Abstract
PURPOSE OF REVIEW:
We present a current perspective of epigenetic alterations that can lead to cardiovascular disease (CVD) and the potential of dietary factors to counteract their actions. In addition, we discuss the challenges and opportunities of dietary treatments as epigenetic modifiers for disease prevention and therapy.
RECENT FINDINGS:
Recent epigenome-wide association studies along with candidate gene approaches and functional studies in cell culture and animal models have delineated mechanisms through which nutrients, food compounds and dietary patterns may affect the epigenome. Several risk factors for CVD, including adiposity, inflammation and oxidative stress have been associated with changes in histone acetylation, lower global DNA methylation levels and shorter telomere length. A surplus of macronutrients such as in a high fat diet or deficiencies of specific nutrients such as folate and other B-vitamins can affect the activity of DNA methyltransferases and histone modifying enzymes, affecting foetal growth, glucose/lipid metabolism, oxidative stress, inflammation and atherosclerosis. Bioactive compounds such as polyphenols (resveratrol, curcumin) or epigallocatechin may activate deacetylases SIRTs, histone deacetylases or acetyltransferases and in turn the response of inflammatory mediators. Adherence to cardioprotective dietary patterns, such as the Mediterranean diet (MedDiet), has been associated with altered methylation and expression of genes related to inflammation and immuno-competence.
SUMMARY:
The mechanisms through which nutrients and dietary patterns may alter the cardiovascular epigenome remain elusive. The research challenge is to determine which of these nutriepigenetic effects are reversible, so that novel findings translate into effective dietary interventions to prevent CVD or its progression.
Keywords: cardiovascular, chromatin, diet, epigenetics, histone acetylation, methylation, nutrition
INTRODUCTION
Cardiovascular disease (CVD) is a major global cause of morbidity and mortality [1], whose prevalence is predicted to further increase in both developing and developed countries [2]. The pathophysiology of CVD is characterised by endothelial dysfunction, vascular inflammation, atherosclerosis, fibrosis and thrombosis, with multiple pathways simultaneously perturbed [3]. The progressive remodelling and loss of flexibility that occurs during ageing of the vasculature contributes to the onset of CVD. Telomere attrition, epigenetic alterations and mitochondrial dysfunction are, among others, hallmarks of molecular damage that accumulate during ageing [4]. Many environmental factors that lead to CVD are mediated by epigenetic changes, which include posttranslational modifications to histone tails of nucleosomes (such as methylation, acetylation and phosphorylation), DNA methylation and non-coding RNA regulation of chromatin remodelling [5]. The role of diet and physical activity on CVD outcomes has been extensively studied [6][7], but evidence for the mechanisms by which dietary components might manipulate the epigenome of ageing vascular cells reversibly, perhaps, remains poor.
Mitochondria: a cellular powerhouse of epigenetic modifiers
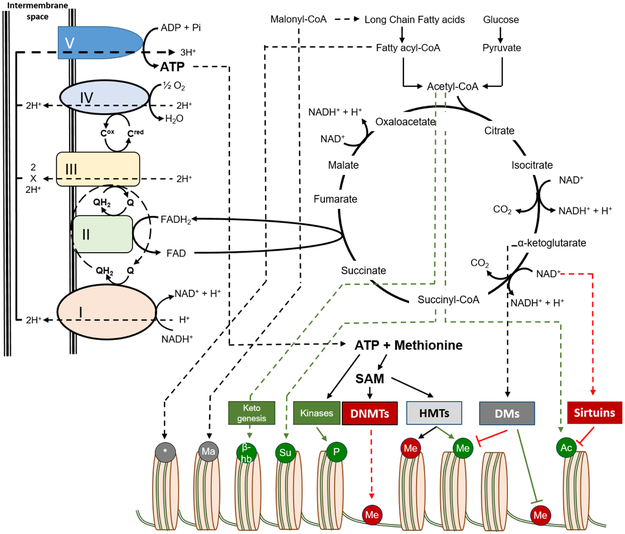
Epigenetics describes the reversible and heritable changes in genes, which alter gene functions without affecting the DNA sequence [5]. A key component of the effect of the epigenome on gene expression is the assembly of DNA and proteins into chromatin. Genomic DNA in eukaryotic cells is wrapped around histone proteins to form nucleosomes, which comprise the basic building blocks of chromatin [8]. Various post-translational modifications of the histones affect its 3D-structure and lead either to exposure of DNA, which makes it accessible to the transcriptional machinery, or have the opposite effect, which results in gene silencing. In addition to their primary role as the cellular ATP producers, mitochondria constitute the platform where most of the epigenetic modifiers or other metabolites that affect epigenetic modifications are formed [8]. Thus, changes in mitochondrial function are closely associated with availability of epigenetic modifiers (Figure 1).
Figure 1.
Sources of epigenetic modifiers: Availability of epigenetic modifiers, which are used for modification of histones or DNA, is determined by alterations in synthesis or catabolism of metabolites that are derived via glucose and fatty acid oxidation pathways, including the Krebs/TCA cycle and oxidative phosphorylation. Ac: Acetyl group, Me: Methyl group, P: Phosphoryl group, Su: Succinyl group, β-hb: β-hydroxybutyryl group, Ma: Malonyl group, *: Other acyl groups, DNMTs: DNA Methyl-transferases, HMTs: Histone Methyl-transferases, DMs: Demethylases, SAM: S-adenosylmethionine. Green color depicts modifications that activate transcription and red color indicates inhibitory effects.
Histone acetylation
Acetyl-CoA is used as an acetyl group donor by histone acetyltransferases (ΗΑΤ), which mediate acetylation of nucleosomal histones, chromatin remodeling and activation of gene transcription. Acetyl-CoA is the main donor for acetylation of chromatin and of proteins involved in nDNA transcription and replication [8]. It is formed in mitochondria from glycolysis (catabolism of pyruvate) or from β-oxidation of fatty acids. De-acetylation of chromatin by histone deacetylases (HDACs) leads to transcriptional repression. Sirtuins (SIRTs) are class III HDACs that couple lysine deacetylation to NAD+ hydrolysis and lead to formation of nicotinamide, which signals back to SIRTs by inhibiting their activity [5]. Thus, the levels of NAD+, NADH or NAM affect SIRT-mediated deacetylation of chromatin. NAD+, which is formed in mitochondria, is an obligatory cofactor for the activity of SIRT1, SIRT6 and SIRT7.
Histone and DNA methylation
S-adenosylmethionine (SAM) is the methyl group donor for both histone methyltransferases (HMTs) and DNA methyltransferases (DNMTs) generated by methionine and ATP, which is formed in mitochondria. Methylation of lysine, arginine or histidine of histones has been associated with both repression and activation of transcription. On the other hand, DNA methylation of cytosine residues in CpG dinucleotides (CpG sites) is a repressive gene regulation mechanism, regulating chromatin architecture and gene transcription [9]. S-adenosyl homocysteine (SAH), which is formed following demethylation of SAM, functions as a repressor of both DNMTs and histone lysine demethylases (KDMs).
Histone phosphorylation
ATP is critical for phosphorylation of core histones that affects gene expression, as well as cell division and DNA damage repair. Phosphorylation of histones promotes chromatin condensation, which happens during apoptosis, DNA fragmentation, and cell death [10].
Other chromatin modifications
Mass spectrometry analyses have identified additional non-acetyl histone acylations with hydrophobic (propionylation, crotonylation butyrylation), polar (2-hydroxyisobutyrylation and β-hydroxybutyrylation) or acidic (succinylation, malonylation, glutarylation) modifiers [11]. Histone propionylation, crotonylation, butyrylation, 2-hydroxyisobutyrylation, β-hydroxybutyrylation, and succinylation have been associated with increased transcription (Figure 1). On the other hand, the effect of malonylation and glutarylation on gene transcription remains elusive. Availability of non-acetyl epigenetic modifiers is regulated by synthesis of short chain fatty acids, which is mediated by Acyl-CoA synthetase short-chain family member 2 (ACSS2) that are eventually oxidized and provide the acyl modifiers. Ketogenesis, a mitochondrial process that occurs in response to low blood glucose and hepatic glycogen, accounts for the formation of β-hydroxybutyrate.
The expression levels of many of the enzymes that are involved in acetylation and methylation processes, along with the impact of genetic changes, such as single nucleotide polymorphisms (SNPs), represent components of an interactive network that orchestrates changes in chromatin structure [8]. In that context, cellular metabolic functions that generate ATP, acetyl-CoA, NADH and ROS from carbohydrates and fats drive a range of epigenetic modifications.
The epigenome and cardiovascular disease
The interrelationship between subendothelial accumulation of apo-B containing lipoproteins and inflammation is central for the pathogenesis of atherosclerosis [3]. The association of inflammatory markers (IL6, IL1β, IL8, high-sensitivity C-reactive protein, vascular cell adhesion molecule-1) and dyslipidaemias with DNA methylation in CVD has been recently reviewed [12]. Epigenetic changes affect endothelial function, vasomotor tone, cytokine signaling and fibrosis at the site of plaque formation [6][13]. Hypomethylation of genomic DNA, which is generally associated with increased gene expression, has been observed in several tissues, including vascular smooth-muscle cells, atherosclerotic lesions and peripheral blood leukocytes, of patients with atherosclerosis [14]. Muka et al. [13] looked at studies that examined DNA methylation of CpG sites in or near CVD-related genes. They reported that lower levels of methylation at long-interspersed nuclear elements (LINE-1), a surrogate for global genomic DNA methylation levels), and higher methylation of Alu repeats were inversely associated with CVD risk, independent of established cardiovascular risk factors [13]. In addition, altered methylation patterns were found in genes involved in foetal growth, glucose/lipid metabolism, oxidative stress, inflammation and atherosclerosis in blood cells related with CVD [13].
A recent epigenome-wide association study (EWAS) in Japanese individuals reported that 10 CpG sites were hypermethylated and 16 hypomethylated in genomic DNA extracted from the aortic intima of atheromatous plaque lesions compared to control plaque-free intima [15]. One of the hypermethylated CpG sites was located in the long non-coding HOX transcript antisense RNA (HOTAIR) gene, which may be activated by thymic stromal lymphopoietin (TSLP), a vascular smooth muscle cell protein with anti-inflammatory and anti-oxidant effects [16]. Some of the hypomethylated sites were in genes encoding proteins in the Wnt signaling pathway, which is known to be involved in atherosclerosis [17]. Individuals with established CVD have higher promoter methylation levels of various metabolism-related genes including those that encode metalloproteinases, gene products involved in homocysteine and one carbon-metabolism and in ATP production [13]. In population cohorts from Sweden [18] and Japan [19], EWAS have found differential DNA methylation at individual CpG sites (211 sites vs 3 sites respectively) to be independently associated with MI risk. However, these studies were carried out using DNA from peripheral blood mononuclear cells (PBMCs) due to the practical difficulties in obtaining myocardium or blood vessel wall biopsies, so their relevance to cardiac cell function remains to be established.
There are only few studies associating histone modifications with CVD risk or disease mechanisms. A review of animal and cell studies concluded that histone acetylation in endothelial cells can either prevent (c-jun) or induce (RNAP II) transcription factor binding or specificity [20]. Other chromatin-modifying enzymes may affect transcription factor recruitment. A recent in vivo study in human hearts reported that during postnatal development there is a fine cooperation of active CpG methylation and histone marks in regions enriched for CVD-associated variants to shape the cardiomyocyte transcriptome [21]. In contrast, when cardiomyocytes from failing hearts were compared to those from non-failing hearts, active histone marks of several pathological genes were affected, including connective tissue growth factor (CTGF) and natriuretic peptides A and B (NPPA, NPPB), while CpG methylation was stable [21].
Telomeres
Telomeres are nucleoprotein structures that protect the ends of linear chromosomes from DNA degradation and fusion. Vertebrate telomeres are composed of multiple copies of the sequence TTAGGG bound to the protein complex shelterin [22]. Highly heterochromatic, heavily methylated adjacent regions called subtelomeres also help to preserve telomere integrity [23].
In most somatic tissues, telomeres become shorter with every cell division; a process accelerated by oxidative stress [23]. Chronic inflammation is presumed to shorten telomeres in leukocytes, due to increased cell turnover. Thus, in addition to mitochondrial dysfunction, dysregulated nutrient sensing and epigenetic alterations, telomere length is a well-established ‘hallmark’ of cellular ageing [4]. Mean leukocyte telomere length (LTL) is an independent predictor of several age-related conditions, including coronary heart disease and other forms of CVD [24]. Furthermore, recent studies suggest that in addition to telomere length, the integrity of telomeres decreases with age and with development of hypertension in vascular tissue [25].
Diet as an epigenetic intervention point in cardiovascular disease
Most metabolic pathways affect a range of cellular functions and often act as a network, with a single gene product affecting several distant parts of a pathway. Thus, it becomes difficult to identify particular targets for epigenetic interventions. Additional challenges are the identification of direct epigenetic effects of any single dietary component and the inability of epigenetic biomarkers to distinguish between causal, consequential and coincidental relationships. With these caveats, dietary components associated with particular exposures and epigenetic changes are reviewed below, in an effort to identify potential intervention points.
One of the important factors to consider is the timing of epigenetic alterations triggered by dietary factors which often defines the magnitude of the epigenetic impact, such as in the case of nutritional deficiencies being critical during early gestation [26]. An example is folate deficiency, which when corrected preconceptually can prevent neural tube defects, but has no effect if given after the first trimester of pregnancy, as the development and closure of the neural tube is completed 28 days after conception [27]. The persistence of the epigenetic phenotype for more than a few generations was observed early on in malnourished animals [28]. The associations between epigenetic perturbations in utero, maternal dietary changes and developmental programming of CVD has been previously reviewed [29].
Human studies of dietary effects on the cardiovascular epigenome come from ‘natural’ experiments in populations of mothers and babies conceived or born during famine periods. These include investigations carried out in the Dutch Famine birth cohort [30] and studies in the Gambia, where pregnant women in rural areas experience seasonal nutritional changes due to weather conditions [31]. New data from the Dutch Famine birth cohort reveals that prenatal famine exposure during early gestation is associated with higher risk for CAD and hypertension in adulthood, and with altered DNA methylation within the INSR and CPT1A loci, which encode proteins involved in prenatal growth and fatty acid oxidation, respectively [26].
Macronutrients
Nutrient excess has been causally linked to the pathogenesis of CVD, metabolic syndrome and insulin resistance [7]. Chronic carbohydrate and lipid excess may elevate acetyl-CoA, which changes chromatin structure, increases acetylation of DNA-binding proteins, suppresses autophagy and accelerates age-associated pathologies [11]. Circulating fatty acids such as α-linolenic acid, EPA and DHA affect changes in DNA methylation sites for genes such as APOE, IL6 and ABCA1 which are correlated with CVD traits [12]. Two recent human studies reported that obese participants had an increased CpG-methylation and reduced expression of PLIN1 [32] which facilitates among others lipolysis of triglycerides (TG) and, in a genome-wide methylation study, hypermethylation of PPARG, HAND2, HOXC6, SORBS2, CD36, and CLDN1 gene promoters which may be associated with reduced adipogenesis, impaired triglyceride uptake and insulin sensitivity [33].
Obesogenic high-fat diets in experimental animals were also associated with increased DNA methylation in the Leptin and PPARG2 gene promoters [34]. In a recent clinical trial, overfeeding by high-fat diets independent of the fat composition (saturated or polyunsaturated fat) increased DNA methylation in adipose tissue, especially in promoter regions affecting adipocyte differentiation, lipid metabolism, and adipose tissue expandability pathways [35]. Interestingly, overfeeding with saturated fatty acids changed the mean methylation of a set of proinflammatory signals (eg. FTO, IL6, POMC) which were different from the ones triggered by polyunsaturated fatty acids (eg. ADIPOQ) [35]. Furthermore, Hussay et al. [36] reviewed the inconclusive evidence on the effects of ω-3 fatty acids supplementation on DNA methylation, pointing out that males and females not only metabolise and store ω-3 fatty acids in a different way, but they also differ in their global DNA methylation patterns.
In vascular and inflammatory cells, hyperglycaemia leads to chromatin changes, which in turn affect gene transcription [37]. In an attempt to integrate genetic (GWAS) and epigenetic (EWAS) data, the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study researchers challenged study participants with a high-fat meal and compared changes in plasma concentration of 35 fatty acids and 11 sterols in the fasting and postprandial state [38]. The authors proposed that integration of clinical data with genomic and epigenomic data on sterols and fatty acids may lead to identification of novel CVD biomarkers [38]. Postprandial TG responses to a high-fat dietary challenge increased methylation at sites of genes such as APOA5, SREBF1 and ABCG1 that regulate lipogenesis and lipoprotein metabolism, while it lowered methylation of CPT1A [39] which promotes hepatic fatty acid β-oxidation. These findings were supported by other cohorts (KORA and InCHIANTI), which reported in two European populations that newly identified loci associated with methylation of four CpG sites in CPT1A, APOA5, SREBF1, and ABCG1 might regulate disturbed blood lipid levels and thus contribute to the development of complex lipid-related diseases [40][41].
Fasting conditions have the opposite effect to overnutrition. In mouse hepatoma cells, depletion of glucose and/or amino acids from culture media led to increased DNA methylation and chromatin accessibility at the transcription start site of SIRT genes [42]. Similar to starvation and fasting, caloric restriction has been linked to a reduced risk of a cardiovascular disease [43], possibly due to its effect on weight and adiposity. A recent animal study that attempted to investigate the relationship between caloric restriction, lipid metabolism and vascular risk reported some transient beneficial effects on total cholesterol, HDL, and TG levels without any effect on DNA methylation patterns [44].
Folic acid, B-vitamins, other methyl donors
The epigenetic effects of “methyl-donor” nutrients on vascular ageing and cardiometabolic risk, are better understood. Dietary intake of methyl-group donors and co-factors during pregnancy has been reported to affect foetal growth and development, thus establishing a major link between early environmental exposure and chronic disease development in the offspring later in life [45][46]. Dietary methyl-groups derived from methionine, choline/betaine and methyl/folate/vitamin B12 directly influence DNA and histone methylases, as they are the precursors leading to the formation of SAM [47]. Pauwels et al. [48] reported that maternal dietary and supplemental intake of methyl-group donors, in the periconception period only, increased infant buccal DNA methylation in genes related to growth (IGF2 DMR), metabolism (RXRA), and appetite control (LEP), all affecting cardiovascular health.
Dietary intake of carotenoids and B-vitamins has been associated with longer telomeres [49][50], which are linked with lower CVD risk [51]. A recent prospective study reported longer LTL and reduced LINE-1 methylation accompanied by higher SAM in elderly subjects after one year of supplementation with B-vitamins [52]. In contrast, shorter LTL is associated with lower levels of folate, both in adults and in newborns [53]. The underlying mechanism is currently unknown, although a decrease in DNA methylation of the subtelomeres and subsequent loss of telomere integrity is one plausible explanation. The increased incorporation of uracil into the telomeric sequence itself may also account for the observed changes, since the conversion of dUMP to dTTP requires folate. This could compromise shelterin binding, affect histone methylation, or lead to repeated telomere breakages [22].
Bioactive compounds
Polyphenols including flavonoids, curcuminoids, and stilbenes contained in fruits, vegetables, and other dietary components including green tea, red wine, and cocoa compose the largest ubiquitous group of bioactive compounds with well-documented anti-inflammatory and cardioprotective actions [7]. Several studies have confirmed their beneficial effects on vascular structure and function, inflammation and multiple cardiovascular risk factors [54]. Their effects on the epigenome have been studied extensively in the cancer field, but their role in the “cardiovascular epigenome” is still largely unexplored. In a recent in vitro study, cocoa polyphenols down-regulated key genes involved in the epigenetic process (DNMTs, MTHFR and MTRR) of PBMCs [55]. Studies on resveratrol, a polyphenol found in grapes, berries, peanuts and red wine, have shown that it affects chromatin segregation [56] but it also has the capacity to activate the deacetylases SIRTs [57]. This affects high-glucose-induced cardiac oxidative stress, mitochondrial dysfunction, myocardial fibrosis and vascular ageing. A recent trial on PBMCs of type 2 diabetes patients reported that boosting SIRT-1 expression with resveratrol supplements lowered H3K56ac levels and alleviated oxidative stress [58].
Cruciferous vegetables are rich in the anti-inflammatory isothiocyanate sulforaphane, which suppresses NF-κB signalling, as well as TNF-α-induced monocyte adhesion, circulating adhesion molecules and chemokines in C57BL/6 mice [59]. Though cancer studies showed that sulforaphane downregulates histone deacetylase activity directly and affects methylation indirectly [60], its role on epigenetics of vascular disease has not been explored.
The epigenetic effects of other bioactive compounds have been explored sparsely in a few recent cell culture and animal studies. In spontaneously hypertensive rats, dietary curcumin, the principal curcuminoid of turmeric and member of the ginger family, suppressed degradation of extracellular matrix following abnormal changes in vasomotor tone [61]. The animals fed curcumin also demonstrated decreased expression levels of HDAC1, and of the inflammatory markers matrix metalloproteinase-2 (MMP-2) and transforming growth factor β (TGFβ) in their coronary arteries, as well as TIMP1 transcription activation by increasing histone H3 acetylation at its promoter [61]. Similarly, epigallocatechin-3-gallate (EGCG), found in green tea, has been recognised as a histone acetyltransferase inhibitor [55]. In endothelial cells, in vitro EGCG induced hypoacetylation of H3 and suppressed HDAC1 expression and blocked the response of inflammatory mediators [62].
Thus, a wide range of dietary factors may alter epigenetic signatures. However, whether these changes play a causative role in CVD or can be used as biomarkers for early prevention and interventions remains to be clarified.
Dietary patterns
In recent years, nutrition epidemiology studies have used dietary patterns instead of isolated nutrients as a more accurate tool to study eating habits. Among the dietary patterns, dietary patterns rich in fruits and vegetables as well as the Mediterranean diet (MedDiet) rich in olive oil, legumes, fruits and vegetables [63], have been associated with longer LTL [64] and with lower risk of CVD incidence and mortality, including CHD and MI [65]. Higher adherence to the diet protects from hypertension, inflammation and other complications related to excessive adiposity and epigenetic modifications may mediate some of these effects. Adherence to MedDiet in a subset of the PREDIMED-Navarra study was associated with methylation and thus suppression of the expression of eight genes related to inflammation and immunocompetence [66]. However, further longitudinal studies and randomised controlled trials are needed before firm conclusions can be drawn regarding the causal direction of the relationships between dietary patterns, epigenetic changes, telomere length and disease risk.
CONCLUSION
Recent advances in study design combined with novel dietary assessment methods, high throughput technologies, and big data analytics are contributing to the development of the field of nutritional epidemiology, which will be invaluable in furthering our understanding of the relationship between diet and cardiovascular health. Epigenetics and epigenomics research can help to identify novel causal biomarkers. Dietary patterns that favour cardiovascular health, such as the MedDiet, are based on the combined action of meals high in antioxidant- and anti-inflammatory- compounds and on olive oil, a rich source of monounsaturated fats. However, the MedDiet is more than just a diet. One of its significant components is a lifestyle characterised by frequent social interactions and associated positive effects on mood - factors that are well recognised to modulate disease outcomes. The extent to which pathological epigenetic changes can be prevented or manipulated by dietary factors in order to prevent human cardiovascular disease remains to be established. However, the exploration of this novel field represents a rich seam of data that can now be mined in order to answer this timely question.
KEY POINTS.
Epigenetic alterations help elucidate the mechanisms by which diet and nutrients may affect gene expression and regulation. Dysregulation of the epigenome, including changes in DNA methylation and histone acetylation, plays a major role in CVD.
A surplus of macronutrients (fatty acids and carbohydrates) or nutrient deficiencies (folate and other B-vitamins), as well as bioactive compounds such as polyphenols (resveratrol, curcumin) or epigallocatechin may affect the expression levels of methylation and acetylation enzymes and thus affect the expression of genes that are linked to vascular dysfunction.
Identification of the causal directions of the relationships between dietary patterns, epigenetic changes, telomere length and disease risk will facilitate the development of treatment and intervention measures.
ACKNOWLEDGEMENTS
All authors contributed in writing and editing the manuscript and generating the figures. They all proofread the manuscript.
FINANCIAL SUPPORT AND SPONSORSHIP: A. Z. Kalea received support by the British Heart Foundation (BHF) as a chair scholar and from NIHR UCLH BRC Cardiometabolic Programme; K. Drosatos was supported by the National Heart, Lung and Blood Institute (NIH-HL130218) and the W.W. Smith Charitable Trust.
COMMON ABBREVIATIONS:
- AMPK
Adenosine monophosphate-activated protein kinase
- ATP
Adenosine Triphosphate
- CAD
coronary artery disease
- CHD
coronary heart disease
- CpG
Cytosine-phosphate-guanine
- CVD
cardiovascular disease
- DNMT
DNA methyltransferase
- EGCG
epigallocatechin-3-gallate
- EWAS
epigenome-wide association study
- GWAS
genome-wide association study
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HMT
histone methyltransferase
- KDM
lysine demethylases
- LTL
leukocyte telomere length
- MedDiet
Mediterranean diet
- MI
myocardial infarction
- MMP
matrix metalloproteinase
- mRNA
Messenger ribonucleic acid
- miRNA
Micro ribonucleic acid
- mTOR
mammalian target of rapamycin
- NAD
Nicotinamide adenine dinucleotide
- NADH
Nicotinamide adenine dinucleotide-reduced form
- NAM
Nicotinamide
- NF-κB
nuclear factor κB
- nDNA
nuclear DNA
- PBMCs
peripheral blood mononuclear cells
- PUFA
polyunsaturated fatty acid
- ROS
reactive oxygen species
- SAH
S-adenosyl homocysteine
- SAM
S-adenosylmethionine
- SFA
saturated fatty acid
- SIRT
Sirtuin
- SNP
single nucleotide polymorphisms
- TIMP
tissue inhibitor of metalloproteinases
- TG
triglycerides
- TGFβ
transforming growth factor β
- TNF-α
tumor necrosis factor alpha
- TF
Transcription factor
- UTR
untranslated region
Footnotes
CONFLICTS OF INTEREST: The authors declare no competing financial interests in relation to the work described.
REFERENCES
- 1.World Health Organization. WHO Cardiovascular Disease Facts Sheet [Internet]. Available from: http://www.who.int/mediacentre/factsheets/fs317/en/
- 2.Vasan RS, Benjamin EJ. The Future of Cardiovascular Epidemiology: Table. Circulation. 2016;133(25):2626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabas I 2016 Russell Ross Memorial Lecture in Vascular Biology: Molecular-Cellular Mechanisms in the Progression of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol, 2017; 37(2): 183–189.• Great paper on the latest research on the natural progression of atherosclerosis and its link with inflammation
- 4.López-Otín C, Blasco MA, Partridge L et al. The Hallmarks of Aging. Cell 2013; 153(6): 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corella D, Ordovas JM. Basic Concepts in Molecular Biology Related to Genetics and Epigenetics. Rev Espanola Cardiol Engl Ed 2017;70(9):744–53.• A general perspective on the basic concepts of molecular biology related to epigenetics
- 6.Wallace RG, Twomey LC, Custaud M-A et al. The role of epigenetics in cardiovascular health and ageing: A focus on physical activity and nutrition. Mech Ageing Dev [Internet]. 2017. November; Available from: http://linkinghub.elsevier.com/retrieve/pii/S0047637417302336• A review on lifestyle factors linked to epigenetic mechanisms pre-disposing individuals to cardiovascular disease
- 7.Bowen KJ, Sullivan VK, Kris-Etherton PM, Petersen KS. Nutrition and Cardiovascular Disease—an Update. Curr Atheroscler Rep [Internet]. 2018; 20(2):8 Available from: http://link.springer.com/10.1007/s11883-018-0704-3• Updated review on the links between nutritional factors and CVD
- 8.Aon MA, Cortassa S, Juhaszova M, Sollott SJ. Mitochondrial health, the epigenome and healthspan. Clin Sci Lond Engl 1979. 2016;130(15):1285–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics & Chromatin. 2017;10(1). Available from: http://epigeneticsandchromatin.biomedcentral.com/articles/10.1186/s13072-017-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15(11):703–8. [DOI] [PubMed] [Google Scholar]
- 11.Costello KR, Schones DE. Chromatin modifications in metabolic disease: Potential mediators of long-term disease risk. Wiley Interdiscip Rev Syst Biol Med. 2018; 25;e1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y and Ordovas JM. The integration of epigenetics and genetics in nutrition research for CVD risk factors. Proc Nutr Soc 2017; 76(3): 333–346.• A review on integration of genomic and epigenomic data in nutrition research as related to CVD
- 13.Muka T, Koromani F, Portilla E et al. The role of epigenetic modifications in cardiovascular disease: A systematic review. Int J Cardiol. 2016;212:174–83.• A review on epigenetic mechanisms associated with CVD
- 14.Findeisen HM, Kahles FK, Bruemmer D. Epigenetic regulation of vascular smooth muscle cell function in atherosclerosis. Curr Atheroscler Rep. 2013;15(5):319. [PubMed] [Google Scholar]
- 15.Yamada Y, Horibe H, Oguri M et al. Identification of novel hyper- or hypomethylated CpG sites and genes associated with atherosclerotic plaque using an epigenome-wide association study. Int J Mol Med 2018;41(5):2724–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y, Meng K, Jiang L et al. Thymic stromal lymphopoietin-induced HOTAIR activation promotes endothelial cell proliferation and migration in atherosclerosis. Biosci Rep. 2017;37(4):BSR20170351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albanese I, Khan K, Barratt B, Al‐Kindi H, Schwertani A. Atherosclerotic Calcification: Wnt Is the Hint. J Am Heart Assoc. 2018;7(4):e007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rask-Andersen M, Martinsson D, Ahsan M et al. Epigenome-wide association study reveals differential DNA methylation in individuals with a history of myocardial infarction. Hum Mol Genet. 2016;ddw302. [DOI] [PubMed] [Google Scholar]
- 19.Nakatochi M, Ichihara S, Yamamoto K et al. Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin Epigenetics [Internet]. 2017;9(1). 2017 May15;9:54.• Epigenome-wide association study on CVD
- 20.Bauer AJ, Martin KA. Coordinating Regulation of Gene Expression in Cardiovascular Disease: Interactions between Chromatin Modifiers and Transcription Factors. Front Cardiovasc Med 2017. April 6;4:19. doi: 10.3389/fcvm.2017.00019. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilsbach R, Schwaderer M, Preissl S et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat Commun 2018. January 26;9(1):391. doi: 10.1038/s41467-017-02762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pisano S, Gilson E, Giraud-Panis M-J. Dynamics under the Telomeric Bridge. Mol Cell. 2017;68(4):643–4. [DOI] [PubMed] [Google Scholar]
- 23.Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Wnuk M. Downregulation of methyltransferase Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J Cell Physiol. 2017;232(12):3714–26 [DOI] [PubMed] [Google Scholar]
- 24.D’Mello MJJ, Ross SA, Briel M et al. Association between shortened leukocyte telomere length and cardiometabolic outcomes: systematic review and meta-analysis. Circ Cardiovasc Genet. 2015;8(1):82–90. [DOI] [PubMed] [Google Scholar]
- 25.Morgan RG, Donato AJ, Walker AE. Telomere uncapping and vascular aging. Am J Physiol Heart Circ Physiol. 2018. March 16. doi: 10.1152/ajpheart.00008.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobi EW, Goeman JJ, Monajemi R et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014;5:5592.• New analysis from the Dutch Hunger Winter cohort
- 27.Haider BA, Bhutta ZA. Multiple-micronutrient supplementation for women during pregnancy. Cochrane Pregnancy and Childbirth Group, editor. Cochrane Database Syst Rev [Internet]. 2017. April 13;4:CD004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart RJ, Sheppard H, Preece R, Waterlow JC, The effect of rehabilitation at different stages of development of rats marginally malnourished for ten to twelve generations. Br J Nutr 1980; 43(3): 403–412. [DOI] [PubMed] [Google Scholar]
- 29.Sun C, Burgner DP, Ponsonby A-L et al. Effects of early-life environment and epigenetics on cardiovascular disease risk in children: highlighting the role of twin studies. Pediatr Res. 2013;73(4-2):523–30. [DOI] [PubMed] [Google Scholar]
- 30.Painter RC, de Rooij SR, Bossuyt PM et al. Early onset of coronary artery disease after prenatal exposure to the Dutch famine. Am J Clin Nutr. 2006;84(2):322–7. [DOI] [PubMed] [Google Scholar]
- 31.Moore SE. Early life nutritional programming of health and disease in The Gambia. J Dev Orig Health Dis. 2016;7(2):123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bialesova L, Kulyté A, Petrus P et al. Epigenetic Regulation of PLIN 1 in Obese Women and its Relation to Lipolysis. Sci Rep 2017; 7(1):10152. doi: 10.1038/s41598-017-09232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller M, Hopp L, Liu X et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol Metab. 2017;6(1):86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwamborn RAJ, Slieker RC, Mulder PCA et al. Prolonged high-fat diet induces gradual and fat depot-specific DNA methylation changes in adult mice. Sci Rep. 2017;7:43261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perfilyev A, Dahlman I, Gillberg L et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105(4):991–1000.• RCT on the epigenetic effects of different types of dietary fatty acids
- 36.Hussey B, Lindley MR, Mastana SS. Omega 3 fatty acids, inflammation and DNA methylation: an overview. Clin Lipidol. 2017;12(1):24–32.• Updated review discussing the limitations of nutriepigenetic studies with omega-3
- 37.Block T, El-Osta A. Epigenetic programming, early life nutrition and the risk of metabolic disease. Atherosclerosis. 2017;266:31–40. [DOI] [PubMed] [Google Scholar]
- 38.Irvin MR, Zhi D, Aslibekyan S et al. Genomics of Post-Prandial Lipidomic Phenotypes in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. PLoS ONE. 2014;9(6):e99509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai C-Q, Wojczynski MK, Parnell LD et al. Epigenome-wide association study of triglyceride postprandial responses to a high-fat dietary challenge. J Lipid Res. 2016;57(12):2200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irvin MR, Zhi D, Joehanes R et al. Epigenome-Wide Association Study of Fasting Blood Lipids in the Genetics of Lipid-Lowering Drugs and Diet Network Study. Circulation. 2014;130(7):565–72.• Intervention study integrating genome-wide and epigenome-wide data
- 41.Pfeiffer L, Wahl S, Pilling LC et al. DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet. 2015;8(2):334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zullo A, Simone E, Grimaldi M et al. Effect of nutrient deprivation on the expression and the epigenetic signature of sirtuin genes. Nutr Metab Cardiovasc Dis. 2018;28(4):418–24. [DOI] [PubMed] [Google Scholar]
- 43.Weiss EP, Albert SG, Reeds DN et al. Effects of matched weight loss from calorie restriction, exercise, or both on cardiovascular disease risk factors: a randomized intervention trial. Am J Clin Nutr. 2016;104(3):576–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowacka-Woszuk J, Madeja ZE, Chmurzynska A. Prenatal caloric restriction alters lipid metabolism but not hepatic Fasn gene expression and methylation profiles in rats. BMC Genet [Internet]. 2017. December;18(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franke K, Gaser C, de Rooij SR, Schwab M, Roseboom TJ. Premature brain aging in humans exposed to maternal nutrient restriction during early gestation. NeuroImage 2017;173:460–471 [DOI] [PubMed] [Google Scholar]
- 46.Van Dijk SJ, Zhou J, Peters TJ et al. Effect of prenatal DHA supplementation on the infant epigenome: results from a randomized controlled trial. Clin Epigenetics. 2016;8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeisel S, Choline Other Methyl-Donors and Epigenetics. Nutrients. 2017;9(12):445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauwels S, Ghosh M, Duca RC et al. Maternal intake of methyl-group donors affects DNA methylation of metabolic genes in infants. Clin Epigenetics 2017. February 7;9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Min K-B, Min J-Y. Association between leukocyte telomere length and serum carotenoid in US adults. Eur J Nutr. 2017;56(3):1045–52. [DOI] [PubMed] [Google Scholar]
- 50.Mazidi M, Kengne A-P, Banach M. Mineral and vitamin consumption and telomere length among adults in the United States. Pol Arch Intern Med. 2017;127(2):87–90. [DOI] [PubMed] [Google Scholar]
- 51.Baragetti A, Palmen J, Garlaschelli K et al. Telomere shortening over 6 years is associated with increased subclinical carotid vascular damage and worse cardiovascular prognosis in the general population. J Intern Med 2015; 277(4): 478–487. [DOI] [PubMed] [Google Scholar]
- 52.Pusceddu I, Herrmann M, Kirsch SH et al. Prospective study of telomere length and LINE-1 methylation in peripheral blood cells: the role of B vitamins supplementation. Eur J Nutr. 2016;55(5):1863–73. [DOI] [PubMed] [Google Scholar]
- 53.Lee J-Y, Shin C, Baik I. Longitudinal associations between micronutrient consumption and leukocyte telomere length. J Hum Nutr Diet. 2017;30(2):236–43. [DOI] [PubMed] [Google Scholar]
- 54.Santhakumar AB, Battino M, Alvarez-Suarez JM. Dietary polyphenols: Structures, bioavailability and protective effects against atherosclerosis. Food Chem Toxicol 2018;113:49–65. [DOI] [PubMed] [Google Scholar]
- 55.Vahid F, Zand H, Nosrat-Mirshekarlou E, Najafi R, Hekmatdoost A. The role dietary of bioactive compounds on the regulation of histone acetylases and deacetylases: a review. Gene. 2015;562(1):8–15. [DOI] [PubMed] [Google Scholar]
- 56.Lee JH, Wendorff TJ, Berger JM. Resveratrol: A novel type of topoisomerase II inhibitor. J Biol Chem. 2017;292(51):21011–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang W-J, Wang C-J, He Y et al. Resveratrol alleviates diabetic cardiomyopathy in rats by improving mitochondrial function through PGC-1α deacetylation. Acta Pharmacol Sin. 2018;39(1):59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bo S, Togliatto G, Gambino R et al. Impact of sirtuin-1 expression on H3K56 acetylation and oxidative stress: a double-blind randomized controlled trial with resveratrol supplementation. Acta Diabetol. 2018;55(4):331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nallasamy P, Si H, Babu PVA, et al. Sulforaphane reduces vascular inflammation in mice and prevents TNF-α-induced monocyte adhesion to primary endothelial cells through interfering with the NF-κB pathway. J Nutr Biochem. 2014;25(8):824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaufman-Szymczyk A, Majewski G, Lubecka-Pietruszewska K, Fabianowska-Majewska K. The Role of Sulforaphane in Epigenetic Mechanisms, Including Interdependence between Histone Modification and DNA Methylation. Int J Mol Sci. 2015;16(12):29732–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu J, Shen T, Xie J, et al. Curcumin modulates covalent histone modification and TIMP1 gene activation to protect against vascular injury in a hypertension rat model. Exp Ther Med. 2017;14(6):5896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu D, Perkins JT, Hennig B. EGCG prevents PCB-126-induced endothelial cell inflammation via epigenetic modifications of NF-κB target genes in human endothelial cells. J Nutr Biochem. 2016;28:164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grosso G, Marventano S, Yang J et al. A comprehensive meta-analysis on evidence of Mediterranean diet and cardiovascular disease: Are individual components equal? Crit Rev Food Sci Nutr. 2017;57(15):3218–32. [DOI] [PubMed] [Google Scholar]
- 64.Rafie N, Golpour Hamedani S et al. Dietary patterns, food groups and telomere length: a systematic review of current studies. Eur J Clin Nutr. 2017;71(2):151–8.• Updated systematic review on diet and telomere length
- 65.Rosato V, Temple NJ, La Vecchia C et al. Mediterranean diet and cardiovascular disease: a systematic review and meta-analysis of observational studies. Eur J Nutr. 2017. November 25. doi: 10.1007/s00394-017-1582-0. [DOI] [PubMed] [Google Scholar]
- 66.Arpón A, Riezu-Boj JI, Milagro FI et al. Adherence to Mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J Physiol Biochem. 2016;73(3):445–55.• Nutriepigenetic data from the PREDIMED trial