Abstract
Background and Aim:
The genetic relationship among serotypes of Salmonella enterica from food animals, food of animal origin, and human is of interest as the data could provide an important clue for the source of human infection. This study aimed to determine the genetic relatedness of S. enterica from pig production and human in Thailand–Laos border provinces.
Materials and Methods:
A total of 195 S. enterica serotypes isolated from pig and pork (n=178) and human (n=17) including four serotypes (Typhimurium, Rissen, Derby, and Stanley) were randomly selected to examine their genetic relatedness using highly conserved sequence of three genes (fim A, man B, and mdh).
Results:
The results showed that 195 Salmonella isolates of four different serotypes were grouped into five different clusters, and members of the same Salmonella serotypes were found in the same cluster. Salmonella isolated from pig production and human in Thailand–Laos border provinces represented overlapping population and revealed a high degree of similarity, indicating close genetic relationship among the isolates.
Conclusion:
The results support that the determination of Salmonella serotyping combined with analysis of phylogenetic tree can be used track the clonal evolution and genetic diversity of Salmonella serotypes in different host species.
Keywords: Laos, pig, Salmonella, Thailand
Introduction
Due to the regional economic integration (known as ASEAN Economic Community), rapid growth of cross-border activities between Thailand and their regional neighbors is expected. These cross-border activities including the movement of people, goods, and services, in either legal or illegal manners, could lead to greater risk of cross-border transmission of infectious diseases, especially foodborne pathogens [1]. Thailand and Laos share a common border of approximately 1800 km in length with up to 36 crossing points [2]. Livestock, such as pigs and cattle, are common commodities [3],movement and unhygienic slaughter of livestock occur in and around Thailand–Lao PDR border. Thus, causing major concern among public health authorities on either side of the border. Pigs are smuggled in and out between Thailand and Laos every day, and these animals are illegally slaughtered without recourse to best hygienic practices [4]. Therefore, these could be a critical source for spreading of foodborne pathogens.
Salmonellosis represents a growing threat to global public health. It was estimated that Salmonellosis accounts for about 155,000 deaths and 9,380,000 infections in humans worldwide [5]. Salmonella surveillance program using serotyping for subtyping and strain differentiation has been reported [6]. This technique examined Salmonella O and H antigen based on immune reaction technique. Even if around 2500 Salmonella serotypes can be differentiated, this technique has limited value in determining the genetic relationship of strains within the same or different serotype [7]. More discriminatory subtyping methods, such as phage typing and pulsed-field gel electrophoresis, are thus commonly used to subtype Salmonella, particularly as part of national and international salmonellosis surveillance systems [8,9].
Information on the main food animal reservoirs for Salmonella and the modes of transmission are extremely limited in Thailand.
This study was conducted to gain a better understanding of the ability of Salmonella to be transmitted to humans from pigs. Therefore, this study aimed to determine the genetic relatedness of Salmonella enterica isolates obtained from pig production and human in Thailand–Laos border provinces.
Materials and Methods
Ethical approval
Research protocols involving human subjects were approved by the Ethics Committee of the Faculty of Medicine, Khon Kaen University (the authorization ID, HE572136).
Samples collection
A total of 195 S. enterica isolates collected from pig and pork (n=178), and human (n=17) were selected and used in this study. All isolates were obtained from samples collected in three provinces in Thailand including Nong Khai, Mukdahan, and Ubon Ratchathani and two provinces in Laos including Vientiane and Savannakhet (Table-1). The study locations included one municipal pig slaughterhouses, one municipal fresh market, and one municipal hospital in each province. These slaughterhouses and markets were selected so that the animals could be tracked from the slaughterhouses to the markets. The samples were collected 3 times with 3-4-month interval from each sampling site. Due to logistical constraints, random sampling was not possible. The fecal samples were collected from pigs by rectal evacuation after bleeding but before the scalding process. Carcass samples were collected before delivery to the market by swabbing an area of approximately 50 cm2 on each carcass using sterile gauze. The same pigs were tracked and sampled again at fresh markets. At fresh markets, the samples were collected from pig carcasses by swabbing with gauze at the corresponding retail shop.
Table-1.
Number of Salmonella isolates examined in this study (n=195).
| Sample type | Number of Salmonella isolates | Total | ||||
|---|---|---|---|---|---|---|
| Thailand | Laos | |||||
| NK | MH | U | VT | SV | ||
| Pig | 11 | 11 | 6 | 13 | 19 | 60 |
| Pig carcasses | 10 | 11 | 7 | 10 | 20 | 58 |
| Pork | 12 | 9 | 9 | 20 | 10 | 60 |
| Human | 3 | 9 | 0 | 2 | 3 | 17 |
| Total | 36 | 40 | 22 | 45 | 52 | 195 |
NK=Nong Khai, MH=Mukdahan, U=Ubon Ratchathani, VT=Vientiane, SV=Savannakhet
Human stool samples were collected from workers at slaughterhouses, butchers, and workers at a retail meat shop in fresh markets and patients with diarrhea at hospitals. The slaughterhouse workers, butchers, and retail meat workers were requested to provide their own stool samples. Nurses or technicians collected samples from patients. Due to a low number of possible human samples, efforts were made to obtain most possible participants.
Salmonella isolation and identification
The Salmonella strains were isolated using the standard protocol ISO6579:2002 (E). Three colonies were initially collected from each positive sample and subjected to serotyping by slide agglutination based on the Kauffman–White scheme at the Center of Antimicrobial Resistance in Foodborne Pathogens (in cooperation with the World Health Organization), Faculty of Veterinary Science, Chulalongkorn University.
Polymerase chain reaction (PCR) and DNA sequencing
All Salmonella isolates were subjected to molecular typing based on fimA, manB, and mdh genes with specific primer pairs as reported [10]. PCR reactions were carried out in 25 μL reaction volume containing 12.5 μL of 2× Reddymix™ PCR Master Mix, 1 μL of each forward and reverse primers, 5 μL of DNA template, and 5.5 μL of sterile distilled water. Amplification conditions for manB, fimA, and mdh gene targets were as follows: Initial denaturation at 95°C for 9.5 min, followed by 40 cycles of 95°C for 45 s, annealing for 45 s at either 55°C (manB) or 58°C (fimA and mdh), extension at 72°C for 1 min, and a final extension step of 7 min at 72°C. Amplified PCR products were analyzed by electrophoresis through 1.2% agarose gels. All PCR products were purified using Nucleospin® Gel and PCR clean up (Macherey-Nagel, Düren, Germany) and submitted for sequencing (First Base Laboratories, Selangor Darul Ehsan, Malaysia). All sequences were assembled and proofread using SeqMan and aligned using the MUSCLE [11] in MEGA 6.0.
Phylogenetic analyses
DNA sequence alignments of fimA, manB, and mdh were used for the construction of phylogenetic trees. The phylogenetic tree was built using the neighbor-joining (NJ) method. The evolutionary history was inferred using the NJ method [12]. The evolutionary distances were computed using the Maximum Composite Likelihood method [13] and are in the units of the number of base substitutions per site. The analysis involved 195 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1989 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [14].
Results and Discussion
Salmonellosis presents a significant public health challenge due to the number of cases per year and due to the ease of transmission to humans through the food chain [15]. Molecular typing methods that utilize the sequences of highly conserved housekeeping and virulence gene offer a robust and versatile approach for the detection of foodborne salmonellosis. The technique helps to accurately identify routes of transmission and sources of infection, especially in the detection of bacterial population (Salmonella Enteritidis and Salmonella Typhimurium) with a very large clonal lineage and limited heterogeneity [16]. In addition, despite the extensive strategy invested in surveillance, the burden of foodborne Salmonellosis due to S. enterica in humans is on the increase. Moreover, since S. enterica serotypes are made up of several sublineages with divergent antigenic representation, antimicrobial resistance phenotypes, and virulence. Understanding the evolution of these species is crucial in identifying sources, routes of transmission as well asprediction and prevention of outbreaks [17].
In this study, an NJ phylogenetic tree was constructed by concatenated fimA, manB, and mdh sequences (1989 bp). The tree showed that Salmonella isolates of four different serotype were separated into five clusters (Clusters A, B, C, D, and E) (Table-2). Members of the same serotypes clustered into distinct groups. The percentage of similarity within each cluster ranged from 99.6% to 100% and the average genetic distance of Salmonella isolates within each cluster ranged from 0.001 to 0.003 (Figure-1). This study examined S. enterica isolated from pig production and human to determine the genetic relatedness of four different serotypes from various hosts. 195 Salmonella isolates of four serovars (Typhimurium, Rissen, Derby, and Stanley) in Thailand–Laos border provinces were assessed using molecular typing based on the sequences of three highly conserved genes (fimA, manB, and mdh). The molecular typing schemes utilized the combination of housekeeping genes and virulence or flagellin genes and offered more discriminatory power in determining the evolutionary relationship of S. enterica serovars [18,19]. The four serotypes used in this study were commonly found in pig production and human in Thailand–Laos border provinces. They have been incriminated in several cases of salmonellosis in humans that occur due to the consumption of contaminated pork or ingestion of water contaminated by pig feces. Phylogenetic analysis revealed that Salmonella isolates were grouped into five different clusters and members of the same serotypes were found in the same cluster (Table-1 and Figure-1). This showed that Cluster A consists of 56 S. enterica serotype Rissen, including pig fecal sample (n=17), pig carcass (n=16), pork (n=17), and human isolates (n=6) from Thailand–Laos border provinces. Cluster B contained S. enterica ser. Stanley (n=34) and S. enterica ser. Typhimurium (n=3), Cluster C contained S. enterica ser. Derby (n=17), Cluster D contained one S. enterica ser. Rissen and one S. enterica ser. Typhimurium, and Cluster E contained S. ser. Typhimurium (n=73) (Table-2). Salmonella isolated from pig production and human in Thailand–Laos border provinces represents overlapping population. This is because consumption of contaminated pork and other pig product can increase the risk of salmonellosis in humans in addition to significant economic losses. Contamination of pork can occur during processing through improper handling of the intestinal content at the slaughterhouse [20]. This can, in turn, increase the chances of transmission of Salmonella serotypes from humans to animals and vice versa.
Table-2.
Distribution of Salmonella serotype among pig, pig carcass, pork, and human isolates.
| Cluster | Salmonella serovars | Sample type (Number of isolates) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Thailand | Laos | ||||||||||
| Pig | Pig carcass | Pork | Human | Total | Pig | Pig carcass | Pork | Human | Total | ||
| A | S. Rissen | 10 | 9 | 10 | 5 | 34 | 7 | 7 | 7 | 1 | 22 |
| S. Typhimurium | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| S. Derby | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| S. Stanley | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | |
| B | S. Stanley | 1 | 5 | 3 | 0 | 9 | 9 | 7 | 8 | 1 | 25 |
| S. Typhimurium | 0 | 0 | 1 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | |
| C | S. Derby | 0 | 0 | 3 | 0 | 3 | 3 | 1 | 8 | 2 | 14 |
| S. Typhimurium | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | |
| S. Rissen | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| D | S. Rissen | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| S. Typhimurium | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| E | S. Typhimurium | 15 | 13 | 10 | 5 | 43 | 11 | 13 | 6 | 0 | 30 |
| S. Derby | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| S. Stanley | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| S. Rissen | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
Figure-1.
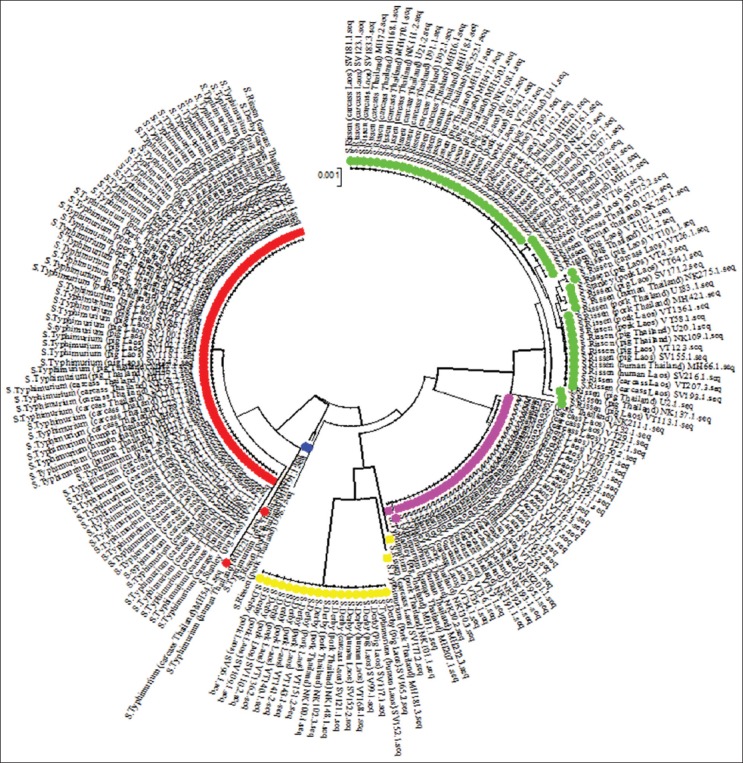
Phylogenetic relationships of 195 Salmonella isolates based on concatenated of the three MLST genes (fimA, manB, and mdh) sequenced were generated by MEGA6 using the neighbor-joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Each color represents different clusters (A - green, B - pink, C - yellow, D - blue, and E - red).
Most of pig production and human isolates in Thailand–Laos border provinces in each cluster showed a high degree of similarity, indicating close relationship among the isolates. Exceptions were S. enterica ser. Rissen (n=3), S. enterica ser. Typhimurium (n=6), S. enterica ser. Stanley (n=2), and S. enterica ser. Derby (n=2). Three S. enterica ser. Rissen were found in different branches and one S. enterica ser. Rissen was isolated from pig carcass in Laos clustered with S. enterica ser. Derby (Cluster C). In addition, one S. enterica ser. Rissen isolates in Thailand from pig carcass clustered with S. enterica ser. Typhimurium (Cluster E) and one S. enterica ser. Rissen isolates in Thailand from pork clustered with S. enterica ser. Typhimurium (Cluster E). Six S. enterica ser. Typhimurium were in different branches; one S. enterica ser. Typhimurium isolates in Thailand from pig clustered with S. enterica ser. Rissen (Cluster A). Three S. enterica ser. Typhimurium clustered with S. enteric a ser. Stanley (Cluster B), two of the isolates were from humans in Thailand, while the other is from pig carcass from Thailand. Two S. enterica ser. Typhimurium clustered with S. enterica ser. Derby (Cluster B) contained one isolate in Laos which was from human and one isolate in Thailand which was from pork, and one S. enterica ser. Typhimurium isolate in Thailand was from pig clustered with S. enterica ser. Rissen (Cluster D). In addition, two S. enterica ser. Stanley were found in different branches, one of these from pork isolated in Laos clustered with S. enterica ser. Rissen (Cluster A) and the other clustered with S. enterica ser. Typhimurium (Cluster E). Similarly, two other S. enterica ser. Derby were on different branches of the phylogenetic tree, one of these clustered with S. enterica ser. Rissen (Cluster A) a pig isolates from Laos and the other clustered with S. enterica ser. Typhimurium (Cluster E). The clustering of the isolates with similar genotypes from pig, pork, and human in both Thailand and Lao could be attributed to factors such as the environment, transportation, and cross-contamination during handling and processing of pork [21]. Altogether, the results also indicate that pig and pork may be a reservoir for the Salmonella serovars in humans. Similar studies among Salmonella isolates from pig, pork, and human have been reported in Germany, France, and Mexico [22-25]. The combination of molecular typing technique and epidemiological studies plays a pivotal role in authenticating the relationship between Salmonella strains isolated from different regions and sources. These approaches also helped in providing valuable information on various contamination routes and differentiate strains obtained from sporadic and outbreak cases [26].
Conclusion
In this study, sequence of the three conserved housekeeping genes of Salmonella serotypes isolated from pigs, pork, and humans from Thailand and Laos border provinces showed a high degree of similarity except for few isolates that cluster on different branches of the phylogenetic tree. Hence, giving room for the likelihood of animal to human transmission, increased dissemination and maintenance of resistance determinants and the risk of human infection.
Authors’ Contributions
RC conceived and designed the study. RP and SA collected the samples. RP and NS carried out the experiment. AAB and RP analyzed the data. RP and AAB wrote the first draft of the manuscript. RC revised the manuscript. All authors read and approved the final draft of the manuscript.
Acknowledgments
We thank Genetic Strains Research Center, National Institute of Genetics, Japan, for providing E. coli K12 MG1655. We deeply thank Wattana Theppangna, Department of Livestock and Fisheries, Ministry of Agriculture and Forestry and Dethaloun Meunsene, Faculty of Agriculture, National University of Lao for facilitating sample collection in Lao PDR. This work was financially supported by the Research Grant for Mid-Career University Faculty RSA5680051 co-funded by Thailand Research Fund (TRF), the Faculty of Veterinary Science, and Chulalongkorn University. It was partially supported by the 90th anniversary of Chulalongkorn University fund, Thailand.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Käferstein F.K, Motarjemi Y, Bettcher D.W. Foodborne disease control:A transnational challenge. Emerg. Infect. Dis. 1997;3(4):503. doi: 10.3201/eid0304.970414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paitoonpong S. Thailand's cross-border trade in the Greater Mekong Subregion:Some issues never solved. TDRI Q. Rev. 2006;21(1):11–19. [Google Scholar]
- 3.KNIPS V. Review of the Livestock Sector in the Mekong Countries(Livestock Information, Sector Analysis and Policy Branch Report), Rome, Food and Agriculture Organization of the United Nations. 2004. [[Last accessed on 19-09-2018]]. Available from: http://www.fao.org/WAICENT/FaoInfo/Agricult/againfo/resources/es/publications/sector_reports/lsr_mekong.pdf .
- 4.Cocks P, Abila R, Bouchot A, Benigno C, Morzaria S, Inthavong P, Van Long N, Bourgeois-Luthi N, Scoizet A, Sieng S. FAO ADB and OIE SEAFMD Study on Cross-Border Movement and Market Chains of Large Ruminants and Pigs in the Greater Mekong Sub Region. Thailand, Bangkok: Funded by the Government of PR China and Australian Standz Initiative; 2009. pp. 1–63. [Google Scholar]
- 5.Li K, Ye S, Alali W.Q, Wang Y, Wang X, Xia X, Yang B. Antimicrobial susceptibility, virulence gene and pulsed-field gel electrophoresis profiles of Salmonella Enterica serovar Typhimurium recovered from retail raw chickens, China. Food Control. 2017;72(A):36–42. [Google Scholar]
- 6.CDC 2004. Centers for Disease Control and Prevention. Atlanta, Ga: US Department of Health and Human Services; 2003. Salmonella Surveillance Study. [Google Scholar]
- 7.Selander R.K, Beltran P, Smith N.H, Helmuth R, Rubin F.A, Kopecko D.J, Musser J.M. Evolutionary genetic relationships of clones of Salmonella Serovars that cause human typhoid and other enteric fevers. Infect. Immun. 1990;58(7):2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher I.S. The Enter-net international surveillance network-how it works. Eur. Commun. Dis. Bull. 1999;4(5):52–55. doi: 10.2807/esm.04.05.00073-en. [DOI] [PubMed] [Google Scholar]
- 9.Swaminathan B, Barrett T.J, Hunter S.B, Tauxe R.V, Force C.P.T. The molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 2001;7(3):382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukhnanand S, Alcaine S, Warnick L.D, Su W.L, Hof J, Craver M.P.J, Wiedmann M. DNA sequence-based subtyping and evolutionary analysis of selected Salmonella Enterica serotypes. J. Clin. Microbiol. 2005;43(8):3688–3698. doi: 10.1128/JCM.43.8.3688-3698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar R.C. MUSCLE:Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naruya M.N. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Koichiro T.M, Nei S.K. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palhares J.C.P, Kich J.D, Bessa M.C, Biesus L.L, Berno L.G, Triques N.J. Salmonella and antimicrobial resistance in an animal-based agriculture river system. Sci. Total Environ. 2014;472(15):654–661. doi: 10.1016/j.scitotenv.2013.11.052. [DOI] [PubMed] [Google Scholar]
- 16.Ranjbar R, Ahmadi M, Memariani M. Multiple-locus variable-number tandem repeat analysis (MLVA) for genotyping of Salmonella Enterica subspecies enterica serotype infantis isolated from human sources. Microb. Pathog. 2016;100(1):299–304. doi: 10.1016/j.micpath.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Fricke W.F, Mammel M.K, McDermott P.F, Tartera C, White D.G, LeClerc J.E, Cebula T.A. Comparative genomics of 28Salmonella Enterica isolates:Evidence for CRISPR-mediated adaptive sublineage evolution. [Last accessed on 25-09-2018];J. Bacteriol. 2011 JB-00297 doi: 10.1128/JB.00297-11. Available from: https://www.jb.asm.org/content/early/2011/05/20/JB.00297-11.short . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tankouo-Sandjong B, Sessitsch A, Liebana E, Kornschober C, Allerberger F, Hächler H, Bodrossy L. MLST-v, multilocus sequence typing based on virulence genes, for molecular typing of Salmonella Entericasubsp. enterica serovars. J. Microbiol. Method. 2007;69(1):23–36. doi: 10.1016/j.mimet.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Foley S.L, White D.G, McDermott P.F, Walker R.D, Rhodes B, Fedorka-Cray P.J, Zhao S. Comparison of subtyping methods for differentiating Salmonella Enterica serovar Typhimurium isolates obtained from food animal sources. J. Clin. Microbiol. 2006;44(10):3569–3577. doi: 10.1128/JCM.00745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wannaprasat W, Padungtod P, Chuanchuen R. Class 1 integrons and virulence genes in Salmonella Enterica isolates from pork and humans. Int. J. Antimicrob. Agents. 2011;37(5):457–461. doi: 10.1016/j.ijantimicag.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Rönnqvist M, Välttilä V, Ranta J, Tuominen P. Salmonella risk to consumers via pork is related to the Salmonella prevalence in pig feed. Food Microbiol. 2018;71(1):93–97. doi: 10.1016/j.fm.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Zaidi M.B, McDermott P.F, Fedorka-Cray P, Leon V, Canche C, Hubert S.K, Tollefson L. Nontyphoidal Salmonella from human clinical cases, asymptomatic children, and raw retail meats in Yucatan, Mexico. Clin. Infect. Dis. 2006;42(1):21–28. doi: 10.1086/498508. [DOI] [PubMed] [Google Scholar]
- 23.Hauser E, Tietze E, Helmuth R, Junker E, Blank K, Prager R, Malorny B. Pork contaminated with Salmonella Enterica serovar 4,[5], 12:i:An emerging health risk for humans. Appl. Environ. Microbiol. 2010;76(14):4601–4610. doi: 10.1128/AEM.02991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser E, Hebner F, Tietze E, Helmuth R, Junker E, Prager R, Malorny B. Diversity of Salmonella Enterica serovar Derby isolated from pig, pork and humans in Germany. Int. J. Food Microbiol. 2011;151(2):141–149. doi: 10.1016/j.ijfoodmicro.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Kerouanton A, Rose V, Weill F.X, Granier S.A, Denis M. Genetic diversity and antimicrobial resistance profiles of Salmonella Enterica serotype derby isolated from pigs, pork, and humans in France. Foodborne Pathog. Dis. 2013;10(11):977–984. doi: 10.1089/fpd.2013.1537. [DOI] [PubMed] [Google Scholar]
- 26.Campioni F, Davis M, Medeiros M.I.C, Falcão J.P, Shah D.H. MLVA typing reveals higher genetic homogeneity amongSEnteritidis strains isolated from food, humans and chickens in Brazil in comparison to the North American strains. Int. J. Food Microbiol. 2013;162(2):174–181. doi: 10.1016/j.ijfoodmicro.2013.01.008. [DOI] [PubMed] [Google Scholar]


