Abstract
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma of childhood with an unmet clinical need for decades. A single oncogenic fusion gene is associated with treatment resistance and a 40 to 45% decrease in overall survival. We previously showed that expression of this PAX3:FOXO1 fusion oncogene in alveolar RMS (aRMS) mediates tolerance to chemo- and radiotherapy and that the class I–specific histone deacetylase (HDAC) inhibitor entinostat reduces PAX3:FOXO1 protein abundance. Here, we established the antitumor efficacy of entinostat with chemotherapy in various preclinical cell and mouse models and found that HDAC3 inhibition was the primary mechanism of entinostat-induced suppression of PAX3:FOXO1 abundance. HDAC3 inhibition by entinostat decreased the activity of the chromatin remodeling enzyme SMARCA4, which in turn de-repressed the microRNA miR-27a. This re-expression of miR-27a led to PAX3:FOXO1 mRNA destabilization and chemotherapy sensitization in aRMS cells in culture and in vivo. Furthermore, a phase I clinical trial (ADVL1513) has shown that entinostat is tolerable in children with relapsed or refractory solid tumors and is planned for phase I-b cohort expansion or phase II clinical trials. Together, these results implicate an HDAC3–SMARCA4–miR-27a–PAX3:FOXO1 circuit as a driver of chemoresistant aRMS and suggest that targeting this pathway with entinostat may be therapeutically effective in patients.
Introduction
Soft tissue sarcomas are amongst the most common and deadliest childhood cancers (1, 2). Despite improved survival for other childhood cancers, progress for particularly metastatic sarcoma has been minimal and therapeutic options remain limited (2, 3). Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children (2). Patients with diagnosed metastatic RMS have dismal prognoses for survival that have not improved in years, if not several decades (4–6). Even though ~85% of local lesions can be excised by surgery, undetectable circulating tumor cells and recurrence at the primary site from occult microscopic residual disease are presumed to act as a gateway to relapse and subsequent regional and distal metastasis (7).
Alveolar rhabdomyosarcoma (aRMS) is the more aggressive RMS subtype and is generally not survivable long-term once metastatic (<8%)(4, 6, 8). This disease does respond to chemotherapy in most cases, but the clinical challenge is preventing or overcoming recurrence after chemotherapy and/or radiation (9). Unique to aRMS is the expression of a fusion protein called PAX3:FOXO1, which results from a chromosomal translocation and comprises the transcription factors PAX3 (paired box 3) and FOXO1 (forkhead box 1), PAX3:FOXO1, is seen in some patients with aRMS. We have previously reported that native, non-constitutive, G2 cell cycle phase-enriched expression of the PAX3:FOXO1 oncogene in aRMS facilitates checkpoint adaptation (meaning a high tolerance of DNA breaks or mitotic catastrophe) [(10), and reviewed (11)]. In terms of clinical impact, this PAX3:FOXO1-mediated treatment resistance causes a notable 40–45% difference (drop) in survival over 10 years compared to PAX3:FOXO1-negative cases (9). In experimental studies, genetic knockdown of PAX3:FOXO1 abundance improves chemotherapy and radiation sensitivity and reduces tumor re-establishment (10), but drugging transcription factors like PAX3:FOXO1 in patients is a daunting task. However, in this study, we found that PAX3:FOXO1 can be pharmacologically silenced at both the mRNA and protein levels by entinostat, a class I histone deacetylase (HDAC) inhibitor (with weak HDAC10/HDAC11 inhibition) that was granted FDA breakthrough designation for certain breast cancers in adults.
In combination with the chemotherapy vincristine, entinostat had strong anti-tumor activity in a genetically-engineered mouse model (GEMM) of aRMS and in both orthotopic allograft and patient-derived xenograft (PDX) mouse models of aRMS. Pretreatment with entinostat also improved radiation sensitivity. We further established the comparative efficacy of entinostat to other HDAC inhibitors at reducing the expression of PAX3:FOXO1 and that of another, related fusion protein also found in aRMS, PAX7:FOXO1. Mechanistically, we determined that in aRMS cells, HDAC3 inhibition by entinostat decreases SMARCA4, which in turn de-represses miR-27a, which then silences PAX3:FOXO1; by blocking HDAC3, entinostat suppressed the abundance and activity of PAX3:FOXO1 and the growth of fusion-positive aRMS tumors in mice. Together, these preclinical and mechanistic studies have led to the Children’s Oncology Group pediatric phase I clinical trial, ADVL1513, and a planned phase IB cohort expansion for RMS patients for this same trial.
Results
Entinostat in combination with vincristine slows aRMS tumor growth in vivo
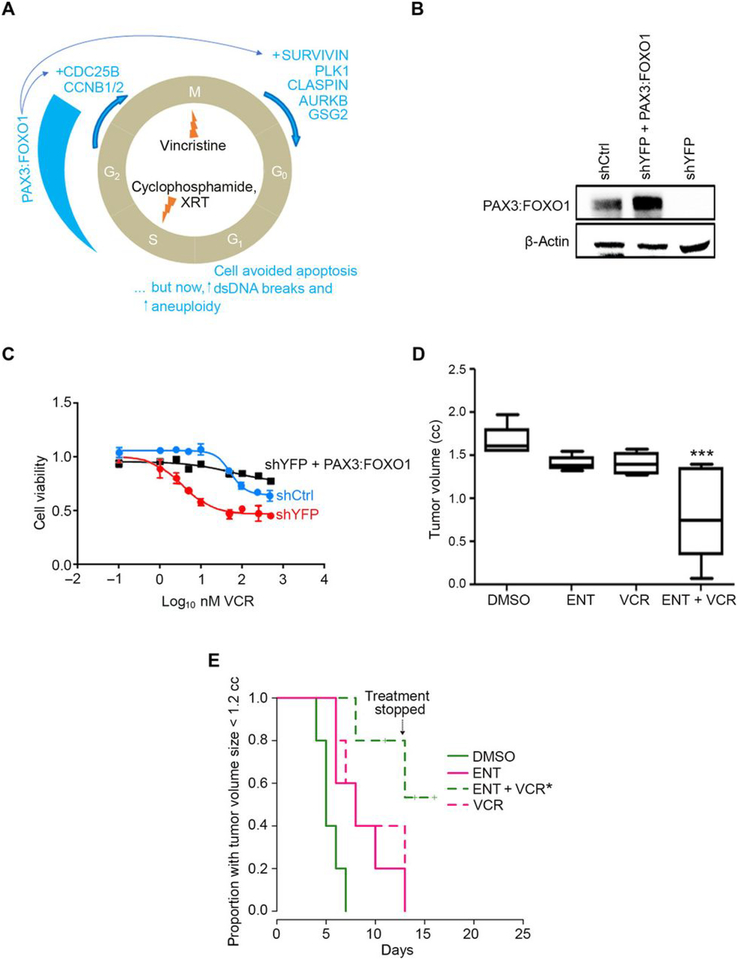
In preclinical studies using RNA interference, depletion of PAX3:FOXO1 abundance in aRMS cells is shown to be critical to overcome checkpoint adaptation, a process whereby tumor cells survive chemotherapy and radiation in late phases of the cell cycle (10) (summarized in Fig. 1A). To affirm genetically that silencing PAX3:FOXO1 increases chemosensitivity of aRMS to vincristine (VCR) in vitro, we first re-validated two derivative cell lines from the aRMS GEMM-derived primary tumor culture U23674 that did or did not have stable knockdown of PAX3:FOXO1 (shYFP or shCtrl, respectively) (10). PAX3:FOXO1 was then also restored by retroviral-mediated transduction into the shYFP-U23674 line. Immunoblot analysis confirmed stable knockdown of PAX3:FOXO1 and/or restored expression of PAX3:FOXO1, respectively (Fig. 1B). When treated with VCR for 24 hours, shYFP (PAX3:FOXO1 knockdown) cells alone had an absolute IC50 of 33 nM. However, control cells did not reach an absolute IC50 (Fig. 1C). Restoring expression of PAX3:FOXO1 in the stable knockdown cells (shYFP + PAX3:FOXO1) rescued the cells from the cytotoxic effects of VCR, in that no absolute IC50 was reached (Fig. 1C). Notably, although PAX3:FOXO1 was tumor-protective against chemotherapy, PAX3:FOXO1 knockdown was only cytostatic, which is consistent with prior reports of murine and human aRMS (10, 12).
Fig. 1. Entinostat treatment of aRMS in vivo.
(A) Diagrammatic representation of checkpoint adaptation, a process whereby tumor cells survive chemotherapy and radiation in late phases of the cell cycle. XRT, X-ray telescope radiotherapy; PLK1, polo-like kinase 1; AURKB, aurora kinase B; GSG2, genomic structure of haspin; CDC25B, cell division cycle 25b; CCNB1/2, cyclin B1/2; DS, double-stranded. (B) Basal PAX3:FOXO1 protein expression in murine U23674 aRMS cells transfected with control shRNA (shCtrl), PAX3:FOXO1-targeted shRNA (shYFP), or targeted shRNA plus a lentivirus expressing PAX3:FOXO1 (shYFP + PAX3:FOXO1). Blot is representative of N=3 biological replicates. (C) Viability of the U23674 cells described in (B) exposed to vincristine (VCR) at 4 nM for 24 hours. Graph plotted using GraphPad Prism. Data are mean ± S.D. of N= 3 independent experiments. (D) Box-and-whisker plot showing the tumoristatic efficacy of entinostat (ENT) or vincristine (VCR), alone and in combination, in aRMS mice at day 13 (DMSO vs ENT+VCR). Treatment at a daily dose of 5 mg/kg of ENT by intraperitoneal (IP) injection, VCR at a dose of 1 mg/kg weekly by IP injection, or a combination of both. Data are means ± SEM (N=5 mice per cohort), ***P < 0.001 by log-rank test. (E) Kaplan-Meier plot of the proportion of mice with tumors smaller than 1.2 cubic cm after treatment with ENT and/or VCRat a daily dose of 5 mg/kg of ENT by intraperitoneal (IP) injection, VCR at a dose of 1 mg/kg weekly by IP injection, or a combination of both. Treatment was stopped after day 13 because body weight loss approached 10–15%. Data are means ± SEM (N=5 mice per cohort), *P < 0.05 by log-rank test. In this experiment, treatment was stopped for all mice after day 13 because body weight loss approached 10–15%.
Our previous studies examined whether cell of origin influenced PAX3:FOXO1 expression, and whether expression could be pharmacologically altered by HDAC inhibitors or other epigenetic agents (13). To test whether the narrow-spectrum Class I HDAC inhibitor entinostat (ENT) could sensitize PAX3:FOXO1-positive aRMS to the chemotherapies most often used in high-risk or relapsed disease, we first tested the efficacy of ENT and VCR as single agents and in combination in orthotopic allograft mouse models of aRMS. This aRMS model was generated by injecting murine aRMS primary cell cultures into the cardiotoxin-preinjured gastrocnemius muscle of SHO mice. The tumor bearing mice were treated with ENT at a daily dose of 5 mg/kg administered intraperitoneally, VCR at 1 mg/kg administered intraperitoneally once a week, or in combination. Unlike monotherapy of either agent, the combination of ENT and VCR was effective in delaying tumor growth in the aRMS mice (Fig. 1, D and E). Histologically, no cytodifferentiation was seen (table S1).
To determine whether pre-treatment with ENT potentiates the effect of radiation on RMS cells, we pre-treated a murine primary tumor cell culture (U23674) with ENT or DMSO for 24 hours followed by irradiation (10 Gy) (fig. S1A). After irradiation, 500,000 viable cells were injected into the cardiotoxin-preinjured gastrocnemius muscle of SCID hairless outbred (SHO) mice. Tumor cells treated with DMSO alone or ENT alone were used as controls. After injection of tumor cells, mice were monitored for tumor development. Mice that received tumor cells pretreated with ENT prior to irradiation showed increased latency of tumor development compared to mice that received cells pretreated with DMSO only (fig. S1B), suggesting that ENT treatment sensitizes RMS cells to radiation. With respect to chemotherapy, we also confirmed that the biochemical effect of PAX3:FOXO1 silencing occurs – and with synergy – at clinically achievable concentrations of ENT and VCR in human RMS cultures (fig. S2, A–C).
To further assess the clinical feasibility of ENT in a clinical setting, we investigated whether ENT had a dose-dependent effect, and/or whether ENT was unique among HDAC inhibitors for its suppressive effect on PAX3:FOXO1. First, we observed a dose-dependent effect of ENT on PAX3:FOXO1 expression in Rh30 cells in response to doses from 0.1 to 2 μM, when examining the mRNA abundance at 24 hours (fig. S3A) and the protein abundance at 72 hours (fig. S3B). A PDX-derived explant aRMS culture, CF-001, also exhibited a dose-dependent response (fig. S3C), as did the murine aRMS culture, U23674 (fig. S3D).
To examine the relative efficacy of other HDAC inhibitors at reducing PAX3:FOXO1 when compared to ENT (an HDAC1–3 inhibitor), we tested SAHA, panobinostat (PAN), CUDC-907, and CUDC-101 in human and murine aRMS using the highest reported maximum drug serum concentration (Cmax) in humans. Where Cmax was unknown, we treated the cell cultures at the measured drug IC25. ENT at its highest achievable Cmax (1000 nM) (14) significantly reduced PAX3:FOXO1 expression in human aRMS (Rh30 and Rh41) cells (fig. S4A), which was reflected by near complete reduction at the protein level in the human aRMS cells and in murine aRMS (U23674) cells (fig. S4B). In comparison, PAN (a broad spectrum HDAC inhibitor that targets HDAC1–11) at its Cmax (45 nM) (15) did not consistently diminish PAX3:FOXO1 at either the mRNA or protein level (fig. S4, C and D), and SAHA (an HDAC1/2/3/6/8 inhibitor) was only somewhat effective at reducing PAX3:FOXO1 abundance at its Cmax (1000 nM) (16) (fig. S4, E and F), but to a lesser degree than ENT. Of note, near-complete cytotoxicity was observed in Rh41 cells with SAHA at the end of 72 hours treatment – preventing inclusion of Rh41 in these biochemical studies. In addition to PAN and SAHA, CUDC-907 (a dual PI3K and HDAC1/2/3/10 inhibitor) and CUDC-101(Class I and II HDACs inhibitor) were tested but found to be less efficient at reducing PAX3:FOXO1 compared to ENT at their IC25 (fig. S4, G and H). To examine whether these HDAC inhibitors (HDACi’s) can have an effect on the related fusion protein, PAX7:FOXO1, the human aRMS cell line CW9019 harboring the t(1;13) translocation was treated with HDACi’s at their respective Cmax or IC25 concentration. ENT and all other HDACi’s tested were effective at reducing PAX7:FOXO1 protein abundance to an almost undetectable level (fig. S4I).
Entinostat plus vincristine has efficacy in aRMS patient-derived xenografts
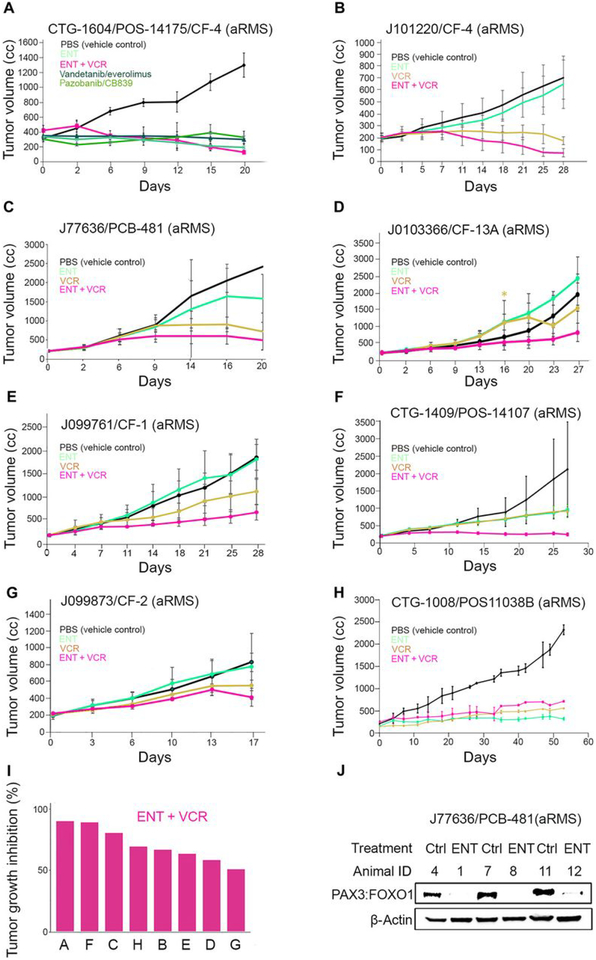
We next investigated the anti-tumor efficacy of ENT and VCR as single agents or in combination for various biologically-independent patient-derived xenografts models of aRMS. The dosing details are given in table S2, and PDX model characteristics are given in tables S3 and S4. Seven of the eight models were from recurrent and/or metastatic tumors taken from biopsy or autopsy. All these contemporary models were established after 2010. Seven of eight models carried the PAX3:FOXO1 oncogene, whereas one model (J99873/CF-2) was established from an autopsy tumor harboring PAX7:FOXO1. In each case, tumor growth inhibition by combined ENT plus VCR was superior to either drug alone (Fig. 2, A–G) except for one model (CTG-1008), in which tumor growth inhibition by ENT alone was better than combined treatment (Fig. 2H). Waterfall plots summarizing the tumor growth inhibition (%) for the eight models (Fig. 2I and fig. S5, A–C) showed that in 5 of 8 cases, ENT had single-agent activity relative to the control. No difference was seen between different treatment groups in terms of rhabdomyoblast differentiation (fig. S5, D and E, and table S5). Biochemical analysis of tumor lysates from J77636 PDXs treated with ENT showed pharmacodynamic down-regulation of PAX3:FOXO1 protein abundance, which was consistent with results seen in 2-dimensional cultures of human and murine aRMS cell lines (Fig. 2J). Statistical summaries of the 8 aRMS PDX models are provided in tables S6–13.
Fig. 2. In vivo evaluation of entinostat and vincristine in aRMS patient derived xenograft.
(A-H) Graphical analysis of tumor volume inhibition by either ENT alone or in combination with VCR in 8 different PDX aRMS mouse models (Champions Oncology and Jackson Lab) established from clinical biopsies, recurrent aRMS, or autopsies. Demographic features of these models are in tables S3 and S4; treatment schedules are in table S2. Statistical analyses are given in tables S6–S13. (I) Waterfall plot showing tumor growth inhibition (%) for combination treatment (ENT+VCR) in the 8 models shown in panels (A-H), as labeled. (J) Pharmacodynamic assessment of tumor lysates from J77636 treated with ENT for PAX3:FOXO1 protein expression. Blot shows N=3 biological replicates (N represents tumor lysates from 3 mice per cohort).
Mechanisms underlying entinostat treatment converge on known downstream genes
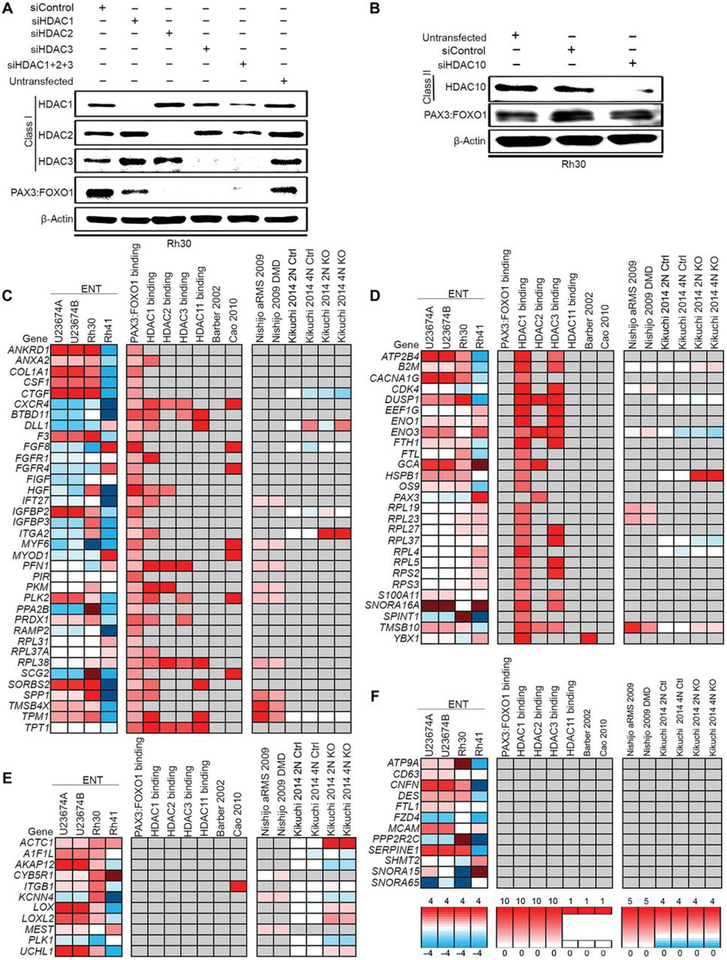
ENT (entinostat) targets class I HDACs (HDAC1, HDAC2 and HDAC3) but that at higher concentrations also inhibits the class II member HDAC10. To determine further the mechanism by which ENT silences PAX3:FOXO1 expression, we reduced the endogenous expression of HDAC1, HDAC2, and/or HDAC3 in Rh30 cells using siRNA (siHDAC1/2/3) alongside a scrambled siRNA control (Fig. 3A). PAX3:FOXO1 abundance was decreased to some extent by each individual HDAC siRNA, but depletion of HDAC2 or HDAC3 were the most potent (Fig. 3A). Depleting Rh30 cells of HDAC10, albeit somewhat less efficient than HDAC1/2/3 depletion, had no detectable impact on the protein abundance of PAX3:FOXO1 (Fig. 3B). These results indicated the importance of Class I HDACs, specifically HDAC2 and HDAC3, for PAX3:FOXO1 expression and suggests that inhibition of specifically these HDACs is the mechanism through which ENT represses the fusion protein.
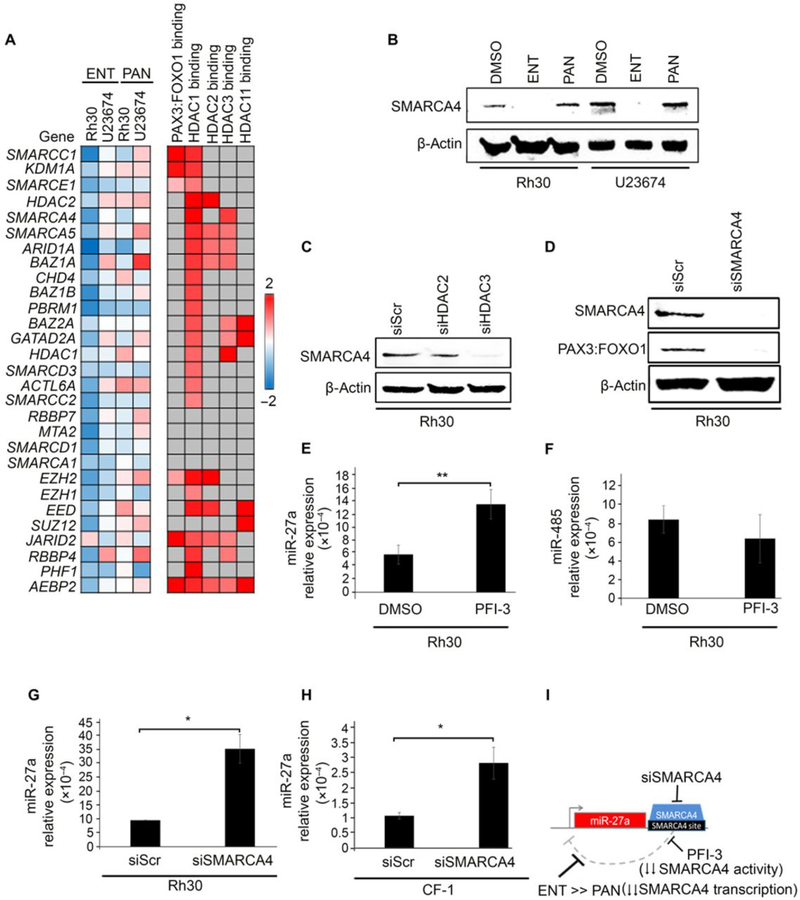
Fig. 3. siRNA-mediated knockdown of HDAC and gene expression and PAX3:FOXO1 binding data for key aRMS gene targets.
(A and B) PAX3:FOXO1 expression in Rh30 cells transfected with siRNA at 100 nM for 72 hours targeting HDAC1, HDAC2, and/or HDAC3 (A), as well as HDAC10 (B). Blots are representative of N=3 independent experiments. (C-F) Three aRMS samples [U23674 in two replicates (U23674A and U23674B), Rh30, and Rh41] were sequenced after treatment with ENT at 2 μM for 72 hours or DMSO. Featured genes were decreased (log2 ratio of ENT-induced expression divided by control expression > 1) in all samples. Additional expression data for these key targets were curated from previous publications (10, 33) and PAX3:FOXO1 binding data was curated from the literature as indicated (31, 32) or was generated through ChIP-Seq experiments. Four subclasses of ENT-induced gene expression were identified as (C) PAX3:FOXO1 binding with or without HDAC binding, (D) HDAC1, 2, 3 or 11 binding only, (E) indirect targets of PAX3:FOXO1, and (F) otherwise-regulated genes.
Our RNA-sequencing (RNA-seq) and chromatin immunoprecipitation-exonuclease (ChIP-exo) da6ta identified four classes of cell-autonomous gene expression changes related to ENT treatment: these subclasses were PAX3:FOXO1 binding with or without HDAC binding (Fig. 3C); HDAC1, HDAC2, HDAC3 or HDAC11 binding only (Fig. 3D); indirect targets of PAX3:FOXO1 (Fig. 3E), and otherwise-regulated genes (13)(Fig. 3F). Whereas HDAC-related and HDAC-unrelated changes in expression of the known PAX3:FOXO1 direct and indirect targets were expected, neither these nor those of the otherwise-regulated genes could explain altered transcription of PAX3:FOXO1 with respect to PAX3 regulatory elements.
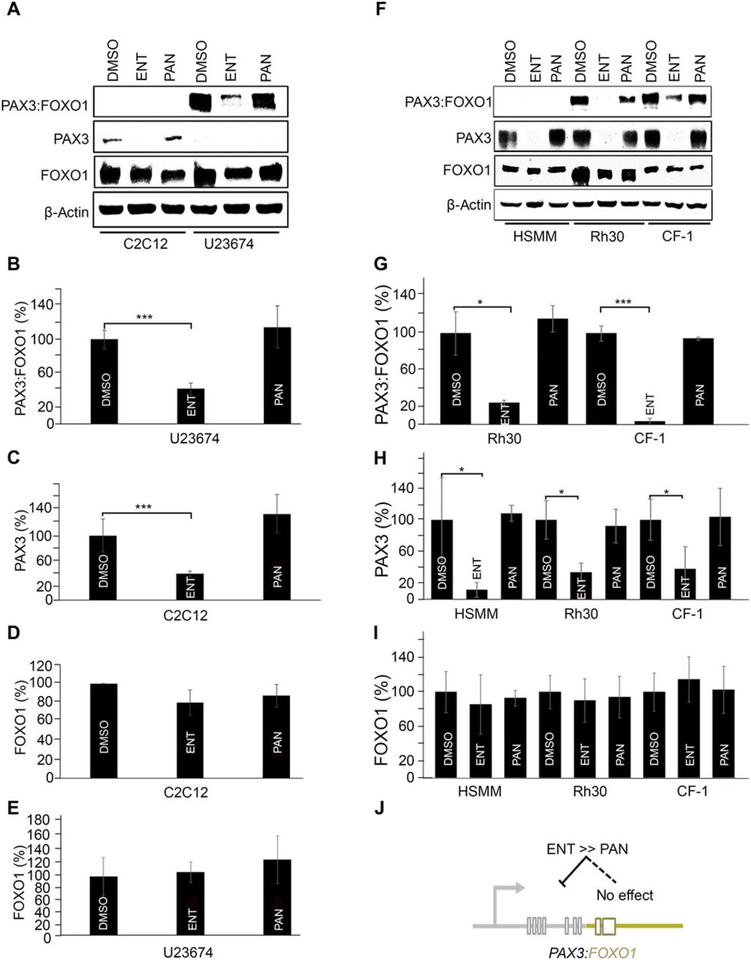
To understand the mechanisms of action for ENT and PAN, we also analyzed the expression of wild-type PAX3 and FOXO1 in U23674 and Rh30 aRMS cells as well as C2C12 mouse myoblasts, human skeletal myoblasts (HSMM), and CF-1 mouse embryonic fibroblasts cells, each treated separately with ENT and PAN. Consistent with previous results (13), PAX3:FOXO1 was significantly reduced by ENT but not by PAN in U23674 cells (Fig. 4, A and B). These cells do not express detectably sufficient wild-type PAX3 for analysis (Fig. 4A); however, in C2C12 cells, which do, ENT significantly reduced wild-type PAX3 but not wild-type FOXO1 abundance (Fig. 4, A and C), and in both cells lines neither ENT nor PAN altered PAX3 or FOXO1 (Fig. 4, A, D, and E). Similar results were observed in Rh30, CF-1, and HSMM cells (Fig. 4, F to I). These results suggest that for PAX3:FOXO1 in aRMS tumor cells, the PAX3 cis-regulatory elements are affected differently by ENT treatment versus PAN treatment, perhaps attributable to secondary effects on PAX3 cis-elements or PAX3 mRNA stability factors (Fig. 4J).
Fig. 4. Wild-type PAX3 expression altered by entinostat and not by panobinostat.
(A) Immunoblots of PAX3:FOXO1, wild-type PAX3, and wildtype FOXO1 in murine C2C12 myoblasts and U23674 aRMS cells upon 72 hours’ treatment with entinostat (ENT; 1 μM) and panobinostat (PAN; 45 nM). Blots are representative of N=3 independent experiments. Note that in the murine aRMS GEM culture U23674, both wild-type PAX3 alleles are homozygously replaced by PAX3:FOXO1, but both FOXO1 alleles are intact (homozygous wild-type). (B-E) Densitometric analysis of (A) PAX3:FOXO1, wild-type PAX3 and wild-type FOXO1expression upon treatment with ENT and PAN. Data are means ± SD, N=3 independent experiments; ***P < 0.001 by two-sided Student’s t test. (F) Western blots of PAX3:FOXO1, wild-type PAX3, and wild-type FOXO1 in HSMM, Rh30 and CF-1 cells upon 72 hours’ treatment with ENT (1μM) or PAN (45 nM). Blots are representative of N=3 biological replicates. In contrast to murine aRMS cultures, PAX3 is heterozygous (wild-type/PAX3:FOXO1) and FOXO1 is heterozygous (wild-type/null) in human aRMS cultures, and both are homozygous wild-type in human HSMM myoblasts. (G-I) Densitometric analysis of PAX3:FOXO1, wild-type PAX3 and wild-type FOXO1 protein expression upon treatment with ENT. Data are means ± SD, N=3 independent experiments; * P < 0.05, ***P < 0.001 by two-sided Student’s t tests. (J) A diagrammatic representation attributing the effect of ENT on PAX3:FOXO1 expression to PAX3 cis-regulatory elements.
Loss of SMARCA4 expression or activity increases, or de-represses, the expression of the microRNA miR-27a
To further understand how ENT silences the PAX3:FOXO1 gene, while PAN does not, RNA-seq analysis of ENT-treated versus PAN-treated human and murine aRMS cells were carried out with an emphasis on assessing the expression of transcription factors and chromatin remodeling complexes. Differential expression across samples after ENT treatment versus after PAN treatment identified key upregulated and downregulated genes in the ENT-specific effects (Fig. 5A). Many epigenetic targets, including chromatin modifying enzymes and remodeling complexes, were indeed decreased by ENT treatment and increased by PAN treatment in Rh30 and U23674 cells (Fig. 5A). Specifically, ENT treatment had a strong effect on the expression of many of the SWI/SNF (switch/sucrose non-fermentable) complexes, which included that of SMARCA4/BRG1, which was markedly reduced by ENT but not PAN treatment (Fig. 5A). At the protein level, only ENT reduced SMARCA4 protein abundance in both cells, notably to an undetected level (Fig. 5B). Consistent with ENT-mediated pharmacological inhibition of class I HDACs, siRNA-mediated knockdown of HDAC3 (siHDAC3) decreased SMARCA4 protein abundance to an almost undetectable level; knockdown of HDAC2 had markedly less of an effect (Fig. 5C). This finding concurs with a previous report indicating a non-canonical role of HDAC3 as transcriptional activator (17). Furthermore, the abundance of the PAX3:FOXO1 fusion protein was decreased in Rh30 cells upon reduction of SMARCA4 by siRNA (siSMARCA4) (Fig. 5D), altogether suggesting an HDAC3-SMARCA4 circuit in promoting PAX3:FOXO1 fusion protein abundance in aRMS cells.
Fig. 5. Loss of SMARCA4 expression or activity de-represses miR-27a.
(A) Differential expression analysis by RNA-seq for genes associated with chromatin-modifying complexes in Rh30 and U23674 cells after treatment for 72 hours with ENT (1 μM) or panobinostat (45 nM) relative to cells treated with DMSO. SMARCA4 average transcripts per million for DMSO-treated cells: 86.86; for ENT-treated cells: 56.15; and for PAN-treated cells: 72.19. (B) SMARCA4 protein abundance in Rh30 and U23674 aRMS cultures treated with ENT or PAN. Blots are representative of N=3 independent experiments. (C) SMARCA4 protein abundance in Rh30 cells transfected with HDAC2- or HDAC3-targeted siRNA (100 nM for 72 hours). Blots are representative of N=3 independent experiments. (D) PAX3:FOXO1 protein abundance in Rh30 cells transfected with SMARCA4-targeted siRNA (100 nM for 72 hours). Blots are representative of N=3 independent experiments. (E) Quantitative PCR (qPCR) analysis of miR-27a expression in Rh30 cells treated with the SMARCA4 bromodomain inhibitor PFI-3 (10 μM for 24 hours). (F) qPCR analysis of miR-485 in Rh30 cells treated with PFI-3. (G and H) qPCR analysis of miR-27a expression in Rh30 and CF-1 cells transfected with SMARCA4-targeted siRNA (100 nM for 72 hours). Data were normalized to the expression of U6snRNA. Gene expression was quantified using the 2^-dCt method. Data are means ± SD, N=3 independent experiments each in triplicate; * P < 0.05, **P < 0.01 by a two-sided Student’s t test. (I) Diagrammatic representation of how ENT and PFI-3 disrupt SMARCA4-mediated interference of miR-27a transcription.
Upon searching the miR search engine miRBase for miRNA that bound the PAX3 promoter or cis-elements, we observed that miR-27a was unique in that in both mouse and human cells it bound PAX3 upstream (5’) of the PAX3:FOXO1 breakpoint (in intron 4). Notably, miR-27a-5p reportedly targets PAX3, as well as PAX7, in developing and adult muscle cells (18). For comparison, we chose miR-485–5p as a conserved miRNA binding the PAX3 3’ untranslated region (UTR). Referencing the University of California Santa Cruz (UCSC) genome browser visualization of ChIP-seq data from the Encyclopedia of DNA Elements (ENCODE) predicted that SMARCA4 binds to the 3’ UTR of miR-27a. We then measured miR-27a expression upon pharmacological inhibition of SMARCA4 bromodomain activity using the small molecule inhibitor PFI-3 at a concentration of 10 μM. PFI-3 inhibits 95% of the SMARCA4 bromodomain activity at this concentration according to analysis by time-resolved fluorescence energy transfer (TR-FRET; fig. S6). Inhibition of SMARCA4 bromodomain activity increased miR-27a expression (Fig. 5E) but not that of miR-485 (Fig. 5F). We then carried out siRNA-mediated knockdown of SMARCA4 in Rh30 cells and CF-1 cells, which showed an increase in miR-27a compared to scrambled siRNA control, consistent with the effect seen on miR-27a upon pharmacological inhibition of SMARCA4 (Fig. 5, G and H). Together, these results suggested that a SMARCA4-miR27a regulatory circuitry controls the expression of PAX3:FOXO1 (Fig. 5I).
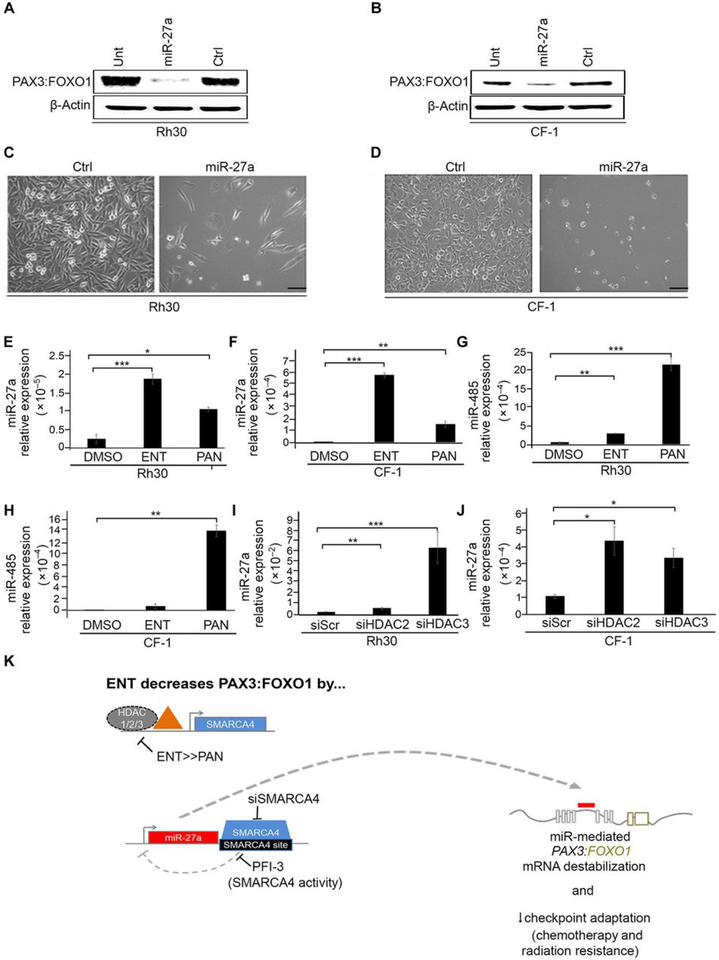
Entinostat-induced re-expression of miR-27a silences PAX3:FOXO1
To understand whether ENT silences PAX3:FOXO1 transcript via miR-27a-mediated transcription interference, we re-expressed miR-27a in Rh30 and CF-1 cells by transient transfection of miR-27a mimics and examined PAX3:FOXO1 protein expression. Compared to a control oligonucleotide (a “negative mimic”), miR-27a re-expression decreased PAX3:FOXO1 abundance in both Rh30 and CF-1 cells (Fig. 6, A and B), which also exjibited altered cell morphology (Fig. 6, C and D). qPCR analysis then showed a more potent increase in the expression of miR-27a in ENT-treated than PAN-treated Rh30 and CF-1 cells (Fig. 6, E and F). However, PAN increased miR-485 expression more potently than did ENT (Fig. 6, G and H). Because our earlier studies showed that ENT reduced PAX3:FOXO1 expression by inhibiting HDAC2/3 (Fig. 3A), we silenced HDAC2/3 by siRNA in Rh30 and CF-1 cells, which resulted in increased miR-27a expression relative to that in scrambled-siRNA control cells (Fig. 6, I and J). These results altogether suggest that, by inhibiting HDAC3 and maybe in part HDAC2, ENT blocks epigenetic suppression of an miRNA, thereby enabling its restriction of PAX3:FOXO1 expression and reducing the growth of aRMS tumors (Fig. 6K).
Fig. 6. miR-27a over-expression silences PAX3:FOXO1.
(A and B) Western blot of PAX3:FOXO1 protein abundance in Rh30 and CF-1 cells transfected with mimics of miR-27a (10 μM for 72 hours) or a negative control (ctrl). Also shown is blotting in lysates from untransfected control cells (unt). Blots are representative of N=3 independent experiments. (C and D) Light microscopy images of Rh30 and CF-1 cells transfected with mimics of miR-27a. Images are representative of N=3 independent experiments. Scale bar, 50 μM. (E to H) qPCR analysis of miR-27a (E and F) or miR-485p (G and H) expression in Rh30 and CF-1 cells treated for 72 hours with ENT (1 μM) or PAN (45 nM). (I and J) qPCR of miR-27a expression in Rh30 and CF-1 cells transfected with HDAC2- or HDAC3-targeted siRNA (100 nM). Data were normalized to U6snRNA expression. Gene expression was quantified using the 2^-dCt method. Data are means ± SD, N=3 independent experiments each in triplicate.; * P < 0.05, **P < 0.01, and ***P < 0.001 by a two-sided Student’s t tests. (K) Summary of the HDAC3–SMARCA4–miR-27a–PAX3:FOXO1 regulatory circuit targeted by ENT.
Discussion
Our previous studies uncovered that cell-of-origin conveys an epigenetic memory to tumor cells, leading to differences in histological phenotype and drug sensitivity, and that cell-of-origin also epigenetically influences the transcription of the PAX3:FOXO1 oncogene in tumor cells (13). Apropos to those findings, mechanistic studies we performed here suggest that HDAC2/HDAC3 inhibition is the primary mechanism of entinostat-mediated repression of PAX3:FOXO1 in aRMS. We also found that entinostat and panobinostat, despite both being HDAC inhibitors, are very different drugs with different global actions and specifically different activity with respect to SMARCA4, the inhibition of which we have now shown is critical to miR-27a expression and PAX3:FOXO1 mRNA destabilization.
De-regulated expression and function of miRNAs has been reported in many cancers, including RMS. HDACi’s and entinostat specifically have been previously reported to alter miRNA expression (19, 20). In this study, we found an miRNA that inhibited PAX3:FOXO1 mRNA translation and/or its stability. Our focus was on miRNAs that could bind either the PAX3 promoter and exon 1–8, or the FOXO1 exons 1–3 and its 3’ UTR, as well as those that were conserved across humans and mice, given that entinostat reduced both human and mouse PAX3:FOXO1. Notably, miR-27a-5p targets both PAX3 and PAX7 (18), miR-27a’s own promoter is regulated in muscle by the myogenic transcription factors MYF6, MYOD and MEF2C (21) (which aligns with RMS features), and SMARCA4 is predicted by ENCODE to bind the 3’ UTR of miR-27a. Indeed, knockdown of SMARCA4 or pharmacological inhibition of it bromodomain activity increased the expression of miR-27a. Although SMARCA4 is typically characterized as a transcriptional activator, reports suggest that SMARCA4 can also act as a repressor (22, 23). How precisely SMARCA4 represses miR-27a requires further investigation and may reveal insight on its function in other contexts.
From a clinical point of view, entinostat in combination with the chemotherapy agent vincristine showed strong anti-tumor activity in aRMS orthotopic mouse models at clinically achievable adult drug concentrations. For the purposes of clinical trial planning, we have also established the comparative efficacy of entinostat to other HDAC inhibitors at reducing PAX3:FOXO1 and PAX7:FOXO1 expression. Not only did entinostat reduce PAX3:FOXO1 abundance across species in both murine and human aRMS cell lines/cultures and patient-derived xenograft explant cell cultures, but entinostat also inhibited PAX3:FOXO1 levels more effectively than the other HDACi’s tested. We note that in the time since our studies were performed, Malempati et al. (24) have reported the Phase I clinical trial results for entinostat in pediatric patients, showing higher drug exposures and decreased clearance without any additional toxicity for children versus adults (Cmax of 140.8 nM at a dose of 4 mg/m2 and half-life 45 hours in children). Thus, our preclinical studies may be overly-stringent with respect to clinical modeling.
In summary, our preclinical data with strong mechanical evidence suggest that targeting HDAC3-SMARCA4-miR27a-PAX3:FOXO1 regulatory circuitry in aRMS may provide real therapeutic benefit for patients with aRMS. The preclinical efficacy in vivo and overall biochemical performance at PAX3:FOXO1 suppression make entinostat a promising drug candidate for combining with chemotherapy and/or radiation in clinical trials for aRMS. Our findings directly support the pediatric phase IB clinical trial ADVL1513 concept under review for the use of entinostat as single agent in rhabdomyosarcoma and possible phase II chemotherapy-entinostat trials to follow.
Materials and Methods
Cell culture
Murine primary tumor cell cultures (U23674) were generated as described previously (25). Human aRMS cell lines Rh30 and Rh41 were cultured in growth medium (GM) RPMI 1640 (11875–093; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (26140079; Thermo Fisher Scientific) and 1% penicillin/streptomycin (15140-122; Thermo Fisher Scientific). CW9019 cultured in Dulbecco’s Modified Eagle Medium (DMEM) (11965-084; Sigma) supplemented with 10% FBS and 1% penicillin/streptomycin. C2C12, mouse myoblast cell line obtained from ATCC and cultured in DMEM supplemented with 20% FBS and 1% penicillin/streptomycin. Primary human skeletal mouse myoblast (HSMM) (CC-2580; Lonza Inc.) cultured in GM (151-500, Cell Applications). Phoenix-AMPHO cells (a second-generation retrovirus producer cell line) (CRL-3213; ATCC) cultured in DMEM supplemented with 10% FBS and 1% antibiotics. Patient-derived xenograft explant cell culture CF-1 was cultivated in RPMI 1640 growth supplemented with 10% FBS and 1% antibiotics. All cells were incubated at 37°C and 5% CO2. Rh30 and Rh41 were obtained from the Children’s Oncology Group (COGcell.org) and were authenticated by STR validation assay through Biosynthesis. The murine cell culture U23674 was authenticated by PCR validation by Transnetyx. CF-001 was authenticated by analysing the expression of PAX3:FOXO1.
Vincristine chemotherapy, HDAC inhibition, and SMARCA4 inhibition
The chemotherapy agent vincristine sulfate, used here in the treatment of Rh30 and U23674 cells, was obtained from Sigma (V8879). HDAC inhibitors (HDACi used in this study were were purchased from SelleckChem: entinostat (also known as MS-275, a class I HDACi; cat. no. S1053), panobinostat (a broad-spectrum HDACi; S1030), SAHA (suberoylanilide hydroxamic acid, a class and class II HDACi; S1047), CUDC-907 (a PI3K inhibitor and class I and II HDACi; S2759), and CUDC-101 (a class I and II HDACi and inhibitor of epidermal growth factor receptor family members EGFR and HER2; S1194). aRMS cell lines and primary tumor cell cultures were treated with these drugs at their clinically relevant Cmax (maximum plasma concentrations) or, where Cmax is not yet reported, their determined IC25 (25% inhibiting concentration).
TR-FRET assay of the and treatment of cells SMARCA4 inhibitor
The SMARCA4 bromodomain activity inhibitor, PFI-3 (S7315), was purchased from SelleckChem. Rh30 cells were treated with PFI-3 at a concentration of 10 μM. PFI-3 inhibits 95% of the SMARCA4 bromodomain activity at 10 μM based on TR-FRET assay carried out by a commercial vendor, BPS bioscience. Briefly, the assay was performed by TR-FRET technology using recombinant bromodomain and BET Ligand. The TR-FRET signal from the assay is correlated with the amount of Ligand binding to the bromodomain. Binding experiments were performed in duplicate at each concentration. The TR-FRET data were analyzed using the computer software, GraphPad Prism (GraphPad Software Inc.). In the absence of the compound in wells containing BET ligand, the TR-FRET signal (Ft) in each data set was defined as 100% activity. Select wells where control inhibitors were more than 100-fold the IC50 were used to define the TR-FRET signal (Fb) as 0% activity. The percent activity in the presence of each compound was calculated according to the following equation: % activity = [(F-Fb)/(Ft - Fb)]×100, where F= the TR-FRET signal in the presence of the compound. The percent inhibition was calculated according to the following equation: % inhibition = 100 - % activity.
RNA-extraction and RT-PCR
Rh30 (human aRMS) cell line and murine aRMS primary tumor cell cultures (U23674) were treated with 0.1, 0.2, 0.4, 0.8, 1 and 2 μM entinostat for 24 hours. DMSO treatment was used as a control. After treatment with entinostat, total RNA was extracted and cDNA was synthesized as previously described (13). Expression of PAX3:FOXO1 was determined by RT-PCR using custom Taqman primers and probe (cat# 4304970 and 4316034) on a StepOnePlus real-time PCR machine (Applied Biosystems). Rh30 and Rh41 cell lines were treated with 1 μM of entinostat, 45nM of panobinostat and 1 μM of SAHA for 24 hours. DMSO treated cells were used as control. Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Complementary DNA was prepared from RNA using high capacity cDNA reverse transcription kit (Applied Biosystems) with RNase inhibitor. Quantitative PCR (qPCR) was performed using TaqMan Universal Master Mix, no AmpErase UNG on ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). Primers used were Gapdh-Hs02758991_g1 and PAX3:FOXO1-Hs03024825_ft. Gene expression was quantified using the 2^-dCt method.
Immunoblotting
Human Rh30 and Rh41 cell lines and murine aRMS primary tumor cell cultures (U23674) were treated with 1 μM entinostat, 45 nM panobinostat, 1 μM SAHA, 150 nM CUDC907 or 150nM CUDC-101 for 72 hours. DMSO (vehicle) treated cells were used as control. After 72 hours, lysates were collected using RIPA lysis buffer with Halt Protease and Phosphatase Inhibitor Cocktail (78440;Thermo Fisher Scientific) and analyzed for PAX3:FOXO1 expression using an anti-PAX3 antibody (MAB2457, 1:400; R & D Systems), PAX7:FOXO1 expression using an anti-FKHR antibody (H-128, 1:300; Santa Cruz Biotechnology), SMARCA4 expression using anti-SMARCA4 antibody (G-7, 1:300; Santa Cruz Biotechnology) and matched for protein expression using an anti-β-actin antibody (ab8227, 1:10,000; Abcam). All blots were visualized using a FluorChemQ system (ProteinSimple). For analyzing endogenous wild-type FOXO1 expression, recombinant FOXO1 (NM_002015) (Origene) was used. FOXO3 (NM_001455) was used as negative control to choose the appropriate antibody (fig. S7). Anti-FOXO1 antibody (clone 2H8.2, 1:500; Millipore) detects recombinant FOXO1 without background compared to other antibodies and subsequently used for all immunoblots (fig. S7).
Orthotopic allograft studies
All animal studies were conducted with IACUC approval at the Oregon Health & Science University. Orthotopic allograft mouse model of aRMS (U23674, genotype Myf6Cre, PAX3:FOXO1, p53) was generated as described (13). Mice were treated with entinostat at a daily dose of 5 mg/kg by intraperitoneal (IP) injection, vincristine sulfate at a dose of 1 mg/kg weekly by IP injection, or a combination of both. Treatment was started once the tumors reached 0.25 cc and ended when the tumors reached 1.5 cc. During treatment, mice becoming with body weight loss (10–15%) were euthanized early. For the radiation studies, murine aRMS primary tumor cell cultures (U23674) were either pretreated with DMSO or 2 μM entinostat and then subjected to 10 Gy radiation before injecting 500,000 viable cells into each mouse. Failure was defined as an event for tumor size greater than or equal to 1.2 cc.
Patient-derived xenograft (PDX) models at Champions Oncology
The Champions Personalized Tumorgraft™ chemosensitivity tests were conducted using a TumorGraft model established from two independent rhabdomyosarcoma biopsy specimens. The explants were received and immediately implanted into immunodeficient mice. The antitumor activity of entinostat and vincristine was tested in a low-passage immune-compromised female mice (Harlan; nu/nu) between 5–8 weeks of age housed on irradiated papertwist-enriched 1/8” corncob bedding (Sheperd) in individual HEPA ventilated cages (Innocage® IVC, Innovive USA) on a 12-hour light-dark cycle at 68–74°F (20–23°C) and 30–70% humidity. Animals were fed water ad libitum (reverse osmosis, 2 ppm Cl2) and an irradiated Test rodent diet (Teklad 2919) consisting of 19% protein, 9% fat, and 4% fiber. All compounds were formulated according to manufacturer’s specifications. Beginning Day 0, tumor dimensions were measured twice weekly by digital caliper and data including individual and mean estimated tumor volumes (Mean TV ± SEM) recorded for each group; tumor volume was calculated using the formula (1): TV= width2 x length × 0.52. All studies were done with the approval of Champions Oncology IACUC.
PDX models at The Jackson Laboratory
To establish each PDX model, NSG (NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ) mice were obtained by The Jackson Laboratory. Tumor explants were obtained from the patients and immediately implanted into the rear flanks of recipient female NSG (JAX # 5557) mice using a trochar. Once tumors reached about 2000 mm3, they were collected and passaged for serial transplantation in NSG mice to create low-passage fragments or cohort for future studies. The tumor volume range for enrollment were 150–250 mm3. All studies were done with the approval of The Jackson Laboratory IACUC. Mice were treated with vehicle or ENT/VCR as a single agent or combination at dose and route of administration provided in table S2 until tumors reached 2000 mm3 or reached study day 28. The antitumor activity of entinostat and vincristine was tested. All compounds were formulated according to manufacturer’s specifications. Beginning Day 0, tumor dimensions were measured twice weekly by digital caliper and data including individual and mean estimated tumor volumes (Mean TV ± SEM) recorded for each group; tumor volume was calculated using the formula (1): TV= (width)2 × length/2.
siRNA-mediated silencing of Class I and Class II HDACs
For silencing of Class I HDACs (1, 2 and 3), Rh30 cells were transfected with 100 nM of HDAC1 (L-003493-00-0005 5 nmol), HDAC2 (L-003495-02-0005 5 nmol), HDAC3 (L-003496-00-0005 5 nmol) or HDAC10 (L-004072-00-0005 5 nmol) SMARTpool siRNA reagent (a pool of four siRNA duplexes all designed to target distinct sites within the specific gene of interest) (Dharmacon) versus scrambled siRNA (Dharmacon) using Lipofectamine RNAimax (13778030; Invitrogen). siRNA Cell lysates were subjected to western blotting using anti-HDAC1 (H-51, 1:400; Santa Cruz Biotechnology), anti-HDAC2 (C-8, 1:400; Santacruz Biotechnology), anti-HDAC3 (B-12, 1:400; Santa Cruz Biotechnology) antibody, anti-HDAC10 (E-2, 1:400; Santa Cruz Biotechnology) and PAX3:FOXO1 expression using anti-PAX3 antibody (R & D Systems).
Retroviral infection
To restore expression of PAX3:FOXO1 in U23674 with stable knockdown of PAX3:FOXO1 (U23674 shYFP), retroviral vector pk1-PAX3:FOXO1 transfected into Phoenix-AMPHO packaging cells using Lipofecatmine Plus reagent (15338100; Invitrogen). Medium was harvested after 24, 48 and 72 hours and filtered with 0.45 μm syringe filter. Retroviral supernatant with polybrene was used for transduction of U23674 shYFP culture.
siRNA-mediated silencing of SMARCA4
For silencing of SMARCA4, Rh30 cells were transfected with 100 nM of SMARCA4 (L-010431-00-0005 5 nmol) SMARTpool siRNA reagent (Dharmacon) versus scrambled siRNA (Dharmacon) using Lipofectamine RNAimax (Invitrogen). Cell lysates were subjected to western blotting using anti-SMARCA4 (G-7, 1:300; Santa Cruz Biotechnology).
miRNA over-expression
For over-expression of miR-27a-5p, Rh30 and CF-1 cells were transfected with 10 μM of miR-27-a-5p mimics (assay name: hsa-miR-27a-5p; accession number: MI0000085) or miR-27a-5p negative control (4464058 5 nmol) (Thermo Fisher Scientific) using Lipofectamine RNAimax (Invitrogen). Cell lysates collected at the end of 72 hours subjected to western blotting for analysis of PAX3:FOXO1 expression using an antibody against PAX3 (R & D systems).
miRNA isolation and RT-PCR
miRNA isolation was performed as per the manufacturers protocol (K157001; Invitrogen). Briefly, Rh30 and CF-1 cells were seeded at a density of 0.3 million cells and the next day, treated with entinostat (1 μM) or DMSO (vehicle). At the end of 24 hours treatment, miRNA was isolated per manufacturer standard protocol. The recovery tube with RNA was then quantified using Bio-Tek microvolume plate. Complementary DNA was prepared from 5 ng of miRNA using TaqMan miRNA reverse transcriptase kit (4366596; Thermo Fisher Scientific). Quantitative PCR was performed using TaqMan Universal Master Mix, no AmpErase UNG (4440040; Thermo Fisher Scientific) on Bio-Rad Thermocycler Real-Time PCR System. Primers used were hsa-miR-27a and hsa-miR-485 (TaqMan miRNA assays). U6snRNA (TaqMan miRNA assays) used as loading control. miRNA expression was quantified using the 2^-dCt method.
RNA-seq
For the identification of transcriptional changes (Fig. 3, C–F) in aRMS cells after ENT treatment, each of the three aRMS cultures (human cell lines Rh30 and Rh41, mouse cell culture U23674) was treated with ENT alongside a paired untreated sample for a fixed time period. All samples were treated with ENT for 72 hours, except for Rh41 which was treated for 24 hours due to higher sensitivity to entinostat. All cells were cultured on 10cm dishes and treatment began when plates were 60% confluent. Passages lower than 7 were used for all mouse cultures. RNA isolation and sequencing was performed by the commercial service provider Beijing Genomics Institute (BGI). Differential expression for a single sample we defined as post-ENT treatment expression divided by vehicle-treated expression. These criteria identified 348 overexpressed and 358 under-expressed genes in aRMS. Differential expression of genes associated with chromatin modification and remodeling following ENT and PAN treatment (72 hours) (Fig. 5A) was analyzed by one more set of RNA-seq with RNA isolation and sequencing performed by BGI. Log2 scaled ratios of entinostat-treated vs. vehicle-treated RH30 and U23674 and panobinostat-treated vs. vehicle-treated RH30 and U23674 organized by change in regulation.
Bioinformatic analysis of RNA-seq
The paired end raw reads for all RNA-seq data samples were aligned using TopHat version 2.0.9. Up to two mismatches in the alignment were permitted before a read alignment was discarded. The reads for human samples were aligned to the UCSC hg36 human reference genome. The reads for mouse samples were aligned to the UCSC mm19 mouse reference genome. The aligned reads were assembled into transcripts using Cufflinks version 2.2.1. Differential comparisons were performed by the Cuffdiff function of Cufflinks, version 2.2.1. Differential comparisons were made between samples treated with entinostat and untreated control samples. For the differential analysis of treated vs. untreated, the standard Cuffdiff parameters were used. Reported in all figures are log2 scaled differential values. Genes for which treated samples had non-zero expression and untreated samples had zero expression had expression fixed were set to the minimum of the treated expression and 1024, before log2 scaling, to prevent divide by zero errors during downstream analysis. Only genes with quantified expression across all samples (total of 14,575 genes) were considered. Log2 scaled mean differential expression across all samples of a common subtype was used to identify over- and under-expression.
ChIP-exo
For identification of ENT-related gene expression changes for the known targets of PAX3:FOXO1, ChIP-exo studies were performed by Peconic Genomics using anti-PAX3 antibody (cat# MAB2457, R & D Systems) with the murine U23674 aRMS culture as previously described (26). Paired-end, 40 base pair reads were generated. Putative binding sites (peaks) were identified using the MACS program, version 2.1.0.20140616, with default parameters without a control sample (in which case MACS uses regional read levels for background) (27). Overall data quality and peak quality were verified by a combination of methods, including cross-correlation of positive- and negative-strand reads (which showed a peak expected from punctate binding at a plausible offset of approximately 15bp) (26) and the R package ChIPQC (which showed good numbers of reads-in-peaks (28). ChIPseq data for additional ENT targets HDAC1, HDAC2, HDAC3 and HDAC11 were taken from published sources (29, 30). HDAC11 was used as a surrogate for HDAC10, since HDAC10 data was not available. Additional data was gathered from published sources for PAX3:FOXO1 binding identified by cyclic amplification and selection of targets (CASTing) (31), and by previously published PAX3:FOXO1 ChIP-seq data (32). Published expression data (33) as well as cell-cycle dependent expression data (10) were also integrated to infer indirect targets of PAX3:FOXO1. Differential expression across samples following ENT treatment was used to identify key upregulated and downregulated genes in aRMS. Bioinformatic analysis of PAX3:FOXO1 and HDACs binding sites was then cross-referenced for RNAseq data for ENT-treated cells.
Statistical analysis
Bioinformatics and computational methods are described above. Continuously distributed outcomes were summarized with the mean plus or minus one standard deviation. Treatment groups were contrasted on the mean with analysis of variance in log units. Time to event distributions were summarized with Kaplan-Meier curves and significance of variation with treatment group was assessed with log rank tests. Corrections for multiple comparisons were made with the Dunnett method for ANOVA and the Bonferroni method for log rank testing. Statistical testing on means and time to event was two-sided with a nominal significance level of 5%, carried out with R. For PDX mouse models, the significance of variation in tumor volume with treatment was assessed with a repeated measures linear model with an autoregressive order 1 autocorrelation matrix and a Tukey correction for multiple comparisons in terms of treatment, day, and the treatment by day interaction. All analyses were carried out in log10 units and all statistical testing was two sided with a 5% experiment wise significance level. SAS Version 9.4 for Windows (SAS Institute) was used throughout. GraphPad Prism was used for statistical analysis of cell viability assay and TR-FRET assay. For densitometric analysis (% change), significance was determined by a two-tailed Student’s t test and p values of <0.05 considered to be statistically significant. Statistical significance was set at * P < 0.05, **P < 0.01 and ***P < 0.001. Error bars indicate mean ± SD or SEM.
Supplementary Material
Supplementary Materials
Fig. S1. ENT treatment of aRMS in vivo.
Fig. S2. ENT reduces PAX3:FOXO1 abundance at a clinically relevant dose and time of exposure and reduces cell viability synergistically with vincristine.
Fig. S3. PAX3:FOXO1 mRNA expression is reduced by ENT in aRMS cell lines.
Fig. S4. Better than other HDAC inhibitors, ENT reduces PAX3:FOXO1 abundance in murine and human aRMS cells.
Fig. S5. Representative histology of PDX mouse aRMS tissue.
Fig. S6. PFI-3–mediated inhibition of SMARCA4 bromodomain activity
Fig. S7. Validation of commercially available antibodies for detecting endogenous FOXO1 abundance in aRMS cell lines.
Table S1. Histological markers of differentiation in aRMS orthotopic allograft model mice.
Table S2. Treatment schedules for CF-4/PCB-513 PDX model mice.
Table S3. Patient history of the Champions Oncology PDX aRMS models.
Table S4. Patient history of the Jackson Laboratory PDX aRMS models.
Table S5. Histological scoring of markers of differentiation in PDX aRMS mice
Table S6. Statistical analysis of CTG-1604/POS-14175 data.
Table S7. Statistical analysis of J101220/CF-4 data.
Table S8. Statistical analysis of J77636/PCB-481 data.
Table S9. Statistical analysis of J0103366/CF-13A data.
Table S10. Statistical analysis of J099761/CF-1 data.
Table S11. Statistical analysis of CTG-1409/POS 14107 data.
Table S12. Statistical analysis of J099873/CF-2 data.
Table S13. Statistical analysis of CTG-1008 data.
Acknowledgements:
This work was supported by the NIH grants 5R01CA189299,1R01CA143082 P30CA006973, UL1TR001079, UL1TR001079; St. Baldrick’s Foundation; Braver, Stronger, Smarter Foundation; Michelle Paternoster Foundation for Sarcoma Research; dancingwithbrad.com; Christina Renna Foundation; Friends of Doernbecher Foundation; Friends of T.J. Foundation; Clarke Gilles Foundation; and a private anonymous gift in memory of Nanette. Additional funding was provided to FGB by the Intramural Research Program of the National Cancer Institute. We are grateful to the families who shared the PDX model data. We thank Ken Kikuchi for assistance.
Footnotes
Publisher's Disclaimer: “This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Competing Interests: C.K. received an unrestricted grant from Syndax Pharmaceuticals that supported third-party testing of entinostat in patient-derived xenografts. P.O. is an employee of Syndax Pharmaceuticals. J.E.W. is an employee of AbbVie Pharmaceuticals. Unrelated to this study, C.K. has had sponsored research agreements with Eli Lilly and Roche-Genentech, and N.E.B. is a scientific officer at First Ascent Biomedical Corp. C.R.V. is an advisor to KSQ Therapeutics and receives research funding from Boehringer-Ingelheim. D.S.H. has received reimbursement for travel to Medical Advisory Board meetings for Loxo Oncology, Bristol Myers Squibb, Celgene, and Bayer, but no compensation otherwise.
Data & Materials Availability: RNAseq data have been deposited in the NCBI Gene Expression Omnibus Database (GEO Accession number GSE115698). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Davis LE, Keller C, Integrative biology of rhabdomyosarcoma using genetic murine models. AACR 2012 Annual Meeting Educational Book, (2012). [Google Scholar]
- 2.Smith MA, Altekruse SF, Adamson PC, Reaman GH, Seibel NL, Declining childhood and adolescent cancer mortality. Cancer 120, 2497–2506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. I. o. H. U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES, National Cancer Institute., L. Y. A. Alliance., “Closing the Gap: Research and Care Imperatives for Adolescents and Young Adults with Cancer Report of the Adolescent and Young Adult Oncology Progress Review Group.,” NIH Publication No. 06–6067 (National Cancer Institute, Bethesda, MD, 2006). [Google Scholar]
- 4.Breneman JC et al. , Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma--a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol 21, 78–84 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Malempati S, Hawkins DS, Rhabdomyosarcoma: review of the Children’s Oncology Group (COG) Soft-Tissue Sarcoma Committee experience and rationale for current COG studies. Pediatric blood & cancer 59, 5–10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams BA et al. , Metastatic rhabdomyosarcoma: a retrospective review of patients treated at the hospital for sick children between 1989 and 1999. J Pediatr Hematol Oncol 26, 243–247 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Pappo AS et al. , Survival After Relapse in Children and Adolescents With Rhabdomyosarcoma: A Report From the Intergroup Rhabdomyosarcoma Study Group. Journal of Clinical Oncology 17, 3487–3493 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Rudzinski ER et al. , Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: A report from the Children’s Oncology Group. Pediatric blood & cancer 64, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Missiaglia E et al. , PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol 30, 1670–1677 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi K et al. , Cell-cycle dependent expression of a translocation-mediated fusion oncogene mediates checkpoint adaptation in rhabdomyosarcoma. PLoS genetics 10, e1004107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen EY et al. , Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc Natl Acad Sci U S A 111, 5349–5354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kikuchi K et al. , Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun 365, 568–574 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Abraham J et al. , Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes & development 28, 1578–1591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yardley DA et al. , A phase 1 study to assess the food effect on the pharmacokinetics (PK) of entinostat. Journal of Clinical Oncology 32, 591–591 (2014). [Google Scholar]
- 15.Bauer S et al. , Phase I study of panobinostat and imatinib in patients with treatment-refractory metastatic gastrointestinal stromal tumors. British journal of cancer 110, 1155–1162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin EH et al. , A study to determine the effects of food and multiple dosing on the pharmacokinetics of vorinostat given orally to patients with advanced cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 12, 7039–7045 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Lu XF et al. , Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transducer and activator of transcription 3 signaling pathway. Cell death & disease 9, 398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crist CG et al. , Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci U S A 106, 13383–13387 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S et al. , Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis 4, e556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampath D et al. , Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 119, 1162–1172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Torres F, Aranega AE, Franco D, Identification of regulatory elements directing miR-23a-miR-27a-miR-24–2 transcriptional regulation in response to muscle hypertrophic stimuli. Biochim Biophys Acta 1839, 885–897 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Zhang X et al. , Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem cell reports 3, 460–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ, BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. The Journal of biological chemistry 281, 38974–38980 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malempati S et al. , ADVL1513: Results of a phase 1 trial of entinostat, an oral histone deacetylase inhibitor, in pediatric patients with recurrent or refractory solid tumors. Journal of Clinical Oncology 36, 10556–10556 (2018). [Google Scholar]
- 25.Aslam MI et al. , PDGFRbeta reverses EphB4 signaling in alveolar rhabdomyosarcoma. Proceedings of the National Academy of Sciences of the United States of America 111, 6383–6388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramachandran P, Palidwor GA, Porter CJ, Perkins TJ, MaSC: mappability-sensitive cross-correlation for estimating mean fragment length of single-end short-read sequencing data. Bioinformatics (Oxford, England) 29, 444–450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y et al. , Model-based Analysis of ChIP-Seq (MACS). Genome Biology 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll TS, Liang Z, Salama R, Stark R, de Santiago I, Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet 5, 75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Negre N et al. , A comprehensive map of insulator elements for the Drosophila genome. PLoS genetics 6, e1000814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z et al. , Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barber TD et al. , Identification of target genes regulated by PAX3 and PAX3-FKHR in embryogenesis and alveolar rhabdomyosarcoma. Genomics 79, 278–284 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Cao L et al. , Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res 70, 6497–6508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishijo K et al. , Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer research 69, 2902–2911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Fig. S1. ENT treatment of aRMS in vivo.
Fig. S2. ENT reduces PAX3:FOXO1 abundance at a clinically relevant dose and time of exposure and reduces cell viability synergistically with vincristine.
Fig. S3. PAX3:FOXO1 mRNA expression is reduced by ENT in aRMS cell lines.
Fig. S4. Better than other HDAC inhibitors, ENT reduces PAX3:FOXO1 abundance in murine and human aRMS cells.
Fig. S5. Representative histology of PDX mouse aRMS tissue.
Fig. S6. PFI-3–mediated inhibition of SMARCA4 bromodomain activity
Fig. S7. Validation of commercially available antibodies for detecting endogenous FOXO1 abundance in aRMS cell lines.
Table S1. Histological markers of differentiation in aRMS orthotopic allograft model mice.
Table S2. Treatment schedules for CF-4/PCB-513 PDX model mice.
Table S3. Patient history of the Champions Oncology PDX aRMS models.
Table S4. Patient history of the Jackson Laboratory PDX aRMS models.
Table S5. Histological scoring of markers of differentiation in PDX aRMS mice
Table S6. Statistical analysis of CTG-1604/POS-14175 data.
Table S7. Statistical analysis of J101220/CF-4 data.
Table S8. Statistical analysis of J77636/PCB-481 data.
Table S9. Statistical analysis of J0103366/CF-13A data.
Table S10. Statistical analysis of J099761/CF-1 data.
Table S11. Statistical analysis of CTG-1409/POS 14107 data.
Table S12. Statistical analysis of J099873/CF-2 data.
Table S13. Statistical analysis of CTG-1008 data.