ABSTRACT
Lineage-restricted transcription factors, such as the intestine-specifying factor CDX2, often have dual requirements across developmental time. Embryonic loss of CDX2 triggers homeotic transformation of intestinal fate, whereas adult-onset loss compromises crucial physiological functions but preserves intestinal identity. It is unclear how such diverse requirements are executed across the developmental continuum. Using primary and engineered human tissues, mouse genetics, and a multi-omics approach, we demonstrate that divergent CDX2 loss-of-function phenotypes in embryonic versus adult intestines correspond to divergent CDX2 chromatin-binding profiles in embryonic versus adult stages. CDX2 binds and activates distinct target genes in developing versus adult mouse and human intestinal cells. We find that temporal shifts in chromatin accessibility correspond to these context-specific CDX2 activities. Thus, CDX2 is not sufficient to activate a mature intestinal program; rather, CDX2 responds to its environment, targeting stage-specific genes to contribute to either intestinal patterning or mature intestinal function. This study provides insights into the mechanisms through which lineage-specific regulatory factors achieve divergent functions over developmental time.
KEY WORDS: Lineage-specifying, Transcription factor, Development, Patterning, Intestine, Chromatin
Summary: Temporal shifts in chromatin accessibility lead to CDX2 activating distinct target genes in developing versus adult mouse and human intestinal cells, explaining the differential phenotypes of CDX2 loss over developmental time.
INTRODUCTION
Lineage-specifying transcription factors are required for a cellular lineage to develop in embryos, and their loss can lead to a selective failure of the respective tissue to develop in mutant embryos. Examples include MYOD1, which establishes muscle development and induces muscle trans-differentiation in fibroblasts (Davis et al., 1987; Tapscott et al., 1988), and PDX1, which is essential for pancreas development (Jonsson et al., 1994; Offield et al., 1996). Study of such transcription factors is informative during early development, when tissues are specified; however, the same factors often have additional roles in subsequent tissue maturation, organogenesis and adult homeostasis. For example, Foxa2 plays a crucial role in liver specification in the embryo, but also regulates bile acid transport in the adult liver (Bochkis et al., 2008; Lee et al., 2005). ChIP-seq of FOXA2 in embryonic and adult hepatocytes revealed distinct sets of target genes in these distinct contexts. Adult enhancer chromatin structure in the liver, as assayed by H3K4me1 ChIP-seq, indicated that FOXA2-bound regions specific to the adult took on an active chromatin structure, whereas sites bound by FOXA2 only in the embryo were occupied by a nucleosome in the adult hepatocytes (Alder et al., 2014). Chromatin-binding activity of the intestine-specifying factor CDX2 has not been explored in distinct developmental contexts.
The homeodomain transcription factor CDX2 is expressed from the onset of intestinal development through adult life. In mice, early deletion of Cdx2 in the visceral endoderm precludes formation of the intestinal epithelium, which undergoes an esophagus-like homeotic conversion (Gao et al., 2009). Squamous epithelial conversion also occurs upon spontaneous loss of CDX2 heterozygosity in mouse development, leading to harmatomas in the colon (Chawengsaksophak et al., 1997; Tamai et al., 1999). Moreover, expression of certain gastric markers is observed in the intestinal epithelium upon early embryonic inactivation of Cdx2 by a Villin-Cre driver (Grainger et al., 2010). In contrast, ablation of Cdx2 in the intestinal epithelium late in fetal or adult life compromises vital digestive function without inducing histological features of the rostral gut (Verzi et al., 2010, 2011). Conversely, rostral gut genes are ectopically expressed when adult intestinal stem cells lacking CDX2 are cultured in the presence of factors conducive to gastric differentiation (Simmini et al., 2014) or upon prolonged loss of CDX2 in a subset of adult intestinal tissues (Hryniuk et al., 2012; Stringer et al., 2012).
The distinct consequences of CDX2 loss at different developmental stages prompt questions about the mechanistic basis of lineage-specifying transcription factor activities: do these factors bind their chromatin targets from the outset or do their transcriptional targets differ along the developmental continuum? How do they interact with chromatin across developmental time? Is expression of a lineage-specific transcription factor sufficient to activate appropriate target genes, even in the absence of extracellular signals? Additionally, although many insights have been garnered from studies in mice, CDX2 function in human tissue specification remains untested.
Here, we employ mouse models and human pluripotent stem cell-derived models, coupled with investigation of chromatin accessibility, to show that CDX2 binds distinct chromatin sites in embryonic and adult intestines; this distinction is conserved in mice and humans. We find that CDX2 is incapable of instructing major shifts in the chromatin to direct its own binding, and is thus not functioning as a pioneer factor in the developing gut. Rather, dynamic chromatin-binding properties of CDX2 correlate with the dynamic transitions in chromatin accessibility that occur in the course of intestine development. Importantly, CDX2 is required to sustain newly accessible chromatin regions in mature tissues. Our identification of distinct CDX2-binding sites and regulatory functions during tissue specification versus in adults may explain why tissue plasticity is only observed upon CDX2 inactivation during a distinct window during embryonic development. These findings support a model in which lineage-specifying transcription factors operate in context-dependent roles, shaped by a dynamic developmental chromatin landscape.
RESULTS
CDX2 is required for human intestinal development
In mouse embryos with conditional deletion of Cdx2 in the developing endoderm, the intestinal epithelium exhibits morphological and molecular characteristics of foregut identity. Depending on the timing of CDX2 loss using different Cre drivers, esophageal, gastric corpus or gastric antral identities are observed, with earlier onset of Cdx2 inactivation producing more anterior (rostral) identity shifts (Gao et al., 2009; Grainger et al., 2010). It is unknown whether CDX2 has a conserved role in human intestine development, and new methods to direct differentiation of human pluripotent stem cells (hPSCs) allow us to ask this question (D'Amour et al., 2005; Finkbeiner et al., 2015; McCracken et al., 2011; Spence et al., 2011).
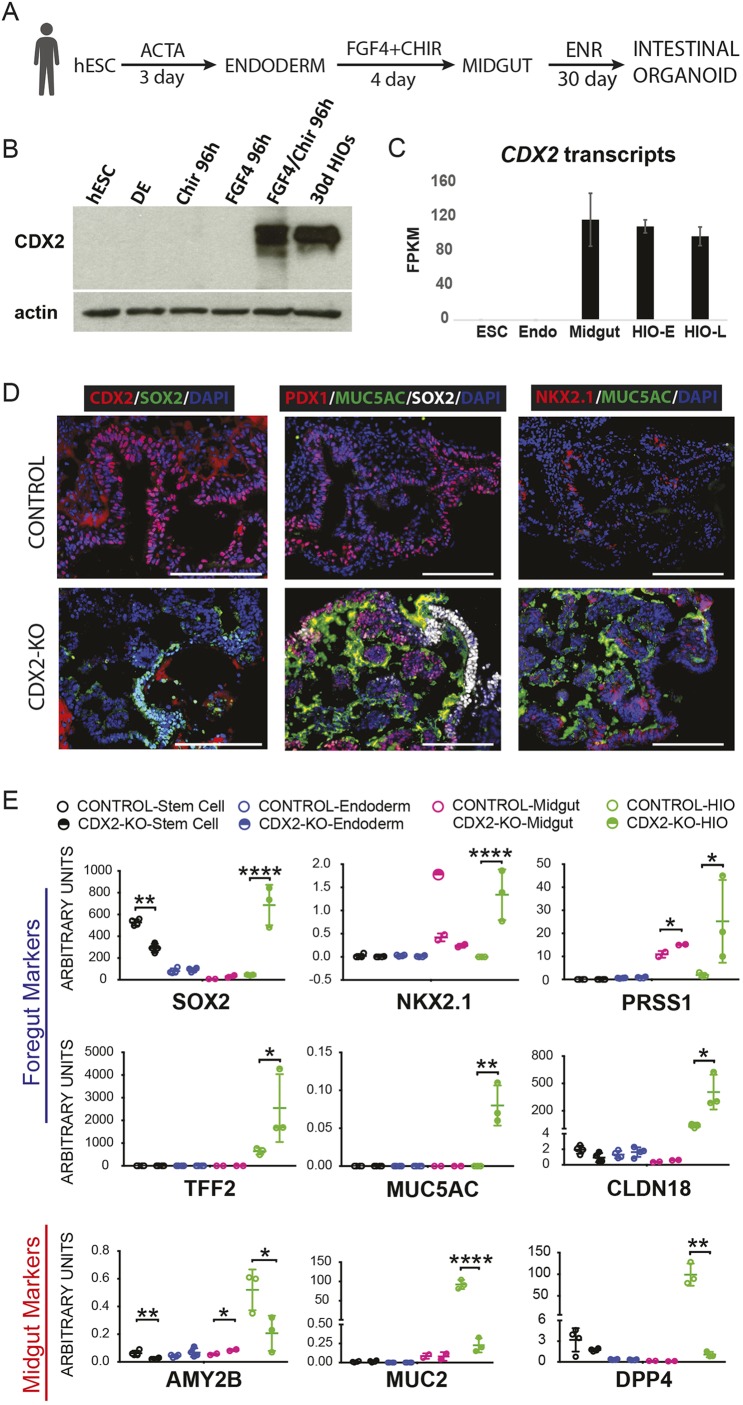
CDX2 expression is first observed upon specification of the midgut by WNT and FGF signaling (Wells and Melton, 2000). Immunoblotting confirmed absence of CDX2 in human embryonic stem cells (hESCs) or definitive endoderm, but combined activation of WNT (using the GSK3β inhibitor CHIR99021 to stabilize β-catenin) and FGF (using recombinant FGF4) signaling triggered robust CDX2 expression, which was sustained in three-dimensional human intestinal organoid (HIO) cultures months later (Fig. 1A-C). To determine if CDX2 is required to induce the intestinal lineage, we used CRISPR/Cas9 to engineer CDX2-null hESCs in the H9 line and differentiated the cells into HIOs. Sequencing confirmed mutation of CDX2, and CDX2 immunoreactivity was lost in mutant cells directed toward intestine differentiation (Fig. 1D, Fig. S1). Whereas control hESCs gave rise to tissues that expressed CDX2 (Fig. 1D) and the intestine-restricted markers AMY2B, MUC2 and DPP4 (Fig. 1E), CDX2-null hESCs failed to develop tissues with these characteristic markers. Instead, they gave rise to organoids expressing transcripts and proteins of CDX2-negative foregut lineages, including stomach, esophagus, pancreas and gall bladder (Fig. 1D,E). These findings are consistent with mouse models in which CDX2 loss in the developing epithelium leads to the development of a tissue with foregut properties (Gao et al., 2009). In contrast to the mouse genetic models, the HIO system contains both epithelial and mesenchymal components, and CDX2 expression is lost in both tissues in this model. Still, the resulting foregut features developing in CDX2-negative human organoids is consistent with a conserved role for CDX2 in initiating human intestine development, and underscores the importance of pursuing the mechanisms through which CDX2 imparts intestinal fate.
Fig. 1.
CDX2 is required for human intestinal development. (A) Differentiation schema used to differentiate hESCs into human intestinal organoids (HIOs). ACTA, activin A treatment, 100 ng/ml; FGF4+CHIR, 500 ng/ml FGF4 + 2 µM CHIR99021; ENR, 100 ng/ml Egf, 5% Noggin-conditioned media and 5% R-SPONDIN2-conditioned media. (B,C) Immunoblot (B) and RNA-seq (E-MTAB-4168; Tsai et al., 2017) (C) show that CDX2 expression is only induced robustly after differentiation of ESCs into endoderm with activin and then induction of midgut specification via treatment with FGF4 (500 ng/ml) and the WNT agonist CHIR99021 (2 µM; Chir). HIO-E, HIO early, pre-transplant; HIO-L, HIO late, after transplant into mouse kidney capsule for further intestinal maturation. DE, definitive endoderm. (D,E) HIOs result from treating ESCs with this differentiation protocol, but ESCs lacking CDX2 as a result of CRISPR targeting (CDX2-KO, Fig. S1) do not give rise to HIOs, and instead give rise to organoids expressing genes characteristic of foregut lineages, as seen by immunofluorescence staining (D, representative of at least three independent experiments) and qRT-PCR (E) on organoids collected after 30 days in ENR (EGF, Noggin, RSPO) culture conditions, with each independent biological sample comprising three to five organoids pooled together. *P<0.05, **P<0.01, ****P<0.0001 (unpaired t-test). Scale bars: 200 µm.
CDX2 regulates distinct transcriptional targets across human intestinal development
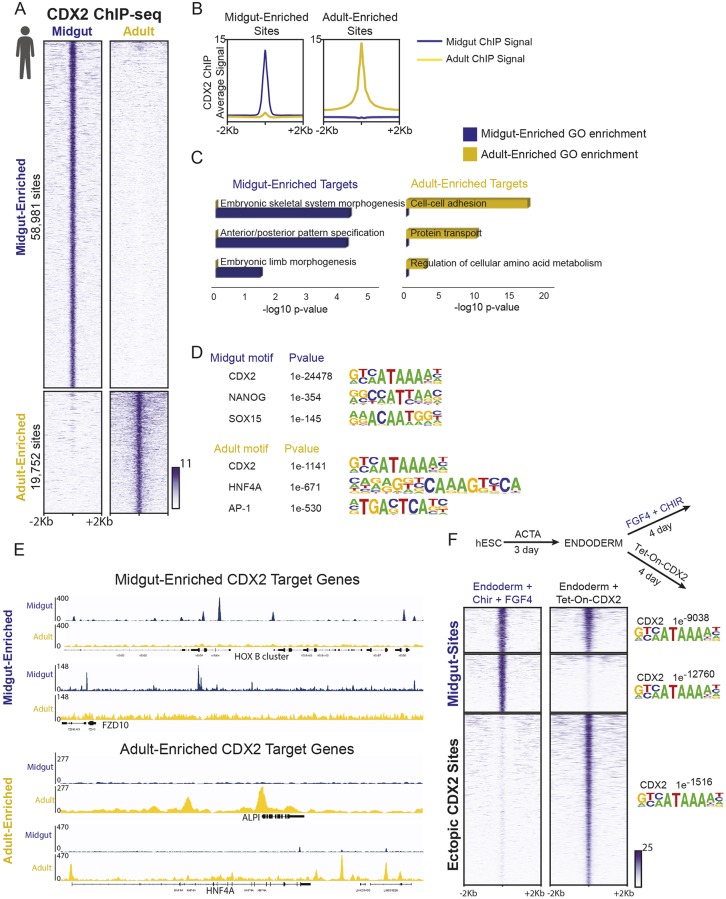
To explore these mechanisms, we performed ChIP-seq on biological replicates (Fig. S2A) of intestine-specified endoderm (herein referred to as ‘midgut’), using the differentiation protocol outlined above (Fig. 1A), which has been demonstrated to generate intestine with duodenal properties (Tsai et al., 2017). We compared CDX2 ChIP-seq in these midgut tissues with ChIP-seq on primary adult human duodenal epithelium (herein referred to as ‘adult’). We used Model-based Analysis of ChIP-Seq (MACS; Zhang et al., 2008) to identify ChIP-seq binding sites and k-means clustering to define genomic regions with differential CDX2 occupancy (Fig. 2A,B). Ontology analysis of genes linked to CDX2-binding sites revealed that midgut-enriched CDX2 binding occurred near genes that were enriched for functions in development, patterning and morphogenesis, whereas genes near adult-enriched CDX2-binding sites were associated with adult intestinal physiological functions (Fig. 2C). DNA sequence motifs enriched at midgut- and adult-enriched regions were also distinct, suggesting that CDX2 works in different regulatory complexes in embryos and adults. The CDX2 motif was strongly enriched in both groups of binding regions, but whereas midgut-enriched sites were enriched in NANOG and SOX15 motifs, consistent with a role for SOX factors supporting endoderm development (Kanai-Azuma et al., 2002), adult-enriched regions were most enriched in HNF4A motifs (Fig. 2D, Table S1), consistent with the function of HNF4 in the mature intestine (Babeu et al., 2009; Cattin et al., 2009; San Roman et al., 2015; Stegmann et al., 2006). Examples of midgut-enriched target genes included the HOXB cluster genes (Fig. 2E, Fig. S2B) known for their roles in tissue patterning and axial elongation (Young et al., 2009), whereas adult-enriched target genes included the enterocyte differentiation marker ALPI and the differentiation-promoting transcription factor gene HNF4A (Fig. 2E, Fig. S2C). These results reveal that human CDX2 binds distinct genomic regions in a stage-specific manner and regulates stage-specific genes.
Fig. 2.
CDX2 has distinct transcriptional targets in the developing midgut versus the adult intestine. Human icon indicates data from human tissues. (A,B) CDX2 ChIP-seq performed in in vitro differentiated midgut (4 day FGF4+CHIR-treated endoderm, derived from human ESCs, as described in Fig. 1A) and in primary adult human duodenum epithelium (collected from duodenal tissue adjacent to the bile duct from patients undergoing Whipple procedures) show distinct binding profiles, as defined by k-means clustering of the identified genomic regions interacting with CDX2 (A), and compared using average signal traces at all indicated sites (B). Data from two merged biological replicates each, see Fig. S2A. (C) Distinct binding sites correspond to different sets of nearby target genes, as indicated by gene ontology (GO) analysis using DAVID (genes with their transcriptional start sites within 5 kb of CDX2-binding sites were used for analysis). (D) DNA-motif enrichment analysis finds that CDX2-binding motifs are present in both categories of binding regions, but other transcription factor-binding motifs are differentially enriched in the midgut-enriched (58,981 sites) or adult-enriched (19,752 sites) CDX2-binding regions, as defined using HOMER. (E) Examples of stage-specific CDX2-binding sites are shown using IGV to visualize merged replicates of the ChIP-seq experiments. (F) Ectopic expression of CDX2 in endoderm, using a doxycycline-inducible hESC line (Endoderm+4 days doxycycline treatment), is not sufficient to recapitulate CDX2 binding to the majority of its normal midgut target sites (Endoderm+4 days CHIR+FGF). Approximately 52% of CDX2 midgut sites (MACS P<1e−10) were not detected by MACS in the doxycycline-inducible condition (second cluster of sites in heatmap), even at a lower peak-calling stringency (P<1e−3), and 48% of the midgut regions were bound by expression of CDX2 in endoderm (first cluster of sites). Lack of binding cannot be attributed to differences in CDX2-binding motifs, which were robustly detected in each category of binding sites (HOMER). Also see Fig. S2 and Table S2.
Both WNT and FGF signals are required to drive robust differentiation of human endoderm into midgut (Spence et al., 2011) and activation of these signals induces CDX2 expression (Fig. 1A). It is unclear whether CDX2 expression is sufficient for its binding at midgut regulatory regions or whether CDX2 also requires other sequelae of dual signaling pathway activation to bind to its midgut-enriched binding regions. To address this question, we engineered hESCs for doxycycline-inducible CDX2 expression. ChIP-seq after forced CDX2 expression in our ESC-derived endoderm cells yielded more than twice as many binding sites as were present in CHIR- and FGF4-treated endoderm (135,194 sites compared with 58,981 sites, Fig. 2F), suggesting that many of these sites are off-target from the endogenous intestinal program driven by WNT and FGF. Both groups of regions were enriched in the CDX2 consensus motif (Fig. 2F, Table S2), consistent with bona fide CDX2 binding to these regions. Surprisingly, despite robust expression (Fig. S2E) and occupancy of significantly more genomic sites, ectopic CDX2 failed to bind more than half of the midgut-enriched targets occupied in the presence of CHIR and FGF4 (Fig. 2F), even though these regions exhibited a strong presence of consensus CDX2-binding sequences (HOMER, P=1e−12760; Fig. 2F, Table S2). Thus, although CDX2 expression is necessary for specification of human intestine (Fig. 1C,D), CDX2 cannot reach its full complement of midgut chromatin targets in the absence of WNT- and FGF-induced alterations to regulatory mechanisms governing CDX2 binding.
Dynamic CDX2 activity across intestinal development is conserved in mice
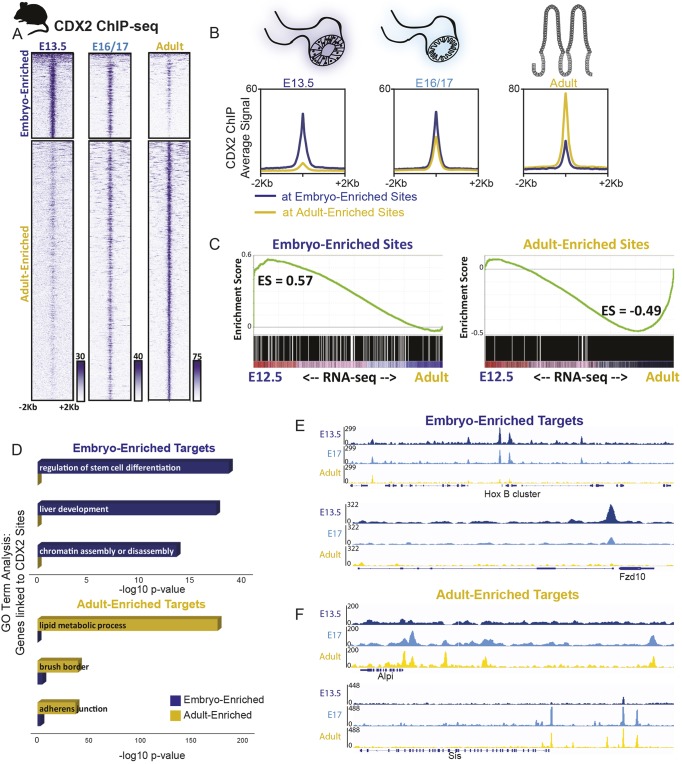
To refine and corroborate this model of CDX2 binding and activity in vivo, we turned to mice. First, we identified CDX2-binding sites in intestinal cells isolated at embryonic days (E) 13.5, 16.5 and 17.5, and from adult intestines. k-means clustering identified four major groups: 4123 genomic regions were bound more robustly in embryos, 12,551 sites were more specific to the adult (Fig. 3A,B), and the remaining sites were bound similarly at all stages (1791 sites with robust CDX2 binding and 15,368 sites with modest signals; clusters 1 and 4 in Fig. S3A and Table S3). CDX2 binding showed a notable transition late in gestation: binding at embryo-enriched sites declined by E16.5, as occupancy of adult-enriched regions became evident (Fig. 3A). To test stage-specific binding functions in gene regulation, we purified control and CDX2-depleted epithelium after embryonic knockout (Shh-Cre; Cdx2f/f, collected at E12.5) or adult knockout (Villin-CreERT2; Cdx2f/f) and used RNA-seq to identify CDX2-dependent transcripts. Consistent with CDX2's activity as a direct transcriptional activator, the genes nearby CDX2-binding sites showed reduced expression upon CDX2 loss at the corresponding developmental stages (Fig. S3B). Developmental stage-specific CDX2-binding regions also correlated with stage-specific gene expression, as genes linked to adult-enriched binding sites were expressed more robustly in adult than in the developing intestine, and the converse was true for embryo-enriched binding targets (Fig. 3C). Thus, stage-specific CDX2 binding predicts nearby gene expression in each context.
Fig. 3.
Temporal-specific shifts in CDX2 binding is conserved between mice and humans, and corresponds to temporal shifts in intestinal gene expression. Mouse icon indicates experiments performed in mouse tissues. (A) CDX2 ChIP-seq performed at the indicated stages of intestinal development on isolated epithelium show distinct, stage-dependent binding profiles (two biological replicates each, see Fig. S3; for E13.5, 12 embryos were pooled per replicate; for E16.5, three embryos pooled per replicate; for E17.5, two embryos pooled per replicate). The heatmap depicts CDX2 ChIP-seq intensity at each of the regions defined as embryo- or adult-enriched via k-means clustering of all binding sites identified in adult or embryonic conditions (Fig. S3). (B) The average ChIP-seq signals at the binding regions shown in A is plotted. (C) Embryo-enriched and adult-enriched binding sites, as identified in A, correspond to different sets of nearby target genes, which show stage-specific gene expression. RNA-seq was conducted on E12.5 or adult intestinal epithelium and genes linked to embryo-enriched or adult-enriched CDX2 binding (within 5 kb) were analyzed for their distribution along the E12.5-to-adult expression continuum using GSEA analysis. The GSEA plots indicate that embryo-enriched sites are nearby genes that are expressed at higher levels in the embryo than in the adult (leftward shift), whereas the opposite trend is observed for expression of genes nearby CDX2 sites that are stronger in the adult. CDX2-bound genes are also dependent upon CDX2 for expression, as revealed by RNA-seq analysis in Cdx2 knockout tissues (Fig. S3B). ES, enrichment score. (D) Embryo- and adult-enriched CDX2-binding regions are nearby genes with distinct gene ontologies, corresponding to embryo-specific or adult-specific functions, as indicated in GO term analysis. (E,F) Representative examples of embryo-enriched (E) or adult-enriched (F) CDX2-binding sites are shown using IGV with merged biological replicates. Also see Fig. S3.
Transcription factor motifs enriched at stage-specific enhancers also suggest dynamic regulatory activity. Although CDX2-binding motifs were enriched in both adult- and embryo-enriched binding sites, other motifs were selectively enriched. For example, embryo-enriched CDX2 binding was associated with HOX and FOXA motifs, whereas adult-enriched binding sites were associated with HNF4, FRA1 and KLF5 motifs (Fig. S3C, Table S3). This divergence points to different transcriptional regulatory networks in the embryonic gut, where developmental competence and tissue patterning predominate (Wang et al., 2015), versus in the adult intestine, where cell identity is fixed and CDX2 and partner factors control physiological functions (Verzi et al., 2010, 2011). Indeed, ontology analysis indicated that genes near embryo-enriched binding sites associate with developmental processes, such as liver development and regulation of stem cell differentiation, whereas genes near adult-enriched regions associate with mature intestinal functions, including lipid metabolism and brush border localization (Fig. 3D). Condition-specific binding at developmental genes (HoxB, Fzd10) or those expressed exclusively in the mature tissue (Alpi, Sis) illustrates these findings (Fig. 3E,F, Fig. S3D,E). Together, these results reveal that CDX2 has distinct genomic targets at different developmental stages, regulates target genes in a stage-dependent manner, and likely partners with different factors in each context to accomplish stage-dependent gene regulation. These results generally parallel observations observed in the human model (Fig. 2), suggesting that the dynamic functions of CDX2 across intestinal development are conserved between these mammalian species.
Temporal dissection of Cdx2 requirements for intestinal identity
We next sought to correlate our observed changes in CDX2's genomic occupancy with observations of homeotic conversion phenotypes upon CDX2 loss. CDX2 loss triggers profound homeotic transformation of the intestine to esophagus when it occurs at E8.5 (FoxA3-Cre; Gao et al., 2009). When Cdx2 is deleted at E13.5, however, certain stomach-specific genes are ectopically expressed without overt squamous cell differentiation (Grainger et al., 2010).
We examined the timing of CDX2 dependency more closely, with additional time points of CDX2 inactivation. Mouse endoderm engineered to delete Cdx2 at ∼E9.5 using the Shh-Cre driver (Harfe et al., 2004) exhibited stomach characteristics anteriorly in the jejunum [ATP4B and foveolar Periodic acid-Schiff (PAS) staining] and stratified squamous esophageal features posteriorly (Fig. 4A, Fig. S4A). Thus, loss of CDX2 ∼E9.5 results in a homeotic phenotype of intermediate severity compared with when CDX2 is lost earlier (E8.5; Gao et al., 2009) or later (E13.5; Grainger et al., 2010). In contrast, we observed no evidence of transformation when we inactivated Cdx2 at later embryonic stages by tamoxifen treatment of dams carrying Villin-CreERT2; Cdx2f/f embryos at E13.5 or E15.5 (Fig. 4A; see also figure 4A of Banerjee et al., 2018). Thus, these late-stage embryos, like the adult epithelium upon CDX2 loss (Verzi et al., 2011), retain features of intestinal identity. RNA-seq analysis of these mutants provides a more global perspective of lineage-specific gene expression upon CDX2 loss at distinct developmental time points. Consistent with our histological analysis, early loss of CDX2 via Shh-Cre triggered a substantial increase in transcripts normally enriched in the hind stomach, though not to levels typically found in the hind stomach (see figure 4C in Banerjee et al., 2018). Elevated expression of Sox2 transcripts in the Shh-Cre; Cdx2f/f mutant exemplifies these findings, as Sox2 transcripts in E12 epithelium increase upon loss of CDX2, but are less robustly detected, or undetectable, upon CDX2 loss at later developmental or adult stages, respectively (Fig. S4C). Whereas rostral transcripts were elevated upon CDX2 loss at the earliest time points examined, transcripts normally enriched in the intestinal epithelium were reduced to levels similar to those found in the hind stomach, though not to levels as low as those of intestinal genes in esophagus samples (see supplementary figure 4B in Banerjee et al., 2018). It is interesting to note that whereas the ileal-esophageal transition observed upon Shh-Cre-driven CDX2 loss is accompanied by squamous histology, the gastric characteristics present in the jejunal epithelium of this model exist within an undulating, villus-like epithelial morphology.
Fig. 4.
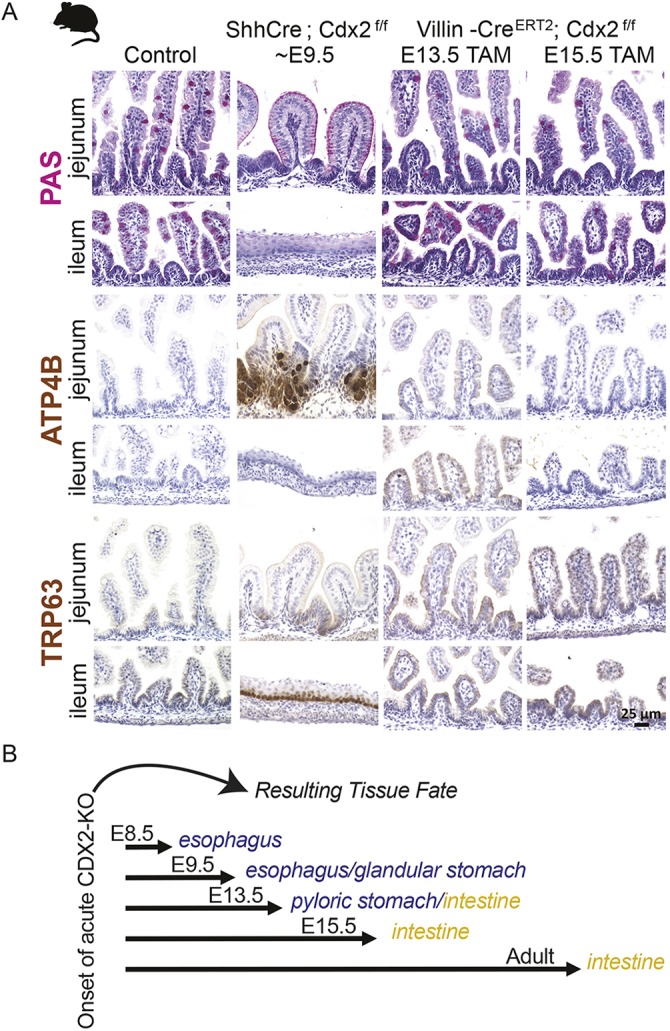
Developmental stage dictates the consequences of CDX2 loss. (A) Analysis of regional tissue identity on samples from control, Shh-Cre; CDX2f/f or Villin-CreERT2; CDX2f/f embryos, treated with tamoxifen by oral gavage [5 mg of tamoxifen (Sigma, T5648) dissolved in 0.5 ml of corn oil] to induce CDX2 loss as indicated, and collected at E18.5 from the indicated intestinal region. PAS stain marks mucosubstances, including intestinal Goblet cells, which are identified by a punctate staining and are characteristic in the intestinal epithelium (Control or Villin-CreERT2), whereas broad PAS staining at the apical cell surface is indicative of gastric foveolar cells, as observed in the jejunum of Shh-Cre; Cdx2f/f embryos. TRP63 marks esophageal cells and is not typically found in the intestine, but appears ectopically in the ileum of Shh-Cre; Cdx2f/f embryos, but not in control embryos, or embryos induced to delete Cdx2 in the later stages. The parietal cell marker ATP4B is typically expressed in the glandular stomach, but is ectopically expressed in the CDX2-negative jejunum of Shh-Cre; Cdx2f/f embryos, but not in embryos deficient in intestinal CDX2 at the later stages tested. Each immunostain or histological stain is representative of at least three experiments on different embryos. (B) Summary of CDX2 loss-of-function phenotypes from the data shown in A and previous studies. Temporal-specific CDX2 loss at stages earlier than E12.5 induces homeotic transformations, as observed by Gao and colleagues using the Foxa3-Cre driver (∼E8.5) (Gao et al., 2009), in this study using the Shh-Cre driver (∼E9.5), or at ∼E13.5 using the VIllin-Cre driver (Grainger et al., 2010). However, acute loss of CDX2 at later developmental stages (>E13.5) and in the adult compromises intestinal digestive functions, but intestinal identity is preserved (Villin-CreERT2 with tamoxifen treatment at E13.5, E15.5 or adult), as documented in these studies in the embryo (Gao et al., 2009; Grainger et al., 2010), and previously in the adult (Verzi et al., 2011).
Taken together, our analysis of CDX2 inactivation in the intestine at different developing time points, along with previously published Cdx2 mouse knockout studies, suggests that a critical window for intestinal plasticity exists prior to E13.5. After this E13.5 time point, intestinal identity is stabilized and less susceptible to transformation to anterior structures (Fig. 4B). These temporal phenotypes roughly correspond to the dynamic CDX2-binding patterns we observed in ChIP-seq assays of early versus late embryonic intestinal epithelia (Fig. 3A) and are consistent with observations that CDX2 target genes partition into functions associated more with developmental processes (embryo-enriched sites) versus mature intestinal physiological functions (adult-enriched sites, Fig. 3D).
CDX2 follows, then stabilizes, a developmentally dynamic chromatin landscape
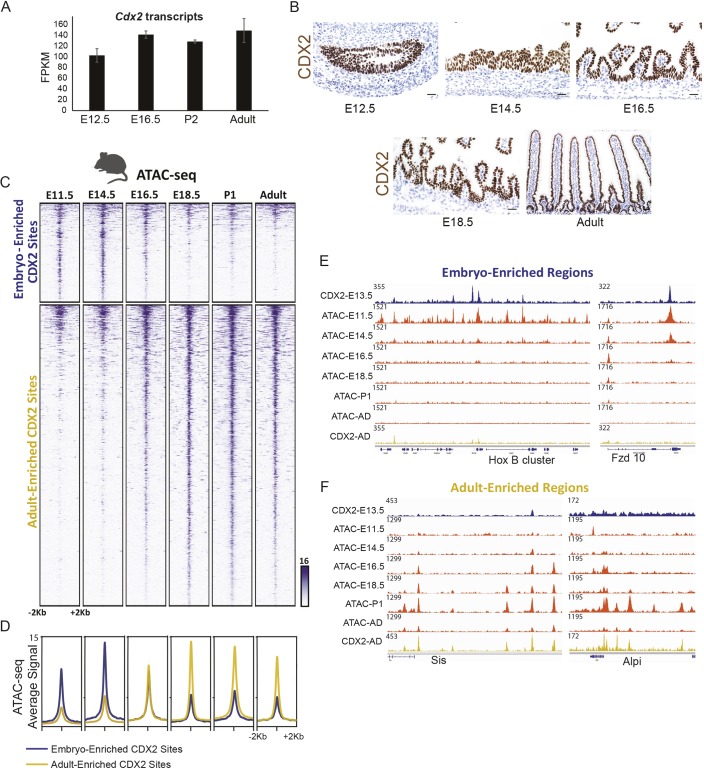
CDX2 expression is nearly constant from the onset of intestine development through adult life in both humans (Fig. 1B,C) and mice (Fig. 5A,B). To resolve why CDX2 could function in different capacities despite relatively consistent levels of expression (Figs 2, 3), we examined the possibility that a dynamic chromatin environment could underlie stage-specific CDX2 genomic binding. Chromatin accessibility was measured at all embryonic and adult binding sites using ATAC-seq (assay for transposase-accessible chromatin using sequencing) at E11.5, E14.5, E16.5, E18.5, postnatal day (P) 1 and adult. We observed a clear pattern at CDX2-bound regions, with embryo-enriched sites losing accessible chromatin after E14.5, and adult-enriched sites gaining chromatin access thereafter (Fig. 5C,D). Four features are notable in this regard. First, 74% of genomic regions with strong adult-enriched CDX2 binding showed poor accessibility in early embryos, lacking ATAC signals at E11.5 (P<10−5; Fig. 5C). Second, open chromatin emerged at these sites in adults (Fig. 5C), where CDX2 binding was clearly absent in the embryo (Fig. 3A); the converse was apparent for the embryo-enriched CDX2-binding sites. Third, chromatin dynamics were similar for the active histone mark H3K27ac (Kazakevych et al., 2017), with stage-specific enhancer marks corresponding to stage-specific CDX2-binding regions (Fig. S5A,B). Fourth, stage-specific binding in mouse endoderm resembled the profound shift in binding between human ESC-derived gut endoderm and adult human intestine (Fig. 2A). Thus, despite constant expression across time, CDX2 binding closely follows a temporal wave of accessible chromatin from embryo-enriched to adult-enriched sites. Examples of these findings are illustrated at genes selectively expressed in developing or adult intestine (Fig. 5E,F).
Fig. 5.
CDX2 temporally enriched binding patterns follow a temporally dynamic chromatin landscape that requires CDX2 for maintenance. (A,B) CDX2 transcripts (from RNA-seq on isolated epithelium; A) and protein levels (via immunostaining; B) are consistent across mouse developmental time, yet CDX2 occupies distinct sets of genomic regions at different developmental stages (Fig. 3). (C) CDX2 chromatin binding shifts may be due to a shifting chromatin landscape across developmental time, as indicated by the ATAC-seq assay for accessible chromatin in isolated intestinal epithelial cells. Regions shown are the CDX2 condition-enriched binding regions (as defined in Fig. 3A). (D) Average ATAC-seq signal is robust at embryo-enriched CDX2-binding sites prior to E16.5, after which chromatin accessibility at these regions diminishes. The converse is true for adult-enriched CDX2-binding sites. (E,F) Specific examples of condition-enriched chromatin accessibility at CDX2-binding sites, as visualized using IGV. CDX2-binding regions are also dependent upon CDX2 binding to remain accessible to the ATAC-seq assay, as seen in CDX2 knockout epithelium (Fig. S5).
The contemporaneous transitions we observed after E14.5 in chromatin accessibility (Fig. 5C,D), CDX2 binding (Fig. 3A) and histone marks (Fig. S5A,B) coincide roughly with the restricted temporal window in which Cdx2 loss is permissive to ectopic expression of more rostral gut features in the intestine (Fig. 4A,B). Importantly, although CDX2 cannot access adult-enriched target sites in the developmental context (Figs 2A and 3A), ATAC-seq in Shh-Cre; CDX2f/f embryos at E16.5 revealed diminished chromatin accessibility at the vast majority of regions occupied by CDX2 at that developmental time point (Fig. S5C,D). Reduced chromatin access was specific to CDX2-bound regions, as chromatin accessibility at all annotated promoters was unchanged (Fig. S5D,E). Together, these data reveal that although chromatin accessibility directs CDX2 binding during endoderm development, CDX2 (likely in partnership with other activators) is required to sustain access at regions associated with adult intestinal genes. This finding is consistent with CDX2 control of enhancer chromatin structures in mature mice (Verzi et al., 2013). Thus, CDX2 acts in distinct developmental contexts, constrained in part by temporal chromatin landscapes, to promote tissue specification in embryos and essential digestive functions in adults.
DISCUSSION
Pioneer factors help shape accessible chromatin (Iwafuchi-Doi and Zaret, 2016), and developmental signaling pathways and partner transcription factors prime chromatin structure to create a state of ‘developmental competence’ (Wang et al., 2015). Our findings fit within this model, in that CDX2 is unable to bind to selected genomic regions outside of a proper developmental context. Our findings demonstrate that CDX2 is re-purposed across the transitions in developmental competence: first CDX2 binds genes associated with developmental and specification functions, and subsequently CDX2 binds genes that promote the mature differentiated state in adults. The correlation between chromatin accessibility (Fig. 5), active histone modifications (Fig. S5), and CDX2 binding suggests that the chromatin environment is likely a crucial factor in dictating condition-specific binding of CDX2. Our experiments with forced CDX2 expression in hESC-derived endoderm are similar to our findings in the mouse, with the observation that CDX2 is not sufficient to access many of its primary midgut-specific targets independently of the proper signaling and/or chromatin environment. These findings are consistent with a previous study in which CDX2 was ectopically expressed in ESCs, endoderm, or motor neuron progenitors. In these distinct contexts, CDX2 could access some binding sites that are common to each tissue, but depends on accessible chromatin at sites that are specifically occupied in only one of these three lineages (Mahony et al., 2014). In addition to regulation of chromatin accessibility, post-translational modification of CDX2, expression of transcriptional co-regulators, or other regulatory mechanisms likely function downstream of WNT and FGF signaling to create an environment in which CDX2 can access its midgut target sites and impart intestinal identity. A similar set of regulatory mechanisms could govern the transition of CDX2 from embryo-enriched binding sites to adult-enriched binding sites (Figs 2, 3) across crucial developmental transitions (Fig. 4B). DNA-binding motif analyses (Figs 2, 3) suggest that CDX2 has different binding partners in embryonic versus mature intestine, and that these partners are likely to be at least partially conserved, with HNF4 and AP-1 factor motifs enriched at CDX2-binding sites in mature intestines of both mice and humans. These findings should be corroborated by testing the roles of these candidate partners in genetic models and by assaying for direct protein-protein interactions. CDX2 binding during intestinal specification is likely to rely upon pioneer factors, and motif analysis points to FoxA, a known pioneer factor (McPherson et al., 1993), as a likely candidate (Fig. S3C). As-yet-unidentified chromatin regulators likely generate the corresponding competence for CDX2 occupancy and intestinal maturation as the intestinal chromatin landscape is altered in late gestation (Fig. 5C). Expression of these chromatin regulators could even be under the control of, or partner with, CDX2.
Our observations that CDX2 chromatin binding and chromatin accessibility shifts during development provide new insights into the molecular basis of intestinal maturation, and indicate that mere expression of lineage regulators is insufficient to achieve desired tissue fates, at least for CDX2 in the intestine. Interestingly, expression of CDX2 in ESCs is both required (Strumpf et al., 2005) and sufficient (Niwa et al., 2005) to induce trophectoderm fate. CDX2 expression activates trophectoderm target genes and indirectly represses activity of pluripotency transcription factors, as CDX2 binding is not observed nearby genes regulated by pluripotency transcription factors, in spite of the observation that the pluripotency factors no longer bind chromatin robustly after expression of CDX2 (Nishiyama et al., 2009). One potential mechanism of trophectoderm lineage induction by CDX2 is through a direct interaction with Oct3/4 (POU5F1), as measured by co-immunoprecipitation and fluorescence resonance energy transfer (Niwa et al., 2005). The trophectoderm and intestinal systems offer an interesting contrast in the ability of one transcription factor to induce lineage differentiation, and it will be intriguing to define potential protein interactors of CDX2 in the embryonic intestine to identify potential negative regulatory interactions akin to what is observed in trophectoderm development.
In models of Barrett's esophagus, a predisposing condition of esophageal adenocarcinoma, CDX2 is unable to induce intestinal metaplasia simply by its ectopic expression in the adult squamous foregut via a keratin 14 promoter (Kong et al., 2011). Intestinal metaplasias are only triggered upon CDX2 expression within the proper cellular context – either in the fetal stomach, or in a subset of junctional cells at the border of the squamous and columnar epithelium in the stomach (Jiang et al., 2017; Silberg et al., 2002). The inability of CDX2 to induce Barrett's metaplasia in most adult contexts is consistent with the observations we report. CDX2 only seems to influence tissue patterning at early developmental stages, corresponding to stages in which chromatin accessibility is more broadly similar across all regions of the developing gastrointestinal tract (Banerjee et al., 2018). After the epigenome is re-structured late in gestation, CDX2 accesses adult-specific binding regions, but its loss no longer leads to ectopic expression of foregut markers. Consistent with the observation that the intestinal chromatin landscape shifts late in development, loss of CDX2 in the adult mouse epithelium leads to a fetus-like transcriptome (Mustata et al., 2013), but not the early embryonic state permissive to homeotic transformation. CDX2 loss in early embryos permits such transformation in both mice and humans (Gao et al., 2009; Grainger et al., 2010; Fig. 4A,B), likely because a permissive chromatin environment is present for a limited time. It will be interesting to define how chromatin accessibility is altered in instances of homeotic transformation upon prolonged loss of CDX2 in adult tissues (Simmini et al., 2014; Stringer et al., 2008). Transformation of adult tissues will likely require reversal of repressive chromatin features, such as H3K9me3 domains that require re-structuring, as observed during induced pluripotent stem cell reprogramming (Soufi et al., 2012), or the loss of PRC2 function that is required for expression of alternate lineage genes in the intestine (Saxena et al., 2017). Future studies investigating these epigenomic processes in the context of CDX2 loss in the early embryonic intestine or upon ectopic CDX2 expression leading to Barrett's metaplasia will be important next steps in understanding the mechanisms of lineage plasticity. In addition, defining the factors that craft embryonic chromatin landscapes toward their mature forms will bolster efforts of regenerative medicine to develop these tissues accurately in vitro.
MATERIALS AND METHODS
Mice
Shh-cre mice (Harfe et al., 2004) were purchased from The Jackson Laboratory and male Shh-cre; Cdx2f/+ were bred to Cdx2 f/f female mice for inducing Cdx2 knockout (KO) in the developing intestine beginning at ∼E9.5. Cre-negative embryos were used as littermate controls. Villin-CreERT2 (el Marjou et al., 2004) and CDX2f/f mice (Verzi et al., 2010) were previously described. Wild-type CD1 mice were obtained from Charles River Laboratories. All mouse protocols and experiments were approved by the Rutgers Institutional Animal Care and Use Committee.
Human pluripotent stem cell culture and differentiation, and human adult tissue collection
Human embryonic stem cells (line H9, WiCell Research Institute, NIH registration number 0062) (tested for mycoplasma contamination and validated with STR profiling) were maintained and differentiated into endoderm, midgut and intestinal organoids as published previously (Cruz-Acuña et al., 2017; Tsai et al., 2016, 2017). hESCs were maintained in mTESR1 (Stem Cell Technologies) on hESC qualified Matrigel (Corning) in 6-well dishes. Prior to differentiation, hESCs were passaged as small clumps into 24-well Nunc Cell-Culture Treated multi-well plates with the Nunclon Delta surface treatment. When cultures were approximately 60% confluent, hESC media was replaced with endoderm differentiation media (RPMI1640+100 ng/ml Activin A; R&D Systems) for 3 days supplemented with 0%, 0.2% and 2% HyClone FBS on subsequent days. Following endoderm differentiation, cells were exposed to midgut differentiation media for 4-6 days, which consisted of RPMI1640+2% HyClone FBS+2 µM CHIR99021+500 ng/ml FGF4 (made in-house, as previously described; Leslie et al., 2015). Free-floating midgut spheroids formed on the 4th-6th days, and were subsequently placed in a droplet of Matrigel and were grown/expanded into HIOs for 28-35 days, in HIO media as previously described (Tsai et al., 2016), which consisted of Advanced DMEM/F12 media supplemented with 1× Pen/Strep, 1× B27 (Gibco, Life Technologies), 100 ng/ml EGF, 5% NOGGIN conditioned media (Heijmans et al., 2013) and 5% R-SPONDIN2 conditioned media (Bell et al., 2008).
For human patients, de-identified duodenal biopsies were collected with Rutgers IRB approval, cut longitudinally and epithelial cells were obtained by scrapping the luminal surface. Cells were washed twice with PBS, fixed with 1% formaldehyde (15 min at 4°C followed by 30 min at room temperature), washed twice and cell pellets were flash frozen for subsequent chromatin pulldown.
Histology, immunofluorescence and immunohistochemistry
Human organoids derived from stem cells (Spence et al., 2011) for 30 days were processed for immunofluorescence using antibodies (CDX2, Bio-Genex, MU392-UC, 1:500; MUC5AC, Abcam, ab79082, 1:500; NKX2.1, ThermoFisher, 8G7G3, 1:50; PDX1, Epitomics, 3470-1 1:300; SOX2, Santa Cruz Biotechnologies, sc-27603, 1:100). Mouse immunostaining was performed with the following antibodies: CDX2, Cell Signaling, 12306, 1:200; ATP4B, MBL International Corp., D032-3, 1:200; TRP63, Santa Cruz Biotechnologies, sc-8343, 1:500. Human organoids were collected and fixed in 4% paraformaldehyde (PFA) for 1 h, followed by cryoprotection with 30% sucrose overnight. On the next day, tissues were embedded with OCT (Fisher, Tissue-Plus, 4585) and stored at −80°C. Tissues, were sectioned at 3 µm and immunofluorescence staining was carried out using standard methods (Spence et al., 2011) by staining primary antibody overnight and 1 h secondary antibody on the next day. For midgut immunofluorescence staining, the monolayer of cells was fixed by 4% PFA for 20 min, followed by the same methods as organoids. Images were taken using an Olympic microscope IX71.
For mouse immunostaining, intestinal tissue was dissected and fixed overnight in 4% PFA at 4°C, washed with PBS, passed through increasing concentrations of an ethanol series and paraffin. Intestinal sections (5 µm) were cut from paraffin blocks, processed for immunostaining with the indicated primary antibodies, developed using the Vectastain ABC Kit (Vector Laboratories, PK6101) and counterstained with Hematoxylin. An antigen retrieval step of 1 h in 10 mM sodium citrate solution under 15 psi pressure was used for all stains. Slides were incubated with primary antibody overnight at 4°C (CDX2, Cell Signaling, 12306, 1:200; ATP4B, MBL International Corp., D032-3, 1:200; TRP63, Santa Cruz Biotech, sc-8343, 1:500). PAS staining was conducted by treating slides with 0.5% periodic acid and staining with Schiff's Reagent (Alfa Aesar, J612171). Images were taken using a Retiga 1300CCD (Q-Imaging) camera and a Nikon Eclipse E800 microscope with QC-Capture imaging software. Adjustments in contrast and sharpness, when made, were applied to complete figure panels in Adobe Photoshop.
qRT-PCR
Briefly, RNA isolation was performed using MagMAXTM-96 total RNA isolation kit (Ambion, AM1830). SuperScript VILO cDNA synthesis kit (ThermoFisher, 11754250) was used to make cDNA from 200 ng RNA. cDNA levels were detected using QuantiTect SYBR Green (Qiagen, 608056). Relative gene expression was plotted as arbitrary units, using the following formula: [2^(housekeeping gene Ct-gene Ct)]×10,000.
Generating CDX2 hESC knockout cell line using CRISPR/Cas9
The CDX2-KO was generated using the pCas-Guide-EF1a-GFP plasmid (Blue Heron Biotech, GE100018) using the CDX2 sgRNA of targeting sequence (CCTCTCAGAGAGCCCCAGCGTGG) at the human CDX2 exon2 DNA-binding domain. Plasmid was introduced into hESCs using electroporation (NEPA21 Electro-Kinetic Transfection System). Total plasmid DNA (10 µg) was prepared and mixed with 1×106 hESCs in transfection medium (OPIT-MEM, Gibco, 31985-062) to a total volume of 100 µl and added into an electroporation cuvette (Bulldog Bio, 12358-346). Cells plus DNA were electroporated using the following settings: poring pulse: 175 volt; transfer pulse: 20 volt. Immediately, the transfected cells were transferred to one well of a Matrigel coated 6-well plate with 1.5 ml mTeSR1 medium plus ROCK inhibitor (Reagents Direct, Y27632, 53-B85). After 24 h, GFP-positive cells were sorted by fluorescence-activated cell sorting (FACS) and plated in Matrigel-coated 6-well plates with mTeSR1 medium plus ROCK inhibitor at low density. After 5-7 days culture, single colonies were manually picked and transferred to individual wells of a 24-well plate, and were expanded. Clones were screened by DNA sequencing and were differentiated to interrogate CDX2 protein expression by immunofluorescence. DNA-sequencing primer sequences were GCATCCTCCTGCTTCAGTCT and GCAGTTCTCAGCCCTCACTT. Control cells were treated the same way, but were electroporated with a GFP plasmid lacking a sgRNA.
Generating doxycycline-inducible CDX2 expression in hESCs
The pInducer20 vector is an inducible Lentiviral vector system, which carries both rtTA3 and neomycin-resistance genes under the UBC promoter as well as a cDNA of interest under the control of a tetracycline-responsive promoter (Meerbrey et al., 2011). pInducer plasmids are available from Addgene (44012). Generation of the pInducer-GFP inducible control cell line has been described elsewhere (Chen et al., 2014). The human CDX2 clone HsCD00045643 was purchased from the Arizona State DNA Plasmid Repository (Dnasu.org), and was introduced into the pInducer20 plasmid using Gateway recombination cloning technology (ThermoFisher Scientific) following standard manufacturer protocols. Lentiviral particles were generated at the University of Michigan Viral Vector Core, and inducible hESCs lines were generated as previously described (Tsai et al., 2016). Following antibiotic selection (G418), GFP or CDX2 gene expression was induced by adding 2 µg/ml doxycycline to the culture medium.
Western blotting
The cells at indicated differentiation stages were collected, homogenized and lysed in lysis buffer. Lysis buffer consisted of 50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1.5 mM MgCl2, 5 mM EGTA, 1% glycerol and protease and phosphatase inhibitors [30 mM sodium pyrophosphate (Na4P2O7), 50 mM sodium fluoride (NaF), 0.1 mM sodium orthovanadate (Na3O4V) and Complete, EDTA-free (Roche, 11873580001)]. Cells were lysed for 30 min at 4°C with rotation and cleared by centrifugation at 16,000 g. Protein concentrations were determined by Bio-Rad protein assay (Bio-Rad, 500-0006). Samples containing 20 μg of protein lysate in Laemmli sample buffer were separated on 15% SDS-PAGE gel and transferred (semi-dry) to nitrocellulose membrane. Membrane was probed with rabbit mAb CDX2 (D11D10) (Cell Signaling Technology, 12306, 1:500), then re-probed with mouse mAb β-actin (Sigma, A1978). HRP-conjugated rabbit or mouse secondary antibodies (Cell Signaling Technology, 7074 or 7076) were used and the membrane was developed by using chemiluminescence (Western Blotting Luminol Reagent, sc-2048).
Purification of intestinal epithelial cells from mice
Pregnant dams were sacrificed and dissected embryos were kept in ice-cold PBS. Embryo tail tissue was used for genotyping using KAPA Mouse Genotyping Kits (Kapa Biosystems, KK7352). For E12.5 RNA-seq entire gut tubes (caudal of stomach), and for E16.5 ATAC-seq midguts (caudal stomach to rostral caecum), were dissected out of the body cavity. Intestinal tissues were treated with pre-warmed 0.25% Trypsin for 8-10 min at 37°C on a vortex station, neutralized with 10% FBS, and passed through a 70 µm filter to obtain single cells. Single cells were incubated with phycoerythrin (PE)-conjugated anti-CD326 (EpCam clone G8.8, eBiosciences, 12-5791-81) for ∼30 min on ice. PE-stained cells were then incubated for ∼30 min with magnetic conjugated anti-PE antibody (Miltenyi Biotec Anti-PE MicroBeads, 130-048-801). Subsequently, upon the availability of anti-CD326 (Epcam) magnetic microbeads antibody (Miltenyi Biotec 130-105-958), cells were stained with the anti-Epcam magnetic microbeads antibody for ∼40 min on ice. Stained cells were passed through a column (Miltenyi Biotec, MS Columns, 130-042-201) in a magnetic field to obtain magnetic antibody-conjugated, EpCam-positive epithelial cells. The purity of magnetic cell isolation was compared with FACS-sorted EpCam-positive cells and found to be comparable. Cells were dissolved in Trizol for RNA processing or used immediately for ATAC-seq.
ChIP-seq
For embryonic CDX2 ChIP-seq (E13.5, 12 embryos pooled per replicate; E16.5, 3 embryos pooled per replicate; and E17.5, 2 embryos pooled per replicate) midguts (caudal stomach to rostral caecum) were cut into small pieces, pooled and fixed. Briefly, tissues were PBS washed twice and flash frozen for subsequent chromatin pulldown assays. Cell pellets were thawed on ice and dissolved in 3× volume of lysis buffer: 1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1, Protease Inhibitor 1× (Complete inhibitors, Roche) or mammalian Protearrest (G-Biosciences). Cells were sonicated (∼25-30 mins) to shear chromatin in 200-600 bp fragments in a Diagenode bioruptor. Chromatin was incubated overnight with CDX2 antibody (6 µl, Bethyl A300-691A, lot 1) conjugated to protein A/G beads (15 µl each). Chromatin-bound beads were washed five times with RIPA wash buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% sodium deoxycholate, 1% NP-40; 0.5 M LiCl) and rinsed once with TE buffer (10 mM Tris, 0.1 mM EDTA). Chromatin-bound beads were re-suspended in reverse crosslinking buffer (1% SDS, 0.1 M NaHCO3) and incubated at 65°C for 6 h to release ChIP DNA. DNA was column purified using a PCR purification column (Qiagen) and quantified using Picogreen (Life Technologies). ChIP DNA was used to prepare ChIP-seq libraries using Rubicon Genomics ThruPLEX DNA-seq Kit (R400427/R400428/R40048), fragment size selected using Pippin Prep and sequenced on Illumina HiSeq (50 or 75-bp reads; single end; ∼25 M reads).
ATAC-seq
For ATAC-seq, 25,000-50,000 isolated epithelial cells, from midguts (caudal stomach to rostral caecum, up to 2 embryos per sample) were used, as described previously (Buenrostro et al., 2015) with minor modifications. Briefly, cells were centrifuged at 500 g for 5 min, and lysed in ice-cold ATAC-lysis buffer [10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% (v/v) NP-40], followed by centrifugation at 500 g for 10 min at 4°C to isolate nuclear pellets, which were treated in a 50 μl reaction with Nextera Tn5 Transposase (Illumina, FC-121-1030) for 25-30 min at 37°C. The transposed chromatin was purified with QIAquick PCR Purification Kit (Qiagen) and PCR amplified with high-fidelity 2× PCR Master Mix (New England Biolabs) using universal forward primer and unique reverse primers in a 50 µl reaction for five cycles. After five cycles, 5 µl of the reaction mix was amplified by qPCR for 20 cycles to determine the optimum number of additional cycles, based on one-quarter of the maximum fluorescence intensity. The PCR-amplified libraries were column purified, fragment size selected using Pippin Prep and sequenced on Illumina HiSeq (50/75-bp reads; single end; ∼25 M reads).
RNA-seq
For E12.5 RNA-seq, total RNA was extracted from Epcam-enriched epithelial cells (as described above) from gut tissue caudal to the stomach (2-5 embryos pooled per sample) in Trizol using RNeasy micro kit (Qiagen, 74004). RNA-seq libraries were prepared using the SMARTer-Seq v4 Low Input mRNA library kit (Clonetech SMARTer, 634888). Adult RNA data were curated from GSE70766 and GSE98724 (San Roman et al., 2015; Saxena et al., 2017).
Computational analysis
Raw sequencing reads (fastq) were quality checked using fastQC (v0.11.3), aligned to mouse (mm9) or human (hg19) genomes using TopHat2 (v2.1.0, RNA-seq) or bowtie2 (v2.2.6, ChIP- and ATAC-seq) to generate bam files. For ChIP- and ATAC-seq, bam files were merged using samtools merge (0.1.19) for downstream analysis. Bigwigs were generated from bam files using deeptools bamCoverage (v2.4.2; Ramírez et al., 2014), duplicate reads ignored, RPKM normalized and reads extended for Chip- and ATAC-seq visualization using Integrated Genomics Viewer (IGV; Robinson et al., 2011).
RNA-seq: Cxb files were generated from BAM files using Cuffquant (v2.2.1, frag-bias-correct, multi-read-correct). Normalized expression count tables were generated from cxb files using Cuffnorm (v2.2.1, library-norm-method quartile). Differential gene expression tables were computed from cxb files using Cuffdiff (v2.2.1, --multi-read-correct --frag-bias-correct, --dispersion-method per-condition --library-norm-method quartile) and log2 fold change (FPKM+1) used for analysis.
For ChIP-seq and ATAC-seq, peaks (bed file) were identified from aligned reads (bam) using MACS (1.4.1; Zhang et al., 2008). Two biological replicate ChIPs were performed for each condition, and replicates inspected by measuring Pearson correlation coefficients. Genes associated with ChIP peaks were identified using BETA minus (genes within 5 kb of binding sites, using CTCF boundaries to filter peaks around a gene). Enriched motifs at ChIP peaks were identified using HOMER findMotifsGenome.pl (v4.8.3, knownResults; Heinz et al., 2010). Enriched ontologies were identified from genomic regions (bed file) using GREAT analysis (v3.0.0) using the Proximal- 5 kb upstream and 1 kb downstream and Distal 200 kb setting (McLean et al., 2010) or DAVID (Huang da et al., 2009).
For comparing open chromatin regions (ATAC-seq) or differential CDX2 binding (ChIP-seq), bigwigs were generated (no normalization) and signal intensities were quantile normalized (v0.4.0, bin_size 50, log transformation) using Haystack (Pinello et al., 2018). k-means-clustered heatmaps were generated from haystack normalized bigwigs using computeMatrix and plotHeatmap from deeptools package (v2.4.2). Genomic regions (bed file) associated with desired k-mean clusters were extracted from bed files generated by PlotHeatmap, to obtain selected bed files (genomic regions).
To identify condition-specific binding sites in mouse, MACS called peaks at P-value 10−3 for E13.5 and at P-value 10−3 and 10−5 for adult were concatenated for each biological replicate, yielding a total of 33,834 genomic regions. E13.5 and adult replicate bam files were merged respectively, and quantile normalized bigwigs were used for k-mean clustering (k=4) at these sites. Cluster2 was identified as CDX2's E13.5 enriched genomic targets (4132) and cluster 3 as CDX2’s adult enriched genomic targets (12,551 regions, genomic coordinates provided in Table S3). For human data, replicate bam files were merged for subsequent analysis. MACS peaks at P-value 10−10 were called on merged bam files of midgut-specified endoderm (58,981 peaks) and adult (19,752 peaks). A k-means-clustered heatmap was generated using quantile normalized bigwigs at midgut-specified endoderm (top cluster) and adult sites (bottom cluster). Genes within a 5 kb region of CDX2 summits were identified using BETA minus at midgut-enriched and adult-enriched binding sites, and nearby genes were further used for evaluating gene ontologies (Table S1). For CDX2 ChIP-seq in naive endoderm with doxycycline-induced CDX2, two biological replicates were performed and replicate bam files were merged for subsequent analysis. MACS peaks were called on merged bam files. Genomic sites bound by CDX2 in the doxycycline-induced condition were identified as overlapping sites of midgut-specified endoderm using intersectBed peaks at P-value 10−10 (23,076 peaks). Condition-enriched sites were identified by subtracting lower stringency peaks of one condition (P-value 10−3) from higher stringency peaks of the second condition (P-value 10−10). This approach yielded midgut-only peaks (30,501 sites), not bound by CDX2 in the Tet-On condition, and Tet-On enriched peaks, which were bound upon CDX2 induction without differentiating the cells towards intestine (87,373 sites).
For ATAC-seq, two biological replicates from each developmental stage were performed, bam files were merged and bigwigs were quantile normalized to compare intensities of open chromatin signal. Embryonic and adult ChIP-seq data (Kazakevych et al., 2017) of the active chromatin marker (H3K27ac) was applied to CDX2's E13.5- and adult-genomic targets. Quantile normalized bigwig files of different replicates were merged by BigWigMerge. To evaluate whether CDX2 is required to maintain open chromatin regions where it typically binds, we compared ATAC-seq at E16.5 intestinal epithelium with CDX2 KO (Shh-cre) at E16.5. Two biological replicates were performed, bam files were merged and bigwigs were quantile normalized to compare intensities of open chromatin signal at regions indicated above.
Statistical analysis
For statistical analysis associated with Fig. 1, data are expressed as the median of each sample set. Each data point in the plots represents an independent biological sample. For organoid experiments, each independent biological sample is comprised of three to five organoids pooled together. All organoid experiments were conducted on at least three independent biological replicates. Unpaired t-tests were carried out with GraphPad Prism 5.0 software. Each data point is presented, with the middle line representing the mean, with error bars representing s.e.m. In all figures, *P<0.05. Gene set enrichment analysis (GSEA) was run using the pre-ranked setting on all genes using FPKM+1 values as described (Subramanian et al., 2005).
Supplementary Material
Acknowledgements
We acknowledge members of the Verzi lab and the Rutgers Epigenomics Group for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.K., R.A.S., J.R.S., M.P.V.; Methodology: N.K.; Validation: L.C., J.R.S.; Formal analysis: N.K., Y.-H.T., L.C., A.Z., K.K.B., M.S., S.H., N.H.T., J.X., R.A.S., J.R.S., M.P.V.; Investigation: N.K., Y.-H.T., L.C., A.Z., K.K.B., M.S., S.H., N.H.T., J.X., R.A.S., J.R.S., M.P.V.; Resources: R.A.S., J.R.S.; Data curation: N.K., L.C., A.Z., K.K.B., M.S.; Writing - original draft: N.K., J.R.S., M.P.V.; Writing - review & editing: N.K., L.C., R.A.S., J.R.S., M.P.V.; Visualization: N.K., Y.-H.T., L.C., A.Z., K.K.B., M.S., S.H., N.H.T., J.X., R.A.S., J.R.S., M.P.V.; Supervision: J.X., R.A.S., J.R.S., M.P.V.; Project administration: R.A.S., J.R.S., M.P.V.; Funding acquisition: R.A.S., J.R.S., M.P.V.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institute of Allergy and Infectious Diseases (NIAID) (U01 DK103141 to M.P.V. and J.R.S.); the National Institutes of Health (R01 CA190558 to M.P.V., R01 DK082889 to R.A.S.); the Human Genetics Institute of New Jersey; the Biospecimen Repository Service Shared Resource and Sequencing Facility of the Rutgers Cancer Institute of New Jersey (P30CA072720); and the University of Michigan Center for Gastrointestinal Research (UMCGR) (NIDDK 5P30DK034933). Deposited in PMC for release after 12 months.
Data availability
All RNA-seq, ATAC-seq and ChIP-seq data generated in this study have been deposited in GEO under accession numbers GSE115314 and GSE115541.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.172189.supplemental
References
- Alder O., Cullum R., Lee S., Kan A. C., Wei W., Yi Y., Garside V. C., Bilenky M., Griffith M., Morrissy A. S. et al. (2014). Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Rep. 9, 261-271. 10.1016/j.celrep.2014.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babeu J.-P., Darsigny M., Lussier C. R. and Boudreau F. (2009). Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G124-G134. 10.1152/ajpgi.90690.2008 [DOI] [PubMed] [Google Scholar]
- Banerjee K. K., Saxena M., Kumar N., Chen L., Cavazza A., Toke N. H., O'Neill N. K., Madha S., Jadhav U., Verzi M. P. et al. (2018). Enhancer, transcriptional, and cell fate plasticity precedes intestinal determination during endoderm development. Genes Dev. 32, 1430-1442. 10.1101/gad.318832.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. M., Schreiner C. M., Wert S. E., Mucenski M. L., Scott W. J. and Whitsett J. A. (2008). R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development 135, 1049-1058. 10.1242/dev.013359 [DOI] [PubMed] [Google Scholar]
- Bochkis I. M., Rubins N. E., White P., Furth E. E., Friedman J. R. and Kaestner K. H. (2008). Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat. Med. 14, 828-836. 10.1038/nm.1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Wu B., Chang H. Y. and Greenleaf W. J. (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21-29. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin A.-L., Le Beyec J., Barreau F., Saint-Just S., Houllier A., Gonzalez F. J., Robine S., Pincon-Raymond M., Cardot P., Lacasa M. et al. (2009). Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Biol. 29, 6294-6308. 10.1128/MCB.00939-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawengsaksophak K., James R., Hammond V. E., Köntgen F. and Beck F. (1997). Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84-87. 10.1038/386084a0 [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Finkbeiner S. R., Weinblatt D., Emmett M. J., Tameire F., Yousefi M., Yang C., Maehr R., Zhou Q., Shemer R. et al. (2014). De novo formation of insulin-producing “neo-beta cell islets” from intestinal crypts. Cell Rep. 6, 1046-1058. 10.1016/j.celrep.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Acuña R., Quirós M., Farkas A. E., Dedhia P. H., Huang S., Siuda D., García-Hernández V., Miller A. J., Spence J. R., Nusrat A. et al. (2017). Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326-1335. 10.1038/ncb3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E. and Baetge E. E. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534-1541. 10.1038/nbt1163 [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H. and Lassar A. B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987-1000. 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- el Marjou F., Janssen K.-P., Chang B. H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D. and Robine S. (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186-193. 10.1002/gene.20042 [DOI] [PubMed] [Google Scholar]
- Finkbeiner S. R., Hill D. R., Altheim C. H., Dedhia P. H., Taylor M. J., Tsai Y. H., Chin A. M., Mahe M. M., Watson C. L., Freeman J. J. et al. (2015). Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Reports 4, 1140-1155. 10.1016/j.stemcr.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., White P. and Kaestner K. H. (2009). Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588-599. 10.1016/j.devcel.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger S., Savory J. G. and Lohnes D. (2010). Cdx2 regulates patterning of the intestinal epithelium. Dev. Biol. 339, 155-165. 10.1016/j.ydbio.2009.12.025 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Heijmans J., van Lidth de Jeude J. F., Koo B.-K., Rosekrans S. L., Wielenga M. C. B., van de Wetering M., Ferrante M., Lee A. S., Onderwater J. J. M., Paton J. C. et al. (2013). ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128-1139. 10.1016/j.celrep.2013.02.031 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H. and Glass C. K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576-589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryniuk A., Grainger S., Savory J. G. A. and Lohnes D. (2012). Cdx function is required for maintenance of intestinal identity in the adult. Dev. Biol. 363, 426-437. 10.1016/j.ydbio.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M. and Zaret K. S. (2016). Cell fate control by pioneer transcription factors. Development 143, 1833-1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Li H., Zhang Y., Yang Y., Lu R., Liu K., Lin S., Lan X., Wang H., Wu H. et al. (2017). Transitional basal cells at the squamous-columnar junction generate Barrett's oesophagus. Nature 550, 529-533. 10.1038/nature24269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T. and Edlund H. (1994). Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606-609. 10.1038/371606a0 [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M., Kanai Y., Gad J. M., Tajima Y., Taya C., Kurohmaru M., Sanai Y., Yonekawa H., Yazaki K., Tam P. P. et al. (2002). Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129, 2367-2379. [DOI] [PubMed] [Google Scholar]
- Kazakevych J., Sayols S., Messner B., Krienke C. and Soshnikova N. (2017). Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 45, 5770-5784. 10.1093/nar/gkx167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., Crissey M. A., Funakoshi S., Kreindler J. L. and Lynch J. P. (2011). Ectopic Cdx2 expression in murine esophagus models an intermediate stage in the emergence of Barrett's esophagus. PLoS ONE 6, e18280 10.1371/journal.pone.0018280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Friedman J. R., Fulmer J. T. and Kaestner K. H. (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944-947. 10.1038/nature03649 [DOI] [PubMed] [Google Scholar]
- Leslie J. L., Huang S., Opp J. S., Nagy M. S., Kobayashi M., Young V. B. and Spence J. R. (2015). Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138-145. 10.1128/IAI.02561-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S., Edwards M. D., Mazzoni E. O., Sherwood R. I., Kakumanu A., Morrison C. A., Wichterle H. and Gifford D. K. (2014). An integrated model of multiple-condition ChIP-Seq data reveals predeterminants of Cdx2 binding. PLoS Comput. Biol. 10, e1003501 10.1371/journal.pcbi.1003501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken K. W., Howell J. C., Wells J. M. and Spence J. R. (2011). Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 6, 1920-1928. 10.1038/nprot.2011.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M. and Bejerano G. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495-501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson C. E., Shim E.-Y., Friedman D. S. and Zaret K. S. (1993). An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75, 387-398. 10.1016/0092-8674(93)80079-T [DOI] [PubMed] [Google Scholar]
- Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T. et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA 108, 3665-3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustata R. C., Vasile G., Fernandez-Vallone V., Strollo S., Lefort A., Libert F., Monteyne D., Perez-Morga D., Vassart G. and Garcia M.-I. (2013). Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 5, 421-432. 10.1016/j.celrep.2013.09.005 [DOI] [PubMed] [Google Scholar]
- Nishiyama A., Xin L., Sharov A. A., Thomas M., Mowrer G., Meyers E., Piao Y., Mehta S., Yee S., Nakatake Y. et al. (2009). Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5, 420-433. 10.1016/j.stem.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R. and Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917-929. 10.1016/j.cell.2005.08.040 [DOI] [PubMed] [Google Scholar]
- Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L. and Wright C. V. (1996). PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983-995. [DOI] [PubMed] [Google Scholar]
- Pinello L., Farouni R. and Yuan G.-C. (2018). Haystack: systematic analysis of the variation of epigenetic states and cell-type specific regulatory elements. Bioinformatics 34, 1930-1933. 10.1093/bioinformatics/bty031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B. A. and Manke T. (2014). deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187-W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G. and Mesirov J. P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24-26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Roman A. K., Aronson B. E., Krasinski S. D., Shivdasani R. A. and Verzi M. P. (2015). Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J. Biol. Chem. 290, 1850-1860. 10.1074/jbc.M114.620211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M., Roman A. K. S., O'Neill N. K., Sulahian R., Jadhav U. and Shivdasani R. A. (2017). Transcription factor-dependent ‘anti-repressive’ mammalian enhancers exclude H3K27me3 from extended genomic domains. Genes Dev. 31, 2391-2404. 10.1101/gad.308536.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg D. G., Sullivan J., Kang E., Swain G. P., Moffett J., Sund N. J., Sackett S. D. and Kaestner K. H. (2002). Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology 122, 689-696. 10.1053/gast.2002.31902 [DOI] [PubMed] [Google Scholar]
- Simmini S., Bialecka M., Huch M., Kester L., van de Wetering M., Sato T., Beck F., van Oudenaarden A., Clevers H. and Deschamps J. (2014). Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat. Commun. 5, 5728 10.1038/ncomms6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G. and Zaret K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151, 994-1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J. R., Mayhew C. N., Rankin S. A., Kuhar M. F., Vallance J. E., Tolle K., Hoskins E. E., Kalinichenko V. V., Wells S. I., Zorn A. M. et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105-109. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann A., Hansen M., Wang Y., Larsen J. B., Lund L. R., Ritié L., Nicholson J. K., Quistorff B., Simon-Assmann P., Troelsen J. T. et al. (2006). Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol. Genomics 27, 141-155. 10.1152/physiolgenomics.00314.2005 [DOI] [PubMed] [Google Scholar]
- Stringer E. J., Pritchard C. A. and Beck F. (2008). Cdx2 initiates histodifferentiation of the midgut endoderm. FEBS Lett. 582, 2555-2560. 10.1016/j.febslet.2008.06.024 [DOI] [PubMed] [Google Scholar]
- Stringer E. J., Duluc I., Saandi T., Davidson I., Bialecka M., Sato T., Barker N., Clevers H., Pritchard C. A., Winton D. J. et al. (2012). Cdx2 determines the fate of postnatal intestinal endoderm. Development 139, 465-474. 10.1242/dev.070722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F. and Rossant J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093-2102. 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai Y., Nakajima R., Ishikawa T., Takaku K., Seldin M. F. and Taketo M. M. (1999). Colonic hamartoma development by anomalous duplication in Cdx2 knockout mice. Cancer Res. 59, 2965-2970. [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H. and Lassar A. B. (1988). MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405-411. 10.1126/science.3175662 [DOI] [PubMed] [Google Scholar]
- Tsai Y.-H., Hill D. R., Kumar N., Huang S., Chin A. M., Dye B. R., Nagy M. S., Verzi M. P. and Spence J. R. (2016). LGR4 and LGR5 function redundantly during human endoderm differentiation. Cell Mol. Gastroenterol. Hepatol. 2, 648-662 e648. 10.1016/j.jcmgh.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-H., Nattiv R., Dedhia P. H., Nagy M. S., Chin A. M., Thomson M., Klein O. D. and Spence J. R. (2017). In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development. Development 144, 1045-1055. 10.1242/dev.138453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., Montgomery R. K., Fleet J. C., Brown M., Liu X. S. and Shivdasani R. A. (2010). Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713-726. 10.1016/j.devcel.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., Ho L. L., Liu X. S. and Shivdasani R. A. (2011). Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31, 2026-2039. 10.1128/MCB.01250-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., San Roman A. K., Liu X. S. and Shivdasani R. A. (2013). Intestinal master transcription factor CDX2 controls chromatin access for partner transcription factor binding. Mol. Cell. Biol. 33, 281-292. 10.1128/MCB.01185-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Yue F., Li Y., Xie R., Harper T., Patel N. A., Muth K., Palmer J., Qiu Y., Wang J. et al. (2015). Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16, 386-399. 10.1016/j.stem.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. M. and Melton D. A. (2000). Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development 127, 1563-1572. [DOI] [PubMed] [Google Scholar]
- Young T., Rowland J. E., van de Ven C., Bialecka M., Novoa A., Carapuco M., van Nes J., de Graaff W., Duluc I., Freund J. N. et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516-526. 10.1016/j.devcel.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.