ABSTRACT
The sequence and roles in developmental progression of the microRNA let-7 are conserved. In general, transcription of the let-7 primary transcript (pri-let-7) occurs early in development, whereas processing of the mature let-7 microRNA arises during cellular differentiation. In Caenorhabditis elegans and other animals, the RNA-binding protein LIN-28 post-transcriptionally inhibits let-7 biogenesis at early developmental stages, but the mechanisms by which LIN-28 does this are not fully understood. Nor is it understood how the developmental regulation of let-7 might influence the expression or activities of other microRNAs of the same seed family. Here, we show that pri-let-7 is trans-spliced to the SL1 splice leader downstream of the let-7 precursor stem-loop, which produces a short polyadenylated downstream mRNA, and that this trans-splicing event negatively impacts the biogenesis of mature let-7 microRNA in cis. Moreover, this trans-spliced mRNA contains sequences that are complementary to multiple members of the let-7 seed family (let-7fam) and negatively regulates let-7fam function in trans. Thus, this study provides evidence for a mechanism by which splicing of a microRNA primary transcript can negatively regulate said microRNA in cis as well as other microRNAs in trans.
KEY WORDS: let-7 family microRNAs, lin-28, Trans-splicing, Developmental timing, Caenorhabditis elegans
Summary: RNA-binding protein LIN-28 is necessary for the trans-splicing event that regulates let-7 biogenesis in cis as well as the activity of members of the let-7 seed family in trans.
INTRODUCTION
MicroRNAs are endogenous ∼22 nt RNAs that are enzymatically processed from longer primary transcripts, and that repress protein expression through imperfect base pairing with their target mRNAs. Nucleotides 2-8 of the microRNA, known as the seed, instigate target recognition through essentially complete complementarity, whereas base pairing via the non-seed nucleotides (9-22 of the microRNA) is less constrained than is seed pairing (He and Hannon, 2004). microRNAs that contain an identical seed sequence but differ in their non-seed nucleotides are classified together as a ‘family’ based on their presumed evolutionary relatedness, and their potential to act in combination on the same targets (Bartel, 2009; Ambros and Ruvkun, 2018).
The let-7 gene was initially identified in a screen for developmental defects in Caenorhabditis elegans (Meneely and Herman, 1979), and later found to encode a microRNA that promotes the differentiation of cellular fates (Reinhart et al., 2000). Orthologs of the C. elegans let-7 microRNA are easily identified across animal phyla because of the near perfect conservation of the entire 22 nt sequence (Pasquinelli et al., 2000). In many species, let-7 paralogs encode additional family members, including mir-48, mir-84 and mir-241 in C. elegans, which differ from let-7 in some of their non-seed nucleotides. In C. elegans, let-7 seed family (let-7fam) microRNAs function semi-redundantly to regulate stage-specific larval cell fate transitions, with mir-48, mir-84 and mir-241 primarily promoting the L2-to-L3 transition and let-7 primarily promoting the L4-to-adult transition (Reinhart et al., 2000; Abbott et al., 2005).
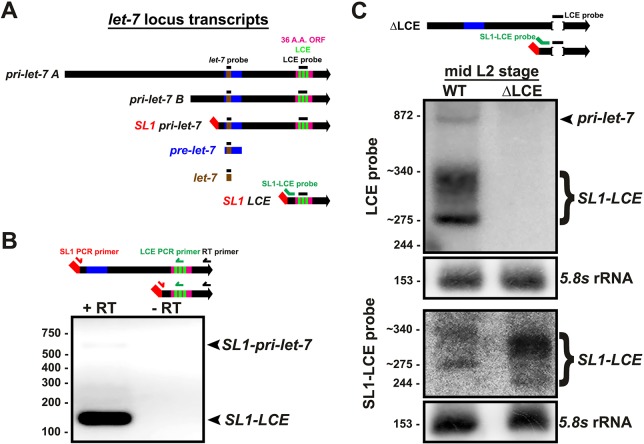
In C. elegans, two major primary transcripts of let-7 (pri-let-7) are produced, pri-let-7 A and pri-let-7 B of 1731 and 890 nucleotides, respectively. The 5′ end of these transcripts can be further processed by trans-splicing with a 22-nucleotide splice leader (SL) RNA (SL1) to produce the 728 nucleotide SL1-pri-let-7 (Bracht et al., 2004; Van Wynsberghe et al., 2011), (Fig. 1A). All three of these pri-let-7 transcripts contain the let-7 precursor hairpin plus additional downstream sequences, which include an element with complementarity to the let-7fam seed sequence that has been shown to associate in vivo with the Argonaute protein, ALG-1 (Zisoulis et al., 2012).
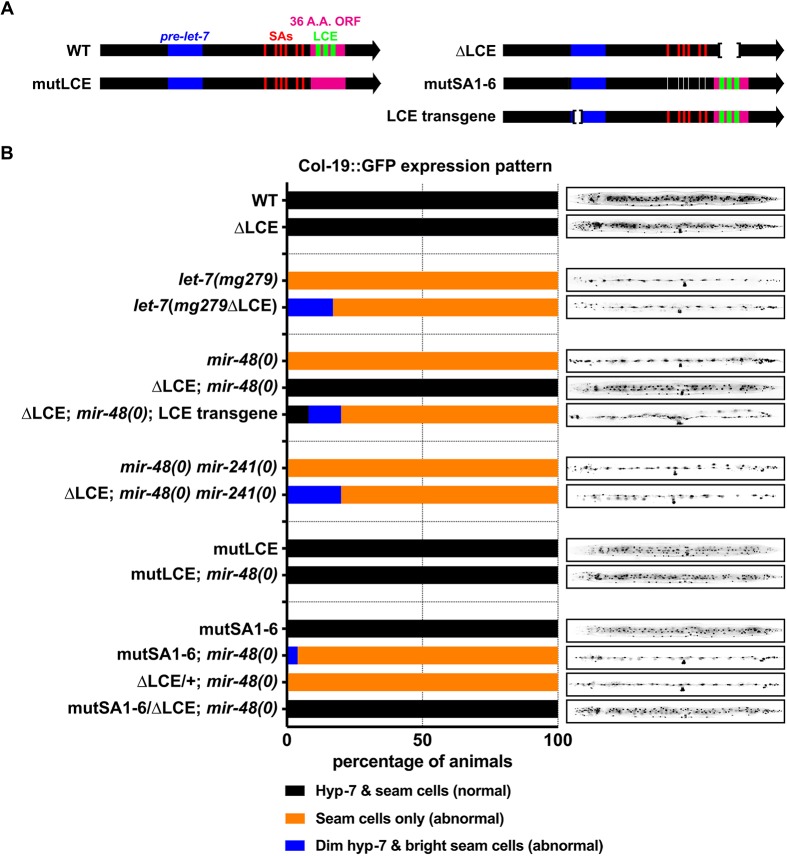
Fig. 1.
The C. elegans let-7 locus produces four transcripts. (A) Major transcripts produced from the C. elegans let-7 locus, and the locations of probes used in the northern blots shown in C, and in subsequent figures. (B) Non-qRT-PCR analysis of total RNA from a mix-population of wild-type (WT) animals, using an SL1 forward primer and LCE reverse primer, with (+) and without (−) reverse transcriptase (RT) in the cDNA synthesis reaction. Numbers mark dsDNA ladder bands in bp. (C) Total RNA from WT and LCE-deletion mid-L2 animals (20 h after plating) analyzed using northern blotting with probes for the LCE and SL1-LCE. Numbers mark known RNA sizes in nt: 872 nt is in vitro transcribed pri-let-7 B, 244 nt is in vitro transcribed SL1-LCE and 153 nt is 5.8s rRNA. Numbers with a tilde before them were estimated from known RNA sizes.
pri-let-7 transcripts are expressed at all four larval stages of C. elegans development (L1-L4), whereas mature let-7 is abundantly expressed only in the L3 and L4 larval stages. Intriguingly, pri-let-7 levels oscillate during each larval stage, peaking mid-stage and dipping during larval molts, likely as a result of underlying pulsatile transcriptional activity of the let-7 locus (Van Wynsberghe et al., 2011; McCulloch and Rougvie, 2014; Perales et al., 2014; Van Wynsberghe et al., 2014). Why pri-let-7 pulses with each larval stage remains unclear. However, the distinct developmental profiles of pri-let-7 and mature let-7, particularly at early larval stages, indicate potent post-transcriptional inhibition of let-7 biogenesis during the L1 and L2 stages.
LIN-28 is a conserved RNA-binding protein that can bind to and regulate a variety of RNAs (Stefani et al., 2015; Wilbert et al., 2012). In C. elegans, LIN-28 is expressed at early larval stages and exerts a strong inhibition of let-7 processing (Van Wynsberghe et al., 2011). Similar to C. elegans, mammalian Lin28 inhibits let-7 processing and is expressed in more pluripotent cells. Extensive studies in mammalian systems have also shown that Lin28 exerts its inhibition of let-7 by binding to the stem-loop of either pri-let-7 or pre-let-7 to directly inhibit processing (reviewed by Tsialikas, 2015). Although CLIPseq data from C. elegans indicated that, similar to in mammals, LIN-28 could bind pri-let-7 in vivo to inhibit accumulation of mature let-7 microRNA, binding occurs downstream of the let-7 stem-loop, which suggests a different mode of regulation on let-7, the precise mechanism(s) of which remains unclear (Stefani et al., 2015).
Trans-splicing is the act of joining two separate RNAs. In C. elegans, ∼70% of all transcripts (including mRNAs and microRNA primary transcripts) are trans-spliced with an SL RNA. The outcomes of trans-splicing include separating the individual mRNAs of polycistronic operon transcripts, shortening the 5′ untranslated regions (5′ UTRs) of RNAs and changing the 5′ RNA cap from monomethyl to trimethyl guanosine. There are two major classes of SL RNAs with distinct sequences: SL1 and SL2. Although there are exceptions, SL1 trans-splicing tends to be the most common 5′ trans-splicing event of a primary RNA transcript and invariably results in rapid degradation of the 5′ ‘outron’. SL2 trans-splicing is generally restricted to downstream open reading frames (ORFs) of operons after SL1 trans-splicing and polyadenylation of the first mRNA (Morton and Blumenthal, 2011; Blumenthal, 2012). Similar to cis-splicing, trans-splicing in C. elegans uses a consensus acceptor sequence of ‘TTTCAG’ (Graber et al., 2007). In mammalian cell culture, Lin28 has been implicated in cis-splicing through its regulation of splicing factor abundance (Wilbert et al., 2012), whereas any involvement of LIN-28 in C. elegans with either cis or trans-splicing remains unknown.
Here, we identify a previously undescribed trans-splicing event in pri-let-7 that occurs downstream of the let-7 stem-loop, and produces a short (∼262 nt) mRNA that contains a 5′ SL1 leader sequence, a short ORF, let-7 complementary sequences (LCSs) and a poly-A tail. We provide evidence that LIN-28 is necessary for this splicing event, and that trans-splicing serves to negatively regulate let-7fam in two ways: First, by preventing precocious let-7 expression through the degradation of the upstream outron which contains the let-7 precursor; second, by inhibiting let-7fam activity via production of an RNA that functions as a let-7fam sponge. Thus, we have characterized a splicing event that involves let-7 primary transcripts and that can regulate let-7 biogenesis in cis as well as let-7fam activity in trans.
RESULTS
The let-7 locus produces a short trans-spliced transcript that contains LCSs
As mentioned above, a previous study identified a region of pri-let-7 in C. elegans that contains a let-7 complementary element (LCE) (Zisoulis et al., 2012). We confirmed that three sites within the LCE element have complementarity to let-7fam microRNAs so as to permit base pairing to the let-7-family seed sequence plus varying degrees of 3′ supplemental pairing (Fig. S1A). We also noted that an additional transcript, C05G5.7, is annotated to be transcribed from the let-7 locus and to contain a 647 nt 5′ UTR that includes the let-7 stem-loop, a 111 nt ORF that contains the LCE, and a 79 bp 3′ UTR. We also identified a potential splice acceptor (SA) sequence that is located upstream of the LCE and downstream of the let-7 stem-loop, which could mediate SL1-trans-splicing and thereby produce a short ∼262 nt transcript that contains the LCE but lacks the upstream let-7 stem-loop (Fig. 1A, Fig. S1A). Analysis of the let-7 locus of other Caenorhabditis species showed that both the trans-SA sequence and the LCE are conserved (Fig. S1A). To determine whether an ∼262 nt SL1-spliced LCE transcript is expressed in vivo, we performed non-quantitative PCR (non-qRT-PCR) using SL1 forward and LCE reverse primers, and we observed two distinct bands. We determined the top band to be SL1-pri-let-7, the previously known SL1-spliced version of pri-let-7 (with the SL1 upstream of the let-7 stem-loop), and the bottom band to be the predicted SL1-spliced LCE transcript (hereafter referred to as SL1-LCE) (Fig. 1B). We also determined that both SL1-LCE and SL1-pri-let-7 are poly-adenylated, by generating cDNA using oligo-dT primer, followed by non-qRT-PCR (Fig. S2).
We confirmed the existence of SL1-LCE by northern blotting with a probe to the LCE sequence and determined that SL1-LCE is more abundant than pri-let-7 at the mid-L2 stage (Fig. 1C). Interestingly, we observed a range of SL1-LCE lengths, from an estimated 275 to 340 nt. After probing specifically to the SL1 splice junction at the 5′ end of SL1-LCE, we observed the same range in length, which indicates that this variation is at the 3′ end, and suggests heterogeneity in poly-adenylation and/or transcriptional stop sites (Fig. 1C). To confirm that the variable length was not at the 5′ end, we also performed 5′ rapid amplification of cDNA ends (RACE) and observed only a single 5′ terminus, at the site of the SL1-LCE trans-splice (Fig. S3). We also confirmed the three alternative 5′ ends of pri-let-7 (Fig. S3 and data not shown). We failed to detect the 5′ end of the annotated transcript C05G5.7.
The location of the trans-SA sequence within the let-7 locus suggests that SL1-LCE could be processed from pri-let-7. Therefore, we tested whether the expression pattern of SL1-LCE is also developmentally regulated, as is the case for pri-let-7. Using quantitative RT-PCR (qRT-PCR), we determined that, similar to pri-let-7, SL1-LCE levels pulsed in phase with the cycle of larval molts, which indicates that SL1-LCE expression could be driven by the same oscillatory transcriptional program as pri-let-7 (Van Wynsberghe et al., 2011; Perales et al., 2014). However, unlike pri-let-7, SL1-LCE stopped pulsing and remained relatively low after the L2 stage (Fig. 2A,B). These findings suggest that SL1-LCE is generated from pri-let-7 by trans-splicing and that it is post-transcriptionally downregulated in conjunction with larval developmental progression.
Fig. 2.
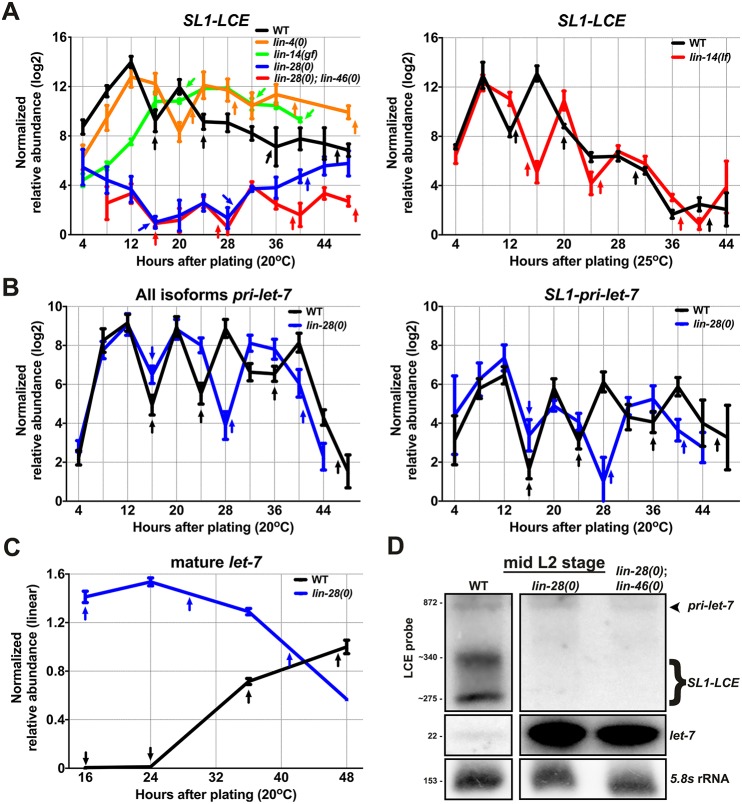
lin-28 is necessary for the expression of SL1-LCE. (A) qRT-PCR analysis of SL1-LCE levels in samples of total RNA from staged populations of synchronously developing WT animals, and the indicated heterochronic mutants at 20°C (left) and at 25°C (right). (B) qRT-PCR analysis of the levels of all pri-let-7 isoforms (left) and SL1-pri-let-7 (right) in samples of total RNA from staged populations of synchronously developing WT and lin-28(0) animals at 20°C. n=3 biological replicates. (C) FirePlex miRNA analysis of let-7 levels in samples of total RNA from staged populations of synchronously developing WT and lin-28(0) animals. n=3 biological replicates. (D) Total RNA from WT, lin-28(0) and lin-28(0); lin-46(0) mid-L2 animals (20 h after plating) analyzed using northern blotting and hybridized with probes for the LCE (top), for let-7 (middle) or for 5.8s rRNA (bottom). Numbers mark known RNA sizes in nt: 872 nt is in vitro transcribed pri-let-7-B, 22 nt is let-7 and 153 nt is 5.8s rRNA. Numbers with a tilde before them were estimated from known RNA sizes. Data are mean±s.d. Arrows mark the times of larval molts.
lin-28 regulates the expression of SL1-LCE
Coordinated with a variety of other factors, let-7 functions within the heterochronic pathway to ensure proper developmental timing of various cell fates, in particular the progression from larval to adult fates during the L4-to-adult transition (Reinhart et al., 2000). The high L1 and L2 expression of SL1-LCE suggested that the heterochronic genes that specify early larval events could promote the expression of SL1-LCE. Therefore, we assessed SL1-LCE expression in heterochronic mutants with altered temporal patterns of early larval cell fates.
lin-4 is a microRNA that is necessary for the transition from the L1 to L2 stages through its targeted repression of the lin-14 3′ UTR. lin-4 null [lin-4(0)] mutations or lin-14 gain-of-function [lin-14(gf)] mutations result in aberrant upregulation of lin-14 that retards developmental progression by continuously specifying the repetition of L1 events (Chalfie et al., 1981; Ambros and Horvitz, 1984; Ambros, 1989; Lee et al., 1993). On the other hand, lin-14 loss-of-function [lin-14(lf)] results in premature developmental progression, which is characterized by a skipping of L1 events and precocious advancement through the L2 to adult stages (Ambros and Horvitz, 1984). We observed that SL1-LCE levels remained high throughout development in lin-4(0) animals and in lin-14(gf) animals, whereas SL1-LCE levels were unchanged in lin-14(lf) animals (Fig. 2A), which indicates that lin-14 promotes, but is not necessary for, SL1-LCE expression.
lin-28 encodes an RNA-binding protein that functions in early larval stages to regulate developmental progression from L2 to later cell fates. Loss of lin-28 results in the skipping of L2 events, precocious advancement to the adult stage and precocious expression of let-7 (Fig. 2C,D) (Ambros and Horvitz, 1984; Van Wynsberghe et al., 2011). We found that lin-28(0) animals exhibited drastically reduced SL1-LCE levels compared with the wild type (Fig. 2A,D). Therefore, in contrast to lin-14, lin-28 appears to be essential for expression of the SL1-LCE transcript.
The fact that lin-14(lf) animals display essentially the same precocious phenotypes as those seen in lin-28(0) animals, yet have normal SL1-LCE expression, suggests that the reduced SL1-LCE expression in lin-28(0) is not an indirect consequence of precocious development. In further support of this conclusion, we observed that in lin-28(0);lin-46(0) animals, which are completely suppressed for precocious phenotypes but not for precocious let-7 levels (Pepper et al., 2004; Vadla et al., 2012) (Fig. 2D), SL1-LCE levels are similarly reduced as in lin-28(0) alone (Fig. 2A,D). Taken together, this suggests that lin-28 has a relatively direct role in promoting LCE trans-splicing.
One possible explanation for why SL1-LCE levels are low in lin-28(0) is that expression of pri-let-7 could be reduced. To test this possibility, we measured the levels of pri-let-7 using qRT-PCR and observed no reduced expression of pri-let-7 in lin-28(0) larvae (Fig. 2B,D). This finding indicates that LIN-28 post-transcriptionally regulates the generation of SL1-LCE. We also noted that, despite the fact that lin-28(0) animals undergo only three larval stages instead of the normal four, the pulses of pri-let-7 still coincided with each larval molt (Fig. 2B).
As mentioned previously, one pri-let-7 isoform is SL1 trans-spliced upstream of the let-7 stem-loop. To test whether lin-28 loss of function also reduced this upstream trans-splicing event, we measured SL1-pri-let-7 levels in lin-28(0) animals and observed no significant reduction (Fig. 2B). Put together, these data indicate that LIN-28 is essential for generating SL1-LCE from pri-let-7 by specifically promoting trans-splicing at the downstream SA.
Mutations of the LCE SA result in the use of cryptic SA sequences
To determine the function of trans-splicing of the LCE transcript, we used CRISPR/Cas9 to introduce mutations of the ‘TTTCAG’ SL1 acceptor sequence of the LCE transcript. To our surprise, deletion of the TTTCAG sequence did not eliminate LCE trans-splicing; non-qRT-PCR and TA-cloning revealed that SL1 trans-splicing still occurred using a cryptic acceptor sequence (TTGTAG) located 27 nt upstream of the canonical TTTCAG (data not shown). qRT-PCR revealed that in wild-type animals use of this cryptic TTGTAG sequence was minimal (Fig. S4A), whereas in the animals in which the canonical TTTCAG was deleted, use of the cryptic TTGTAG was readily detectable and the expression pattern of the resulting (albeit slightly longer) SL1-LCE was similar to that of the normal SL1-LCE in wild-type animals (Fig. S4A). Furthermore, the use of this cryptic TTGTAG SA was dependent upon lin-28 (Fig. S4A), which indicates that LIN-28 can promote LCE-proximal trans-splicing regardless of the acceptor sequence.
With the canonical TTTCAG deleted, northern blots showed a decrease in SL1-LCE levels and an increase in pri-let-7 levels (Fig. 3B), which indicates that SL1-LCE is trans-spliced from pri-let-7, and the non-canonical TTGTAG SL1 acceptor sequence is not as efficient in this context as is the wild-type TTTCAG sequence. Moreover, because this cryptic TTGTAG sequence is upstream to the canonical TTTCAG, all of the corresponding SL1-LCE bands were shifted up, which strengthened our interpretation that all northern blot bands that are associated with this transcript are SL1 trans-spliced (Fig. 3B).
Fig. 3.
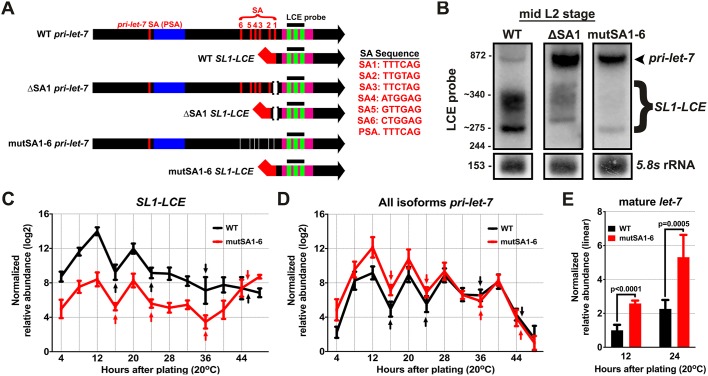
Larvae with SL1-LCE SA site mutations display elevated levels of pri-let-7 and mature let-7. (A) Locations of the NTNNAG motif SL1 SA in the let-7 locus and the SA mutations used in these experiments. PSA refers to the SA sequence in pri-let-7 upstream of the pre-let-7 stem-loop. SA1 is the position of trans-splicing that generates SL1-LCE in the wild type. SA2-SA6 are non-canonical SA sequences, some of which can be utilized when SA1 is mutated. (B) Total RNA from mid-L2 (20 h after plating) WT, SA1-deletion and mutSA1-6 animals, analyzed using northern blotting, hybridized with a probe for the LCE. Numbers mark known RNA sizes in nt: 872 nt is in vitro transcribed pri-let-7-B, 244 nt is in vitro transcribed SL1-LCE and 153 nt is 5.8s rRNA. Numbers with a tilde before them were estimated from known RNA sizes. (C) qRT-PCR analysis of the levels of SL1-LCE in samples of total RNA from staged populations of synchronously developing WT and mutSA1-6 animals. n=3 biological replicates. Note: WT SL1-LCE data are repeated from Fig. 2A as these experiments were performed together and used for comparison purposes. (D) qRT-PCR analysis of the levels of all pri-let-7 isoforms in samples of total RNA from staged populations of synchronously developing WT and mutSA1-6 animals. n=3 biological replicates. Note: WT pri-let-7 data are repeated from Fig. 2B as these experiments were performed together and used for comparison purposes. (E) FirePlex miRNA analysis of let-7 levels in samples of total RNA from staged populations of synchronously developing WT and mutSA1-6 animals at the mid-L1 (12 h after plating) and late-L2 (24 h after plating) stages. n=6 biological replicates. Statistical significance was determined using a two-tailed Student's t-test. Data are mean±s.d. Arrows mark the times of larval molts.
Based on previous genomic analysis of SL1 splice sites, the consensus SL1 acceptor sequence is TTTCAG. Other acceptor sequences can be utilized, but certain nucleotides appear to remain invariant, namely T in the second position, A in the fifth position and G in the sixth position (Graber et al., 2007). There are six occurrences of the corresponding NTNNAG consensus sequence in the region between the let-7 stem-loop and the LCE (Fig. 3A). With the aim of eliminating trans-splicing altogether in this region, we made small deletions in all six SAs using CRISPR/Cas9. Non-qRT-PCR and sequence analysis of LCE transcripts from the sixfold SA mutant (mutSA1-6) revealed that some basal level of trans-splicing still occurred, now using two far-non-canonical acceptor sequences, TTTCGG and TTCGGG, which are 1 nt apart from each other and near the original splice site location (data not shown). Compared with wild type, qRT-PCR and northern blotting of mutSA1-6 showed a reduction of approximately 20-fold in SL1-LCE level, as well as an increase of approximately fivefold in pri-let-7 level in the L1 and L2 stages. (Fig. 3C,D). Interestingly, mutSA1-6 animals had no apparent heterochronic phenotype, which suggests that trans-splicing the SL1-LCE from pri-let-7 is not crucial for normal development under standard laboratory conditions.
SL1-LCE trans-splicing regulates let-7 processing
In C. elegans, the 5′ region of an RNA that is removed by SL1 trans-splicing is called the outron. The outrons of trans-spliced RNAs are rarely detected, which suggests that they are rapidly degraded following trans-splicing (Morton and Blumenthal, 2011). Previously published northern blots of pri-let-7 suggest that the outron is below detectable levels (Bracht et al., 2004; Van Wynsberghe et al., 2011, 2014; Zisoulis et al., 2012). The resolution of our northern blots could not definitively show whether the outron was detectable, so we employed qRT-PCR using RT primers positioned 5′ or 3′ of the SL1-LCE SA, and PCR primer pairs that flanked the SL1-LCE SA. cDNA primed from sequences 5′ of the SA would represent both the pre-spliced pri-let-7 as well as the post-spliced outron, whereas cDNA primed from sequences 3′ of the SA would represent only pre-spliced pri-let-7. Therefore, if the outron were present at detectable levels, cDNA from sequences 5′ of the SA would be more abundant than cDNA from sequences 3′ of the SA. When we performed qRT-PCR to pri-let-7, we observed no difference in the yields of cDNA from sequences 5′ versus 3′ of the SA. This indicates that the outron is undetectable by this assay, and hence relatively unstable compared with unspliced pri-let-7 (Fig. S4B).
If trans-splicing of SL1-LCE produces an unstable outron that contains unprocessed let-7 microRNA, we hypothesized that trans-splicing of SL1-LCE from pri-let-7 could have a net negative effect on the accumulation of mature let-7 microRNA. In L1 and L2 animals, when trans-splicing is normally abundant, let-7 microRNA levels in mutSA1-6 animals were approximately twice as high as those seen in the wild type, which indicates that trans-splicing of SL1-LCE negatively impacts the accumulation of mature let-7 (Fig. 3E, Fig. S4C). Interestingly, at later developmental stages when SL1-LCE trans-splicing is not prevalent, let-7 levels were reduced by ∼60% to 70%, which suggests an additional positive regulatory role for sequences that overlap one or more of the SA elements that are mutated in mutSA1-6 (Fig. S4D).
The LCE functions to negatively regulate the let-7 family
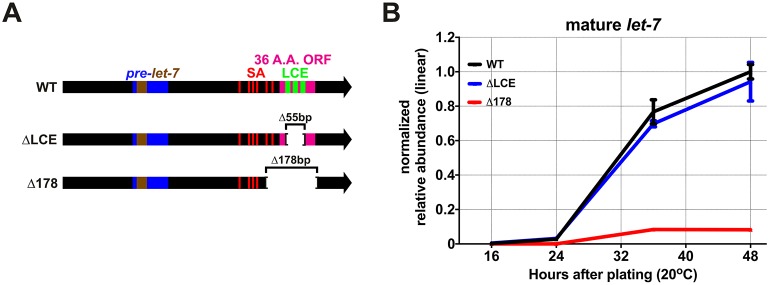
A previous study reported experiments that suggested the LCE region in pri-let-7, in conjunction with ALG-1, could function in cis to facilitate let-7 biogenesis (Zisoulis et al., 2012). In that study, a transgene that carried a modified let-7 locus with a deletion of 178 bp (which removed the LCE and surrounding sequences) expressed decreased levels of mature let-7, which suggested that the LCE, perhaps when bound to the let-7 RNA-induced silencing complex (miRISC), could function to promote microprocessing of pri-let-7. However, because this 178 bp deletion also removed sequences that are upstream of the LCE, including the SL1-acceptor sequence, we used CRISPR/Cas9 to create a 55 bp deletion at the endogenous let-7 locus that removed only the LCE (Fig. 4A). This 55 bp deletion of the LCE did not result in a measurable change in let-7 levels (Fig. 4B). We also used CRISPR/Cas9 to introduce the previously described 178 bp deletion at the endogenous let-7 locus (Fig. 4A), and confirmed the previous (Zisoulis et al., 2012) results: an ∼tenfold reduction in let-7 levels (Fig. 4B). Based on these results, we conclude that the larger 178 bp deletion removes unknown non-LCE positive regulatory elements, and that the LCE sequences themselves do not exert a detectable positive effect on let-7 biogenesis.
Fig. 4.
LCE sequences are dispensable for normal expression of mature let-7 microRNA. (A) Positions of the deletions in the let-7 mutants used in these experiments. (B) FirePlex miRNA analysis of let-7 levels in WT and LCE-deletion (ΔLCE) animals, and in animals containing a 178 bp deletion (Δ178) that has been previously reported to reduce let-7 biogenesis throughout development. Data are mean±s.d. n=3 biological replicates.
Because SL1-LCE contains sequences that are complementary to let-7fam, we hypothesized that SL1-LCE could function as a sponge to negatively regulate let-7fam. To test this, we sought to determine whether mutations that disrupt SL1-LCE could genetically interact with let-7fam in sensitized genetic backgrounds. Of the four major let-7 family genes, only let-7(lf) or mir-48(0) mutants display overt heterochronic phenotypes in normal laboratory conditions. Loss of either mir-48 or let-7 results in retarded hypodermal development and an extra larval molt. In addition, let-7(lf) hermaphrodites burst through an improperly formed vulva (an adult lethality phenotype), and mir-48(0) adult hermaphrodites die because of egg retention, presumably because of their retarded hypodermal development. We hypothesized that if SL1-LCE were to function as a sponge for the let-7fam microRNAs, loss of the LCE sequences would result in increased let-7fam activity, which could be evidenced by suppression of let-7(lf) or mir-48(0) phenotypes.
Col-19::GFP is a reporter that is expressed in hypodermal seam cells and in the hypodermal syncytium (hyp-7) beginning at the L4 molt. In molting L4 animals with either the strong let-7(lf) allele [let-7(mn112)] or the mir-48(0) null allele [mir-48(n4097)], Col-19::GFP expression in hyp-7 is reduced by ∼20-fold compared with the wild type, and is limited to the seam cells. Moreover, let-7(mn112) animals burst through their vulvas as young adults whereas mir-48(n4097) and the weaker let-7(mg279) allele do not burst, but survive to undergo an extra larval molt, and exhibit egg-laying defects and a reduced brood size. Using CRISPR/Cas9, we introduced a deletion of the let-7 hairpin into the LCE deletion background and observed no suppression of the strong let-7(lf) phenotypes (data not shown). Therefore, we hypothesized that the phenotypes that are associated with this substantial reduction of let-7 are too strong to be suppressed by loss of the LCE. To test whether deletion of the LCE could suppress a partial loss of function of let-7 we used CRISPR/Cas9 to delete the LCE of the hypomorphic allele let-7(mg279). The resulting let-7(mg279 ΔLCE) strain exhibited no apparent suppression of the let-7(mg279) retarded phenotypes as measured by the expression pattern of Col-19::GFP, reduced brood size, extra molting phenotype, or adult mortality (Fig. 5B, Fig. S5). Therefore, absence of the LCE did not display genetic interaction with loss of let-7.
Fig. 5.
Inhibition of SL1-LCE function, either by deletion of the LCE, or by mutations of LCE-proximal trans-splicing acceptor sequences, suppresses the retarded hypodermal phenotypes of mir-48(0) animals. (A) Positions of the let-7 LCE mutations (green), SA mutants (red) and LCE transgene used in these experiments. (B) Quantification of the Col-19::GFP phenotypes observed for each genotype in molting late L4 animals (left). From top to bottom: n=9 (WT), 10 (ΔLCE), 23 [let-7(mg279)], 12 [let-7(mg279 ΔLCE)], 9 [mir-48(0)], 22 [ΔLCE; mir-48(0)], 26 [ΔLCE; mir-48(0); LCE transgene], 18 [mir-48(0) mir-241(0)], 15 [ΔLCE; mir-48(0) mir-241(0)], 10 (mutLCE), 15 [mutLCE; mir-48(0)], 10 (mutSA1-6), 10 [mutSA1-6; mir-48(0)], 11 [ΔLCE/+; mir-48(0)] and 16 [mutSA1-6/ΔLCE; mir-48(0)] animals. Images (right) are of a representative molting L4 animal for each genotype.
By contrast, a strong genetic interaction was evident between LCE deletion and mir-48(0). When the LCE deletion mutation was crossed into mir-48(0), Col-19::GFP expression was restored to a wild-type pattern (Fig. 5B). This suggests that removal of the LCE sequences results in upregulation of the activity of one or more members of let-7fam. Consistent with this supposition, when a second family member, mir-241, was also removed, loss of the LCE failed to restore the normal timing of Col-19::GFP expression (Fig. 5B). Deletion of the LCE similarly suppressed the extra molting phenotype, restored survival and restored the brood size of mir-48(0), but only partially suppressed mir-48(0) mir-241(0) double null phenotype (Fig. S5). To determine whether this suppression of mir-48(0) was due to an elevation in let-7fam levels, we measured let-7fam levels in wild-type and LCE deletion animals and observed no difference (Fig. S6). Together, these results indicated that the LCE negatively regulates let-7fam by modulating their activity rather than their levels.
The LCE sequence is predicted to contain a 36 amino acid ORF that is poorly conserved in other Caenorhabditis species (Fig. S1A) and it is therefore unlikely to perform a conserved function. Nevertheless, our 55 bp deletion of the LCE disrupts this putative ORF so it was possible that disruption of the ORF could confound the interpretation of our results. Therefore, we used CRISPR/Cas9 to mutate the three LCSs without altering the amino acid sequence of the ORF. Similar to the LCE deletion, these ‘silent’ LCS mutations did not alter let-7 levels (data not shown), but did restore normal Col-19::GFP expression, normal survival, normal brood size and suppressed the extra molting phenotype in mir-48(0) animals but not in mir-48(0), mir-241(0) double null animals (Fig. 5B, Fig. S5).
Trans-splicing of the LCE is necessary to negatively regulate let-7fam microRNAs
To function as a negative regulator of let-7fam activity by acting as a microRNA sponge, the SL1-LCE transcript would presumably encounter let-7fam miRISC in the cytoplasm. As mentioned above, SL1-LCE contains a putative 36 amino acid ORF. Previously published ribosome-profiling data indicated that ribosomes locate to the LCE sequence (Michel et al., 2014), which suggests that the LCE ORF is translated, and therefore enters the cytoplasm where it could engage let-7fam microRNAs. To confirm that the LCE ORF can be translated in vivo, we generated transgenic animals that carried a transgene with the C-terminus of the LCE ORF fused to GFP and observed fluorescence in cell types that were previously reported to express let-7fam, including head and tail neurons, sensory neurons, ventral and dorsal nerve cords, pharynx, intestine, hypodermis and vulva (Johnson et al., 2003; Kai et al., 2013; Martinez et al., 2008; McCulloch and Rougvie, 2014; Zou et al., 2013; Hayes et al., 2006). GFP expression appeared to be most constant and brightest in neurons, and most dynamic and dimmest in the remaining cell types. Overall, GFP expression recapitulated the temporal expression of SL1-LCE; however, some neurons retained bright GFP expression well into adulthood (Fig. S7).
In addition to being cytoplasmic, to function as a sponge the SL1-LCE transcript should be expressed at levels in molar excess of let-7fam microRNAs. To determine the stoichiometric ratio of SL1-LCE to let-7fam in wild-type larvae, we quantified the amount of SL1-LCE and let-7fam microRNAs in RNA samples from synchronized populations of developing larvae. We calibrated these assays using known amounts of in vitro-transcribed SL1-LCE and pri-let-7, and synthetic let-7fam microRNA oligonucleotides. The results of these quantitative assays indicated that, in whole animals, the SL1-LCE is in molar excess of let-7, mir-48, mir-84 and mir-241 during the L1 and L2 stages (Fig. 6A). Importantly, pri-let-7 levels were not in excess of let-7fam, indicating that pri-let-7, despite containing LCE sequences, is not likely to contribute as significantly to let-7fam sponging as does the SL1-LCE transcript.
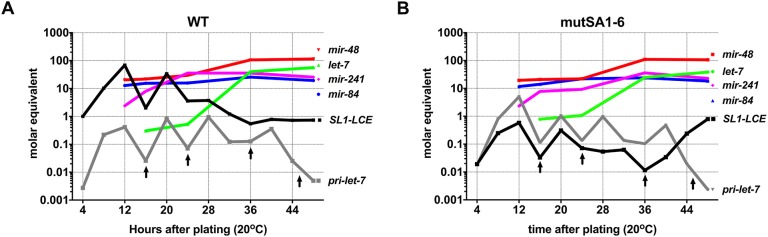
Fig. 6.
The SL1-LCE is in molar excess relative to let-7fam microRNAs during early larval stages. (A,B) Relative levels (molar equivalents, normalized to in vitro transcribed pri-let-7 and synthetic let-7fam) of pri-let-7, SL1-LCE, mir-48, mir-84, mir-241 and let-7, determined using calibrated qRT-PCR (for pri-let-7 and SL1-LCE) or calibrated Fireplex assay (for microRNAs), in samples of total RNA from staged populations of synchronously developing WT (A) and mutSA1-6 (B) animals. n=3 biological replicates. Arrows mark the times of larval molts.
Based on its relative abundance and cytoplasmic location, our results suggest that of the two classes of LCE-containing transcripts that are produced from the let-7 locus (pri-let-7 and SL1-LCE), SL1-LCE is more likely to function as a sponge for let-7fam microRNA. This indicates that the sponging activity of the LCE would depend on trans-splicing of SL1-LCE. To test this supposition, we took advantage of the splice site (mutSA1-6) mutant animals, which have reduced SL1-LCE levels. Using the same calibrated quantitation as above, we determined that the reduced SL1-LCE level in whole mutSA1-6 animals was less than that of let-7fam (Fig. 6B). Moreover, the elevation in pri-let-7 in the mutSA1-6 was not sufficient to put it in excess of all let-7fam (Fig. 6B), although we note that in mutSA1-6, pri-let-7 was in excess of mir-241 and let-7 at the L1 peak and approximately equimolar with let-7 at the L2 peak (Fig. 6B).
We hypothesized that, similar to deletion of the LCE sequences, the reduction in SL1-LCE levels in mutSA1-6 would increase the activity of let-7fam and suppress mir-48(0) phenotypes. However, when mutSA1-6 and mir-48(0) were combined, we observed no suppression (Fig. 5B, Fig. S5). This suggested that, although mutSA1-6 causes a significant decrease in SL1-LCE, the remaining LCE-containing transcripts could nevertheless be functional. Therefore, we aimed to reduce the amount of remaining SL1-LCE by half using mutSA1-6/ΔLCE trans-heterozygous animals. This further reduction in SL1-LCE resulted in suppression of all the heterochronic phenotypes that are associated with mir-48(0), which supports the conclusion that SL1-LCE functions to negatively regulate let-7 family activity (Fig. 5B, Fig. S5).
Because SL1-LCE levels in mutSA1-6 whole-animal RNA extracts were significantly lower than those of let-7fam, we were concerned that the suppression that was observed in the mutSA1-6/ΔLCE trans-heterozygous animals was due to the genetic background of ΔLCE. We therefore examined ΔLCE/+ animals and failed to observe any suppression of mir-48(0) heterochronic phenotypes, which indicates that suppression of mir-48(0) was due to the mutSA1-6/ΔLCE allelic configuration (Fig. 5B, Fig. S5). To further control for a potential genetic background origin for the suppression of mir-48(0) in mutSA1-6/ΔLCE animals, we sought to express transgenically the LCE transcript in mir-48(0) animals that carried the LCE deletion. If the suppression of mir-48(0) were due to bona fide loss of the LCE transcript, then we would expect that restoring expression of the LCE transcript from a transgene should eliminate the suppression. Accordingly, we generated a let-7 extrachromosomal transgene that lacked the mature let-7 sequence but still contained the LCE (Fig. 5A). When expressed in animals that lacked both mir-48 and their endogenous LCE we observed restoration of the heterochronic phenotypes that are associated with mir-48(0). Put together, these results indicate that the suppression of the mir-48(0) phenotypes in ΔLCE homozygotes and ΔLCE/mutSA1-6 trans-heterozygotes is due to a loss or decrease in SL1-LCE levels.
DISCUSSION
A general property of microRNAs across diverse organisms is that they are first transcribed as longer primary transcripts, which are then enzymatically processed to produce the mature 22 nt microRNA. The multiple biogenesis steps required to generate a mature microRNA provide access for a range of transcriptional and post-transcriptional regulation. For example in C. elegans, HBL-1 and LIN-42 can modulate the transcriptional activity of microRNAs including let-7 and lin-4 (Roush and Slack, 2009; McCulloch and Rougvie, 2014; Perales et al., 2014; Van Wynsberghe et al., 2014), and LIN-28 post-transcriptionally regulates let-7 (Van Wynsberghe et al., 2011). The negative regulation of let-7 by LIN-28 is evolutionarily conserved. In mammals, it has been shown that LIN28 can bind to the stem-loop of pri-let-7 and/or pre-let-7 to directly inhibit processing by Drosha and/or Dicer (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008; Heo et al., 2009; Loughlin et al., 2012; Nam et al., 2011; Piskounova et al., 2011). In C. elegans, LIN-28 also appears to inhibit processing of pri-let-7 by Drosha, although apparently not by binding the let-7 stem loop, but rather through binding to sequences ∼170 nt downstream (Stefani et al., 2015). In addition, in both C. elegans and mammals, the 3′ UTR of lin-28 contains sequences that are complementary to the let-7fam, which indicates that let-7fam can repress LIN-28 expression (Reinhart et al., 2000; Rybak et al., 2008). Thus, lin-28 engages in an evolutionarily conserved reciprocal negative feedback with let-7fam microRNAs (Fig. 7). However, the functional significance of these lin-28–let-7fam regulatory interactions, and their precise mechanisms, are not fully understood.
Fig. 7.
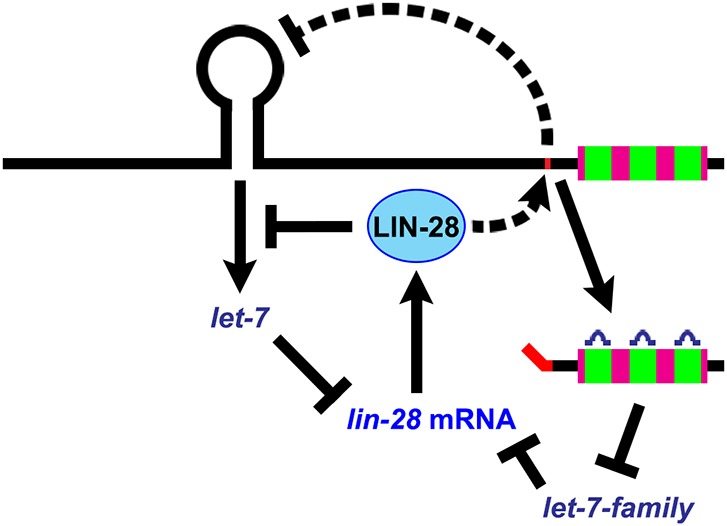
A model of how LIN-28 and let-7fam microRNAs function in a reciprocal negative regulatory network. A summary of the genetic pathway determined by our results in which lin-28 and let-7fam post-transcriptionally exert reciprocal negative feedback on each other.
In this study, we identified a previously undescribed RNA, SL1-LCE, which is trans-spliced from C. elegans pri-let-7 downstream of the pre-let-7 stem-loop, and which contains LCSs followed by 3′ poly-A. We determined that SL1-LCE is highly expressed in the early larval stages, displaying an inverse expression pattern compared with let-7, which suggests that it could be associated with negative regulation of let-7 biogenesis. We find that expression of SL1-LCE coincides with the expression of LIN-28, and is dependent on lin-28 function, which reveals a novel regulatory circuit in which LIN-28 governs trans-splicing of pri-let-7 to negatively impact let-7 microprocessing (Fig. 7). The regulation of SL1-LCE trans-splicing by lin-28 appears to be independent of other phenotypes that are controlled by lin-28, as another precocious mutant lin-14(lf) had no effect on trans-splicing, and SL1-LCE levels were low in lin-28(0);lin-46(lf), in which precocious phenotypes are suppressed.
Interestingly, when we mutated the SL1-LCE trans-SA sequence, even in combination with mutations of nearby putative SAs, trans-splicing persisted using far non-canonical acceptor sequences. This suggests the presence of sequences in pri-let-7 with potent splicing enhancer activity. Although the use of a far non-canonical sequence is unusual, it is not unprecedented in C. elegans (Aroian et al., 1993).
Our findings suggest that LIN-28 inhibits let-7 biogenesis through the combined effects of two mechanisms. On the one hand, LIN-28 binds directly to pri-let-7 to inhibit processing by Drosha/Pasha (Stefani et al., 2015); on the other hand, LIN-28 also promotes SL1-LCE trans-splicing, which results in downregulation of pri-let-7 levels (Fig. 7). LIN-28 could be either directly regulating LCE trans-splicing through its binding to pri-let-7, or indirectly through other means such as regulating splicing components. In fact, in mammalian cells, LIN28 has been shown to indirectly affect alternative splicing by regulating the expression of certain splicing factors (Wilbert et al., 2012).
lin-28(0) animals exhibit greater than 100-fold elevation of let-7 in the L1 and L2 stages, presumably as a result of a release of repression from both trans-splicing and inhibited biogenesis. When we reduced LCE trans-splicing by mutating SA1-6 in a wild-type lin-28 background, mature let-7 was not de-repressed as much as in lin-28(0); although we observed a marked elevation in pri-let-7, which is consistent with a role for LCE trans-splicing in destabilizing pri-let-7, the attendant elevation of mature let-7 was much more modest (only ∼twofold), which apparently reflects a potent inhibition by LIN-28 of pri-let-7 Drosha/Pasha processing.
Interestingly, although we observed an elevation of mature let-7 levels in lin-28(0) larvae that exceeded 100-fold, we observed no corresponding decrease in pri-let-7. Apparently, in the absence of LIN-28, the destabilization of pri-let-7 levels due to increased Drosha/Pasha processing is balanced by a commensurate stabilization of pri-let-7 due to reduced SL1-LCE trans-splicing. These observations further suggest that the level of pri-let-7 in wild-type larvae could be subject to homeostatic regulation.
In mammals, trans-splicing is relatively rare compared with in nematodes (Lei et al., 2016). However, most human microRNA genes, including ten of the 12 genes that encode let-7fam members, are located within introns of mRNAs or non-coding RNAs (Rodriguez et al., 2004; Kim and Kim, 2007). Therefore, it is possible that the spliceosomal machinery could contribute to the regulation of microRNA biogenesis in contexts other than C. elegans let-7. In fact, interplay between microRNA microprocessing and splicing has previously been observed. For example, the microRNA processing machinery Drosha/DGCR8, as well as microRNA primary transcripts, have been observed to be associated with supraspliceosomes, and inhibition of splicing can result in the elevation of microRNAs, including let-7 (Agranat-Tamir et al., 2014). On the other hand, there is evidence of situations in which splicing and the biogenesis of intronic microRNAs can co-occur without apparently influencing each other (Kim and Kim, 2007), which suggests that connectivity between microRNA processing and host gene splicing is likely to be subject to regulation, depending on context and circumstances.
A previous study demonstrated that the C. elegans microRNA Argonaute ALG-1 could bind in vivo to the let-7 locus LCE suggesting that let-7 miRISC could associate with pri-let-7 in the nucleus and regulate let-7 biogenesis (Zisoulis et al., 2012). In support of this idea, it was found that a transgene that contained a mutant let-7 locus that deleted 178 bp spanning the LCE displayed a marked decrease in let-7 biogenesis compared with wild type. When we generated the same 178 bp deletion in the endogenous let-7 locus using genome editing, we also observed that the deletion resulted in decreased let-7 expression, which confirmed that positive regulatory elements are contained in the 178 bp region. However, when we removed only the LCE at the endogenous locus, we observed no difference in let-7 levels, which indicates that the putative positive elements that are contained within the 178 bp deleted region are located outside of the LCE, and that the LCE itself does not exert a detectable positive role in let-7 biogenesis. Our finding that LCE sequences are contained in a cytoplasmic mRNA that is produced by trans-splicing from pri-let-7 has suggested that the LCE likely functions primarily by associating with let-7fam microRNAs in the cytoplasm. However, we cannot rule out the possibility that LCE sequences could also interact with miRISC in the nucleus.
Our results show that LIN-28-dependent trans-splicing of the C. elegans let-7 primary transcript can act in cis to negatively influence let-7 biogenesis, and at the same time produce a trans-acting inhibitory RNA, SL1-LCE, which negatively regulates the activity of let-7fam microRNAs. We propose that SL1-LCE functions as a sponge for let-7fam microRNAs through base pairing to the let-7fam seed sequence. We observed that loss of the LCE suppresses mir-48(0) presumably by boosting the activity of the remaining let-7-family microRNAs. However, loss of the LCE failed to suppress let-7(lf) phenotypes, even in the case of a weak let-7(lf) mutation, mg279. This indicates that perhaps the particular let-7-family microRNA(s) that are hypothetically elevated in activity by loss of the SL1-LCE can substitute for mir-48 but not let-7.
Interestingly, animals with deletions of the LCE or SAs displayed no overt phenotypes except in the sensitized background of mir-48(0). This suggests that regulation of SL1-LCE production is not crucial for normal development under standard laboratory conditions, but may function to modulate let-7 biogenesis and let-7fam activity in the context of ensuring robust developmental timing under stressful physiological or environmental conditions.
Analysis of RNA that was extracted from whole animals with deletions in the LCE's SA sequences indicated that the SL1-LCE was no longer in molar excess compared with let-7fam. Based on this, we would have predicted these animals to exhibit suppressed mir-48(0) phenotypes, but suppression was not evident unless we further reduced SL1-LCE levels by removing one copy of the LCE. This suggests that the change in the molar ratio of SL1-LCE to let-7fam in homozygous mutSA1-6 animals compared with wild type may not be sufficient in specific cell types to suppress the heterochronic phenotypes of mir-48(0). Unfortunately, we do not know which SL1-LCE-expressing cell types contribute to let-7fam regulation, nor do we know the stoichiometry of SL1-LCE and let-7fam within the relevant cells. Furthermore, we note that mutSA1-6 animals displayed reduced levels of let-7 at later larval stages, which could confound the detection of any suppression of mir-48(0). Finally, we cannot also exclude the possibility that the SL1-LCE may directly or indirectly regulate the let-7fam in a non-molar-equivalent manner.
Employing an LCE-ORF::GFP transgenic reporter, we observed expression of the SL1-LCE in a variety of cell types throughout development. All of the cell types in which we observed expression have previously been reported to express members of the let-7fam. Although we observed weak GFP expression in cell types that are associated with the heterochronic phenotypes of let-7fam mutants, namely the hypodermis and vulva, we observed the strongest expression in neurons. This suggests that SL1-LCE-mediated regulation of let-7fam activity could have a neuronal component. Interestingly, all let-7fam members are expressed in neurons throughout development but their roles in these cells are not well understood. One of the hallmarks of let-7fam mutants is the expression of a supernumerary larval molt and, interestingly, neuroendocrine signaling has been shown to regulate molting in arthropods and is also thought to regulate molting in nematodes. Indeed, mutations in the C. elegans neuronal-expressed gene pqn-47 (myrf-1) result in the reiteration of a larval molt, which demonstrates a link between neuronal signaling and the heterochronic pathway (Frand et al., 2005; Russel et al., 2011).
MATERIALS AND METHODS
Nematode methods and phenotypic analysis
C. elegans were cultured on nematode growth medium (NGM) (Brenner, 1974) and fed with E. coli HB101. Synchronized populations of developmentally staged worms were obtained using standard methods (Stiernagle, 2006). Unless otherwise noted, all experiments were performed at 20°C. A list of strains used in this study is in Table S3.
For heterochronic phenotype analysis, early L4 animals were picked from healthy uncrowded cultures, placed onto individual plates seeded with HB101 and observed periodically until the end of the experiment. Fluorescence microscopy was used to score Col-19::GFP expression.
Sequence alignments and target prediction
DNA and ORF alignments were performed using Clustal Omega (www.ebi.ac.uk). let-7fam:LCE target predictions were performed using RNAhybrid (https://bibiserv2.cebitec.uni-bielefeld.de).
Brood size counts
Young adult hermaphrodites were placed individually on plates seeded with HB101 and each animal was transferred daily to a fresh plate. The number of progeny produced on each plate was assessed until the animal stopped producing progeny.
RNA extraction
A population of animals was collected and flash-frozen in liquid nitrogen, and total RNA was extracted using Qiazol reagen (Qiagen) as described by McJunkin and Ambros (2017).
Northern blotting
Northern blotting was adapted from Lee and Ambros (2001). RNA samples were run on 5% urea-PAGE gels and then transferred to GeneScreen Plus Hybridization membranes (PerkinElmer) using electrophoresis. After transfer, the membranes were crosslinked with 120 mjoules of UV (wavelength of 254 nm) and baked at 80°C for 1 h. Oligonucleotide probes (Table S2) were labeled using the Integrated DNA Technologies Starfire Oligos Kit with alpha-32P ATP and hybridized to the membranes at 37°C in 7% SDS, 0.2 M Na2PO4 (pH 7.0) overnight. Membranes were washed at 37°C, twice with 2× SSPE, 0.1% SDS and twice with 0.5× SSPE, 0.1% SDS. The blots were exposed on a phosphorimager screen and imaged with a Typhoon FLA7000 (GE).
Non-qRT-PCR
Samples of total RNA were pre-treated with turbo DNase (Invitrogen) (Pinto and Lindblad, 2010). cDNA was synthesized using SuperScript IV (Invitrogen) following the manufacturer's instructions, using the RT oligonucleotides ‘let-7 RT’ or ‘oligo (dT)’. PCR was then performed using 2× PCR PreMix (Sydlabs) with the primers SL1 F and LCE R for SL1-pri-let-7, SL1-LCE and cryptic SA identifications; SL1-LCE F and LCE R for oligo (dt)-based SL1-LCE identification; and SL1-pri-let-7 F and pri-let-7 R for oligo (dt)-based SL1-pri-let-7 identification following the manufacturer’s instructions using 1 μl of cDNA and an annealing temperature of 55°C. The products were then analyzed using electrophoresis on a 2% agarose gel, imaged, cut out and gel purified using the EZ-10 Spin Column DNA Gel Extraction Minipreps Kit (Bio Basic Canada), TA cloned using the TA Cloning Kit with pCR2.1 Vector (Invitrogen) and subjected to Sanger sequencing using the M13 reverse primer.
Quantitative PCR
cDNA was synthesized as described above using the RT oligonucleotides ‘let-7 RT’ and ‘gpd-1 QPCR R’ (for full-length let-7 locus transcripts) or ‘pri-let-7 R’ and ‘gpd-1 QPCR R’ (for outron detection). qPCR reactions were performed using Qiagen QuantiFast SYBR Green PCR kit following the manufacturer's instructions, using an ABI 7900HT Real Time PCR System (Applied Biosystems). With the exception of the experiments we have reported in Fig. 6, ΔCTs were calculated by normalizing samples to gpd-1 (GAPDH). ΔCTs were then inverted so that greater values reflect greater RNA levels, and were normalized to set the value of the least abundant sample to one. For each biological replicate, the average of three technical replicates was used.
5′ RACE
5′ RACE was adapted from Pinto and Lindblad (2010) and Turchinovich et al. (2014). Samples of total RNA from late L2 animals were pre-treated with Turbo DNase (Invitrogen) (Pinto and Lindblad, 2010). Then 1.6 μl of the RNA (in H2O) was combined with 0.5 μl of 10 μM let-7 RT oligo and 0.4 μl of 25 mM dNTPs. This mixture was incubated at 65°C for 10 min, chilled on ice, and then combined with 1.6 μl 25 mM MgCl2, 0.6 μl 100 mM MnCl2, 4 μl 5× First-Strand Buffer (Invitrogen), 2 μl 0.1 M dithiothreitol, 0.3 μl Ribolock (ThermoFisher) and 8 μl H2O. This mixture was incubated at 42°C for 2 min, then 1.0 μl of SuperScript II (Invitrogen) was added and the mixture was incubated at 42°C for 30 min. Next, 2.0 μl of 5′ RACE template switching oligonucleotide (10 μM) was added and the incubation was continued at 42°C for an additional 60 min. The reaction was heat inactivated at 70°C for 15 min, then diluted 1:10 and used for a standard PCR with the primers Rd1 SP and pri-let-7 R for pri-let-7, and Rd1 SP and LCE R for SL1-LCE. PCR products were TA-cloned (Invitrogen) and subjected to Sanger sequencing.
Quantitative microRNA detection
microRNAs were quantified from total RNA using FirePlex miRNA assay (Abcam) following the manufacturer's instructions. Guava easyCyte 8HT (Millipore) was used for analysis. With the exception of the Calibrated RNA quantitation experiments (below), signals (arbitrary units) were normalized using geNorm (Vandesompele et al., 2002).
Calibrated RNA quantitation
To generate T7 templates for the production of RNA standards that correspond to pri-let-7 and SL1-LCE, the corresponding genomic sequences were PCR amplified from genomic DNA using the oligonucleotides T7 pri-let-7 F and let-7 RT, and T7 SL1-LCE F and let-7 RT, respectively. T7 pri-let-7 added the T7 promoter to the pri-let-7 PCR product, and T7 SL1-LCE added the T7 and SL1 sequences to the SL1-LCE PCR product. RNA from the respective PCR products was in vitro transcribed (IVT) using the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs) following the manufacturer's instructions, and column purified. RNA concentration and quality was measured using an Advanced Analytics Fragment Analyzer. Known amounts of the IVT RNA were then serially diluted. cDNAs from the IVT serial dilutions, and from biological samples, were synthesized as described above and subjected to qPCR. Equal amounts of total RNA were used for each biological sample, and the amounts of pri-let-7 and SL1-LCE in each biological sample were calculated from the standard curve that was generated from the IVT dilutions.
Synthetic oligonucleotides of let-7, mir-48, mir-84 and mir-241 were ordered from Integrated DNA Technologies. Known amounts of these RNA oligonucleotides were serially diluted and subjected to FirePlex miRNA analysis, along with biological samples. Equal amounts of total RNA were used for each biological sample, and the amount of each microRNA in each biological sample were calculated from the standard curve that was generated from the synthetic microRNA dilutions.
Microscopy
Epifluorescence images were obtained using a Zeiss Imager.Z1 with a 10× objective.
Targeted genome editing by CRISPR/Cas9
Mutants were generated using CRISPR/Cas9 methods adapted from Paix et al. (2014, 2015). The germlines of young adult hermaphrodites were injected with a mix of CRISPR RNA (crRNA) that targeted the region of interest in the let-7 locus and the ‘co-CRISPR’ marker dpy-10, trans-activating crRNA, a single-stranded oligonucleotide homologous recombination template, Cas9 protein that was prepared as described in Paix et al. (2015) and water. F1 animals that exhibited the co-CRISPR phenotype were picked, allowed to lay eggs and then genotyped using PCR. F2s were cloned from F1s that scored positively by PCR genotyping for the desired let-7 locus modification. Homozygous F2s were then selected by PCR genotyping and subjected to Sanger sequencing for validation. All mutants were backcrossed to wild type at least thrice. See Table S1 for a list of alleles that were generated for this study along with the crRNAs used to generate them. Some crRNAs were generated using IVT (see below). crRNAs that were generated by IVT are noted in their names.
In vitro transcription of crRNAs
To produce templates for the production of crRNAs by T7 in vitro transcription, equal amounts of two DNA oligonucleotides (100 μM) were mixed together: The sequence of the first oligonucleotide was the reverse compliment to the crRNA of interest followed by the reverse compliment of the T7 promoter; the second oligonucleotide (oCN183) was complementary to the T7 promoter sequence of the first oligonucleotide. The oligonucleotide mixture annealed by rapidly heating to 95°C followed by cooling to 15°C over 10 min. Then 1.0 μl of the annealed oligonucleotide mixture was added to an IVT reaction, and transcription was carried out using the HiScribe T7 high Yield RNA Synthesis Kit following the manufacturer's instructions (New England Biolabs). The RNA product was then column purified.
Transgenic constructs
The pCN30 construct, which contains the let-7 locus ORF tagged with GFP on its C-terminus, was constructed by cloning GFP into the let-7 genomic rescue plasmid pZR001 (Ren and Ambros, 2015). The pCN33 construct, which contains the let-7 locus minus the 22-nucleotide mature let-7 sequence, was constructed from the genomic rescue plasmid pZR001 (Ren and Ambros, 2015).
Supplementary Material
Acknowledgements
We thank the members of the Ambros and the Mello laboratories for helpful discussions and the sharing of resources, especially Takao Ishidate for help with the northern blotting.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.N., V.A.; Methodology: C.N., V.A.; Formal analysis: C.N., V.A.; Investigation: C.N.; Resources: V.A.; Data curation: C.N.; Writing - original draft: C.N.; Writing - review & editing: C.N., V.A.; Supervision: V.A.; Project administration: V.A.; Funding acquisition: C.N., V.A.
Funding
This work was supported in part by a Translational Cancer Biology Training Grant funded by the National Institutes of Health (T32CA130807-06A1), and by National Institutes of Health grants (R01GM34028 and 5R01GM104904). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.172031.supplemental
References
- Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P., Horvitz H. R. and Ambros V. (2005). The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, 403-414. 10.1016/j.devcel.2005.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranat-Tamir L., Shomron N., Sperling J. and Sperling R. (2014). Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 42, 4640-4651. 10.1093/nar/gkt1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. (1989). A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49-57. 10.1016/0092-8674(89)90171-2 [DOI] [PubMed] [Google Scholar]
- Ambros V. and Horvitz H. R. (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409-416. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Ambros V. and Ruvkun G. (2018). Recent molecular genetic explorations of Caenorhabditis elegans microRNAs. Genetics 209, 651-673. 10.1534/genetics.118.300291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian R. V., Levy A. D., Koga M., Ohshima Y., Kramer J. M. and Sternberg P. W. (1993). Splicing in Caenorhabditis elegans does not require an AG at the 3′ splice acceptor site. Mol. Cell. Biol. 13, 626-637. 10.1128/MCB.13.1.626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. (2012). Trans-splicing and operons in C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1-11. WormBook. 10.1895/wormbook.1.5.2 [DOI] [PMC free article] [PubMed]
- Bracht J., Hunter S., Eachus R., Weeks P. and Pasquinelli A. E. (2004). Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 10, 1586-1594. 10.1261/rna.7122604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R. and Sulston J. E. (1981). Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24, 59-69. 10.1016/0092-8674(81)90501-8 [DOI] [PubMed] [Google Scholar]
- Frand A. R., Russel S. and Ruvkun G. (2005). Functional genomic analysis of C. elegans molting. PLoS Biol. 3, e312 10.1371/journal.pbio.0030312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber J. H., Salisbury J., Hutchins L. N. and Blumenthal T. (2007). C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA 13, 1409-1426. 10.1261/rna.596707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G. D., Frand A. R. and Ruvkun G. (2006). The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133, 4631-4641. 10.1242/dev.02655 [DOI] [PubMed] [Google Scholar]
- He L. and Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522-531. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Heo I., Joo C., Cho J., Ha M., Han J. and Kim V. N. (2008). Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276-284. 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Heo I., Joo C., Kim Y.-K., Ha M., Yoon M.-J., Cho J., Yeom K.-H., Han J. and Kim V. N. (2009). TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696-708. 10.1016/j.cell.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Johnson S. M., Lin S. Y. and Slack F. J. (2003). The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 259, 364-379. 10.1016/S0012-1606(03)00202-1 [DOI] [PubMed] [Google Scholar]
- Kai Z. S., Finnegan E. F., Huang S. and Pasquinelli A. E. (2013). Multiple cis-elements and trans-acting factors regulate dynamic spatio-temporal transcription of let-7 in Caenorhabditis elegans. Dev. Biol. 374, 223-233. 10.1016/j.ydbio.2012.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-K. and Kim V. N. (2007). Processing of intronic microRNAs. EMBO J. 26, 775-783. 10.1038/sj.emboj.7601512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C. and Ambros V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862-864. 10.1126/science.1065329 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L. and Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lei Q., Li C., Zuo Z., Huang C., Cheng H. and Zhou R. (2016). Evolutionary insights into RNA trans-splicing in vertebrates. Genome Biol. Evol. 8, 562-577. 10.1093/gbe/evw025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin F. E., Gebert L. F. R., Towbin H., Brunschweiger A., Hall J. and Allain F. H.-T. (2012). Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 19, 84-89. 10.1038/nsmb.2202 [DOI] [PubMed] [Google Scholar]
- Martinez N. J., Ow M. C., Reece-Hoyes J. S., Barrasa M. I., Ambros V. R. and Walhout A. J. M. (2008). Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 18, 2005-2015. 10.1101/gr.083055.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch K. A. and Rougvie A. E. (2014). Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression. Proc. Natl. Acad. Sci. USA 111, 15450-15455. 10.1073/pnas.1414856111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K. and Ambros V. (2017). A microRNA family exerts maternal control on sex determination in C. elegans. Genes Dev. 31, 422-437. 10.1101/gad.290155.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely P. M. and Herman R. K. (1979). Lethals, steriles and deficiencies in a region of the X chromosome of Caenorhabditis elegans. Genetics 92, 99-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A. M., Fox G., Kiran A. M., De Bo C., O'connor P. B. F., Heaphy S. M., Mullan J. P. A., Donohue C. A., Higgins D. G. et al. (2014). GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 42, D859-D864. 10.1093/nar/gkt1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J. J. and Blumenthal T. (2011). RNA processing in C. elegans. Methods Cell Biol. 106, 187-217. 10.1016/B978-0-12-544172-8.00007-4 [DOI] [PubMed] [Google Scholar]
- Nam Y., Chen C., Gregory R. I., Chou J. J. and Sliz P. (2011). Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080-1091. 10.1016/j.cell.2011.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. A., Thomson J. M. and Hammond S. M. (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539-1549. 10.1261/rna.1155108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C. Y., Calidas D., Lu T., Smith J., Schmidt H., Krause M. W. and Seydoux G. (2014). Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198, 1347-1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D. and Seydoux G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47-54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., Maller B., Hayward D. C., Ball E. E., Degnan B., Müller P. et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86-89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Pepper A. S.-R., McCane J. E., Kemper K., Yeung D. A., Lee R. C., Ambros V. and Moss E. G. (2004). The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development 131, 2049-2059. 10.1242/dev.01098 [DOI] [PubMed] [Google Scholar]
- Perales R., King D. M., Aguirre-Chen C. and Hammell C. M. (2014). LIN-42, the Caenorhabditis elegans PERIOD homolog, negatively regulates microRNA transcription. PLoS Genet. 10, e1004486 10.1371/journal.pgen.1004486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto F. L. and Lindblad P. (2010). A guide for in-house design of template-switch-based 5′ rapid amplification of cDNA ends systems. Anal. Biochem. 397, 227-232. 10.1016/j.ab.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Piskounova E., Polytarchou C., Thornton J. E., Lapierre R. J., Pothoulakis C., Hagan J. P., Iliopoulos D. and Gregory R. I. (2011). Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066-1079. 10.1016/j.cell.2011.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., Rougvie A. E., Horvitz H. R. and Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901-906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Ren Z. and Ambros V. R. (2015). Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl. Acad. Sci. USA 112, E2366-E2375. 10.1073/pnas.1422858112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J. L. and Bradley A. (2004). Identification of mammalian microRNA host genes and transcription units. Genome Res. 14, 1902-1910. 10.1101/gr.2722704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S. F. and Slack F. J. (2009). Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev. Biol. 334, 523-534. 10.1016/j.ydbio.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel S., Frand A. R. and Ruvkun G. (2011). Regulation of the C. elegans molt by pqn-47. Dev. Biol. 360, 297-309. 10.1016/j.ydbio.2011.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R. and Wulczyn F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987-993. 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- Stefani G., Chen X., Zhao H. and Slack F. J. (2015). A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA 21, 985-996. 10.1261/rna.045542.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. (2006). Maintenance of C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas J. and Romer-Seiber J. (2015). LIN28: roles and regulation in development and beyond. Development 142, 2394-2404. 10.1242/dev.117580 [DOI] [PubMed] [Google Scholar]
- Turchinovich A., Surowy H., Serva A., Zapatka M., Lichter P. and Burwinkel B. (2014). Capture and Amplification by Tailing and Switching (CATS). An ultrasensitive ligation-independent method for generation of DNA libraries for deep sequencing from picogram amounts of DNA and RNA. RNA Biol. 11, 817-828. 10.4161/rna.29304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadla B., Kemper K., Alaimo J., Heine C. and Moss E. G. (2012). lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 8, e1002588 10.1371/journal.pgen.1002588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. and Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, Research0034 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P. M., Kai Z. S., Massirer K. B., Burton V. H., Yeo G. W. and Pasquinelli A. E. (2011). LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 18, 302-308. 10.1038/nsmb.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P. M., Finnegan E. F., Stark T., Angelus E. P., Homan K. E., Yeo G. W. and Pasquinelli A. E. (2014). The Period protein homolog LIN-42 negatively regulates microRNA biogenesis in C. elegans. Dev. Biol. 390, 126-135. 10.1016/j.ydbio.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q. and Gregory R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97-100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert M. L., Huelga S. C., Kapeli K., Stark T. J., Liang T. Y., Chen S. X., Yan B. Y., Nathanson J. L., Hutt K. R., Lovci M. T. et al. (2012). LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell 48, 195-206. 10.1016/j.molcel.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis D. G., Kai Z. S., Chang R. K. and Pasquinelli A. E. (2012). Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486, 541-544. 10.1038/nature11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Chiu H., Zinovyeva A., Ambros V., Chuang C.-F. and Chang C. (2013). Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340, 372-376. 10.1126/science.1231321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.