Abstract
Previously we showed that alveolar macrophages(AMs) from patients with chronic beryllium disease(CBD) and beryllium sensitization(BeS) demonstrated significantly greater cell surface CD16(encoded by the FCGR3A gene) than controls. We hypothesized that these differences were related to polymorphisms in the FCGR3A gene. This study was to determine the association between FCGR3A polymorphisms in CBD, BeS versus controls as well as clinical data, providing potential information about disease pathogenesis, risk and activity. 189 CBD/154 BeS /150 controls (92 Be-exposed-non-diseased and 58 healthy controls) were included in this study. Sequence specific primers polymerase chain reaction(PCR-SSP) was used to determine FCGR3A 158V/F polymorphisms. We found significantly higher frequencies of the 158V allele (OR:1.60 (CI: 1.17-2.19),p=0.004) and 158VV homozygotes (OR:2.97 (CI:1.48-5.97) p=0.007) in CBD versus controls. No differences were found in the frequencies of FCGR3A alleles or genotypes between BeS versus controls and CBD versus BeS. Average changes in exercise testing maximum workload(Wlm),maximum oxygen consumption(VO2m) and diffusion capacity of carbon monoxide(DLCO) demonstrated greater decline over time in those CBD cases with the 158VV gene, modelled between 10 and 40 years from first beryllium exposure. The FCGR3A V158F polymorphism is associated with CBD compared to BeS and controls and may impact lung function in CBD.
Introduction
Chronic beryllium disease (CBD) develops in up to 16% of individuals exposed to beryllium (Be) and is characterized by granulomatous inflammation and the accumulation of CD4+ T cells in the lung [1–4]. CBD is preceded by beryllium sensitization (BeS), a beryllium-specific immune response demonstrated by a blood test called the beryllium lymphocyte proliferation test (BeLPT)[5, 6]. BeS and CBD are genetically determined, associated with an HLA class II epitope, primarily a glutamic acid at amino acid position 69 (Glu69) in HLA-DPB1, and to a lesser extent a glutamic acid at amino acid position 71 in HLA-DRB1 [7]. Genetic factors important in disease susceptibility and severity are not well-understood, although it is clear that exposure is also important in development of disease and more severe disease [8]. Studies have shown that beryllium persists within the lungs of individuals many years after exposure has ceased [5, 8, 9], suggesting a failure to clear the Be antigen from the lungs. This retention of Be may perpetuate an ongoing Be-specific immune response in CBD and/or progression from BeS to CBD.
Human FcγRIIIa (CD16) receptor is an extensively glycosylated heterogeneous protein with an apparent molecular weight of 50-80 kDa. FCGR3A gene codes for the FcγRIIIa receptor present in macrophages, NK and γδ T cells, with low-affinity for IgG-containing immune complexes (IC)[10–14]. Cross-linking of FcγRs potently triggers functions such as phagocytosis, respiratory burst, degranulation, antibody-dependent cellular cytotoxicity and antigen presentation[10, 15]. There is a G559T polymorphism in the FCGR3A gene that leads to the substitution of Valine for Phenylalanine at position 158 of the polypeptide chain (158V/F). The FcγRIIIa-158V allotype exhibits higher affinity for IgG1 and IgG3 than does FcγRIIIa-158F, and is capable of binding IgG4[16, 17]. Higher levels of FCGR3A transcripts have been observed among individuals with the FCGR3A-158 V/V versus V/F or F/F genotype; increased cell surface CD16 expression by quantitative flow cytometry on NK cells from individuals expressing at least one valine at FCGR3A-158 versus F/F (P = 0.029); as well as augmented rituximab binding and rituximab-mediated, antibody-dependent cellular cytotoxicity (ADCC)[18]. An over-representation of FcγRIIIa-158F allele has been reported in patients with systemic lupus erythematosus (SLE)[19–21]. Although the FCGR3A-158 V-F polymorphism impacts multiple autoimmune and infectious diseases[15, 16], the clinical significance of FcγRIIIa in CBD is not known.
In our previous studies, we showed that alveolar macrophages (AMs) from CBD and BeS demonstrated significantly greater cell surface CD16 expression than controls[3]. However, Be exposure in vitro decreased CD16 expression and phagocytosis by CBD and BeS AMs. Given the important roles that FcγRIIIa plays in binding, phagocytosis and clearance of immune complexes, we hypothesized that these differences were related to polymorphisms in the FCGR3A gene, which has been associated with other autoimmune disorders, such as sarcoidosis and SLE, as well as protein levels. In this study, we aimed to investigate the association of FCGR3A V158F with the susceptibility and clinical markers of CBD. It is the first analysis demonstrating the association between the V158F polymorphism of FCGR3A gene and CBD and with lung function in CBD.
Results
Subject Demographics
The demographics of the subjects are shown in Table 1. No significant differences in the age, race and smoking status were noted in BeS, CBD and Be-exposed control subjects. The majority of BeS, CBD subjects and Be-exposed controls were male and Caucasian, typical of Be-industries. The Be-non exposed controls were more likely to be female and younger (p<0.05) than Be-exposed controls. Not surprisingly, CBD subjects had a statistically significant increase in total BAL WBC count and BAL lymphocytes % compared with BeS subjects (p < 0.05). The BAL BeLPT peak stimulation index was higher in CBD than in BeS subjects (P < 0.05).
Table 1.
Clinical characteristics of Healthy control, BeS, and CBD subjects.
| CBD (n=189) | BeS (n=154) | Be-exposed controls (n=92) | Be-non exposed controls (n=58) | |
|---|---|---|---|---|
| Age (yr) | 53 (29-73) | 57 (30-84) | 66 (44-84) | 36(20-70)* |
| Gender (M/F) | 154/31/4 | 112/40/2* | 68/23/1 | 17/40/1* |
| Race (W/BLACK/ASIAN/other/no data) | 148/9/1/1/30 | 146/6/0/0/2 | 83/6/1/1/0 | 55/1/2/0/1 |
| Hispanic ethnicity (His/Non His/No data) | 17/157/15 | 17/136/1 | 11/80/1 | 0/57/1* |
| Smoking status (CS/FS/NS/no data) | 13/77/90/9 | 19/52/77/6* | No data | No data |
| BAL cells | 39.5(2.1-154.3) | 22.6(3.8-122)** | NA | NA |
| Total WBC count (millions) | ||||
| BAL cells Lymphocytes (%) | 28.6(0.7-84) | 7.4(0.7-48)** | NA | NA |
| Peak stimulation index | 30.3(0.5-515.3) | 1.9(0-20.5)** | NA | NA |
| Wlm | 175 (50-320) | 15040-280) | NA | NA |
| Vo2m | 2.01(0.79-3.21) | 1.74(0.85-3.37) | NA | NA |
| Pao2r | 71 (36-87) | 73(52-91)** | NA | NA |
| Pao2b | 76 (38-99) | 78(35-97) | NA | NA |
| Aado2r | 11 (0-44) | 8.9(0.3-27.1)** | NA | NA |
| Aado2m | 17.0 (0.3-117) | 15.0(0.2-44)* | NA | NA |
| FEV1(L) | 3.13(1.1-5.9) | 3.13(1.2-5.0)* | NA | NA |
| FEV1% pred | 93(35-126) | 94(35-129) | NA | NA |
| FVC(L) | 4.15(1.22-7.23) | 4.01(1.83-6.42) | NA | NA |
| FVC %pred | 90(31-126) | 92.5(51-129)* | NA | NA |
| TLC(L) | 6.60(3.09-10.65) | 6.82(3.85-9.38)** | NA | NA |
| TLC %pred | 106(46-154) | 111(73-157)** | NA | NA |
| DLCO | 29.94(7.83-57.60) | 27.17(11.95-51.33) | NA | NA |
| DLCO % pred | 89(20-152) | 86(34-138) | NA | NA |
P<0.05
P<0.01 Wlm, Vo2m, Pao2r, Pao2b, Aado2r, Aado2m were adjusted for gender, race, age, while FEV1, FVC, TLC, DLCOU were adjusted for gender, race, age, and height.
Furthermore, compared to BeS, CBD subjects had lower PaO2r, higher Aado2r and higher Aado2m, adjusted for age, gender and race (p < 0.05). FEV1 and TLC have significant differences between CBD and BeS subjects, adjusted for age, gender, race and height (p < 0.01). Furthermore, TLC % predicted has significant differences between CBD and BeS subjects (p < 0.05). These demonstrate the impact of granulomatous inflammation. No significant differences were found in Wlm, vo2m, pao2b, FEV1% pred , FVC, DLCO, and DLCO % pred between CBD and BeS subjects.
FCGR3A 158V/F allele and genotype associate with CBD disease risk
As demonstrated in Supplement Figure 1, both the PCR-SSP and DNA sequencing detected the FCGR3A variants. No differences were found in the frequencies of FCGR3A alleles or genotypes between Be-exposed controls and Be-non exposed controls (Table 2). Therefore, we combined the two controls groups together to enhance power for analysis.
Table 2.
Comparative analysis of the frequency of genotypes and alleles of FCGR3A gene in Be-exposed controls and Be-non exposed controls (%)
| Be-non exposed controls (n=58) | Be-exposed controls (n=92) | X2 | P value | |
|---|---|---|---|---|
| GENOTYPE | ||||
| 158FF | 24(41.4) | 42(45.7) | 2.834 | 0.242 |
| 158FV | 31(53.4) | 39(42.4) | ||
| 158VV | 3(5.2) | 11(11.9) | ||
| ALLELE | ||||
| F | 79 | 123 | 0.225 | 0.362 |
| V | 44 | 61 |
The genotype frequencies for the controls (p=0.46) and CBD subjects (p=0.14) polymorphisms were in the Hardy-Weinberg equilibrium. However those for BeS (p=0.008) were not. Genotype frequencies of FCGR3A SNPs are listed in Table 2. There were significant differences for the FCGR3A 158V/F polymorphisms comparing CBD cases to the controls. The 158V allele was significantly higher (OR:1.60 (CI: 1.17-2.19),p=0.004), 158VV homozygotes (OR:2.97 (CI:1.48-5.97) p=0.007) were also higher in CBD versus controls (Table 3). No differences were found in the frequencies of FCGR3A alleles or genotypes between BeS versus controls and versus CBD.
Table 3.
A comparative analysis of the frequency of genotypes of FCGR3A gene between CBD, BeS and healthy controls
| FCGR3A alleles(%) | CBD (n=189) | BES (n=154) | Health control (n=150) | CBD vs control | BES vs control | ||
|---|---|---|---|---|---|---|---|
| X2 (P) | OR(95%CI) | X2 (P) | OR(95%CI) | ||||
| GENOTYPE | |||||||
| 158FF | 65(34.4) | 80(51.9) | 66(44) | 10.013 (0.007) | 0.336 (0.168-0.675) | 5.724 (0.057) | 0.771(0.366-1.625) |
| 158FV | 83(43.9) | 52(33.8) | 70(46.7) | 0.356 (0.613) | 0.405(0.204-0.803) | 5.263 (0.021) | 0.473(0.221-1.011) |
| 158VV | 41(21.7) | 22(14.3) | 14(9.3) | Ref | 1 | Ref | 1 |
| ALLELE | |||||||
| F | 213(56.3) | 212(68.8) | 202(67.3) | 8.499 (0.004) | 0.626(0.457-0.859) | 0.157 (0.692) | 1.071 (0.762-1.507) |
| V | 165(43.7) | 96(31.2) | 98(32.7) | Ref | 1 | Ref | 1 |
Effect of genotypes of FCGR3A gene on pulmonary function testing
The effect of FCGR3A genotypes on pulmonary function test in the CBD subjects were measured at time of initial CBD diagnosis. The pulmonary function tended to be higher in those with the FCGR3A 158FF genotype. Specifically, FVC %pred (p=0.07) and DLCO % pred (p=0.09) were higher in CBD subjects with the158FF in comparison with those with other genotypes (Table 4).
Table 4.
Effect of genotypes of FCGR3A gene on pulmonary function test in the CBD patients at time of initial CBD diagnosis (FEV1, FVC, TLC, DLCOU were adjusted for gender, race, age, and height).
| FEV1 | FEV1PP | FVC | FVCPP | TLC | TLCPP | DLCOU | DLCOUPP | |
|---|---|---|---|---|---|---|---|---|
| FF | 3.3 (1.6-5.9) | 96 (47-123) | 4.21 (2.37-7.23) | 91 (68-121) | 6.70 (4.63-9.38) | 107.5 (77-137) | 29.91 (15.37-51.41) | 93 (44-140) |
| FV | 3.1 (1.1-4.8) | 89 (35-126) | 4.13 (1.22-6.57) | 85 (31-126) | 6.67 (3.09-9.64) | 104 (46-137) | 31.64 (7.83-57.60) | 89 (20-152) |
| VV | 3.0 (2.0-4.5) | 92 (50-123) | 3.92 (2.60-6.08) | 94 (62-122) | 6.5 (3.99-10.65) | 104 (73-154) | 27.73 (15.53-40.15) | 86 (43-122) |
| p value | 0.19 | 0.415 | 0.09 | 0.249 | 0.55 | 0.136 | 0.09 | 0.207 |
Differences in rates of CBD lung function progression were evaluated for associations with FCGR3A polymorphisms using mixed effects models, along with time from first beryllium exposure.
There are 57 FF, 76 FV and 33 VV subjects in this study. Our results showed the average changes of Wlm (p=0.0217), VO2m(p=0.0097), DLCOU(p=0.0240) over time modeled between 10 and 40 years from first beryllium exposure in those CBD cases with 158VV of FCGR3A gene demonstrated a greater decline (Figure 1) than those with 158 FF (p<0.05). No association was noted between the FCGR3A polymorphisms and FEV1 and aado2m over time (Data not shown).
Figure 1.
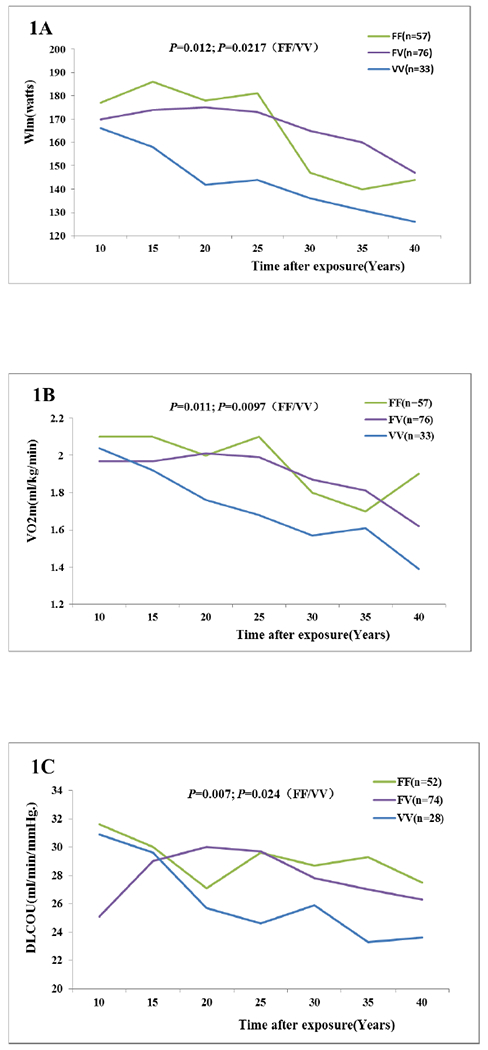
Average changes of Wlm(1A), VO2m(1B), DLCOU(1C) over time modelled between 10 and 40 years from first beryllium exposure in those CBD, adjusted for gender, race and age, demonstrating a greater decline in the 158VV of FCGR3A gene.
Discussion
This is the first study to identify associations between CBD, disease progression over time and FCGR3A polymorphisms (rs396991). We found significantly higher frequencies of the 158V allele, and 158VV homozygotes in CBD versus controls. Furthermore, the FCGR3A polymorphism had clinical relevance in CBD, and pulmonary function tended to be higher in those CBD subjects with the FCGR3A 158FF genotype. Similarly, the average changes in exercise testing and lung function test, such as Wlm, VO2m and DLCO over time modeled between 10 and 40 years from first beryllium exposure in those CBD subjects with 158VV of FCGR3A gene demonstrated a greater decline than those with 158 FF and 158FV.
CBD is a noncaseating granulomatous lung disorder due to beryllium exposure. Genetic susceptibility contributes to the development of BeS and progression of BeS to CBD. Previous studies showed that the granulomatous response in CBD is dictated by functional genetic susceptibility factors in the E69 gene variant in conjuction with exposure[22]. The replication of genes outside the HLA Class II region has been limited using candidate gene studies, although we have defined functional factors associated with more severe CBD and more severe sarcoidosis[9]. Specifically, TGF-β and CCR5 variants were associated with more severe sarcoidosis and CBD lung function and radiographic abnormalities[23, 24]. Similarly, in this study we found 158VV variants associated with more severe CBD lung function, suggesting that these variants are impacting lung inflammation and physiology. However, unlike these variants, we found that the FCGR3A 158VV variant was associated with CBD compared to controls, suggesting that it may play a role in risk for disease as well as disease severity. Similar to our study, FCGR3A polymorphisms have been associated with severity of diseases, including early rheumatoid arthritis (RA) [25], pulmonary tuberculosis [26], and sarcoidosis[27]. The striking finding that 158VV is associated with CBD lung function and gas exchanges decline (lower Wlm, VO2m and DLCOU) over time on average (p=0.0217, 0.0097, 0.0240, respectively), suggested that FCGR3A 158VV homozygous genotype could contribute to the accelerated lung function decline in CBD.
Beryllium persists in the lungs for many years even after exposure stops, raising the question as to whether there are issues with host clearance or beryllium solubility that results in the inability to clear beryllium antigen from the lungs. Furthermore, persistent beryllium exposure has the potential to activate a long-lasting immune response, and result in CBD in workers years after last exposure[28]. Macrophages play an essential role in the process of beryllium clearance and are also the main effector cells during the formation of beryllium induced granuloma. Our previous studies showed that alveolar macrophages from CBD and BeS have significantly higher CD16 expression levels and higher phagocytosis function than controls[3]. However, the biological mechanisms underlying these increased CD16 levels and phagocytosis remain unclear. Many studies have shown that the CD16 V158F polymorphism (rs396991) is related to macrophage phagocytic function; we postulate that this may also be the case in CBD.
Single-nucleotide polymorphism (SNP) and mutations in FCGR3A have been associated with a number of immune mediated diseases , including systemic lupus erythematosus (SLE), rheumatic arthritis (RA), sarcoidosis and other autoimmune diseases[16, 19–21, 29]. Relevant to CBD, previous studies showed that the 158F and 158FF prevalence were much higher in stage I sarcoidosis, another granulomatous lung disease of unknown cause and a milder form of this disease. These same variants have also been associated with decreased FCGRIIIa affinity and clearance of immune complexes (ICs) [27]. Specifically, the V158F polymorphism has been implicated in the binding and phagocytotic function of macrophages; the 158VV genotype presented a higher ICs binding and a more effective macrophage ingestion of ICs. The enhanced phagocytic ability of macrophage followed by increased antigen presentation, may result in significant cytokines release and even an autoimmune response. On the contrary, the 158FF genotype has been associated with lower IC binding ability and fewer ICs ingested by macrophages, leading to lower cytokines production[3, 27]. In our study, we found an increased frequency of the 158V allele and 158VV genotype in CBD subjects compared to controls. Since the 158V polymorphic variant is associated with a very effective uptake of ICs by FCGRIIIa, this may result in excessive antigen presentation, activation of monocytes, macrophages, dendritic and Natural Killer cells, subsequent production of cytokines and initiation of an autoinflammatory process, all of which may contribute to granuloma information. It is plausible that 158V plays an important role in CBD granuloma formation and disease pathogenesis.
There are some limitations to this study. The FCGR3A polymorphisms for BeS (p=0.008) were not in Hardy-Weinberg equilibrium. Deviations from HWE can be very informative. In case subjects, deviation from HWE, assuming sources of error have been eliminated, may indicate the association of a locus with disease. In cases, this deviation could be due to genetic drift, immigration, non-random mating, new mutations, natural selection, or combinations of these effect. It is possible that these factors have impacted HWE in our cases of BeS, as we did not find problems with our genotype calls when we sequenced BeS cases compared to the call for our CBD cases or controls. However, this finding is perplexing and limits our conclusions. The other limitation in our study is whether this variant is functional. We were unable to assess the functional aspects of this variant as we used DNA from cases and controls from prior studies and could not evaluate the function of the FcGR in these same subjects.
In summary, the V158F polymorphism of FCGR3A gene is associated with CBD compared to BeS and controls and may impact the lung function in CBD. We revealed an increase in the occurrence of 158V allele and 158VV homozygotes of FCGR3A gene in CBD versus controls. Furthermore, our data also suggested that FCGR3A 158VV homozygous genotypes could contribute to the accelerated lung function decline in CBD. The FCGR3A polymorphisms may serve as a risk factor of CBD.
Materials and Methods
Study population.
All participants gave informed written consent in accordance with the Declaration of Helsinki, and the study was approved by National Jewish Health (NJH) Institutional Review Board (IRB). 189 CBD subjects, 154 BeS subjects and 150 controls (92 Be-exposed non-diseased and 58 healthy controls) were enrolled in this study, drawn from subjects seen clinically at NJH. The diagnosis of CBD was established using previously defined criteria, including the presence of granulomatous inflammation on lung biopsy, and a positive proliferative response of blood and/or bronchoalveolar (BAL) cells to BeSO4 in vitro. The diagnosis of BeS was established based on a positive proliferative response of blood cells to BeSO4 in vitro on the BeLPT and the absence of granulomatous inflammation on lung biopsy. Individuals were considered to be current smokers if they had smoked daily for at least the last 3 months.
Indices of CBD disease severity
Clinical evaluations were completed on initial evaluation (baseline) and during follow-up over time on the cases, usually annually or biannually. We evaluated data obtained from pulmonary function testing, exercise testing, and chest radiography. The forced expiratory volume in 1 second (FEV1) and forced expiratory vital capacity (FVC) were measured with a pneumotachograph. Total lung capacity (TLC) was measured in a constant pressure body plethysmograph. The single-breath method of Ogilvie et al[30] was used to evaluate the diffusion capacity of carbon monoxide (DLCO). Gas exchange and maximal exercise capacity were determined with a 380 B cycle ergometer (Siemens-Elema) with continuous cardiac rhythm and arterial oxygen content monitoring[31] . An indwelling arterial line measured arterial blood gases at rest and after each minute of exercise. Results are reported as the arterial partial pressure of oxygen, (PaO2) at rest (PaO2r) and during maximal exercise (PaO2m) and the alveolar-to-arterial oxygen pressure difference (AaDO2), at rest (AaDO2r) and during maximal exercise (AaDO2m) and work load at maximum exercise (Wlm). There were 160 CBD cases that had on average 10.3 visits (range 1–14); others did not have additional follow up after diagnosis, as they were recently diagnosed or lost to follow-up.
DNA sample preparation
Genomic DNA was isolated using a Paxgene blood DNA kit according to the manufacturer’s instruction (Qiagen, Valencia, CA). DNA concentration was determined on a NanoDrop ND-1000. The purified DNA samples were stored at −20 °C as stock and we avoided frequently thawing and freezing.
Sequence-specific primer Polymerase Chain Reaction (PCR-SSP) for FCGR3A Genetic Polymorphisms
PCR-SSP was used for the FCGR3A 158V/F polymorphism determination[16]. We used a common reverse primer (5’-CAACTCAACTTCCCAGTGTGAT-3’) that ensured gene specificity for FCGR3A and either a forward 158F-specific primer (5’-TACTTCTGCAGGGGGCTTT-3’) or a primer specific for 158V allele (5’-TACTTCTGCAGGGGGCTTG-3’). Both 158F and 158V PCR product were 147 bp. Each allele specific primer contained intentional single-nucleotide mismatches to provide allele specificity. In order to eliminate false negatives, we also added a common forward primer (5’-TCAGGATCTGGGTGGTACG-3’) that could generate a 423 bp product in all PCR reactions. The PCR-SSP reaction for 158F allele was performed with 20 ng of genomic DNA, 0.3 μM 158F-specific forward primers and common forward primers, 0.5 μM common reverse primers, and 10 μL PCR Green master mix (Promega, San Luis Obispo, CA,) in a total volume of 20 μL. A temperature profile for 158F allele comprised 5 minutes at 95 °C, followed by 30 cycles of amplification (30 seconds at 95 °C, 1 min of annealing at 57 °C and 30 seconds at 72 °C), then followed by a final elongation at 72 °C for 7 min. All the PCR products were analyzed on a 2% agarose gel containing 0.05% ethidium bromide (EB). The genotypes were read blinded to subjects’ status. 10% of the samples were genotyped a second time to ensure appropriate genotyping.
Genomic DNA sequencing
To confirm the FcgRIIIA genomic sequence and above genotypes, primers were designed to amplify a portion of exon 4 of FCGR3A which corresponds to EC2. The forward primer (5’-TCAGGATCTGGGTGGTACG-3’, corresponding to nt 4692-4710). The reverse primer (5’-CAACTCAACTTCCCAGTGTGAT-3’, corresponds to nt 5091–5112). The 423 bp PCR product containing the V158F nucleotide polymorphic site was purified from a 2% agarose gel with the QIAquick Gel Extraction Kit. Fluorescence-based automated cycle sequencing of PCR product was performed by Eton Bioscience Inc. Ten subjects from each group were chosen at random to confirm their genotyping with DNA sequencing.
Statistical Analysis:
Chi-square test and odds ratio (OR, 95% CI) were used to evaluate the difference and relative risk in allele and genotype distribution between the groups. The association of FCGR3A genotypes and pulmonary function parameters was evaluated using analysis of variance (ANOVA) and Mixed models. In all the analyses, Wlm, vo2m, pao2r, pao2b, aado2r and aado2m were adjusted by age, gender and race; FEV1 L, FVC L, TLC L, and DLCO were adjusted by age, gender, race and height, but FEV1% pred, FVC % pred, TLC %pred and DLCO % pred were not adjusted. The statistical analysis was performed using commercially available software (SAS software, version 9.4. A p<0.05 was taken as statistically significant and all tests were two-sided).
Supplementary Material
Supplement Figure 1. Sequence and gel analysis of a portion of FCGRIIIA cDNA from three Representatives. FcgRIIIa encoding cDNA was prepared from purified PBMCs and polymerase chain reaction (PCR) with the sequence specific primers (PCR-SSP) was used (see Methods) to determine the genotype of FCGRIIIA 158V/F polymorphism. The PCR product were run on 2% agrose gel. PCR products were analyzed on a 2% agarose gel containing 0.05% ethidium bromide (EB). The long common PCR product were gel extracted and sequenced. The left panels showed the example sequencing results and the right panel showed the gel picture of three genotypes of FCGRIIIA 158V/F polymorphism. Upper panel: 158FF genotype, Middle panel: 158FV panel, and lower panel: 158VV panel. For the PCR gel pictures, the PCR product at 147 bp represent either 158F (left PCR product) or 158V (right PCR product) genotype depending on the primer specificity, and the PCR product at 423 bp represent the common product which is used to eliminate false negatives.
Acknowledgements
This research was supported by National Institutes of Health 1R01ES025722-01A1, 1K01ES020857-01, P01ES11810, UL 1TR001082 and ES023826-01A1.
Footnotes
Conflict of interest:
The authors declare that they have no conflict of interest.
References:
- 1.Saltini C, Winestock K, Kirby M, Pinkston P, Crystal RG. Maintenance of alveolitis in patients with chronic beryllium disease by beryllium-specific helper T cells. N Engl J Med 1989,320:1103–1109. [DOI] [PubMed] [Google Scholar]
- 2.Rossman MD, Kern JA, Elias JA, Cullen MR, Epstein PE, Preuss OP, et al. Proliferative response of bronchoalveolar lymphocytes to beryllium. A test for chronic beryllium disease. Ann Intern Med 1988,108:687–693. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Hamzeh N, Gillespie M, Elliott J, Wang J, Gottschall EB, et al. Beryllium increases the CD14(dim)CD16+ subset in the lung of chronic beryllium disease. PLoS One 2015,10:e0117276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreiss K, Mroz MM, Zhen B, Martyny JW, Newman LS. Epidemiology of beryllium sensitization and disease in nuclear workers. Am Rev Respir Dis 1993,148:985–991. [DOI] [PubMed] [Google Scholar]
- 5.Henneberger PK, Cumro D, Deubner DD, Kent MS, McCawley M, Kreiss K. Beryllium sensitization and disease among long-term and short-term workers in a beryllium ceramics plant. Int Arch Occup Environ Health 2001,74:167–176. [DOI] [PubMed] [Google Scholar]
- 6.Newman LS, Mroz MM, Maier LA, Daniloff EM, Balkissoon R. Efficacy of serial medical surveillance for chronic beryllium disease in a beryllium machining plant. J Occup Environ Med 2001,43:231–237. [DOI] [PubMed] [Google Scholar]
- 7.Maier LA, McGrath DS, Sato H, Lympany P, Welsh K, Du Bois R, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol 2003,171:6910–6918. [DOI] [PubMed] [Google Scholar]
- 8.Balmes JR, Abraham JL, Dweik RA, Fireman E, Fontenot AP, Maier LA, et al. An official American Thoracic Society statement: diagnosis and management of beryllium sensitivity and chronic beryllium disease. Am J Respir Crit Care Med 2014,190:e34–59. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Silveira LJ, Hamzeh N, Gillespie M, Mroz PM, Mayer AS, et al. Beryllium-induced lung disease exhibits expression profiles similar to sarcoidosis. Eur Respir J 2016,47:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Winkel JG, Capel PJ. Human IgG Fc receptor heterogeneity: molecular aspects and clinical implications. Immunol Today 1993,14:215–221. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto TT, Inoue M, Shimomura T, Fujimura K. Involvement of Fc gamma receptor polymorphism in the therapeutic response of idiopathic thrombocytopenic purpura. Br J Haematol 2001,115:125–130. [DOI] [PubMed] [Google Scholar]
- 12.Hargreaves CE, Rose-Zerilli MJ, Machado LR, Iriyama C, Hollox EJ, Cragg MS, et al. Fcgamma receptors: genetic variation, function, and disease. Immunol Rev 2015,268:6–24. [DOI] [PubMed] [Google Scholar]
- 13.Braakman E, van de Winkel JG, van Krimpen BA, Jansze M, Bolhuis RL. CD16 on human gamma delta T lymphocytes: expression, function, and specificity for mouse IgG isotypes. Cell Immunol 1992,143:97–107. [DOI] [PubMed] [Google Scholar]
- 14.Vance BA, Huizinga TW, Wardwell K, Guyre PM. Binding of monomeric human IgG defines an expression polymorphism of Fc gamma RIII on large granular lymphocyte/natural killer cells. J Immunol 1993,151:6429–6439. [PubMed] [Google Scholar]
- 15.van der Pol W, van de Winkel JG. IgG receptor polymorphisms: risk factors for disease. Immunogenetics 1998,48:222–232. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest 1997,100:1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood 1997,90:1109–1114. [PubMed] [Google Scholar]
- 18.Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the Fc{gamma}RIIIa-158 V/V and V/F polymorphism. Blood 2007,110:2561–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seligman VA, Suarez C, Lum R, Inda SE, Lin D, Li H, et al. The Fcgamma receptor IIIA-158F allele is a major risk factor for the development of lupus nephritis among Caucasians but not non-Caucasians. Arthritis Rheum 2001,44:618–625. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Ptacek TS, Brown EE, Edberg JC. Fcgamma receptors: structure, function and role as genetic risk factors in SLE. Genes Immun 2009,10:380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edberg JC, Langefeld CD, Wu J, Moser KL, Kaufman KM, Kelly J, et al. Genetic linkage and association of Fcgamma receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis Rheum 2002,46:2132–2140. [DOI] [PubMed] [Google Scholar]
- 22.Rossman MD, Stubbs J, Lee CW, Argyris E, Magira E, Monos D. Human leukocyte antigen Class II amino acid epitopes: susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med 2002,165:788–794. [DOI] [PubMed] [Google Scholar]
- 23.Sato H, Silveira L, Spagnolo P, Gillespie M, Gottschall EB, Welsh KI, et al. CC chemokine receptor 5 gene polymorphisms in beryllium disease. Eur Respir J 2010,36:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonth AC, Silveira L, Fingerlin TE, Sato H, Luby JC, Welsh KI, et al. TGF-beta 1 variants in chronic beryllium disease and sarcoidosis. J Immunol 2007,179:4255–4262. [DOI] [PubMed] [Google Scholar]
- 25.Kastbom A, Ahmadi A, Soderkvist P, Skogh T. The 158V polymorphism of Fc gamma receptor type IIIA in early rheumatoid arthritis: increased susceptibility and severity in male patients (the Swedish TIRA project). Rheumatology (Oxford) 2005,44:1294–1298. [DOI] [PubMed] [Google Scholar]
- 26.Sadki K, Lamsyah H, Rueda B, Akil E, Sadak A, Martin J, et al. Analysis of MIF, FCGR2A and FCGR3A gene polymorphisms with susceptibility to pulmonary tuberculosis in Moroccan population. J Genet Genomics 2010,37:257–264. [DOI] [PubMed] [Google Scholar]
- 27.Typiak MJ, Rebala K, Dudziak M, Dubaniewicz A. Polymorphism of FCGR3A gene in sarcoidosis. Hum Immunol 2014,75:283–288. [DOI] [PubMed] [Google Scholar]
- 28.Sawyer RT, Day BJ, Fadok VA, Chiarappa-Zucca M, Maier LA, Fontenot AP, et al. Beryllium-ferritin: lymphocyte proliferation and macrophage apoptosis in chronic beryllium disease. Am J Respir Cell Mol Biol 2004,31:470–477. [DOI] [PubMed] [Google Scholar]
- 29.Wu J, Li Y, Guan W, Viken K, Perlman DM, Bhargava M. FCGR3A and FCGR3B copy number variations are risk factors for sarcoidosis. Hum Genet 2016,135:715–725. [DOI] [PubMed] [Google Scholar]
- 30.Blakemore WS, Forster RE, Morton JW, Ogilvie CM. A standardized breath holding technique for the clinical measurement of the diffusing capacity of the lung for carbon monoxide. J Clin Invest 1957,36:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundgren RA, Maier LA, Rose CS, Balkissoon RC, Newman LS. Indirect and direct gas exchange at maximum exercise in beryllium sensitization and disease. Chest 2001,120:1702–1708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Figure 1. Sequence and gel analysis of a portion of FCGRIIIA cDNA from three Representatives. FcgRIIIa encoding cDNA was prepared from purified PBMCs and polymerase chain reaction (PCR) with the sequence specific primers (PCR-SSP) was used (see Methods) to determine the genotype of FCGRIIIA 158V/F polymorphism. The PCR product were run on 2% agrose gel. PCR products were analyzed on a 2% agarose gel containing 0.05% ethidium bromide (EB). The long common PCR product were gel extracted and sequenced. The left panels showed the example sequencing results and the right panel showed the gel picture of three genotypes of FCGRIIIA 158V/F polymorphism. Upper panel: 158FF genotype, Middle panel: 158FV panel, and lower panel: 158VV panel. For the PCR gel pictures, the PCR product at 147 bp represent either 158F (left PCR product) or 158V (right PCR product) genotype depending on the primer specificity, and the PCR product at 423 bp represent the common product which is used to eliminate false negatives.


