Abstract
Transposition and homologous recombination assays are valuable genetic tools to measure the production and integration of cDNA from the long terminal repeat (LTR) retrotransposon Tf1 in the fission yeast (S chizosaccharomyces pombe). Here we describe two genetic assays, one that measures the transposition activity of Tf1 by monitoring the mobility of a drug resistance marked Tf1 element expressed from a multi-copy plasmid and another assay that measures homologous recombination between Tf1 cDNA and the expression plasmid. While the transposition assay measures insertion of full-length Tf1 cDNA mediated by the transposon integrase, the homologous recombination assay measures levels of cDNA present in the nucleus and is independent of integrase activity. Combined, these assays can be used to systematically screen large collections of strains to identify mutations that specifically inhibit the integration step in the retroelement life cycle. Such mutations can be identified because they reduce transposition activity but nevertheless have wild-type frequencies of homologous recombination. Qualitative assays of yeast patches on agar plates detect large defects in integration and recombination, while the quantitative approach provides a precise method of determining integration and recombination frequencies.
Keywords: Tf1, Schizosaccharomyces pombe, Fission yeast, Transposition assay, Homologous recombination assay, Quantitative assay, LTR-retrotransposon
1. Introduction
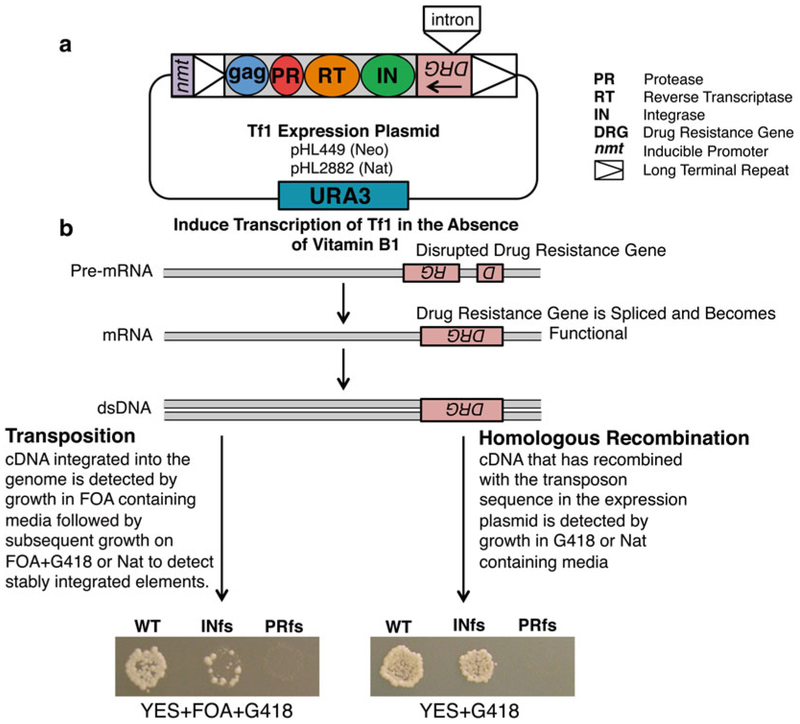
The long terminal repeat (LTR) retrotransposon Tf1 isolated from Schizosaccharomyces pombe encodes four proteins: Gag, protease (PR), reverse transcriptase (RT), and integrase (IN), and coding sequences are flanked by two LTRs (see Fig. 1a). Tf1 mobilizes in a manner similar to retroviruses, through particle formation and reverse transcription followed by integration of the cDNA into the genome of the host cell. As integration into new genomic locations can have deleterious effects on the host by damaging genetic content, determining the targeting mechanism of these elements is of significant importance [1,2]. Previous studies have shown that Tf1 preferentially targets promoters of RNA pol II-transcribed genes, and notably insertions are observed at high levels within promoters of stress response genes. However, it remains largely unclear which host factors play a role in the observed integration preference [3 – 8]. Transposition and homologous recombination assays therefore provide a useful genetic tool to examine these questions and to analyze the role of host factors in mediating transposon integration. These genetic assays function by monitoring integration events into new genomic locations and by measuring homologous recombination of Tf1 cDNA with plasmid sequences [9]. The transposition and recombination activities of Tf1 are measured by expressing a plasmid-encoded copy of Tf1 under the control of a heterologous promoter (nmt1) which is activated in the absence of thiamine. The Tf1 sequence in the expression plasmid contains a drug resistance gene in the opposite orientation with respect to the transposon. This drug resistance gene is interrupted by an artificial intron that due to its orientation is spliced out of the Tf1 transcript but not from the transcript of the resistance gene (see Fig. 1).
Fig. 1.
Genetic assays for transposition and homologous recombination. (a) Tf1, encoding the proteins Gag, PR, RT, and IN, is expressed on a plasmid that is transformed into S. pombe cells. Tf1 expression is under the control of an inducible promoter, nmt1. The expression vector also contains the selectable marker URA3. (b) In the absence of vitamin B1, the Tf1 transcript is expressed and forms a functional drug resistance gene following splicing of the artifi cial intron. Transposition and recombination activity can be detected as patches on the appropriate selection media. Wild-type (WT) Tf1 generates confl uent growth; IN frameshift (INfs) displays signifi cantly reduced growth in the transposition assay (left panel) and approximately twofold less grown in the recombination assay (right panel). The PR frameshift (PRfs) lacks growth on selection media for both assays
Cells with integration and recombination events are selected through the expression of the drug resistance marker [9 – 12]. The qualitative assays of yeast patched on solid media provide a general indication of the transposition and recombination activities in the cell, whereas the quantitative assays provide a highly reproducible measure of cDNA integration and recombination frequencies. The genetic assays outlined here provide a particularly useful genetic tool to study transposon biology.
2. Materials
Here we present details for two genetic assays for Tf1 containing either the neomycin (Neo) or nourseothricin (Nat) drug selection markers. S. pombe can be grown on either complex or synthetic minimal media depending on the needs of the experiment. Complex yeast extract with supplements (YES) medium is made from yeast extract and includes all amino acids, including uracil and adenine supplements. Minimal medium is used to select for auxotrophic markers, and we recommend the use of the pombe glutamate medium (PMG) as it provides high transposition activity and is compatible for selection of the Neo marker by geneticin (G418) resistance. G418 selection is also effective in YES media, and selection of Nat resistance is equally effective in both complex and minimal media. All media and stock solutions should be filter-sterilized (liquid) or autoclaved (liquid and agar solutions) prior to use for only 15 min in order to avoid glucose caramelization [13]. Solid medium is made by the addition of an equal volume of 4 % (w/v) Difco Bacto Agar to 2× concentrated liquid media. Sterile liquid media can be kept for months at room temperature; plates can be stored when wrapped, to protect against drying for up to 4 months at 4 °C.
2.1. Preparationof Fission Yeast Media
YES media (per liter): 5 g yeast extract, 30 g glucose, and 2 g complete dropout powder (see Note 1).
PMG media (per liter): 3 g potassium hydrogen phthalate [14.7 mM], 2.2 g Na 2 PO 4 [15.5 mM], 3.75 g l-glutamic acid, monosodium salt, 20 g glucose (2 % w/v), 20 mL of 50× salt stock, 1 mL of 1000× vitamin stock, 0.1 mL of 10,000× mineral stock, 2 g of dropout powder lacking uracil and leucine (see Note 1).
YES media containing 5-fluoroorotic acid or 5-FOA (YES + 5-FOA): To YES medium add (to a final concentration) 1 mg/mL 5-FOA (US Biologicals) (see Note 2).
YES media containing NAT (YES + NAT): To YES medium, add (to a final concentration) 100 μg/mL nourseothricin (Jena Bioscience).
YES media containing G418 (YES + G418): To YES medium, add (to a final concentration) 500 μg/mL G418 (see Note 3).
YES media containing 5-FOA and nourseothricin (YES + 5-FOA + NAT): To YES medium add (to a final concentration) 1 mg/mL of 5-FOA and 100 μg/mL nourseothricin (see Note 2).
YES media containing 5-FOA and G418 (YES + 5-FOA + G418): To YES medium add (to a final concentration) 1 mg/mL of 5-FOA and 500 μg/mL G418 (see Notes 2 and 3).
PMG containing supplements, 5-FOA and vitamin B1 (PMG + U + L + 5-FOA + B1): To PMG medium add (to a final concentration) 250 μg/mL l-leucine, 50 μg/mL uracil, 10 μM vitamin B1, and 1 mg/mL 5-FOA (see Note 4).
PMG containing supplements leucine and vitamin B1 (PMG-U + L + B1): To PMG medium, add (to a final concentration) 250 μg/mL l -leucine, and 10 μM vitamin B1 (see Note 5).
10. PMG containing the supplement leucine (PMG-U + L-B1): To PMG medium, add (to a final concentration) 250 μg/mL l-leucine (see Note 6).
PMG media containing the supplements leucine, uracil, and vitamin B1 (PMG + U + L + B1): To PMG medium, add (to a final concentration) 250 μg/mL l-leucine, 250 μg/mL uracil, and 10 μM vitamin B1.
2.2. Preparation of Stock Solutions/ Powder
Complete dropout powder: 5 g adenine SO 4 and 2 g each of the following amino acids: alanine, arginine HCl, aspartic acid, asparagine H 2 O, cysteine HCl·H 2 O, glutamic acid, glutamine, glycine, histidine HCl·H 2 O, isoleucine, leucine, lysine HCl, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, uracil, and valine. Mix thoroughly (see Note 7). For addition to PMG media, exclude uracil and leucine components as these will be supplemented or expressed by exogenous plasmid markers (see Note 1).
50× salt stock (per liter): 2 g Na 2 SO 4 [14.1 mM], 50 g KCL [0.67 M], 0.735 g CaCl2 2H2O [4.99 mM], and 52.5 g MgCl2 6H2 O [0.26 M]. Dissolve in deionized water and autoclave.
1000× vitamin stock (per liter): 1 g pantothenic acid [4.20 mM], 10 g nicotinic acid [81.2 mM], 10 g inositol [55.5 mM], and 10 mg biotin [40.8 μM]. Dissolve in deionized water and filter sterilize.
10,000× mineral stock (per liter): 5 g boric acid [80.9 mM], 4 g MnSO 4 [23.7 mM], 4 g ZnSO 4 ·7H 2 O [13.9 mM], 2 g FeCl 2 ·6H 2 O [7.40 mM], 0.4 g molybdic acid [2.47 mM], 1 g Kl [6.02 mM], 0.4 g CuSO 4 ·5H 2 O [1.60 mM], and 10 g citric acid [47.6 mM]. Dissolve in deionized water and filter sterilize.
2.3. Plasmids and Strains
The following plasmids are transformed into S. pombe to assay the NeoAI marked version of Tf1. Wild-type Tf1: pHL449–1, Tf1 with IN frameshift: pHL476–3, and Tf1 with PR frame-shift: pHL490–80.
The following plasmids are transformed into S. pombe to assay the NatAI marked version of Tf1. Wild-type Tf1: pHL2882 Tf1 with IN frameshift: pHL2884, and Tf1 with PR frame-shift: pHL2883.
Strains of S. pombe we commonly use are YHL912 (h −, ura4294, leu1–32) and YHL1101 (h+, ura4-D18, leu1–32, ade6-M210). Strains used with NatAI-expressing Tf1 were derived from the Bioneer collection (h+, ura4-D18, leu1–32, ade6-M210 or ade6-M216). All of these strains were derived from Leupold’s original isolates of S. pombe and therefore lack endogenous copies of Tf1.
3. Methods
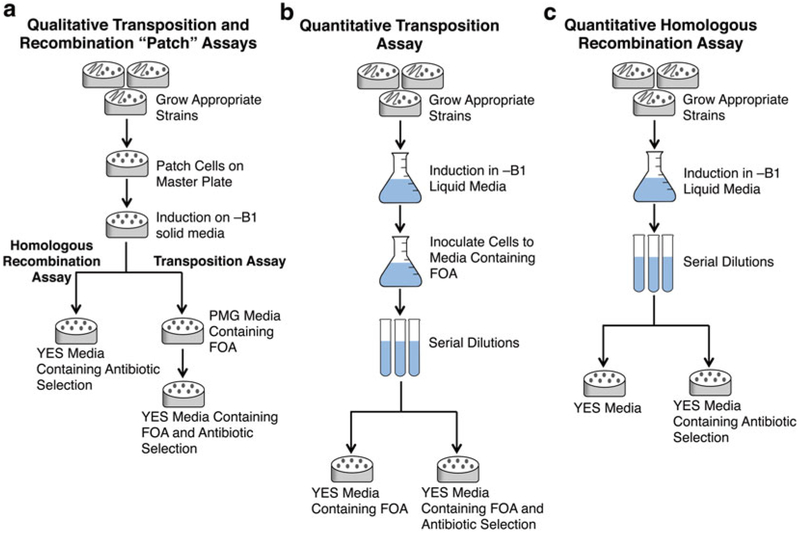
Genetic assays for transposition and recombination can be conducted as a qualitative assay with yeast patched on solid media (see Figs. 1b and 2a) and as a quantitative assay in liquid culture (see Fig. 2b, c). All S. pombe strains should be grown at 32 °C.
Fig. 2.
Flow diagrams for the qualitative and quantitative assays. Qualitative transposition and homologous recombination assays can be conducted with patches of yeast on solid media (a) or as a more accurate quantitative assay in liquid media (b, c)
3.1. Qualitative Transposition Assay (Fig. 2a)
Freshly revive experimental strains (four independent transformants of each Tf1 plasmid are tested with Tf1 controls containing frameshift mutation s) from frozen perm stocks for 3 days on PMG-U + L + B1 solid media (see Note 8).
Establish a master plate containing all experimental strains and controls by removing a small portion of the cell mass with a sterile toothpick from the thick part of the streak and spotting a pinhead-sized amount onto solid PMG-U + L + B1 media (see Note 9). Incubate for 3 days.
activity of the retrotransposon by replica printing the master plate to solid media lacking vitamin B1 (PMG-U + L-B1) using sterile velvets (see Note 10). The master plate can also be propagated at this point by making a second print onto a second non-inducing + B1-containing plate. Incubate the cells on PMG-U + L-B1 for 4 days to ensure sufficient induction of the nmt1 promoter.
Replica print cells to PMG + 5-FOA + U + L + B1 solid media using sterile velvets and incubate for 3 days (see Notes 11 and 12).
Replica print cells to YES + 5-FOA + desired antibiotic-containing media (G418 or Nat) using sterile velvets (see Note 13). Allow growth for 24–48 h (see Note 14 and Fig. 1b).
3.2. Qualitative Homologous Recombination Assay (Fig. 2a)
Freshly revive desired experimental strains (four independent transformants of each Tf1 plasmid are tested with Tf1 controls containing frameshift mutations) from frozen perm stocks by streaking a match-head’s worth of cells onto PMG-U + L + B1 solid media and incubating for 3 days (see Note 8).
Establish a master plate containing all desired experimental and control strains by removing a small portion of the cell mass with a sterile toothpick from the thick part of the streak and spotting a pinhead-sized amount onto solid PMG-U + L + B1 media (see Note 9). Incubate for 3 days.
Induce transposition activity of the retrotransposon by replica printing the master plate to solid media lacking vitamin B1 (PMG-U + L-B1) using sterile velvets (see Note 10). The master plate can also be propagated at this point by making a second print onto a second non-inducing + B1-containing plate. Incubate cells on PMG-U + L-B1 for 4 days to ensure sufficient induction of the nmt1 promoter.
Print cell mass to YES + antibiotic (G418 or Nat)-containing media (see Note 15). Incubate for 24 h (see Note 16 and Fig. 1b).
3.3. Quantitative Transposition Assay (Fig. 2b)
Freshly revive desired experimental strains (each strain is tested in triplicate including controls containing Tf1 frameshift mutation s) from frozen perm stocks by streaking a match-head’s worth of cells onto PMG-U + L + B1 solid media and incubating for 3 days (see Note 8). In addition to testing strains in triplicate, multiple transformants of each Tf1 plasmid must be assayed. It is therefore best to combine these requirements by testing three independent transformants of the Tf1 plasmid in each strain of interest.
Scrape a match-head amount of cell mass from the thick part of the streak and resuspend cells into 1 mL of PMG-U + L-B1 media (see Note 9). Vortex cells for 10 s or until cells are visibly and evenly resuspended in solution.
Harvest cells by centrifugation at (3400 RCF) for 1 min and pour off the supernatant. Blot off excess supernatant on a clean paper towel.
Resuspend cells again in 1 mL PMG-U + L-B1. Vortex, centrifuge, and remove supernatant. Repeat with four washes to ensure adequate removal of the vitamin B1 for efficient promoter induction (see Note 17).
Set up 5 mL induction cultures in PMG-U + L-B1 media using 15 mL disposable glass culture tubes at a starting optical density (OD 600 nm) of 0.05. Grow cultures for 4 days with turning on a roller drum (for optimal aeration of cells).
Measure the OD (600 nm) of the induction cultures and use them to inoculate 5 mL cultures in PMG + 5-FOA + U + L + B1 media using 15 mL disposable class culture tubes with a starting OD 0.1. Grow for 36 h on a roller drum.
Measure the OD (600 nm) of cells after 36 h of growth and dilute samples to OD 1 (2 × 10 7 cells/mL) in PMG + U + L + B1 media.
Prepare tenfold serial dilutions for each sample starting at 2 × 10 7 cells/mL (OD 1) and ending with 2 × 10 4 cells/mL in PMG + U + L + B1 media (see Note 18).
Plate 100 μL of dilutions containing 10 7, 10 6, and 10 5 cells/ mL onto YES + 5-FOA + antibiotic-containing media (see Note 13) using glass beads (~3 mm). Plate 100 μL of dilutions containing 10 6,10 5, and 10 4 cells/mL on YES + 5-FOA-containing media (see Note 19).
Allow cells to grow for exactly 3 days (72 h). Count the number of colonies formed on YES + 5-FOA and YES + 5-FOA + antibiotic-containing plates (see Note 20).
Calculate the frequency of Tf1 transposition by comparing the ratio of cells grown on 5-FOA media to cells grown on 5-FOA + antibiotic-containing media (see Notes 21 and 24).
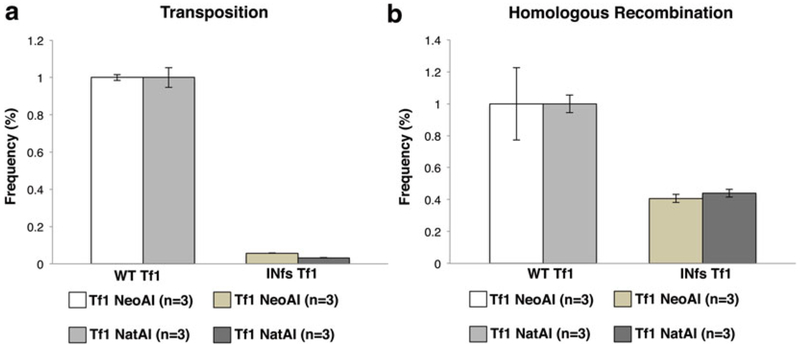
The results of wild-type Tf1 and the frameshift mutation s are shown graphically in Fig. 3a and the quantities are listed in Table 1.
Fig. 3.
Quantitative transposition and homologous recombination assays under G418 and nourseothricin selection. Transposition levels (a) and homologous recombination levels (b) in cells containing the wild-type Tf1 plasmids with either G418 or Nat selection are compared to levels in cells containing an INfs Tf1 expression vector. Transposition levels are reduced roughly 20-fold in the INfs controls (a) and homologous recombination levels are reduced roughly twofold in INfs controls when compared to WT (b). All samples are normalized to WT-Tf1 transposition or recombination levels. Each strain was assayed in triplicate (n = 3) and the results averaged
Table 1.
Raw and normalized transposition frequencies from strains tested in triplicate
| Strain | Transposition frequency (%) |
Standard deviation (%) |
Normalized transposition frequency (%) |
Standard deviation (%) |
|
|---|---|---|---|---|---|
| Tfl NeoAI | WT Tfl | 1.30 | 0.021 | 1.00 | 0.016 |
| INfs Tfl | 0.074 | 0.001 | 0.057 | 0.001 | |
| PRfs Tfl | 0.00 | 0.00 | 0.00 | 0.00 | |
| Tfl NatAI | WTT fl | 0.532 | 0.028 | 1.00 | 0.053 |
| INfs Tfl | 0.017 | 0.001 | 0.033 | 0.002 | |
| PRfs Tfl | 0.00 | 0.00 | 0.00 | 0.00 |
3.4. Quantitative Homologous Recombination Assay (Fig. 2c)
Freshly revive desired experimental strains (each strain is tested in triplicate including Tf1 controls containing frameshift mutation s) from frozen perm stocks by streaking a match-head’s worth of cells onto PMG-U + L + B1 solid media and growing for 3 days (see Note 8). In addition to testing strains in triplicate, multiple transformants of each Tf1 plasmid must be assayed. It is therefore best to combine these requirements by testing three independent transformants of the Tf1 plasmid in each strain of interest.
Scrape a small amount match-head’s worth of cell mass from the thick part of the streak and resuspend cells into 1 mL of PMG-U + L-B1 (see Note 9). Vortex cells for at least 10 s or until cells are evenly resuspended in solution.
Harvest cells with centrifugation at (3400 RCF) for 1 min and pour off the supernatant.
Resuspend cells again in 1 mL PMG-U + L-B1. Vortex, centrifuge, and remove supernatant. Repeat with four washes to ensure adequate removal of vitamin B1 for sufficient induction of the nmt1 promoter (see Note 17).
Set up 5 mL induction cultures of PMG-U + L-B1 media in disposable 15 mL glass culture tubes at a starting optical density (OD 600 nm) of 0.05. Incubate for 4 days with rolling for adequate aeration on a roller drum.
Measure OD (600 nm) of cells and dilute samples to OD 1 (2 × 10 7 cells/mL) in PMG-U + L + B1 medium.
Set up tenfold serial dilutions for each sample ranging from 2 × 10 7 cells/mL (OD 1) to 2 × 10 4 cells/mL in PMG-U + L + B1 media (see Note 18).
8. Plate 100 μL of dilutions containing 10 7, 10 6, and 10 5 cells/ mL onto YES + antibiotic-containing media (see Note 15) using glass beads to spread cells (~3 mm). Plate 100 μL of dilutions containing 10 6, 10 5, and 10 4 cells/mL onto YES media (see Note 22).
Incubate cells for exactly 3 days (72 h). Count the number of colonies formed on YES and YES + antibiotic-containing media (see Note 20).
The frequency of homologous recombination is the ratio of colonies grown on YES + antibiotic-containing media to colonies grown on YES-containing media (see Notes 23 and 25).
The results of wild-type Tf1 and the frameshift mutation s are shown graphically in Fig. 3b and the quantities are listed in Table 2.
Table 2.
Raw and normalized homologous recombination frequencies from strains tested in triplicate
| Strain | Recombination frequency (%) |
Standard deviation (%) |
Normalized recombination frequency (%) |
Standard deviation (%) |
|
|---|---|---|---|---|---|
| Tfl NeoAI | WT Tfl | 0.589 | 0.133 | 1.00 | 0.227 |
| INfs Tfl | 0.239 | 0.015 | 0.407 | 0.026 | |
| PRfs Tfl | 0.00 | 0.00 | 0.00 | 0.00 | |
| Tfl NatAI | WT Tfl | 1.96 | 0.109 | 1.00 | 0.056 |
| INfs Tfl | 0.862 | 0.049 | 0.439 | 0.025 | |
| PRfs Tfl | 0.00 | 0.00 | 0.00 | 0.00 |
4. Notes
Dropout mixture contains equal quantities of all amino acids except for adenine, whose levels are increased to 2.5 times the amount of the other components. The complete dropout mixture should be used in YES media. For the experiments described here, the PMG dropout mixture excludes leucine and uracil as these will be supplemented based on selection needs, or with the amino acid selection markers on the Tf1-expressing plasmids. We find that using this dropout mixture yields far better transposition activity than the traditional five amino acid supplements typically recommended for S. pombe.
5-FOA is used to select against URA3 activity which is expressed from our Tf1 plasmids. Because 5-FOA has low solubility in water, it helps to incubate and shake media at 37 °C for 30–45 min following 5-FOA addition. However, 5-FOA should be added to media only after sterilization of liquid media, since it is unstable at high temperatures.
The purity of commercially available G418 ranges between 60 and 90 %. Therefore, to determine accurate concentrations and weighing for stock solutions, it is necessary to correct for purity. Our 500 μg/mL concentration is the corrected value.
When using 5-FOA plates containing uracil, it is necessary to reduce the amount of uracil from 250 to 50 μg/mL to ensure maximal selection efficiency against URA3. The conversion of 5-FOA to a toxic 5-fluorouridine monophosphate compound by the URA3 enzyme is only effective in media lacking excess amounts of uracil, as uracil will dilute toxicity.
Uracil is excluded from media to select for cells containing the URA3 selection marker in the Tf1 expression vector (see Fig. 1a).
Vitamin B1 (thiamine) is excluded from media to induce Tf1 expression via the nmt1 promoter. All measures to reduce even small amounts of contaminating B1 should be taken, such as using disposable plasticware.
Effective methods include mixing via a coffee grinder or in a stone roller with steel balls.
Controls include strains containing wild-type Tf1 plasmids, as well as versions of Tf1 containing a frameshift stop codon in the beginning of the IN-coding region (Tf1 INfs) and in the PR-coding region (Tf1 PRfs). Tf1 INfs mutants will not produce IN protein and will lack transposition activity. PRfs mutants produce no RT or IN proteins and, as a result, are unable to perform reverse transcription or integration resulting in a lack of cDNA production, homologous recombination activity, and transposition activity (see Fig. 1b).
Using cells from the thick part of the streak from a frozen stock is preferable to single colony selection as this will reduce issues associated with suppressor mutations from colony populations. All experimental cells should be revived from frozen stocks that have previously been single-colony purified and frozen in 15 % glycerol prior to use.
It is helpful after printing plates to sterile velvet, to print a sterile plate lid to the cell mass before printing the velvet to new media. This technique will reduce excess carryover of cell mass and creates more homogeneously phenotypic patches.
Between printings, unwanted cell mass or contamination can occur. These can easily be removed using a sterile scalpel prior to printing to new media.
This intermediate PMG + 5-FOA + U + L + B1 step selects against cells with the expression plasmid as demonstrated by the reduction of the INfs control in experiments. When using nourseothricin as a selection marker, two sequential rounds of 5-FOA selection are required, one round with 3 days of incubation time, followed by a round of 2 days of incubation. G418 is more effective under 5-FOA selection and will need to undergo only a single round of 5-FOA selection for the 3 days (as written in the protocol).
Antibiotics used for selection are either G418 (Neo) or nourseothricin (Nat). These drugs in combination with 5-FOA will allow for selection of cells that have acquired new genomic integration s and have also lost the Tf1 expression plasmid.
The optimal incubation time is determined by observing cell growth; normally this occurs at the point when there is the greatest difference between the wild-type Tf1 and INfs controls. At this point, INfs control patches will exhibit approximately 1/20th the growth/density of WT patches. PRfs patches should exhibit no growth (see Fig. 1b). If incubated for too long, background levels of homologous recombination can occur between Tf1 cDNA and genomic copies of Tf2. This causes the INfs to have similar levels of growth to the wild-type Tf1.
Antibiotic selection is either G418 (Neo) or nourseothricin (Nat). This step will select for cells where Tf1 cDNA has successfully recombined with the expression plasmid, thereby replacing the non-spliceable drug resistance gene with a version that has undergone transcription, splicing, and reverse transcription, creating an active and spliced copy of the drug selection gene on the plasmid.
Confluent growth should be evident after 24 h. A slight but notable (twofold) reduction in growth on the IN frameshift control patches should be evident when compared to WT patches. Single-amino acid mutations in IN that block integration do not exhibit this twofold reduction in homologous recombination indicating that the recombination process is stimulated twofold by IN protein. The PRfs control patches should exhibit no growth (see Fig. 1b).
This step removes residual B1 which will inhibit nmt1 induction of Tf1, resulting in significantly reduced transposition and recombination activity [9].
To ensure the most accurate serial dilutions, we find it ideal to perform dilutions in 1 mL volumes. All samples should be vortexed well (20 s) between each serial dilution. This step is critical to ensure reproducibility between replicates.
This media will allow for selection of cells that have successfully lost the URA3 containing Tf1-expression plasmid.
Micro-colonies are abundant on YES + 5-FOA + antibiotic-containing media. To ensure reproducibility in transposition and recombination frequencies, it is necessary to establish a limit based on colony size. We find it easiest to count all large-to midsize colonies that are well formed and round. Including very small colonies in the count generally does not negatively impact the analysis or decrease experimental reproducibility as long as size limits are consistent through all steps of the experiment.
- Calculate the transposition frequency with the following equation:
Plating cells to YES will determine concentration of viable cells in the culture.
- Calculate the homologous recombination frequency with the following equation:
The frequency of transposition should be roughly 20-fold reduced in INfs samples when compared to WT controls (see Fig. 3a and Table 1).
The frequency of homologous recombination should be roughly twofold reduced in INfs when compared to WT controls (see Fig. 3b and Table 2).
References
- 1.Levin HL, Moran JV (2011) Dynamic interactions between transposable elements and their hosts. Nat Rev Genet 12(9):615–627. doi: 10.1038/nrg3030, nrg3030 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck CR, Garcia-Perez JL, Badge RM, Moran JV (2011) LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12:187–215. doi: 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singleton TL, Levin HL (2002) A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eukaryot Cell 1:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowen NJ, Jordan I, Epstein J, Wood V, Levin HL (2003) Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements derived from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13(Sept):1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leem YE, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, Zhang K, Grewal SIS, Hoffman CS, Levin HL (2008) Retrotransposon Tf1 is targeted to pol II promoters by transcription activators. Mol Cell 30(1):98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Levin HL (2010) High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res 20(2):239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng G, Leem YE, Levin HL (2013) Transposon integration enhances expression of stress response genes. Nucleic Acids Res 41(2):775–789. doi: 10.1093/nar/gks1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee AG, Esnault C, Guo Y, Hung S, McQueen PG, Levin HL (2014) Serial number tagging reveals a prominent sequence preference of retrotransposon integration. Nucleic Acids Res 42(13):8449–8460. doi: 10.1093/nar/gku534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atwood A, Choi J, Levin HL (1998) The application of a homologous recombination assay revealed amino acid residues in an LTR retrotransposon that were critical for integration. J Virol 72(2):1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin HL (1995) A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol Cell Biol 15:3310–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin HL, Boeke JD (1992) Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J 11:1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boeke JD, Garfinkel DJ, Styles CA, Fink GR (1985) Ty elements transpose through an RNA intermediate. Cell 40(491):491–500 [DOI] [PubMed] [Google Scholar]
- 13.Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23(3):173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]