, PTEN, and p53 (160). The differential effects of Dmtf1 for human BC and mouse mammary tumors in tumor aggressiveness can be explained by the fact most of the BC patients without LOH for hDMTF1 showed the involvement of the TP53 and/or INK4a/ARF locus; the deletion of both of which is associated with poor prognoses of patients (161–164). By contrast, involvement of the p53 or Ink4a/Arf locus is rare in mammary tumors from MMTV-neuNT mice (68). Instead, these mouse tumors often overexpressed Ink4a/Arf repressors such as Tbx2 or Pokemon (76) to inactivate the Rb and p53 pathways.
The low incidence of p53 or Ink4a/Arf mutation in MMTV-neuNT tumors does not mean that p53 or Ink4a/Arf has no role in HER2/neu-induced mammary tumorigenesis in mice. Indeed, it was reported that MMTV-neu (wild type); WAP-p53–172H double transgenic mice exhibited dramatically shortened survival than in MMTV-neu single-transgenic mice, with tumors showing anaplastic and aneuploid phenotypes indicating strong cooperativity of ErbB2 and mutant p53 in tumor development (165, 166). It was also reported that MMTV-neu; Ink4a/Arf+/− mammary tumors showed increased Ki67 expression, higher expression of cyclin D1, and decreased mammary tumor apoptosis as compared with those from MMTV-neu mice (167). Thus, tumors with inactivation of any components of the Dmtf1-Arf-p53 pathway lead to more aggressive phenotypes than those without involvement of the pathway in mice. In human BCs, phenotypic comparison of tumors with or without the involvement of the DMTF1-ARF-p53 pathway is very difficult since one of these components is almost always inactivated (51/66, 77 % in our study) in clinical samples (71).
Concluding remarks and future prospects
Among Myb-like proteins, c-Myb is by far the most intensively studied for their roles in signal transduction related to cell proliferation, apoptosis, and differentiation. The c-MYB genomic locus has been mapped to the breakpoint that is frequently translocated in human leukemia and solid tumors; hence the gene product is overexpressed in human cancers including hematopoietic malignancies and carcinoma. Since leukemic cells are often dependent on c-MYB for their growth, the gene has been a target for removal of leukemic cells from the bone marrow needed to autologous bone marrow transplantation for CML. Development of antisense ODNs to other leukemias than CML has not even been undertaken, esp. treatment of acute leukemias through continuous intravenous infusion of antisense ODNs. One major obstacle for antisense ODN-mediated cancer therapy is the stability of the ODNs in vivo. Recently morpholino-modified ODNs have shown outstanding stability in vivo, which should be studied further together with collaboration of several antisense ODNs that have anti leukemic or tumor cell activity.
Although these antisense approaches to c-MYB looked very promising in leukemia therapy, it was not pursued clinically to remove leukemic cells in patients. Instead, small molecules to inhibit c-MYB activity, e.g. by disrupting interactions between c-MYB and its co-regulator p300, has been was attempted to destabilizing the protein itself. Since the approaches look very promising for cancer therapy, clinical trials for small inhibitors celastrol and/or blumbagin should begin soon for future cancer therapy.
Recent studies show that c-MYB overexpression itself caused by translocation rather than creation of chimeric protein is essential for the pathogenesis of human cancer (150, 151). Indeed, juxtaposition of super-enhancer regions to the c-MYB locus has been identified as the unifying feature of ACC translocations. Whatever the mechanism is, c-MYB overexpression itself is the most important event in carcinogenesis than creation of fusion protein(s) by translocation, which can be targeted by immune therapy or small molecule inhibitors.
In contrast to c-MYB, the DMTF1 transcription factor is often (30–40 %) underexpressed in human cancers, due to specific LOH of the locus (67, 71). Both gene knockout and transgenic mouse models (62, 88; 168, 169) have been created for DMTF1 to demonstrate the tumor suppressive and oncogenic functions of the splice variants. Interestingly LOH of hDMTF1 has been reported to be mutually exclusive of those of the INK4a/ARF or the p53 locus in lung and breast cancers, suggesting that DMTF1-loss a new disease category of cancer. Since the disease-free survival is longer in BC with LOH for hDMTF1, it is highly possible that tumors that overexpress hDMTF1 have p53 mutation(s) associated with poor prognosis. Indeed, Dmp1 can bind to both wild type and mutant p53. Future research can thus focus on the role of hDMTF1 in tumors with p53 mutation(s).
Acknowledgements
We thank all other members of Dr. Inoue’s lab for sharing unpublished research data.
Financial Support
K. Inoue was supported by NIH/NCI 2R01CA106314, ACS RSG-07–207-01-MGO, and KG080179.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Literature Cited
- 1.Beug H, von Kirchbach A, Döderlein G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 1979;18:375–390. [DOI] [PubMed] [Google Scholar]
- 2.Anderson SM, Chen JH. In vitro translation of avian myeloblastosis virus RNA. J Virol 1981;40:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Duesberg P. Avian erythroblastosis virus E26: only one (myb) of two cell-derived coding regions is necessary for oncogenicity. Proc Natl Acad Sci USA 1994;91:4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zorbas M, Sicurella C, Bertoncello I, Venter D, Ellis S, Mucenski ML, et al. c-Myb is critical for murine colon development. Oncogene 1999;18:5821–5830. [DOI] [PubMed] [Google Scholar]
- 5.Shin DH, Lee HW, Jeon GS, Lee HY, Lee KH, Cho SS. Constitutive expression of c-myb mRNA in the adult rat brain. Brain Res 2001;892:203–207. [DOI] [PubMed] [Google Scholar]
- 6.Pelicci PG, Lanfrancone L, Brathwaite MD, Wolman SR, Dalla-Favera R. Amplification of the c-myb oncogene in a case of human acute myelogenous leukemia. Science 1984;224:1117–1121. [DOI] [PubMed] [Google Scholar]
- 7.Poenitz N, Simon-Ackermann J, Gratchev A, Qadoumi M, Klemke CD, Stadler R, et al. Overexpression of c-myb in leukaemic and non-leukaemic variants of cutaneous T-cell lymphoma. Dermatology 2005;211:84–92. [DOI] [PubMed] [Google Scholar]
- 8.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood 2007;110:1251–1261. [DOI] [PubMed] [Google Scholar]
- 9.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet 2007;39:593–595. [DOI] [PubMed] [Google Scholar]
- 10.O’Rourke JP, Ness SA. Alternative RNA splicing produces multiple forms of c-Myb with unique transcriptional activities. Mol Cell Biol 2008;28:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murati A, Gervais C, Carbuccia N, Finetti P, Cervera N, et al. Genome profiling of acute myelomonocytic leukemia: alteration of the MYB locus in MYST3-linked cases. Leukemia 2009;23:85–94. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood 2009;113:505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou YE, O’Rourke JP, Edwards JS, Ness SA. Single molecule analysis of c-myb alternative splicing reveals novel classifiers for precursor B-ALL. PLoS One 2011;6:e22880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano K, Uchimaru K, Utsunomiya A, Yamaguchi K, Watanabe T. Dysregulation of c-Myb Pathway by Aberrant Expression of Proto-oncogene MYB Provides the Basis for Malignancy in Adult T-cell Leukemia/lymphoma Cells. Clin Cancer Res 2016;22:5915–5928. [DOI] [PubMed] [Google Scholar]
- 15.Kauraniemi P, Hedenfalk I, Persson K, Duggan DJ, Tanner M, Johannsson O, et al. MYB oncogene amplification in hereditary BRCA1 breast cancer. Cancer Res 2000;60:5323–5328. [PubMed] [Google Scholar]
- 16.Nicolau M, Levine AJ, Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci USA 2011;108:7265–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NF1B transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA 2009;106:18740–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Jin K, van Pelt GW, van Dam H, Yu X, Mesker WE, et al. c-Myb Enhances Breast Cancer Invasion and Metastasis through the Wnt/β-Catenin/Axin2 Pathway. Cancer Res. 2016;76:3364–3375. [DOI] [PubMed] [Google Scholar]
- 19.Guérin M, Sheng ZM, Andrieu N, Riou G. Strong association between c-myb and oestrogen-receptor expression in human breast cancer. Oncogene 1990;5:131–135. [PubMed] [Google Scholar]
- 20.Gudas JM, Klein RC, Oka M, Cowan KH. Posttranscriptional regulation of the c-myb proto-oncogene in estrogen receptor-positive breast cancer cells. Clin Cancer Res 1995;1:235–243. [PubMed] [Google Scholar]
- 21.Torelli G, Venturelli D, Colo A, Zanni C, Selleri L, Moretti L, et al. Expression of c-myb protooncogene and other cell cycle-related genes in normal and neoplastic human colonic mucosa. Cancer Res 1987;47:5266–5269. [PubMed] [Google Scholar]
- 22.Ramsay RG, Thompson MA, Hayman JA, Reid G, Gonda TJ, Whitehead RH. Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ 1992;3:723–730. [PubMed] [Google Scholar]
- 23.Biroccio A, Benassi B, D’Agnano I, D’Angelo C, Buglioni S, Mottolese M, et al. c-Myb and Bcl-x overexpression predicts poor prognosis in colorectal cancer: clinical and experimental findings. Am J Pathol 2001;158:1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abaza MS, Al-Attiyah RJ, Al-Saffar AM, Al-Sawan SM, Moussa NM. Antisense oligodeoxynucleotide directed against c-myb has anticancer activity and potentiates the antiproliferative effect of conventional anticancer drugs acting by different mechanisms in human colorectal cancer cells. Tumour Biol 2003;24:241–257. [DOI] [PubMed] [Google Scholar]
- 25.Hugo H, Cures A, Suraweera N, Drabsch Y, Purcell D, Mantamadiotis T, et al. Mutations in the MYB intron 1 regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer 2006;45:1143–1154. [DOI] [PubMed] [Google Scholar]
- 26.Wallrapp C, Muller-Pillasch F, Solinas-Toldo S, Lichter P, Friess H, Buchler M, et al. Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res 1997;57:3135–3139. [PubMed] [Google Scholar]
- 27.Yang K, He M, Cai Z, Ni C, Deng J, Ta N, et al. A decrease in miR-150 regulates the malignancy of pancreatic cancer by targeting c-Myb and MUC4. Pancreas 2015;44:370–379. [DOI] [PubMed] [Google Scholar]
- 28.Welter C, Henn W, Theisinger B, Fischer H, Zang KD, Blin N. The cellular myb oncogene is amplified, rearranged and activated in human glioblastoma cell lines. Cancer Lett 1990;52:57–62. [DOI] [PubMed] [Google Scholar]
- 29.Patt S, Thiel G, Maas S, Lozanova T, Prosenc N, Cervos-Navarro J, et al. Chromosomal changes and correspondingly altered proto-oncogene expression in human gliomas. Value of combined cytogenetic and molecular genetic analysis. Anticancer Res 1993;13:113–118. [PubMed] [Google Scholar]
- 30.Miyazaki T, Pan Y, Joshi K, Purohit D, Hu B, Demir H, et al. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin Cancer Res 2012;18:1268–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dasgupta P, Linnenbach AJ, Giaccia AJ, Stamato TD, Reddy EP. Molecular cloning of the breakpoint region on chromosome 6 in cutaneous malignant melanoma: evidence for deletion in the c-myb locus and translocation of a segment of chromosome 12. Oncogene 1989;4:1201–1205. [PubMed] [Google Scholar]
- 32.Walker MJ, Silliman E, Dayton MA, Lang JC. The expression of C-myb in human metastatic melanoma cell lines and specimens. Anticancer Res 1998;18:1129–1135. [PubMed] [Google Scholar]
- 33.Morey AL, Murali R, McCarthy SW, Mann GJ, Scolyer RA. Diagnosis of cutaneous melanocytic tumours by four-colour fluorescence in situ hybridisation. Pathology 2009;41:383–387. [DOI] [PubMed] [Google Scholar]
- 34.Mitani Y, Li J, Rao PH, Zhao YJ, Bell D, Lippman SM, et al. Comprehensive analysis of the MYB-NF1B gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin Cancer Res 2010;16:4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu H, Wang Y, Huang Y, Shi H, Xue Q, Yang S, et al. Expression and prognostic role of c-Myb as a novel cell cycle protein in esophageal squamous cell carcinoma. Clin Transl Oncol 2013;15:796–801. [DOI] [PubMed] [Google Scholar]
- 36.Xing D, Bakhsh S, Melnyk N, Isacson C, Ho J, Huntsman DG, et al. Frequent NF1B-associated Gene Rearrangement in Adenoid Cystic Carcinoma of the Vulva. Int J Gynecol Pathol 2017;36:289–293. [DOI] [PubMed] [Google Scholar]
- 37.von Holstein SL, Fehr A, Persson M, Therkildsen MH, Prause JU, Heegaard S, Stenman G. Adenoid cystic carcinoma of the lacrimal gland: MYB gene activation, genomic imbalances, and clinical characteristics. Ophthalmology 2013;120:2130–2138. [DOI] [PubMed] [Google Scholar]
- 38.Stenman G, Andersson MK, Andrén Y. New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle 2010;9:2986–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barroga CF, Pham H, Kaushansky K. Thrombopoietin regulates c-Myb expression by modulating micro RNA 150 expression. Exp Hematol 2008;36:1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clappier E, Soulier J. t(6;7)(q23;q34). Atlas Genet Cytogenet Oncol Haematol 2008;12:248–249. [Google Scholar]
- 41.Dastugue N, Duchayne E, Kuhlein E, Rubie H, Demur C, Aurich J, et al. Acute basophilic leukaemia and translocation t(X;6)(p11;q23). Br J Haematol 1997;9:170–176. [DOI] [PubMed] [Google Scholar]
- 42.Quelen C, Lippert E, Struski S, Demur C, Soler G, Prade N, et al. Identification of a transforming MYB-GATA1 fusion gene in acute basophilic leukemia: a new entity in male infants. Blood 2011;117:5719–5722. [DOI] [PubMed] [Google Scholar]
- 43.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NF1B transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA 2009;106:18740–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 2000;95:745–755. [PubMed] [Google Scholar]
- 45.Ramsay RG. c-Myb a stem-progenitor cell regulator in multiple tissue compartments.Growth Factors 2005;23:253–261. [DOI] [PubMed] [Google Scholar]
- 46.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calabretta B, Sims RB, Valtieri M, Caracciolo D, Szczylik C, Venturelli D, et al. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of c-myb antisense oligodeoxynucleotides: an in vitro study relevant to bone marrow purging. Proc Natl Acad Sci USA 1991;88:2351–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamm I, Wagner M. Antisense therapy in clinical oncology: preclinical and clinical experiences. Mol Biotechnol 2006;33:221–238. [DOI] [PubMed] [Google Scholar]
- 50.Millikin D, Meese E, Vogelstein B, Witkowski C, Trent J. Loss of heterozygosity for loci on the long arm of chromosome 6 in human malignant melanoma. Cancer Res 1991;51:5449–5453. [PubMed] [Google Scholar]
- 51.Ponti G, Ruini C, Massi D, Pellacani G, Tomasi A, Paglierani M, et al. Fluorescence in-situ hybridization and dermoscopy in the assessment of controversial melanocytic tumors. Melanoma Res 2013;23:474–480. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y, Ness SA. Myb proteins: angels and demons in normal and transformed cells. Front Biosci (Landmark Ed) 2011;16:1109–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, et al. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res 1988;16:11075–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trauth K, Mutschler B, Jenkins NA, Gilbert DJ, Copeland NG, Klempnauer KH. Mouse A-myb encodes a trans-activator and is expressed in mitotically active cells of the developing central nervous system, adult testis and B lymphocytes. EMBO J 1994;13:5994–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Golay J, Luppi M, Songia S, Palvarini C, Lombardi L, Aiello A, et al. Expression of A-myb, but not c-myb and B-myb, is restricted to Burkitt’s lymphoma, sIg+ B-acute lymphoblastic leukemia, and a subset of chronic lymphocytic leukemias. Blood 1996;87:1900–1911. [PubMed] [Google Scholar]
- 56.Lipsick JS, Manak J, Mitiku N, Chen CK, Fogarty P, Guthrie E. Functional evolution of the Myb oncogene family. Blood Cells Mol Dis 2001;27:456–458. [DOI] [PubMed] [Google Scholar]
- 57.Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol 1996;16:6457–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol 1998;18:1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue K, Sherr CJ, Shapiro LH. Regulation of the CD13/Aminopeptidase N gene by DMP1, a transcription factor antagonized by D-type cyclins. J Biol Chem 1998;273:29188–29194. [DOI] [PubMed] [Google Scholar]
- 60.Bodner SM, Naeve CW, Rakestraw KM, Jones BG, Valentine VA, Valentine MB, et al. Cloning and chromosomal localization of the gene encoding human cyclin D-binding Myb-like protein (hDMP1). Gene 1999;229:223–228. [DOI] [PubMed] [Google Scholar]
- 61.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci USA 1999;96:3993–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inoue K, Wen R, Rehg JE, Adachi M, Cleveland JL, Roussel MF, et al. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes Dev 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev 2001;15:2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol 2005;25:220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its repression by E2Fs. Oncogene 2006;25:7703–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taneja P, Mallakin A, Matise LA, Frazier DP, Choudhary M, Inoue K. Repression of Dmp1 and Arf promoter by anthracyclins: critical roles of the NF-κB subunit p65. Oncogene 2007;26:7457–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mallakin A, Sugiyama T, Taneja P, Matise LA, Frazier DP, Choudhary M, et al. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell 2007;12:381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taneja P, Maglic D, Kai F, Sugiyama T, Kendig RD, Frazier DP, et al. Critical roles of DMP1 in human epidermal growth factor receptor 2/neu-Arf-p53 signaling and breast cancer development. Cancer Res 2010;70:9084–9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mallakin A, Sugiyama T, Kai F, Taneja P, Kendig RD, Frazier DP, et al. The Arf-inducing transcription factor Dmp1 encodes transcriptional activator of amphiregulin, thrombospondin-1, JunB and Egr1. Int J Cancer 2010;126:1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi M, Srour EF. Regulation of murine hematopoietic stem cell quiescence byDmp1. Blood 2011;118:6562–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maglic D, Zhu S, Fry EA, Taneja P, Kai F, Kendig RD, et al. Prognostic value of the hDMP1-ARF-Hdm2-p53 pathway in breast cancer. Oncogene 2013;32:4120–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu S, Mott RT, Fry EA, Taneja P, Kulik G, Sui G, et al. Cooperation between cyclin D1 expression and Dmp1-loss in BC. Am J Pathol 2013;183:1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fry EA, Taneja P, Maglic D, Zhu S, Sui G, Inoue K. Dmp1α inhibits HER2/neu-induced mammary tumorigenesis. PLoS One 2013;8:e77870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frazier DP, Kendig RD, Kai F, Maglic D, Sugiyama T, Taneja P, et al. Dmp1 physically interacts with p53 and positively regulates p53’s stabilization, nuclear localization, and function. Cancer Res 2012;72:1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kendig RD, Kai F, Fry EA, Inoue K. Stabilization of the p53-DNA complex by the nuclear protein Dmp1α. Cancer Invest 2017;35:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene 2007;26:4329–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugiyama T, Frazier DP, Taneja P, Morgan RL, Willingham MC, Inoue K. The role of Dmp1 and its future in lung cancer diagnostics. Expert Rev Mol Diagn 2008;8:435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inoue K, Fry EA, Frazier DP. Transcription factors that interact with p53 and Mdm2. Int J Cancer 2016;138:1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inoue K, Fry EA. Aberrant expression of Cyclin D1 in cancer. Signal Transduction Insights. 2015;4:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inoue K, Fry EA. Aberrant splicing of the DMP1-INK4a/ARF-MDM2-p53 pathway in cancer. Int J Cancer 2016;139:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fry EA, Taneja P, Inoue K. Oncogenic and tumor-suppressive mouse models for breast cancer employing HER2/neu. Int J Cancer 2017;140:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fry EA, Taneja P, Inoue K. Clinical applications of mouse models for breast cancer engaging HER2/neu. Integr Cancer Sci Ther 2016;3:593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inoue K, Fry EA. Haplo-insufficient tumor suppressor genes A book chapter in ‘Advances in Medicine and Biology’, Nova Science Publishers, Inc. Volume 118, Chapter 6, 83–122, 2017. [PMC free article] [PubMed] [Google Scholar]
- 84.Sherr CJ, Bertwistle D, DEN Besten W, Kuo ML, Sugimoto M, Tago K, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol 2005;70:129–137. [DOI] [PubMed] [Google Scholar]
- 85.Maggi LB Jr, Winkeler CL, Miceli AP, Apicelli AJ, Brady SN, Kuchenreuther MJ, et al. ARF tumor suppression in the nucleolus. Biochim Biophys Acta 2014;1842:831–839. [DOI] [PubMed] [Google Scholar]
- 86.Inoue K, Fry EA. Aberrant expression of ARF in human cancer - a new biomarker? Tumor and Microenvironment, [DOI] [PMC free article] [PubMed]
- 87.Inoue K, Fry EA. Expression of p16INK4a in human cancer - a new biomarker? Cancer Rep Rev 2018. March;2(2). doi: 10.15761/CRR.1000145. Epub 2018 Jan 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maglic D, Stovall DB, Cline JM, Fry EA, Mallakin A, Taneja P, et al. DMP1β, a splice isoform of the tumor suppressor DMP1 locus, induces proliferation and progression of breast cancer. J Pathol 2015;236:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tschan MP, Federzoni EA, Haimovici A, Britschgi C, Moser BA, Jin J, et al. Human DMP1β antagonizes DMP1α regulation of the p14(ARF) tumor suppressor and promotes cellular proliferation. Biochim Biophys Acta 2015;1849:1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inoue K, Fry EA. Novel biomarkers for breast cancer. Biomark Cancer 2016,8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tschan MP, Gullberg U, Shan D, Torbett BE, Fey MF, Tobler A. The hDMTF1 tumor suppressor is a new WT1 target in myeloid leukemias. Leukemia 2008;22,1087–1090. [DOI] [PubMed] [Google Scholar]
- 92.Inoue K, Sugiyama H, Ogawa H, Nakagawa M, Yamagami T, Miwa H, et al. WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood 1994; 84:3071–3079. [PubMed] [Google Scholar]
- 93.Yamagami T, Sugiyama H, Inoue K, Ogawa H, Tatekawa T, Hirata M, et al. Growth inhibition of human leukemic cells by WT1 antisense oligonucleotides. Blood 1996;87:2878–2884. [PubMed] [Google Scholar]
- 94.Inoue K, Ogawa H, Yamagami T, Soma T, Tani T, Tatekawa T, et al. Long-term follow-up of minimal residual disease in leukemia patients by monitoring WT1 (Wilms tumor gene) expression levels. Blood 1996;88:2267–2278. [PubMed] [Google Scholar]
- 95.Tamaki H, Ogawa H, Inoue K, Soma T, Yamagami T, Miyake S, et al. Increased expression of the Wilms tumor gene (WT1) at relapse in acute leukemia. Blood 1996;88:4396–4398. [PubMed] [Google Scholar]
- 96.Ogawa H, Tsuboi A, Oji Y, Tamaki H, Soma T, Inoue K, et al. Successful donor leukocyte transfusion at molecular relapse for a patient with acute myeloid leukemia who was treated with allogeneic bone marrow transplantation: importance of the monitoring of minimal residual disease by WT1 assay. Bone Marrow Transplant 1998;21:525–527. [DOI] [PubMed] [Google Scholar]
- 97.Tamaki H, Ogawa H, Ohyashiki K, Ohyashiki JH, Iwama H, Inoue K, et al. The Wilms’ tumor gene WT1 is a good marker for diagnosis of disease progression of myelodysplastic syndromes. Leukemia 1999;13:393–399. [DOI] [PubMed] [Google Scholar]
- 98.Inoue K, Ogawa H, Sonoda Y, Kimura T, Sakabe H, Oka Y, et al. Aberrant overexpression of the Wilms tumor gene (WT1) in human leukemia. Blood 1997;89:1405–1412. [PubMed] [Google Scholar]
- 99.Inoue K, Tamaki H, Ogawa H, Oka Y, Soma T, Tatekawa T, et al. Wilms’ tumor gene (WT1) competes with differentiation-inducing signal in hematopoietic progenitor cells. Blood 1998;91:2969–2976. [PubMed] [Google Scholar]
- 100.Inoue K, Sugiyama T, Taneja P, Morgan RL, Frazier DP. Emerging roles of DMTF1 in lung cancer. Cancer Res 2008;68:4487–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Inoue K, Fry EA. Alterations of p63 and p73 in human cancers in ‘Mutant p53 and MDM2 in cancer’. Editors: Deb Swati Palit & Deb Sumitra. Springer, NY: Chapter 2: 17–40, 2014. Subcell Biochem 2014;85:17–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Inoue K, Fry EA. Aberrant splicing of estrogen receptor, HER2, and CD44 in breast cancer. Genetics & Epigenetics 2015;7:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016;35:2413–2427. [DOI] [PubMed] [Google Scholar]
- 104.Luz FA, Brígido PC, Moraes AS, Silva MJ. Aberrant Splicing in Cancer: Mediators of Malignant Progression through an Imperfect Splice Program Shift. Oncology 2017;92:3–13. [DOI] [PubMed] [Google Scholar]
- 105.Klempnauer KH, Gonda TJ, Bishop JM. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 1982;31(2 Pt 1):453–463. [DOI] [PubMed] [Google Scholar]
- 106.Gonda TJ, Bishop JM. Structure and transcription of the cellular homolog (c-myb) of the avian myeloblastosis virus transforming gene (v-myb). J Virol 1983;46:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Franchini G, Wong-Staal F, Baluda MA, Lengel C, Tronick SR. Structural organization and expression of human DNA sequences related to the transforming gene of avian myeloblastosis virus. Proc Natl Acad Sci USA 1983;80:7385–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gonda TJ, Gough NM, Dunn AR, de Blaquiere J. Nucleotide sequence of cDNA clones of the murine myb proto-oncogene. EMBO J 1985;4:2003–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 1999;18:3017–3033. [DOI] [PubMed] [Google Scholar]
- 110.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ Jr, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991;65:677–689. [DOI] [PubMed] [Google Scholar]
- 111.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 2003;22:4478–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA 2009;106:21689–21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lieu YK 1, Reddy EP. Impaired adult myeloid progenitor CMP and GMP cell function in conditional c-myb-knockout mice. Cell Cycle 2012;11:3504–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Malaterre J, Carpinelli M, Ernst M, Alexander W, Cooke M, Sutton S, et al. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc Natl Acad Sci USA 2007;104:3829–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood 1996;87:2162–2170. [PubMed] [Google Scholar]
- 116.Levin J, Cocault L, Demerens C, Challier C, Pauchard M, Caen J, et al. Thrombocytopenic c-mpl(−/−) mice can produce a normal level of platelets after administration of 5-fluorouracil: the effect of age on the response. Blood 2001;98:1019–1027. [DOI] [PubMed] [Google Scholar]
- 117.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, et al. Suppressor screen in Mpl−/− mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci USA 2004;101:6553–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell 2005;8:153–166. [DOI] [PubMed] [Google Scholar]
- 119.Dalla-Favera R, Franchini G, Martinotti S, Wong-Staal F, Gallo RC, Croce CM. Chromosomal assignment of the human homologues of feline sarcoma virus and avian myeloblastosis virus onc genes. Proc Natl Acad Sci USA 1982;79:4714–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harper ME, Franchini G, Love J, Simon MI, Gallo RC, Wong-Staal F. Chromosomal sublocalization of human c-myb and c-fes cellular onc genes. Nature 1983;304:169–171. [DOI] [PubMed] [Google Scholar]
- 121.Winqvist R, Knuutila S, Leprince D, Stehelin D, Alitalo K. Mapping of amplified c-myb oncogene, sister chromatid exchanges, and karyotypic analysis of the COLO 205 colon carcinoma cell line. Cancer Genet Cytogenet 1985;18:251–264. [DOI] [PubMed] [Google Scholar]
- 122.Janssen JW, Vernole P, de Boer PA, Oosterhuis JW, Collard JG. Sublocalization of c-myb to 6q21-q23 by in situ hybridization and c-myb expression in a human teratocarcinoma with 6q rearrangements. Cytogenet Cell Genet 1986;41:129–235. [DOI] [PubMed] [Google Scholar]
- 123.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 2002;35:97–112. [DOI] [PubMed] [Google Scholar]
- 124.O’Neil J, Tchinda J, Gutierrez A, Moreau L, Maser RS, Wong KK, et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J Exp Med 2007;204:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barletta C, Pelicci PG, Kenyon LC, Smith SD, Dalla-Favera R. Relationship between the c-myb locus and the 6q-chromosomal aberration in leukemias and lymphomas. Science 1987;235:1064–1067. [DOI] [PubMed] [Google Scholar]
- 126.Ohyashiki K, Ohyashiki JH, Kinniburgh AJ, Toyama K, Ito H, Minowada J, et al. myb oncogene in human hematopoietic neoplasia with 6q- anomaly. Cancer Genet Cytogenet 1988;33:83–92. [DOI] [PubMed] [Google Scholar]
- 127.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 2011;25:1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2002;2:451–461. [DOI] [PubMed] [Google Scholar]
- 129.Yeh ES, Yang TW, Jung JJ, Gardner HP, Cardiff RD, Chodosh LA. Hunk is required for HER2/neu-induced mammary tumorigenesis. J Clin Invest 2011;121:866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhu N, Chen M, Eng R, DeJong J, Sinha AU, Rahnamay NF, et al. MLL-AF9- and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J Clin Invest 2016;126:997–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sroczynska P, Cruickshank VA, Bukowski JP, Miyagi S, Bagger FO, Walfridsson J, et al. shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood 2014;123:1870–1882. [DOI] [PubMed] [Google Scholar]
- 132.Gewirtz AM. Myb targeted therapeutics for the treatment of human malignancies. Oncogene 1999;18:3056–3062. [DOI] [PubMed] [Google Scholar]
- 133.Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B. Requirement of c-Myb for p210(BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood 2008;111:4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science 1986;233:212–214. [DOI] [PubMed] [Google Scholar]
- 135.Luger SM, O’Brien SG, Ratajczak J, Ratajczak MZ, Mick R, Stadtmauer EA, et al. Oligodeoxynucleotide-mediated inhibition of c-myb gene expression in autografted bone marrow: a pilot study. Blood 2002;99:1150–1158. [DOI] [PubMed] [Google Scholar]
- 136.Kalota A, Karabon L, Swider CR, Viazovkina E, Elzagheid M, Damha MJ, et al. 2’-deoxy-2’-fluoro-beta-D-arabinonucleic acid (2’F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res 2006;34:451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kakizuka A, Miller WH Jr, Umesono K, Warrell RP Jr, Frankel SR, Murty VV, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 1991;66:663–674. [DOI] [PubMed] [Google Scholar]
- 138.Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 1992;80:1825–1831. [PubMed] [Google Scholar]
- 139.Uttarkar S, Dassé E, Coulibaly A, Steinmann S, Jakobs A, Schomburg C, et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood 2016;127:1173–1182. [DOI] [PubMed] [Google Scholar]
- 140.Uttarkar S, Piontek T, Dukare S, Schomburg C, Schlenke P, Berdel WE, et al. Small-Molecule Disruption of the Myb/p300 Cooperation Targets Acute Myeloid Leukemia Cells. Mol Cancer Ther 2016;15:2905–2915. [DOI] [PubMed] [Google Scholar]
- 141.Coulibaly A, Haas A, Steinmann S, Jakobs A, Schmidt TJ, Klempnauer KH. The natural anti-tumor compound Celastrol targets a Myb-C/EBPβ-p300 transcriptional module implicated in myeloid gene expression. PLoS One 2018;13:e0190934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jakobs A, Steinmann S, Henrich SM, Schmidt TJ, Klempnauer KH. Helenalin Acetate, a Natural Sesquiterpene Lactone with Anti-inflammatory and Anti-cancer Activity, Disrupts the Cooperation of CCAAT Box/Enhancer-binding Protein β (C/EBPβ) and Co-activator p300. J Biol Chem 2016;291:26098–26108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Uttarkar S, Frampton J, Klempnauer KH. Targeting the transcription factor Myb by small-molecule inhibitors. Exp Hematol 2017;47:31–35. [DOI] [PubMed] [Google Scholar]
- 144.Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPbeta. Mol Cell Biol 1997;17:6609–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell 2015;58:1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mitani Y, Liu B, Rao PH, Borra VJ, Zafereo M, Weber RS, et al. Novel MYBL1 Gene Rearrangements with Recurrent MYBL1-NF1B Fusions in Salivary Adenoid Cystic Carcinomas Lacking t(6;9)Translocations. Clin Cancer Res 2016;22:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wysocki PT, Izumchenko E, Meir J, Ha PK, Sidransky D, Brait M. Adenoid cystic carcinoma: emerging role of translocations and gene fusions. Oncotarget 2016;7:66239–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gonda TJ, Ramsay RG. Adenoid Cystic Carcinoma Can Be Driven by MYB or MYBL1 Rearrangements: New Insights into MYB and Tumor Biology. Cancer Discov 2016;6:125–127. [DOI] [PubMed] [Google Scholar]
- 149.Brayer KJ, Frerich CA, Kang H, Ness SA. Recurrent Fusions in MYB and MYBL1 Define a Common, Transcription Factor-Driven Oncogenic Pathway in Salivary Gland Adenoid Cystic Carcinoma. Cancer Discov 2016;6:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Persson M, Andrén Y, Moskaluk CA, Frierson HF Jr, Cooke SL, Futreal PA, et al. Clinically significant copy number alterations and complex rearrangements of MYB and NFIB in head and neck adenoid cystic carcinoma. Genes Chromosomes Cancer. 2012;51:805–817. [DOI] [PubMed] [Google Scholar]
- 151.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet 2016;48:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation Science 1987;237:1473–1476. [DOI] [PubMed] [Google Scholar]
- 153.Thompson MA, Flegg R, Westin EH, Ramsay RG. Microsatellite deletions in the c-myb transcriptional attenuator region associated with over-expression in colon tumour cell lines. Oncogene 1977;14:1715–1723. [DOI] [PubMed] [Google Scholar]
- 154.Drabsch Y 1, Hugo H, Zhang R, Dowhan DH, Miao YR, Gewirtz AM, Barry SC, Ramsay RG, Gonda TJ. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci USA 2007;104:13762–13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jin W, Liu Y, Chen L, Zhu H, Di GH, Ling H, et al. Involvement of MyoD and c-myb in regulation of basal and estrogen-induced transcription activity of the BRCA1 gene. Breast Cancer Res Treat 2011;125:699–713 [DOI] [PubMed] [Google Scholar]
- 156.Gasco M, Yulug IG, Crook T. TP53 mutations in familial breast cancer: functional aspects. Hum Mutat 2003;21:301–306. [DOI] [PubMed] [Google Scholar]
- 157.Masood S Prognostic/predictive factors in breast cancer. Clin Lab Med 2005;25:809–25,viii. [DOI] [PubMed] [Google Scholar]
- 158.Taneja P, Maglic D, Kai F, Zhu S, Kendig RD, Fry EA, et al. Classical and novel molecular prognostic markers for human breast cancer and their clinical significance. Clin Med Oncol 2010;4:15–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wiesner FG, Magener A, Fasching PA, Wesse J, Bani MR, Rauh C, Jud S, et al. Ki-67 as a prognostic molecular marker in routine clinical use in breast cancer patients. Breast 2009;18:135–141. [DOI] [PubMed] [Google Scholar]
- 160.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther 2006;13:115124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Silva J, Domínguez G, Silva JM, García JM, Gallego I, Corbacho C, et al. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene 2001;20:4586–4590. [DOI] [PubMed] [Google Scholar]
- 162.Milde-Langosch K, Bamberger AM, Rieck G, Kelp B, Lning T. Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype. Breast Cancer Res Treat 2001;67:61–70. [DOI] [PubMed] [Google Scholar]
- 163.Silva J, Silva JM, Domínguez G, García JM, Cantos B, Rodríguez R, et al. Concomitant expression of p16INK4a and p14ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol 2003;199:289–297. [DOI] [PubMed] [Google Scholar]
- 164.Ladanyi M Implications of P16/CDKN2A deletion in pleural mesotheliomas.Lung Cancer. 2005;49 Suppl 1:S95–S98. [DOI] [PubMed] [Google Scholar]
- 165.Li B, Rosen JM, McMenamin-Balano J, Muller WJ, Perkins AS. neu/ERBB2 cooperates with p53–172H during mammary tumorigenesis in transgenic mice. Mol Cell Biol 1997;17:3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Taneja P, Frazier DP, Kendig RD, Maglic D, Sugiyama T, Kai F, Taneja NK, Inoue K. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev Mol Diagn 2009;9:423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.D’Amico M, Wu K, Di Vizio D, Reutens AT, Stahl M, Fu M, et al. The role of Ink4a/Arf in ErbB2 mammary gland tumorigenesis. Cancer Res 2003;63:3395–3402. [PubMed] [Google Scholar]
- 168.Taneja P, Zhu S, Maglic D, Fry EA, Kendig RD, Inoue K. Transgenic and knockout mice models to reveal the functions of tumor suppressor genes. Clin Med Oncol 2011;5:235–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Inoue K, Fry EA, Taneja P. Recent progress in mouse models for tumor suppressor genes and its implications in human cancer. Clin Med Oncol 2013;7:103–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.George OL, Ness SA. Situational awareness: regulation of the myb transcription factor in differentiation, the cell cycle and oncogenesis. Cancers (Basel) 2014;6:2049–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fu SL, Lipsick JS. FAETL motif required for leukemic transformation by v‑Myb. J Virol 1996;70:5600–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dash AB, Orrico FC, Ness SA. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev 1996;10:1858–1869. [DOI] [PubMed] [Google Scholar]