Abstract
Activation of the transcription factor, the aryl hydrocarbon receptor (AHR), in normal human epidermal keratinocytes (NHEKs) increased AHR binding in the promoters of the glucose transporter, SLC2A1, and the glycolytic enzyme, enolase 1 (ENO1). This increased chromatin binding corresponded with AHR-dependent decreases in levels of SLC2A1 and ENO1 mRNA, protein and activities. Studies of the ENO1 promoter showed activation of the AHR decreases the transcription of ENO1. Glycolysis was lowered by activation of the AHR as measured by decreases in glucose uptake and the production of pyruvate and lactate. Levels of ATP were also decreased. Down-regulation of glucose metabolism, either by activation of the AHR, inhibition of glycolysis, inhibition of glucose transport, or inhibition of enolase, increased SIRT1 protein levels in NHEKs and the immortalized keratinocyte cell line, N/TERT-1. This increase in SIRT1 was abrogated by the addition of exogenous pyruvate. Moreover, keratinocyte differentiation in response to downregulation of glycolysis, either by activation of the AHR, inhibition of glucose transport, or inhibition of enolase, was dependent on SIRT1. These results indicate that regulation of glycolysis controls keratinocyte differentiation, and that activation of the AHR, by lowering the expression of SLC2A1 and ENO1, can determine this fate.
INTRODUCTION
The epidermis of mammals consists mainly of keratinocytes undergoing a sequence of cellular changes to form an epidermal barrier that controls water loss, and limits exposure to infectious and chemical agents (Candi et al., 2005). As keratinocytes migrate outward from the basement membrane they change shape, from cuboidal to flat, alter their lipid composition, extrude a lipid matrix, increase adherens and tight junctions, lose cellular organelles such as the nucleus and mitochondria, and replace their outer cell membrane with a proteinaceous cornified envelope, before being sloughed. The regulation of these changes is of intense scientific interest because abnormalities, such as increased or decreased thickening of the epidermis or breaches in the epidermal barrier, can have serious human health consequences.
Activation of the aryl hydrocarbon receptor (AHR), a basic helix-loop-helix/Per-Arnt- Sim transcription factor, accelerates terminal differentiation of human keratinocytes (Greenlee et al., 1985), increases the thickness of the stratum corneum (Loertscher et al., 2001), and increases the expression of numerous genes important to keratinocyte differentiation, such as members of the epidermal differentiation complex as well as ceramide biosynthetic genes (Kennedy et al., 2013, Sutter et al., 2011, Sutter et al., 2009). In utero studies of mice demonstrate that activation of the AHR by exposure to the potent and selective AHR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), accelerates the formation of the epidermal barrier (Sutter et al., 2011). Consistently, models of either genetic loss or chemical inhibition of the AHR demonstrate the importance of the AHR in the regulation of markers of differentiation, formation of normal human skin equivalents (van den Bogaard et al., 2015) and transepidermal water loss (Haas et al., 2016). Limiting dietary AHR ligands has a similar deleterious effect on the barrier function as knocking out the AHR, either in skin or gut (Haas et al., 2016, Li et al., 2011), arguing that the physiological actions of the AHR require ligand activation. Together, these studies show that the AHR plays an important role in keratinocyte differentiation and the formation of the epidermal barrier.
Glycolysis is an essential metabolic pathway that provides energy and reducing equivalents to sustain cell division. While the link between increased glycolysis and cell proliferation is widely supported, a link between decreased glycolysis and cell differentiation, particularly in epithelial cells is emerging (Hamanaka and Mutlu, 2017, Kennedy et al., 2013). As previously reported, activation of the AHR not only increases many of the critical proteins and lipids involved in the differentiation of keratinocytes, but also diminishes the glycolytic and mitochondrial function of these cells and increases mitochondrial reactive oxygen species (ROS). Decreases in glycolytic intermediates and end products paralleled the decreases in many of the mRNA transcripts encoding the proteins of the glycolytic pathway (Kennedy et al., 2013), indicating possible regulation by the AHR. Here we investigate whether alterations in glucose uptake and glycolysis by activation of the AHR play a role in keratinocyte differentiation.
RESULTS
The AHR targets SLC2A1 and ENO1 gene expression in normal human epidermal keratinocytes (NHEKs)
To see if AHR activation induced chromatin binding in the promoter region of any of the glycolytic genes, we performed ChIP-Seq analysis. Induced binding of the AHR was significant in three genes, SLC2A1, ENO1 (Figure 1a) and GAPDH (data not shown). ChIP-PCR could only confirm the sites of SLC2A1 (SLC2A1-1, SLC2A1-2 and SLC2A1-3) and ENO1 (Figure 1b).
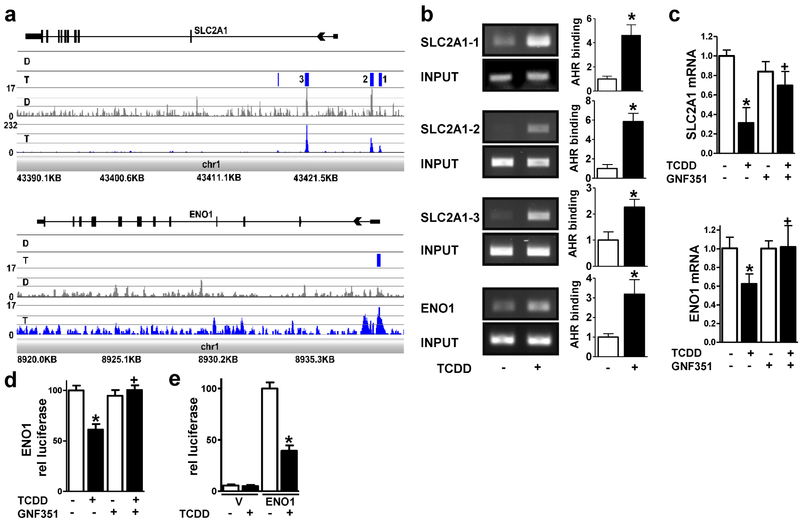
Figure 1. The AHR decreases SLC2A1 and ENO1 expression in NHEKs.
(a) ChIP-Seq analyses identify SLC2A1 and ENO1 as direct targets of the AHR. From the bottom to the top of each browser image, the first track displays the chromosomal location of the mapped reads. The next track shows the peak height in the TCDD (T, blue)- and DMSO (D, grey)-treated samples. The third track displays the statistical analyses, where statistically significant peaks are identified as a blue box. For the SLC2A1 gene, four significant peaks were identified (p < 1e-045 – p < 2.015e-017) in the TCDD-treated sample compared to the DMSO control. The peaks labeled as 1-3 were within +/− 5 kilobases of the transcriptional start site of SLC2A1. For ENO1, one statistically significant peak was identified (p < 9e-004) in the TCDD-treated sample compared to the DMSO control. The final track at the top of each image displays the gene structure. (b) ChIP-PCR verifies SLC2A1 and ENO1 as AHR targets. Representative images (left) and quantitation (right) corresponding to PCR of the three peaks within +/− 5 kilobases of the transcriptional start sites in SLC2A1 and one peak in ENO1 identified in (a). AHR binding [mean (n=3) +/− SD] was determined by densitometry. Levels of AHR binding were normalized with input and expressed in units relative to the vehicle control. *indicates a significant difference versus vehicle control, p < 0.05 by two-tailed Student’s t-test. (c) SLC2A1 and ENO1 transcripts are regulated in an AHR-dependent manner. Expression of SLC2A1 and ENO1 mRNA following treatment with or without (+/−) the AHR ligand, TCDD (24 hours), and +/− the AHR antagonist, GNF351 (100 nM, 24 hours) as indicated. Relative levels of mRNA [mean (n =4) +/− SD] were normalized with TUBA1C. (d) Transcription from the ENO1 promoter (1550 base pairs) is repressed by TCDD and is dependent on the AHR. Stable ENO1-luc integrant N/TERT-1 cells were treated with or without (+/−) the AHR ligand, TCDD (48 hours), and +/− the AHR antagonist, GNF351 (100 nM, 48 hours) as indicated. Relative levels of luciferase (mean +/−SEM) were determined from two replicate experiments, each with n=6. (e) Transcription from the ENO1 promoter (189 base pairs) is repressed by TCDD. NHEKs transfected with the vector (V) or ENO1 promoter (ENO1) were treated with or without (+/−) the AHR ligand, TCDD (72 hours) as indicated. Relative levels of luciferase (mean +/−SEM) were determined from four replicate experiments, each with n=3-6. * indicates a significant difference versus the vehicle control, p < 0.05 and + indicates a significant difference versus TCDD only, p 0.05 by two-way ANOVA followed by Tukey post hoc tests.
Activation of the AHR decreased SLC2A1 and ENO1 mRNA expression. This decrease was reversed by an antagonist of the AHR, GNF351, demonstrating a requirement for AHR activation in mediating these effects of TCDD (Figure 1c). Here we focused our efforts on the promoter of ENO1 since the region of AHR binding overlapped its promoter and TSS site. The initial study was designed to integrate an ENO1 promoter-luciferase construct into the genome to mimic chromatin context. The transcriptional activity of a 1550 bp fragment of ENO1 was repressed by activation of the AHR. This decrease was prevented by GNF351, demonstrating a requirement for AHR activation in mediating these effects of TCDD (Figure 1d). To determine if the region of the ENO1 promoter corresponding to AHR binding (Figure 1a) could repress transcription, a 189 bp region of the ENO1 promoter was inserted into the luciferase reporter vector, pGL3-Basic. Following activation of the AHR with TCDD, the transcriptional activity of this promoter was repressed (Figure 1e).
Ligand-activation of the AHR decreases facilitated glucose transport and glycolysis in human keratinocytes
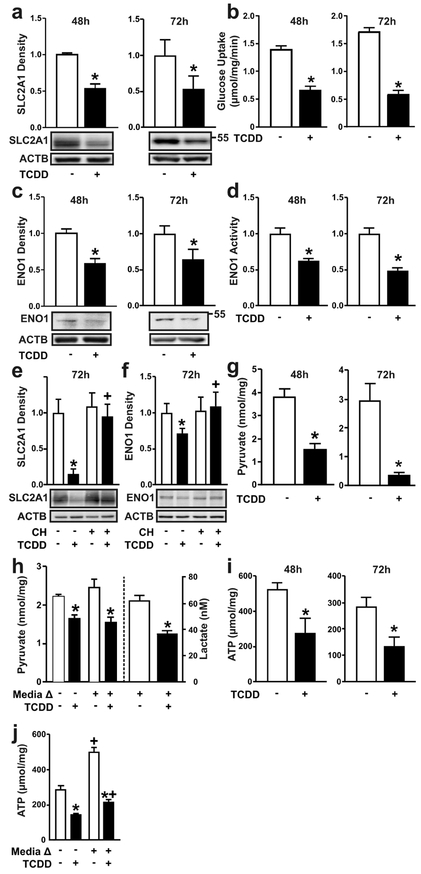
Parallel to mRNA, SLC2A1 and ENO1 proteins were decreased in NHEKs (Figure 2a and 2c) following 48 or 72 hours of TCDD exposure. Lower levels of SLC2A1 and ENO1 were accompanied by decreased rates of cellular glucose uptake (Figure 2b) and phosphopyruvate hydratase (ENO1) activity (Figure 2d). The dependency of the AHR on the effects of TCDD on protein expression of SLC2A1 and ENO1 was confirmed in cells co-treated with the highly selective AHR antagonist, CH223191 for 72 hours (Figures 2e, 2f). Consistent with these effects, levels of intracellular pyruvate, the end product of the glycolytic pathway, was lower after 48 hours of treatment with TCDD, and continued to decrease at 72 hours post treatment (Figure 2g). Similarly, levels of lactate were decreased and the decreases in pyruvate (Figure 2h, left axis) and lactate (Figure 2h, right axis) following TCDD treatment were maintained even when fresh media was provided at 48 hours, indicating that the observed effects on glycolytic metabolism were not due to glucose limitation in the media. Consistent with decreased glycolytic flux and the production of pyruvate, decreases of ATP were also observed (Figure 2i); this endpoint was unaffected by fresh media replacement (Figure 2j).
Figure 2. Ligand-activation of the AHR decreases facilitated glucose transport and glycolysis in human keratinocytes.
(a) SLC2A1 protein and (b) glucose uptake were measured in NHEKs treated with TCDD for 48 or 72 hours, as indicated. (c) ENO1 protein and (d) enzyme activity were measured in NHEKs treated with TCDD for 48 or 72 hours, as indicated. (e) SLC2A1 and (f) ENO1 proteins were measured in N/TERT-1 cells treated with TCDD +/− CH223191 (1 μM) for 72 hours. * indicates a significant difference versus the vehicle control, p 0.05; + indicates a significant difference versus TCDD only, p < 0.05. (g) Levels of pyruvate or (i) ATP were measured in NHEKs treated with TCDD for 48 or 72 hours, as indicated. (h, left y-axis) Levels of pyruvate, (h, right y-axis) lactate and (j) ATP were measured in NHEKs treated with TCDD for a total of 72 hours. After the first 48 hours, treatment media was replaced with fresh media as indicated (+ Media Δ) for an additional 24 hours. * indicates a significant difference versus the vehicle control within each group (media change or no media change), p < 0.05; + indicates a significant difference versus no media change within each treatment group, p 0.05. For all immunoblots, representative images are shown and levels of protein [mean (n =4) +/− SD] are normalized with ACTB and expressed in units relative to the vehicle control. Quantitative results (mean +/− SD) for glucose uptake assay (n=6), ENO1 enzymatic assay (n=6), pyruvate (n=5), lactate (n=6) and ATP (n=5-6) are graphed. For single comparisons, two-tailed Student’s t-test was used. For multiple comparisons, data were analyzed using two-way ANOVA followed by Tukey post hoc tests.
Ligand-activation of the AHR increases SIRT1 by nutrient deprivation, similar to chemical inhibition of glycolysis or glucose transport
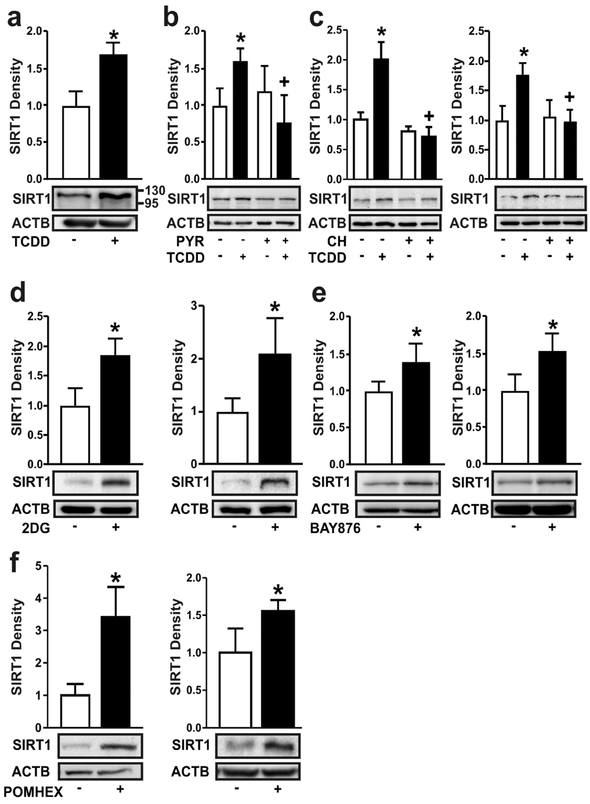
Decreased glycolytic flux is associated with nutrient stress and the up-regulation of SIRT1 (Kanfi et al., 2008). Treatment of NHEKs with TCDD for 72 hours increased SIRT1 protein levels (Figure 3a). This increase in SIRT1 protein was abrogated by co-treatment with exogenous pyruvate (Figure 3b). The AHR antagonist, CH223191, prevented the ligand-mediated increases of SIRT1 protein in both NHEKs and N/TERT-1 cells (Figure 3c), providing additional evidence of AHR dependency. Treatment of cells with 2-deoxyglucose (2DG), a glucose analog and competitive inhibitor of glucose metabolism (Wick et al., 1957) resulted in increased SIRT1 protein in both NHEK and N/TERT-1 cells (Figure 3d). Similar results were observed in cells treated with BAY876, a highly selective inhibitor of SLC2A1 (Siebeneicher et al., 2016) (Figure 3e) or POMHEX, an inhibitor of enolase (Lin et al., 2018) (Figure 3f).
Figure 3. Ligand-activation of the AHR increases SIRT1 by nutrient deprivation, similar to chemical inhibition of glycolysis, glucose transport or enolase.
(a) SIRT1 protein was measured in NHEKs treated with TCDD for 72 hours. (b) SIRT1 protein was measured in NHEKs treated with TCDD +/− sodium pyruvate (PYR, 5 mM) for 72 hours. (c) SIRT1 protein was measured in NHEKs (left) and N/TERT-1 cells (right) treated with TCDD +/− CH223191 (1 μM) for 48 hours. (d) SIRT1 protein was measured in NHEKs (left) and N/TERT-1 cells (right) treated with 2DG (20 mM) for 48 hours. (e) SIRT1 protein was measured in NHEKs (left) and N/TERT-1 cells (right) treated with the select SLC2A1 inhibitor, BAY876 (5 μM) for 48 hours. (f) SIRT1 protein was measured in NHEKs (left) and N/TERT-1 cells (right) treated with the enolase inhibitor, POMHEX (25 μM) for 48 hours. For all immunoblots, representative images are shown and levels of protein [mean (n =4) +/− SD] were normalized with ACTB. * indicates a significant difference versus the vehicle control, p < 0.05; + indicates a significant difference versus TCDD only, p < 0.05. For single comparisons, a two-tailed Student’s t-test was used. For multiple comparisons, data were analyzed using two-way ANOVA followed by Tukey post hoc tests.
AHR-induced keratinocyte terminal differentiation is SIRT1-dependent
Genetic knockdown of SIRT1 was used to determine its importance to AHR-regulated differentiation and its epistatic relationship to the AHR. NHEKs and N/TERT-1 cells were transduced with lentiviral SIRT1 shRNA particles, resulting in an approximately 70% decrease in SIRT1 protein (Figure 4a and 4b, left panel). A decrease in SIRT1 inhibited TCDD-mediated increases of SIRT1 in both N/TERT-1 cells and NHEKs (Figures 4a and 4b, left panels). Interestingly, the knock down of SIRT1 blocked the AHR-mediated increase of FLG in both NHEKs and N/TERT-1 cells Figures 4a and 4b, right panels), a protein required for normal epidermal barrier function and induced during keratinocyte differentiation. To determine whether SIRT1 acted upstream of AHR transcriptional activation, two genes, whose transcription is regulated by the AHR, were measured. Induction of CYP1B1 and CYP1A1 mRNA was not altered by SIRT1 knock down (Figure 4c and 4d, left panels). This is also reflected in the levels of CYP1B1 protein, where protein induction in response to ligand was not affected by SIRT1 levels (Figure 4c, right panel). Additionally, the reduction in the levels of SLC2A1 and ENO1 mRNA following activation of the AHR are independent of SIRT1 (Figure 4d, middle and right panels). An increase of FLG was also observed in cells treated with 2DG, BAY876, or POMHEX. In each case the increase was SIRT1-dependent, as it was completely blocked by knock down of SIRT1 (Figure 4e, 4f and 4g). TCDD-mediated increases in additional keratinocyte differentiation proteins, IVL, SPRR2 and TGM1, were also dependent on the expression of SIRT1 (Figure 4h), as was cornified envelope formation, an index of terminal cell differentiation (Figure 4i).
Figure 4. AHR-induced keratinocyte terminal differentiation is SIRT1-dependent.
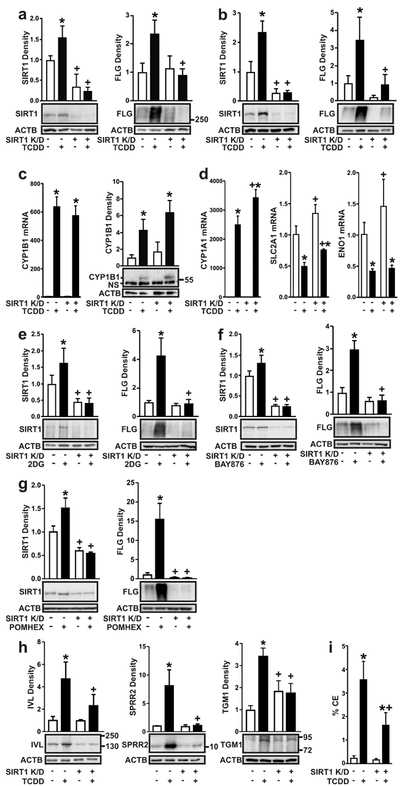
Keratinocytes were stably transduced with the empty vector or the vector containing a shRNA to knock down expression of SIRT1 (SIRT1 K/D). (a) SIRT1 and FLG proteins were measured in NHEKs, vector control or SIRT1 K/D, treated with TCDD for 72 h. (b) SIRT1 and FLG proteins were measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with TCDD for 72 hours. (c) CYP1B1 mRNA (left) was measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with TCDD for 24 hours. CYP1B1 protein (right) was measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with TCDD for 72 hours. (d) CYP1A1, SLC2A1 and ENO1 mRNA were measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with TCDD for 24 hours. (e) SIRT1 and FLG proteins were measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with 2DG (20 mM) for 72 hours. (f) SIRT1 and FLG proteins were measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with BAY876 (5 μM) for 72 hours. (g) SIRT1 and FLG proteins were measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with POMHEX (50 μM) for 72 hours. (h) IVL, SPRR2, and TGM1 proteins were measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with TCDD for 72 hours. (i) CE formation was measured in N/TERT-1 cells, vector control or SIRT1 K/D, treated with TCDD for 5 days. For all immunoblots, representative images are shown and levels of protein [mean (n =4) +/− SD] were normalized with ACTB. Relative levels of mRNA [mean (n =4) +/− SD] were normalized with TUBA1C. * indicates a significant difference versus vehicle control of the same cell line, p < 0.05. + indicates a significant difference versus the same treatment in the vector control cell line, p < 0.05. For single comparisons, two-tailed Student’s t-test was used. For multiple comparisons, data were analyzed using two-way ANOVA followed by Tukey post hoc tests.
DISCUSSION
Metabolic reprogramming as a mechanism to regulate cell fate has been described in several systems where changes in a precursor cell give rise to a phenotypically different cell. Glycolysis, the tricarboxylic acid (TCA) cycle, oxidative phosphorylation, and ROS generation play varying yet profound roles in the ability of a cell to maintain a particular cell phenotype or to differentiate (Shyh-Chang and Ng, 2017, Tang and Mauro, 2017).
In the epidermis, glycolysis is regulated during the differentiation process. Proliferative cells of the basal layer have greater glycolytic enzymatic activities than the differentiated keratinocytes of the suprabasal layers (Adachi and Yamasawa, 1966b, Im et al., 1966). SLC2A1 is the predominant glucose transporter expressed in keratinocytes and is regulated throughout differentiation. Its expression is greatest in basal cells, decreasing in suprabasal cells (Bedogni and Powell, 2006). ENO1 activity also decreases from the basal cells outward in the epidermis (Adachi and Yamasawa, 1966a).
Here we report that ligand-activated AHR-chromatin binding in the promoter regions of SLC2A1 and ENO1 preceded decreases in their levels of mRNA, protein, and activity. Furthermore, we show that the region where the AHR binds to the ENO1 promoter drives TCDD-mediated repression of ENO1 transcription. While demonstrating regulation at the level of transcription, these results do not determine whether direct AHR-xenobiotic response element (XRE) binding in the ENO1 promoter causes repression. Evidence of AHR-XRE-mediated transcriptional repression has been reported for other genes, including FOS (Duan et al., 1999) and NLRP3 (Huai et al., 2014). In the context of the ENO1 promoter, direct binding of the AHR could (1) compete for the overlapping DNA response elements of the transcription factor HIF1 (Semenza et al., 1996), (2) compete with HIF1A for the common dimerization partner of ARNT, or (3) recruit corepressors. Additionally, direct DNA binding of the AHR with a novel partner to non-canonical XREs needs to be considered. While this has been reported for transcriptional activation (Wilson et al., 2013), involvement of a novel partner in transcriptional repression has not been described, yet remains a possibility, as do other mechanisms such as indirect DNA binding.
Overall, the rates of glucose uptake, and the production of pyruvate and lactate were all decreased by activating the AHR, indicating an apparent decrease in glycolytic flux. Recent in vitro studies of the glycolytic enzyme, PFKFB3, show the importance of the glycolysis in keratinocyte proliferation and differentiation. PFKFB3 is a target of p63 and both proteins localize to basal epidermal keratinocytes. Their expression decreases during calcium-induced differentiation. Furthermore, decreasing PFKFB3 by chemical or genetic means elevates the expression of differentiation markers and inhibits the proliferation of keratinocytes, while genetic expression of PFKFB3 opposes differentiation (Hamanaka and Mutlu, 2017).
Our results indicate a general role of glycolysis in regulating keratinocyte differentiation as treatment with either 2DG, BAY876 or POMHEX promoted keratinocyte differentiation. As a ligand activated receptor regulating glucose transport and glycolysis, the AHR is a potential therapeutic target. In fact, the treatment effectiveness of coal tar for atopic dermatitis is attributed to its ability to ligand activate the AHR (van den Bogaard et al., 2013). Similarly, chemicals like BAY876, 2DG, or POMHEX may have utility for treating skin abnormalities characterized by hyper-proliferation and reduced differentiation.
In other cell types nutrient deprivation from decreased glycolysis increases SIRT1 protein and activity (Jesko and Strosznajder, 2016, Kanfi et al., 2008). Furthermore, supplemental pyruvate, which is readily taken up by cells, acts to bypass nutrient deprivation and blocks cytotoxicity caused by inhibition of glycolysis (Chung et al., 2004). Here, the addition of pyruvate abrogated the AHR-mediated SIRT1 accumulation, supporting the idea that the mechanism by which AHR activation induces SIRT1 is in response to decreased glycolysis, specifically a decrease of pyruvate, the end product of glycolysis and a major source of cell energy through the TCA cycle. This idea is further supported by our results showing that nutrient deprivation induced by 2DG or BAY876 also increased SIRT1 protein levels, similar to the effects of AHR activation.
A previous study reports that overexpression of SIRT1 in keratinocytes increases the level of the involucrin, a differentiation-specific protein, and several mRNAs associated with keratinocyte differentiation; knockdown or loss of SIRT1 has the opposite effect (Blander et al., 2009, Ming et al., 2015). Here we showed that genetic knock down of SIRT1 by shRNA resulted in a decrease in the AHR-mediated differentiation in both NHEKs and N/TERT-1 keratinocytes, demonstrating that the AHR-mediated keratinocyte differentiation is SIRT1-dependent. This was shown to be true for both the expression of differentiation markers and cornified envelope formation, a well-established functional measure of terminal differentiation (Sun and Green, 1976).
Of interest to AHR signaling, this latter study (Ming et al., 2015) indicates that SIRT1 is upstream of AHR signaling, with loss of SIRT1 preventing AHR-mediated induction of CYP1B1 RNA and protein. Our contrary results showed that SIRT1 knockdown did not affect AHR ligand-mediated induction of CYP1A1 or CYP1B1 nor repression of SLC2A1 or ENO1. This supports the idea that SIRT1 affects keratinocyte differentiation downstream of AHR transcriptional events and it is elevated in response to nutrient deprivation caused by decreased glycolysis. As SIRT1 acts as a deacetylase of histones, signaling proteins and transcription factors to regulate both epigenetic and protein-protein interaction mechanisms (Jesko and Strosznajder, 2016), further study of SIRT1 in differentiation is warranted.
In addition to glycolysis, mitochondria and oxidative metabolism contribute to metabolic reprogramming to affect cell fate by regulating energy production, amino acid and lipid metabolism and ROS production. While the current study only provides insight into the role and regulation of glycolysis, our previous study described much of this biochemistry (Kennedy et al., 2013). As expected, the large decrease in glucose utilization following activation of the AHR is accompanied by lower levels of citrate, generated in the combination of glycolysis-derived acetyl CoA and oxaloacetate. However, levels of other TCA intermediates are much less affected suggesting that the TCA cycle is maintained by other sources such as amino acids through anaplerosis. Despite the maintenance of the TCA cycle, mitochondrial oxidative phosphorylation appears to be affected as ATP levels and mitochondrial membrane potential decrease. Overall, levels of reducing equivalents decrease and elevation of cysteine-glutathione disulfide provides a clear indication of an increasingly oxidative environment. In the mitochondria, the ratio of GSH/GSSG decreases, consistent with lower levels of glutathione reductase activity. This is accompanied by increased production of mitochondrial ROS, specifically the production of H2O2. Notably, treatment with antioxidants inhibits keratinocyte differentiation, indicating the importance of the oxidative environment to the program of differentiation (Kennedy et al., 2013). Supportive of our findings, a subsequent study reported that epidermal deletion of the murine mitochondrial transcription factor A (TFAM) diminishes mitochondrial ROS and inhibits epidermal differentiation (Hamanaka et al., 2013). Together with the results described here, these studies demonstrate that regulated decreases of glycolytic flux along with additional metabolic reprogramming promote keratinocyte differentiation. Moreover, AHR regulation of SLC2A1 and ENO1 expression appears to be an early and contributing mechanism in keratinocytes.
MATERIALS AND METHODS
Cell Culture
Neonatal NHEKs were purchased from Lonza (Walkersville, MD). N/TERT-1 cells, kindly provided by James G. Rheinwald, [(Dickson et al., 2000) Cell Culture Core, Harvard University]. NHEKs and N/TERT-1 cells were grown as previously described for NHEKs (Sutter et al., 2009). Cells were grown to confluence before pretreatment in basal KSFM for 24 hours, followed by treatment in basal KSFM as indicated.
Chemicals
The following chemicals were purchased from MilliporeSigma (Billerica, MA): DMSO (0.1%, D2650); 2DG (20 mM, 25972); BAY876 (5 μM, SML1774); GNF351 (100 nM, 182707); CH223191 (1 μM, C8124). Sodium pyruvate (5 mM, 11360070, Invitrogen). POMHEX (25 or 50 μM) was kindly provided by Florian L. Muller (UT MD Anderson Cancer Center, Houston, TX).
Antibodies
Antibodies used for immunoblotting are described in the Supplementary Materials and Methods.
Protein Quantitation
Protein quantitation is described in the Supplementary Materials and Methods.
Immunoblotting, Enhanced Chemiluminescence, and Densitometry
Immunoblotting, enhanced chemiluminescence, and densitometry are described in the Supplementary Materials and Methods.
ChIP-Seq Library Preparation
ChIP-Seq Library Preparation is described in the Supplementary Materials and Methods.
ChIP-PCR
ChIP-PCR is described in the Supplementary Materials and Methods.
Transcriptional Luciferase Reporter Constructs and Assays
ENO1-promoter (1550 bp), lentiviral transduction of N/Tert-1 cells. Cells were transduced with particles containing a 1550 bp fragment of the ENO1 promoter (−1186 to +364, relative to the TSS at +1) (LPP-HPRM43681-LvPG04-050, GeneCopoeia) using a MOI of 15 and selected for stable integration with puromycin (1μg/ml, Sigma). Cells were treated with TCDD and/or GNF351 for 48 hours. Gaussia luciferase and secreted alkaline phosphatase (normalizer) were measured following manufacturer’s instructions using Secrete-Pair Dual Luminescence Assay Kit (LF031, GeneCopoeia) and the TD-20/20 Luminometer (Promega, Madison, WI). ENO1-promoter (189 bp), transient transfection into NHEKs. The fragment (−34 to +155, relative to the TSS of +1) of the ENO1 promoter (corresponding to the site of AHR binding from ChIP-Seq and ChIP-PCR results) was amplified from human genomic DNA using PfuUltra II Fusion HotStart DNA Polymerase (600670, Agilent) and the primers listed in Table S3. Following amplification, the fragment was digested with KpnI and HindIII (NEB) and ligated into KpnI- and HindIII-digested pGL3-Basic (E1751, Promega). NHEKs were transfected using Lipofectamine 3000 (L3000015, Invitrogen). Each well of a 12-well dish of NHEKs received 4.5 μl P3000, 2 μg of the ENO1-pGL3-Basic luciferase or vector alone, 0.01 μg of renilla luciferase vector (pGL4.74, E6921, Promega) and 0.25 μg of AHR in pcDNA3.1 in basal KSFM. Cells were transfected for 24 hours after which they were treated with TCDD for 72 hours. Luciferase was measured following the manufacturer’s instructions for the Dual-Luciferase Reporter Assay System (E1960, Promega) and the TD-20/20 Luminometer (Promega).
RNA Isolation and Quantitative PCR
RNA isolation and quantitative PCR is described in the Supplementary Materials and Methods. For all RNA studies, cells were treated for 24 hours. Total RNA isolation and quantitative PCR (qPCR) were performed as previously described (Sutter et al., 2009). Samples were normalized to values of tubulin, alpha 1C. Primers used in this study are listed in Table S2.
Glucose Uptake Assay
Following treatment, NHEKs were collected in phosphate buffered saline and the amount of glucose taken into the cells over a 10 minute period was measured using a Glucose Uptake-Glo Kit (J1341, Promega) according to the manufacturer’s instructions.
ENO1 Activity Assay
Following treatments, NHEKs were collected in phosphate buffered saline and enzyme activity was determined by the ENO1 Human Activity Assay Kit (ab117994, Abcam), according to the manufacturer’s instructions.
Intracellular Pyruvate
Following treatments, NHEKs were collected in phosphate buffered saline and intracellular pyruvate was determined by the Fluorescent Pyruvate Assay Kit [ab65342, Abcam (Cambridge, MA)], according to the manufacturer’s instructions. Pyruvate concentrations were calculated from a standard curve of the provided pyruvate standard. When media was changed during a treatment, the last 24 hours of a 72-hour treatment was in fresh media with the same treatment.
Extracellular Lactate
Following 48 hours of treatment, NHEKs were placed in fresh media containing the same treatment for an additional 24 hours. The amount of lactate transported into the media over this 24-hour period was measured using the Lactate Assay Kit (MAK065, MilliporeSigma) according to the manufacturer’s instructions.
Intracellular ATP
Following treatment, intracellular ATP was determined by the Luminescent ATP Detection Assay Kit (ab113849, Abcam), according to manufacturer’s instructions. ATP concentrations were calculated from a standard curve.
CE Assay
The formation of CEs, a measure of terminal differentiation, was quantified essentially as previously described (Banks-Schlegel and Green, 1980). NHEKs were counted before they were spun and resuspended in solution containing 10 mM Tris-HCl pH 7.5, 1% SDS and 1% 2-mercaptoethanol and CEs were counted following a 10-minute incubation at room temperature with DNase (80 Units/ml) and 10-minute incubation at 90°C. The number of CEs were determined per number of cells and expressed as a percentage.
Knockdown of SIRT1
NHEKs and N/TERT-1 cells were transduced with lentiviral particles of SIRT1 shRNA (TRCN0000229631, SigmaMillipore) or the vector control (SHC201V, SigmaMillipore) using a multiplicity of infection of 1 and a 24 hour incubation in the presence of polybrene (5 mg/ml, SigmaMillipore). Puromycin (1 mg/ml, SigmaMillipore) resistant cells were expanded and used for experiments.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7.03. The use of particular tests is described in the legends of each figure.
Supplementary Material
ACKOWLEDGEMENTS
We would like to thank Haley Rainwater for help in performing experiments. This work was funded by the National Institutes of Health (R01 ES017014).
ABBREVIATIONS
- AHR
aryl hydrocarbon receptor
- NHEK
normal human epidermal keratinocyte
- ChIP
chromatin immunoprecipitation
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- ROS
reactive oxygen species
- 2DG
2-deoxyglucose
- CE
cornified envelope
- qPCR
quantitative PCR
- TCA
tricarboxylic acid
- XRE
xenobiotic response element
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adachi K, Yamasawa S. Quantitative histochemistry of the primate skin. 8. Enolase. J Invest Dermatol 1966a;47(4):293–5. [DOI] [PubMed] [Google Scholar]
- Adachi K, Yamasawa S. Quantitative histochemistry of the primate skin. I. Hexokinase. J Invest Dermatol 1966b;46(5):473–6. [DOI] [PubMed] [Google Scholar]
- Banks-Schlegel SP, Green H. Studies on the development of the definitive cell type of embryonic epidermis using the cross-linked envelope as a differentiation marker. Dev Biol 1980;74(2):275–85. [DOI] [PubMed] [Google Scholar]
- Bedogni B, Powell MB. Skin hypoxia: a promoting environmental factor in melanomagenesis. Cell Cycle 2006;5(12):1258–61. [DOI] [PubMed] [Google Scholar]
- Blander G, Bhimavarapu A, Mammone T, Maes D, Elliston K, Reich C, et al. SIRT1 promotes differentiation of normal human keratinocytes. J Invest Dermatol 2009;129(1):41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 2005;6(4):328–40. [DOI] [PubMed] [Google Scholar]
- Chung SJ, Lee SH, Lee YJ, Park HS, Bunger R, Kang YH. Pyruvate protection against endothelial cytotoxicity induced by blockade of glucose uptake. J Biochem Mol Bio 2004;37(2):239–45. [DOI] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 2000;20(4):1436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Porter W, Samudio I, Vyhlidal C, Kladde M, Safe S. Transcriptional activation of c-fos protooncogene by 17beta-estradiol: mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Endocrinol 1999; 13(9): 1511–21. [DOI] [PubMed] [Google Scholar]
- Haas K, Weighardt H, Deenen R, Kohrer K, Clausen B, Zahner S, et al. Aryl Hydrocarbon Receptor in Keratinocytes Is Essential for Murine Skin Barrier Integrity. J Invest Dermatol 2016;136(11):2260–9. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal 2013;6(261):ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Mutlu GM. PFKFB3, a Direct Target of p63, Is Required for Proliferation and Inhibits Differentiation in Epidermal Keratinocytes. J Invest Dermatol 2017;137(6):1267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai W, Zhao R, Song H, Zhao J, Zhang L, Gao C, et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun 2014;5:4738. [DOI] [PubMed] [Google Scholar]
- Im MJ, Yamasawa S, Adachi K. Quantitative histochemistry of the primate skin. III. Glyceraldehyde-3-phosphate dehydrogenase. J Invest Dermatol 1966;47(1):35–8. [DOI] [PubMed] [Google Scholar]
- Jesko H, Strosznajder RP. Sirtuins and their interactions with transcription factors and poly(ADP-ribose) polymerases. Folia Neuropathol 2016;54(3):212–33. [DOI] [PubMed] [Google Scholar]
- Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett 2008;582(16):2417–23. [DOI] [PubMed] [Google Scholar]
- Kennedy LH, Sutter CH, Leon Carrion S, Tran QT, Bodreddigari S, Kensicki E, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated production of reactive oxygen species is an essential step in the mechanism of action to accelerate human keratinocyte differentiation. Toxicol Sci 2013;132(1):235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011;147(3):629–40. [DOI] [PubMed] [Google Scholar]
- Lin Y, Satani N, Hammoudi N, Ackroyd JJ, Khadka S, Yan VC, et al. Eradication of ENO1-deleted Glioblastoma through Collateral Lethality. bioRxiv 2018; 10.1101/331538. [DOI] [Google Scholar]
- Loertscher JA, Sattler CA, Allen-Hoffmann BL. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters the differentiation pattern of human keratinocytes in organotypic culture. Toxicol Appl Pharmacol 2001;175(2):121–9. [DOI] [PubMed] [Google Scholar]
- Ming M, Zhao B, Shea CR, Shah P, Qiang L, White SR, et al. Loss of sirtuin 1 (SIRT1) disrupts skin barrier integrity and sensitizes mice to epicutaneous allergen challenge. J Allergy Clin Immunol 2015;135(4):936–45 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem 1996;271(51):32529–37. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Ng HH. The metabolic programming of stem cells. Genes Dev 2017;31(4):336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebeneicher H, Cleve A, Rehwinkel H, Neuhaus R, Heisler I, Muller T, et al. Identification and Optimization of the First Highly Selective GLUT1 Inhibitor BAY-876. ChemMedChem 2016;11 (20) :2261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TT, Green H. Differentiation of the epidermal keratinocyte in cell culture: formation of the cornified envelope. Cell 1976;9(4 Pt 1):511–21. [DOI] [PubMed] [Google Scholar]
- Sutter CH, Bodreddigari S, Campion C, Wible RS, Sutter TR. 2,3,7,8-Tetrachlorodibenzo-p-dioxin increases the expression of genes in the human epidermal differentiation complex and accelerates epidermal barrier formation. Toxicol Sci 2011;124(1):128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter CH, Yin H, Li Y, Mammen JS, Bodreddigari S, Stevens G, et al. EGF receptor signaling blocks aryl hydrocarbon receptor-mediated transcription and cell differentiation in human epidermal keratinocytes. Proc Natl Acad Sci U S A. 2009;106(11):4266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CY, Mauro C. Similarities in the Metabolic Reprogramming of Immune System and Endothelium. Front Immunol 2017;8:837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest 2013;123(2):917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaard EH, Podolsky MA, Smits JP, Cui X, John C, Gowda K, et al. Genetic and pharmacological analysis identifies a physiological role for the AHR in epidermal differentiation. J Invest Dermatol 2015;135(5):1320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick AN, Drury DR, Nakada HI, Wolfe JB. Localization of the primary metabolic block produced by 2-deoxyglucose. J Biol Chem 1957;224(2):963–9. [PubMed] [Google Scholar]
- Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther 2013;345(3):419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.