ABSTRACT
The metabolism of PI(3,5)P2 is regulated by the PIKfyve, VAC14 and FIG4 complex, mutations in which are associated with hypopigmentation in mice. These pigmentation defects indicate a key, but as yet unexplored, physiological relevance of this complex in the biogenesis of melanosomes. Here, we show that PIKfyve activity regulates formation of amyloid matrix composed of PMEL protein within the early endosomes in melanocytes, called stage I melanosomes. PIKfyve activity controls the membrane remodeling of stage I melanosomes, which regulates PMEL abundance, sorting and processing. PIKfyve activity also affects stage I melanosome kiss-and-run interactions with lysosomes, which are required for PMEL amyloidogenesis and the establishment of melanosome identity. Mechanistically, PIKfyve activity promotes both the formation of membrane tubules from stage I melanosomes and their release by modulating endosomal actin branching. Taken together, our data indicate that PIKfyve activity is a key regulator of the melanosomal import–export machinery that fine tunes the formation of functional amyloid fibrils in melanosomes and the maintenance of melanosome identity.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: PIKfyve; FIG4; VAC14; PI(3,5)P2; Phosphoinositide; Melanosome; Lysosome; PMEL; Amyloid
Summary: Generation of PI(3,5)P2 by the PIKfyve complex regulates membrane remodeling of early melanosomes; this controls the import–export balance that is required for functional amyloid formation and melanosome identity.
INTRODUCTION
The protein complex composed of the kinase PIKfyve (also known as Fab1), the phosphatase FIG4 (also known as Sac3) and the scaffolding protein VAC14 (also known as ArPIKfyve) regulates levels of phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2] (Ikonomov et al., 2009; Jin et al., 2008) and PI(5)P in mammalian cells (Sbrissa et al., 2012; Zhang et al., 2007). These low-abundance signaling lipids of endosomal membranes are key regulators in the homeostasis of the endolysosomal system (McCartney et al., 2014; Viaud et al., 2014). Although FIG4 acts as a PI(3,5)P2 5-phosphatase (Duex et al., 2006a; Rudge et al., 2004), its activity is also required, together with VAC14, for activation of PIKfyve (Bonangelino et al., 2002; Dove et al., 2002; Duex et al., 2006b; Jin et al., 2008). Thus, depletion not only of VAC14 and PIKfyve, but also of FIG4 reduces cellular PI(3,5)P2 levels (Chow et al., 2007; Zhang et al., 2007; Zolov et al., 2012). Inhibition or depletion of the PIKfyve–VAC14–FIG4 complex (hereafter called the PIKfyve complex) impairs endolysosomal functions, such as endosome-to-trans-Golgi network (TGN) transport, endosomal homeostasis and autophagy (Ferguson et al., 2009; McCartney et al., 2014). However, the molecular mechanisms by which the PIKfyve complex controls these processes and their physiological relevance remain to be fully understood. Mouse models with mutations in VAC14 or FIG4 exhibit diluted pigmentation (Chow et al., 2007; Jin et al., 2008; Zhang et al., 2007). This hypopigmentation suggests that, in melanocytes, the PIKfyve complex may be involved in the biogenesis of melanosomes, the organelles in which the pigment melanin is synthesized. Our current understanding of the stepwise process of melanosome biogenesis from endosomes provides a unique opportunity to investigate the molecular origin of hypopigmentation and the role of the PIKfyve complex in the endosomal system (Bissig et al., 2016; Sitaram and Marks, 2012).
Melanogenesis involves two sequential but independent processes, functional amyloid fibril formation and pigment synthesis. Melanosomes are lysosome-related organelles that are derived from early endosomes, which are called stage I melanosomes in pigment cells. During melanosome biogenesis, stage I melanosomes mature into stage II melanosomes, which have acquired specific identity and morphology distinct from lysosomal organelles. Stage I melanosomes have been proposed to be the crossroad at where melanosomal and endolysosomal pathways segregate (Raposo et al., 2001). However, the underlying molecular mechanisms of this segregation are not fully understood. Stage I melanosomes are characterized by possessing a clathrin coat and having a few intralumenal vesicles (ILVs) (Raposo et al., 2001). In their lumen, amyloid fibril generation is initiated from cleavage products of PMEL, a transmembrane protein expressed in melanocytes (Bissig et al., 2016). During maturation into stage II melanosomes, the formation of these fibrils requires concomitant proteolytic processing of PMEL and differential sorting of PMEL cleavage products (Hurbain et al., 2008; Rochin et al., 2013; Theos et al., 2006b; van Niel et al., 2015, 2011). PMEL amyloidogenesis is completed in ellipsoidal shaped stage II melanosomes, where the assembled amyloid lumenal fibrils serve as a matrix for pigment deposition in later stage III and IV melanosomes, which have acquired melanogenic enzymes (Raposo et al., 2001). This matrix is proposed to sequester highly reactive oxidative intermediates of melanin production, which otherwise may oxidize melanosomal content and damage organelle integrity (Fowler et al., 2006; Lee et al., 1996).
Here, we show that PIKfyve activity promotes the formation of membrane tubules emerging from stage I melanosomes and regulates their release by controlling branched actin dynamics. Interference with PIKfyve activity abrogates membrane tubule formation and branched actin-dependent release, which strongly affects stage I melanosomes homeostasis and leads to accumulation of unstructured aggregates of amyloidogenic PMEL fragments, preventing amyloid matrix formation. These defects are accompanied by prolonged kiss-and-run interactions of stage I melanosomes with lysosomal compartments that affect melanosome identity and maturation.
RESULTS
The PIKfyve complex is required for PMEL fibril formation
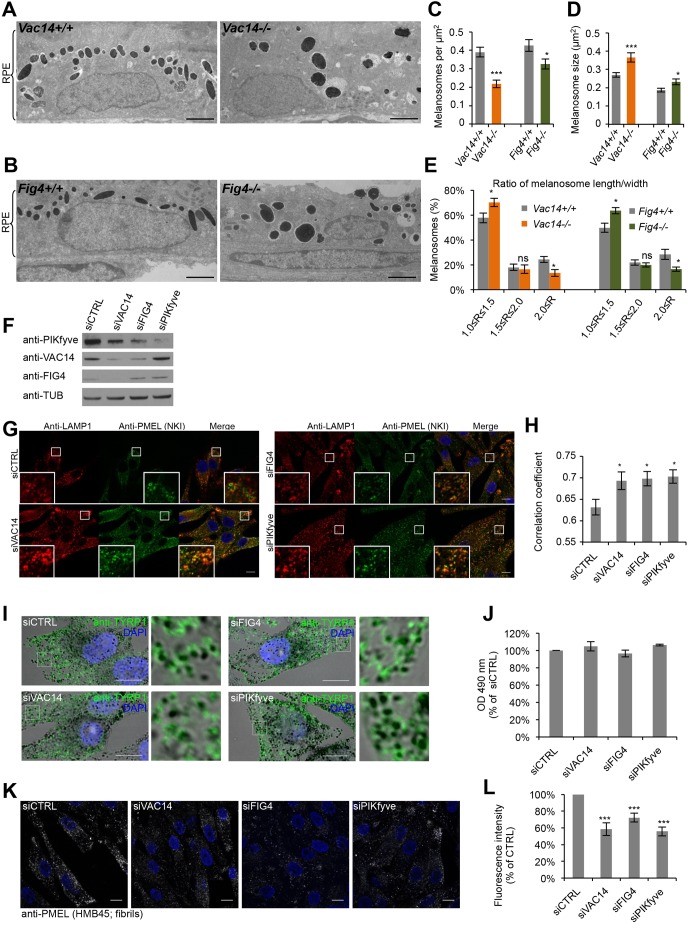
To gain insight into the pigmentation defect caused by loss of VAC14 and FIG4, we used conventional electron microscopy (EM) for morphological analysis of melanosomes in the retinal pigment epithelium (RPE) in eyes of newborn mutant mice (Chow et al., 2007; Zhang et al., 2007). These melanosomes are fully mature and less susceptible to compensatory effects, as they are formed during a brief developmental period (Lopes et al., 2007). Compared to wild-type (WT) RPE, we found fewer, but enlarged, melanosomes per µm2 in the RPE of Fig4−/− and Vac14−/− mice (Fig. 1A–D). The enlargment of melanosomes associated with hypopigmentation is reminiscent of those observed in ocular albinism type 1 (OA1, also known as GPR143)-deficient mice (Incerti et al., 2000), where the melanin-synthesizing enzyme tyrosinase-related protein 1 (TYRP1) is mistrafficked, and LAMP1, a lysosomal protein usually poorly present in early and late melanosomes (Raposo et al., 2001) (Fig. S1, left panels), is enriched in these compartments. The melanosomes in the RPE of Fig4−/− and Vac14−/− mice were also rounder than the elongated WT melanosomes (Fig. 1E). As reported previously for melanosomes in the RPE of mouse Bace2−/− (Rochin et al., 2013), PMEL silver (Dunn and Thigpen, 1930) and Pmel−/− (Hellstrom et al., 2011) mutants, round melanosomes are indicative of a defect in PMEL fibril assembly, since the fibrils give melanosomes their characteristic ellipsoidal shape (Hellstrom et al., 2011; Theos et al., 2006a). Vac14L156R/L156R and Fig4−/− mice also exhibit the diluted coat color characteristic of certain cases of impaired PMEL fibril assembly (Chow et al., 2007; Dunn and Thigpen, 1930; Hellstrom et al., 2011; Jin et al., 2008; Rochin et al., 2013; Zhang et al., 2007).
Fig. 1.
Interference with the PIKfyve complex affects melanosome morphology and identity. (A,B) EM analysis of Epon-embedded RPE of newborn Vac14+/+ and Vac14−/− mice (A) and Fig4+/+ and Fig4−/− mice (B). Scale bar: 2 µm. (C–E) Quantification of melanosome number per µm2 RPE (C), melanosome size (D) and the ratio (R) of maximal width and length of melanosomes (E). (F) MNT-1 cells were treated with control siRNAs or siRNAs against VAC14, FIG4 and PIKfyve and knockdown efficiencies were analyzed by immunoblotting. Antibodies to tubulin (anti-TUB) were used as equal loading marker. (G) MNT-1 cells treated with siRNAs as in F were fixed, permeabilized and immunolabeled using anti-LAMP1 (red) and anti-PMEL-NKI antibodies (green). DAPI was used to stain nuclei. Insets show magnifications of the boxed regions. Scale bars: 10 µm. (H) Quantification of colocalization between LAMP1 and PMEL fluorescence. (I) MNT-1 cells treated with control siRNAs or siRNAs against VAC14, FIG4 or PIKfyve were fixed, permeabilized and immunolabeled using anti-TYRP1 antibody (green) and DAPI (blue) to stain nuclei. Pigmented melanosomes are shown in bright-field images. Panels on the right show magnifications of the boxed regions. Scale bars: 10 µm. (J) Quantification of melanin content of MNT-1 cells treated with control siRNAs or siRNAs against VAC14, FIG4 or PIKfyve. (K) MNT-1 cells treated with control siRNAs or siRNAs against VAC14, FIG4 or PIKfyve were fixed, permeabilized and immunolabeled using DAPI (blue) to stain nuclei and anti-PMEL (HMB45) antibody (gray), which recognizes PMEL fibrils. Scale bars: 10 µm. (L) Quantification of the mean fluorescence intensity per cell normalized to siCTRL for experiments as in K. Means±s.e.m. shown for n≥3 in all panels. *P<0.05; ***P<0.001; ns, not significant (unpaired Student's t-test, unequal variance). See also Fig. S1.
To confirm these findings, we performed RNAi on PIKfyve, VAC14 or FIG4 (denoted siPIKfyve, siVAC14 and siFIG4, respectively) in the human melanocytic cell line MNT-1 (Fig. 1F). In these knocked down cells (Fig. 1G,H) and in MNT-1 cells treated with the PIKfyve inhibitor YM201636 (Fig. S1A–E), we found more lysosomal LAMP1 and PSEN2 localized to stage II melanosomes containing processed PMEL. We also observed miscolocalization of LAMP1 with the melanin-synthesizing protein TYRP1 present on pigmented melanosomes upon YM201636 treatment (Fig. S1A–C). However, TYRP1 was correctly localized to pigmented melanosomes (Fig. 1I) and the melanin content was unchanged (Fig. 1J), excluding abnormal targeting of melanizing enzymes as in OA1-depleted MNT-1 cells (Giordano et al., 2009). As reported in MNT-1 cells displaying a defect in PMEL fibril assembly (Rochin et al., 2013; van Niel et al., 2011), immunolabeling with an antibody recognizing PMEL fibrils (antibody HMB45) was decreased in cells treated with siRNA against the PIKfyve complex protein when compared to control cells (Fig. 1K,L). These findings indicate that the PIKfyve complex is involved in processes regulating PMEL fibril formation during early melanogenesis, which impact melanosome identity and maturation, but not pigment synthesis occurring at later steps of melanosome maturation.
Lumenal PMEL fragments accumulate upon interference with PIKfyve function
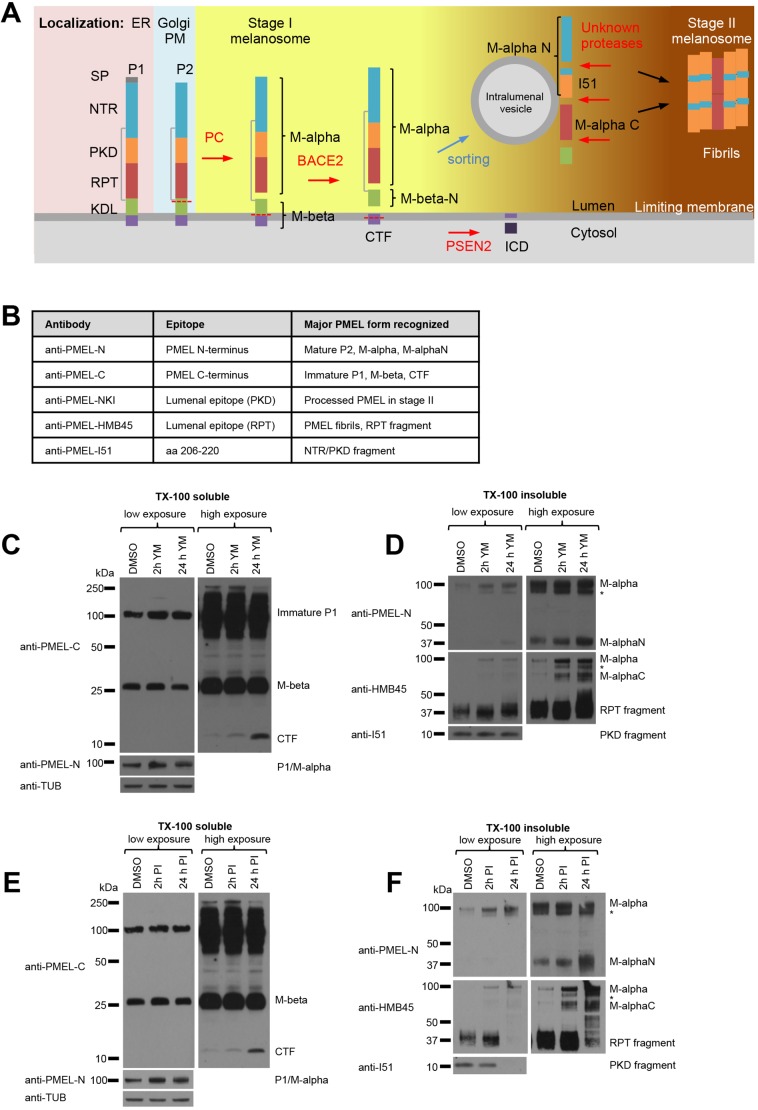
Generation of PMEL fibrils requires stepwise processing of PMEL through the actions of a series of proteases (Fig. 2A). First, the mature P2 form of PMEL is cleaved by proprotein convertase (PC) family enzymes generating a lumenal M-α and transmembrane M-β fragment that remain linked by a disulfide bond (Berson et al., 2003; Leonhardt et al., 2011; Theos et al., 2006b). Then, the M-β fragment is processed by β-site APP-cleaving enzyme 2 (BACE2) generating a membrane-associated C-terminal fragment (CTF) and releasing amyloidogenic M-α into the melanosome lumen (Rochin et al., 2013). Unknown lysosomal proteases then further proteolytically process M-α into amyloidogenic peptides that finally assemble into fibrils (Ho et al., 2016; Kawaguchi et al., 2015; Leonhardt et al., 2013). Given the reported involvement of PIKfyve complex in endolysosome homeostasis (Bissig et al., 2017; Jefferies et al., 2008), we studied whether PIKfyve inhibition or knockdown would affect PMEL processing.
Fig. 2.
M-α fragments accumulate upon interference with PIKfyve function or lysosomal protease activity. (A) Schematic representation of PMEL maturation and proteolytic processing and sorting. PMEL protein domains: signal peptide (SP), N-terminal region (NTR), polycystic kidney disease protein-1-like repeat domain (PKD), repeat domain (RPT), kringle-like domain (KDL), C-terminal fragment (CTF), intracellular domain (ICD). PMEL-processing proteases are indicated in red: proprotein convertase (PC), β-site APP-cleaving enzyme 2 (BACE2) and presenilin 2 (PSEN2). Colored background highlights the intracellular localization of the different PMEL cleavage forms. (B) Table of antibodies used in this study summarizing their epitopes and the major PMEL forms they recognize. (C–F) MNT-1 cells were treated for 2 h or 24 h with 1.6 µM YM201636 (C,D) or with a mixture of protease inhibitors (100 µM leupeptin, 10 µM pepstatin A and 10 µM E-64d) (E,F) and Triton X-100-soluble (C,E) and Triton X-100-insoluble (D,F) lysates were analyzed by immunoblotting using antibodies against the PMEL C-terminus (anti-PMEL-C), the PMEL N-terminus (anti-PMEL-N), the PMEL RPT domain (anti-PMEL-HMB45), the PMEL PKD domain (anti-PMEL-I51) and tubulin (anti-TUB), as an equal loading marker. The different PMEL fragments are annotated on the right. Stars indicate M-α fragments derived from another isoform generated by alternative splicing. Right-hand panels show higher exposures. (See also Fig. S1).
Using a set of antibodies that recognize distinct epitopes and processed fragments of PMEL (Fig. 2B), we found that PIKfyve complex knockdown or inhibition for 2 h and 24 h did not prevent the PC- and BACE2-mediated PMEL cleavages producing M-α, M-β and CTF fragments, but caused an accumulation of the CTF (Fig. 2C; Fig. S1G), which is not part of PMEL fibrils, and of M-α (Fig. S1G). Owing to their amyloidogenic nature, PMEL fibril-associated fragments distribute to the Triton X-100-insoluble fraction in cell lysates (Berson et al., 2003; Leonhardt et al., 2013; Watt et al., 2009). In this fraction, we found accumulation of M-α, M-αN, M-αC (Fig. 2D; Fig. S1H). Treatment of MNT-1 cells for 2 h and 24 h with a mix of lysosomal protease inhibitors (100 µM leupeptin, 10 µM pepstatin A and 10 µM E-64d) led to a similar accumulation CTF, M-α, M-αN and M-αC (Fig. 2E,F), but decreased generation of the polycystic kidney disease-like domain (PKD) and repeat domain (RPT) fragments. These results confirm that lysosomal proteases are required for PMEL processing (Ho et al., 2016; Kawaguchi et al., 2015; Leonhardt et al., 2013), and suggest that accumulation of processed and non-processed fragments of the PMEL lumenal domain under PIKfyve inhibition may be caused by changes of lysosomal activity.
Lysosomal activity is not affected by inhibition of PIKfyve
We assessed the lysosomal functions of melanocytes treated with YM201636. In these cells neither the number nor the size of LAMP1 compartments significantly changed upon inhibition of PIKfyve activity (Fig. 3A–D). By performing ratiometric fluorescence imaging, we detected a similar pH of endolysosomes in mock and YM201636-treated cells (4.4±0.2 and 4.5±0.2, mean±s.e.m., respectively) (Fig. S2A,B) in good agreement with previous reports (Bissig et al., 2017; Ho et al., 2015). Internalization of proteolytically dequenched DQ-BSA showed that treatment with PIKfyve inhibitor slightly increased proteolysis of endocytosed DQ-BSA, although the effect was not statistically significant (Fig. S2C,D). The levels of mature cathepsin D (CatD) decreased and immature CatD accumulated upon treatment with lysosomal protease inhibitors, but not upon YM201636 treatment (Fig. S2E), indicating that PIKfyve inhibition does not impair endolysosomal activity. To reinforce these results, we assessed steady-state levels of the melanocyte-specific transmembrane protein MART1 (also known as MLANA), which is ubiquitylated and degraded in lysosomes (Lévy et al., 2005). PIKfyve inhibition, in contrast to inhibition of lysosomal proteases, did not cause MART1 accumulation (Fig. S2E). The trafficking of a fluid-phase marker [Dextran conjugated to Alexa Fluor 647 (DextranAF647)] (Fig. S2F,G) or a lysosomal transmembrane protein (PSEN2) (Fig. S2H,I) to lysosomes was not affected upon YM201636 treatment, showing that PIKfyve inhibition neither affected transport of endocytosed cargo to endolysosomes nor trafficking of lysosome-resident proteins. This set of data shows that inhibition of PIKfyve does not impair lysosomal activity, excluding lysosomal dysfunction as a cause for the accumulation of PMEL fragments.
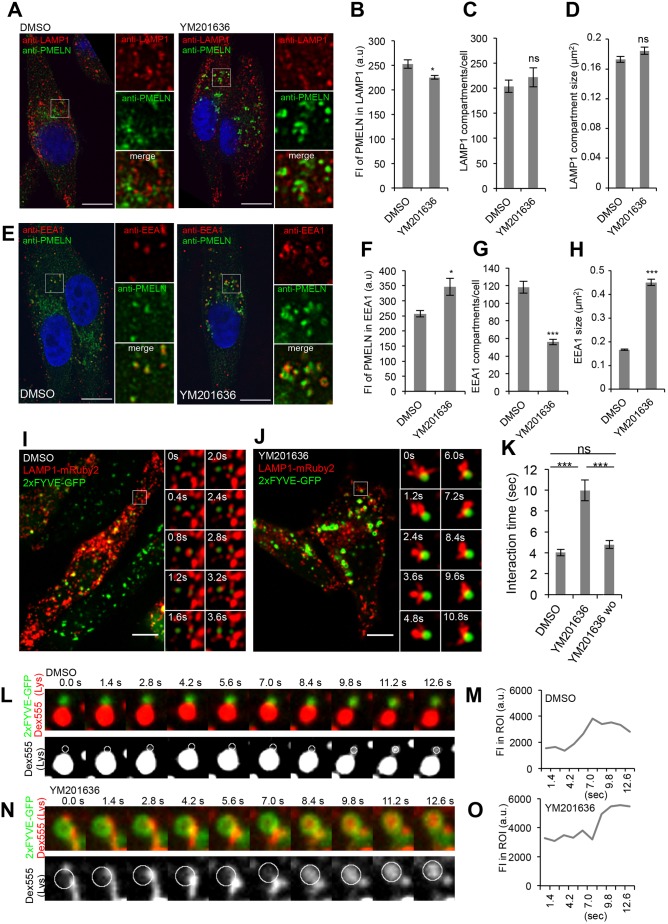
Fig. 3.
Stage I melanosomes and endolysosomes transiently interact and exchange content even upon inhibition of PIKfyve activity. (A) MNT-1 cells treated for 2 h with 1.6 µm YM201636 or DMSO were fixed, permeabilized and immunolabeled using DAPI (blue) to stain nuclei and anti-PMEL-N (green) and anti-LAMP1 (red) antibodies to label mature PMEL/M-α and endolysosomes, respectively. Panels on the right show magnifications of the boxed regions. Scale bars: 10 µm. (B–D) Quantification of the mean fluorescence intensity (FI) of anti-PMEL-N signal in LAMP1 compartments (B), of the number of LAMP1 compartments per cell (C) and of the size of LAMP1 compartments (D). (E) MNT-1 cells treated for 2 h with 1.6 µM YM201636 or DMSO were fixed, permeabilized and immunolabeled using DAPI (blue) to stain nuclei and anti-PMEL-N (green) and anti-EEA1 (red) antibodies to label mature PMEL/M-α and stage I melanosomes, respectively. Panels on the right show magnifications of the boxed regions. Scale bars: 10 µm. (F–H) Quantification of the mean fluorescence intensity (FI) of the anti-PMEL-N signal in EEA1 compartments (F), of the number of EEA1 compartments per cell (G) and of the size of EEA1 compartments (H). (I,J) MNT-1 cells were co-transfected with LAMP1–mRuby2 (red) and 2×FYVE–GFP (green), to mark endolysosomes and stage I melanosomes, respectively. Then cells were treated for 2 h with DMSO (I) or 1.6 µM YM201636 (J) and subsequently YM201636 was washed out (wo) for 1 h (Fig. S3H). Movies were taken at a frame rate of 0.4 s by spinning disc microscopy. The left panels show the first frame of the movies. The right panels show stills of the magnified regions. Scale bars: 10 µm. (K) Quantification of the mean duration time of interactions as shown in I,J and Fig. S3H (for wo). (L–N) DextranAF555 (Dex555) (red) was internalized through a 4 h pulse and 20 h chase into lysosomes of MNT-1 cells overexpressing 2×FYVE–GFP (localized on stage I melanosomes). Then cells were treated for 2 h with DMSO (L) or 1.6 µM YM201636 (N) and movies were taken at a frame rate of 0.4 s by spinning disc microscopy. The upper panels show stills of a movie, where lyososomal DextranAF555 is shown in red and 2×FYVE–GFP localized on stage I melanosomes is in green. Lower panels illustrate DextranAF555 transfer from lysosomes to 2×FYVE–GFP compartments. In these panels, only lysosomal DextranAF555 is shown in gray and circles indicate the positions of the 2×FYVE–GFP compartment. (M–O) Fluorescence intensity of lysosomal DextranAF555 in 2×FYVE–GFP compartments denoted with circles in L and N in cells treated with DMSO (M) and YM201636 (O). Means±s.e.m. shown for n≥3 in all panels. *P<0.05; ***P<0.001; ns, not significant (unpaired Student's t-test, unequal variance). See also Fig. S3. a.u., arbitrary units.
Lysosomal activity is delivered to melanosomes through kiss-and-run interactions
Our data and that of others show that lysosomal protease activity is required for PMEL processing and fibril formation in stage I melanosomes (Ho et al., 2016; Leonhardt et al., 2013). However, in melanocytes, lysosomes and melanosomes co-exist as separate entities with distinct content and morphology (Raposo et al., 2001). In line with this, immunolabeling of M-α species revealed their accumulation in EEA1-, but not LAMP1-labeled compartments at steady state and upon inhibition of PIKfyve activity (Fig. 3A,B,E,F). We speculated that PMEL localized in stage I melanosomes may be processed by lysosomal proteases through transient interactions of stage I melanosomes and endolysosomes, which could be affected by YM201636 treatment.
We tested this hypothesis by live-cell imaging of MNT-1 cells co-expressing LAMP1–mRuby2 and the PI(3)P-binding protein 2×FYVE–GFP, as markers for endolysosomes and stage I melanosomes, respectively. We confirmed, in fixed cells, that 2×FYVE–GFP is found on stage I melanosomes, which correspond to early endosomes in pigment cells and contain mature PMEL and M-α (Fig. S3A,B). In contrast, 2×FYVE–GFP did not colocalize with endolysosomes or with later-stage melanosomes containing fully formed PMEL fibrils or melanin-synthesizing enzymes or endolysosomes (Fig. S3C–F). Our live-cell imaging revealed that in mock-treated cells 2×FYVE–GFP and LAMP1–mRuby2 compartments transiently interact for, on average, 4.0 s without undergoing complete compartment mixing (Fig. 3I,K; Movies 1,2). To assess whether membrane fusion occurs, endolysosomes of 2×FYVE–GFP-expressing cells were loaded by pulse-chase with DextranAF555 and transfer of endolysosomal DextranAF555 to 2×FYVE–GFP compartments was monitored by live-cell microscopy. We found that endolysosomal DextranAF555 can be transferred to 2×FYVE–GFP compartments (Fig. 3L,M; Movie 3), showing that membrane fusion between the two compartments occurs. These findings support our hypothesis that the substrate PMEL, localized to stage I melanosomes, is processed by lysosomal proteases through transient fusions of stage I melanosomes and endolysosomes. These interorganellar kiss-and-run interactions strengthen the role of stage I melanosomes as being the crossroad between melanosomal and endolysosomal pathways, and provide mechanistic insights into the segregation of both pathways.
In YM201636-treated cells, stage I melanosomes and endolysosomes transiently interacted. However, the time of contact doubled when compared to mock (10.0 s versus 4.0 s), but was restored upon YM201636 washout and re-synthesis of PI(3,5)P2/PI(5)P (Fig. 3J,K; Fig. S3G, Movies 4–7). Moreover, upon PIKfyve inhibition, the two compartments still transiently fused, as illustrated by the transfer of endolysosomal DextranAF555 (Fig. 3N,O; Movie 8).
These data show that accumulation of processed and unprocessed PMEL/M-α species in stage I melanosomes upon PIKfyve inhibition did not result from impaired transient fusion between stage I melanosomes and endolysosomes. However, the prolonged duration of contact between stage I melanosomes and endolysosomes upon inhibition of PIKfyve activity may over time cause the observed mislocalization of endolysosomal proteins to melanosomes and loss of melanosome identity.
PMEL fragments accumulate in enlarged EEA1 compartments
The absence of defects in lysosomal activity and morphology upon inhibition of PIKfyve activity led us to investigate potential defects of the EEA1 compartments where PMEL fragments accumulated upon treatment. Interestingly, immunolabeling showed that EEA1-positive compartments that accumulated PMEL/M-α species following YM201636 treatment (Fig. 3E,F) were enlarged and less numerous when compared to those in control cells (Fig. 3G,H). In good agreement with this, after PIKfyve inhibition by YM201636 treatment, we observed enlargement of 2×FYVE–GFP-positive stage I melanosomes, but not LAMP1–mRuby2-positive endolysosomes in our live-cell imaging experiments (Fig. 3J).
It is thus conceivable that PIKfyve activity directly regulates stage I melanosome homeostasis, thereby explaining the lack of PMEL fibrillation when PIKfyve function is impaired.
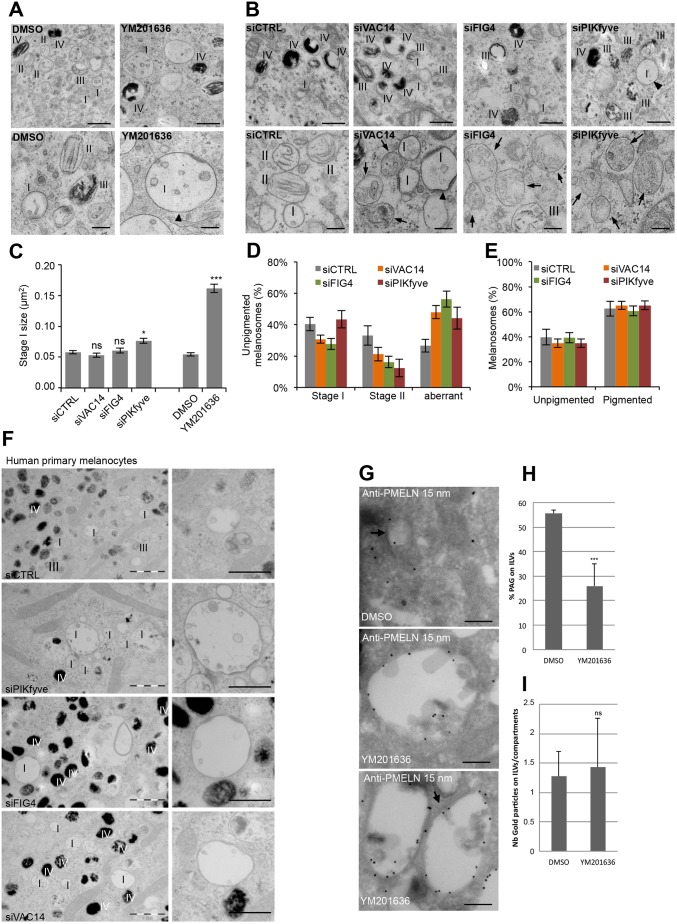
To better characterize the apparent defect in early melanosomes, we performed EM analysis of resin-embedded and chemically fixed MNT-1 cells knocked down for PIKfyve, VAC14 or FIG4 by means of siRNA or treated with the PIKfyve inhibitor YM201636 (Fig. 4A,B). In MNT-1 cells, melanosome biogenesis occurs continuously. Therefore, all four melanosome maturation stages are present and can morphologically be distinguished by EM. We observed enlarged ILV-containing stage I melanosomes decorated with a clathrin coat in inhibitor or siRNA-treated cells (Fig. 4A,B). Of note the mean size increase was only significant for YM201636 treatment and PIKfyve knockdown (Fig. 4C). Similarly, stage I melanosomes were enlarged in human primary melanocytes knocked down for VAC14, FIG4 and PIKfyve (Fig. 4F). In addition, MNT-1 cells treated with siRNA for the PIKfyve complex proteins displayed fewer fibril-containing stage II melanosomes, but more unpigmented melanosomes with unstructured lumenal aggregates (Fig. 4D,E), further confirming our finding that the PIKfyve complex is involved in PMEL fibril formation (Fig. 1K,L). In line with our observations that the PIKfyve complex is not involved in pigment synthesis (Fig. 1I,J), interference with PIKfyve complex did not alter the number and morphology of pigmented melanosomes (Fig. 4A,B,E). These morphological observations show that the PIKfyve complex is required for stage I melanosome homeostasis and PMEL fibril formation, which is initiated in these compartments.
Fig. 4.
PMEL fragments accumulate in enlarged EEA1 compartments. (A,B) EM analysis of Epon-embedded MNT-1 cells treated for 2 h with the PIKfyve inhibitor YM201636 (A) or knocked down for VAC14, FIG4 and PIKfyve (B). Upper panels show low-magnification overviews of unpigmented and pigmented melanosomes, and lower panels show unpigmented melanosomes at higher magnification. Arrowheads point towards enlarged stage I melanosomes and arrows highlight aberrant unpigmented melanosomes containing unstructured aggregates present in VAC14, FIG4 and PIKfyve knockdown cells. I–IV indicates the melanosome stage. Scale bars: 500 nm (upper panels), 200 nm (lower panels). (C) Quantification of stage I melanosome size. (D) Quantification of unpigmented melanosomes grouped into stage I, stage II and aberrant melanosomes. (E) Quantification of pigmented and unpigmented melanosomes expressed as a percentage of the total number of melanosomes. (F) EM analysis of Epon-embedded human primary melanocytes treated with control siRNAs or siRNAs against VAC14, FIG4 and PIKfyve. Right panels show magnifications of stage I melanosomes. I–IV indicates the melanosome stage. Scale bars: 1 µm (left panels), and 500 nm (right panels). (G) Ultrathin cryosections of MNT-1 cells treated for 2 h with 1.6 µM YM201636 or DMSO were immunogold-labeled using anti-PMEL-N antibody followed by protein-A conjugated to 15 nm diameter gold. Arrows indicate clathrin coats. Scale bars: 200 nm. (H) Quantification of the percentage of gold particles on ILVs expressed as a percentage of the total number of gold particles. (I) Quantification of the number of gold particles on ILVs per compartment. Means±s.e.m. shown for n≥3 in all panels. *P<0.05; ***P<0.001; ns, not significant (unpaired Student's t-test, unequal variance).
PMEL fibrillation results from concomitant processing and sorting of the amyloidogenic lumenal domain of PMEL to ILVs of stage I melanosomes (Hurbain et al., 2008; Rochin et al., 2013; Theos et al., 2006 b; van Niel et al., 2015; van Niel et al., 2011). Ultrathin cryosections of MNT-1 cells confirmed the presence of ILV in stage I melanosomes labeled with anti-PMEL-N antibody (Fig. 4G,H), indicating that PIKfyve activity is not critical for ILV formation. These data also revealed that, upon PIKfyve inhibition, mature PMEL/M-α is localized to enlarged clathrin-coat-containing stage I melanosomes, where it was mostly found on the limiting membrane and to a lesser extent on ILVs (Fig. 4G,H). However, a similar amount of gold labeling was found on ILVs between the YM201636-treated and control condition (Fig. 4I), suggesting that PMEL/M-α sorting to ILVs is not impaired. However, accumulation of this fragment on the limiting membrane may indicate a saturation of the sorting machinery.
Taken together, our results suggest that upon interference with the PIKfyve complex, lumenal PMEL fragments accumulate in enlarged stage I melanosomes causing unstructured PMEL aggregation that prevents fibril formation and impairs melanosome maturation.
PIKfyve activity promotes membrane remodeling
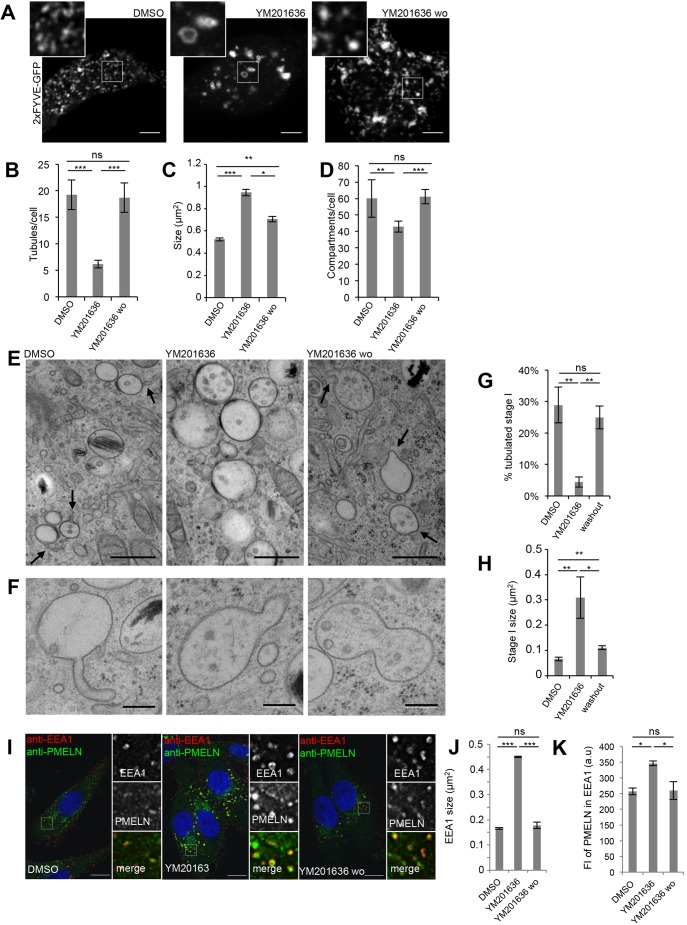
Our observations show that upon PIKfyve inhibition the size of stage I melanosomes doubles, their number decreases and they accumulate PMEL fragments. A similar organelle enlargement induced by inhibition of PIKfyve activity was observed for endolysosomes in HeLa cells. In these cells, PIKfyve activity was shown to control endolysosome homeostasis by promoting membrane remodeling (Bissig et al., 2017). Thus, we hypothesized that PIKfyve exerts a similar function on stage I melanosomes and we studied stage I melanosome homeostasis and membrane dynamics by live-cell microscopy using 2×FYVE–GFP. In mock-treated cells, these compartments were highly dynamic and underwent frequent homotypic fusion and fission (Movie 9). Their shape was heterogeneous, and they were often tubulated (Fig. 5A,B). These features were lost upon inhibition of PIKfyve activity, where stage I melanosomes were fewer, enlarged, spherical and lacked membrane tubules (Fig. 5A–D). In addition, the compartments were less dynamic as compared to control (Movie 10). Upon YM201636 washout and re-synthesis of PI(3,5)P2/PI(5)P, membrane dynamics was restored and tubules and buds were formed and released, resulting in recovery of compartment size and number (Fig. 5A–D; Movie 11). These findings indicate that PIKfyve activity promotes the membrane remodeling of stage I melanosomes required to maintain compartment size, number and homeostasis during the homotypic fusion–fission processes of stage I melanosomes.
Fig. 5.
PIKfyve activity promotes formation of membrane tubules that modulate PMEL accumulation. (A) Snapshots of live 2×FYVE–GFP-transfected MNT-1 cells treated for 2 h with DMSO or 1.6 μM YM201636, and when subsequently YM201636 was washed out (wo) for 4 h. Magnifications of the boxed regions illustrate the absence of membrane tubules in PIKfyve inhibitor-treated cells. Scale bars: 10 µm. (B–D) Quantification of 2×FYVE–GFP-labeled tubules per cell (B), and of the 2×FYVE–GFP compartment size (C) and number per cell (D). (E) EM analysis of HPF/freeze substituted MNT-1 cells treated for 2 h with DMSO or 1.6 µM YM201636, and when subsequently YM201636 was washed out (wo) for 4 h. Arrows point at membrane tubules and deformations. Scale bars: 500 nm. (F) Examples of compartments undergoing membrane remodeling upon 4 h of YM201636 washout. Scale bars: 200 nm. (G,H) Quantification of the percentage of tubulated stage I melanosomes (G) and of stage I melanosome size (H) from experiments as in E. (I) MNT-1 cells treated as indicated were fixed, permeabilized and immunolabeled using DAPI (blue) to stain nuclei and anti-PMEL-N (green) and anti-EEA1 (red) antibodies to label the PMEL/M-α and stage I melanosomes, respectively. Panels on the right show magnifications of the boxed regions. wo, wash out of the YM drugs as performed in A–D. Scale bars: 10 µm. (J,K) Quantification of EEA1 size (J) and the mean fluorescence (FI) of PMELN in EEA1 compartment (K). Means±s.e.m. shown for n≥3 in all panels. *P<0.05; ***P<0.001; ns, not significant (unpaired Student's t-test, unequal variance).
To confirm these findings, we performed EM analyses on high-pressure frozen and freeze-substituted melanocytes, because this technique optimally preserves subtle membrane deformation such as membrane tubulations. In mock-treated cells, multivesicular stage I melanosomes often displayed tubules and showed a clathrin coat on their limiting membrane. In contrast, after YM201636 treatment, these compartments lost their tubular appearance and were enlarged, but still displayed clathrin coats. Upon YM201636 washout, the tubular feature was restored and dividing enlarged compartments were observed (Fig. 5E–H), showing that PIKfyve activity is required for membrane remodeling on stage I melanosomes. We assessed the contribution of this membrane remodeling to PMEL accumulation by performing immunofluorescence microscopy. YM201636 induced accumulation of PMEL fragments in enlarged EEA1-positive stage I melanosomes, and both this accumulation and enlargement were rescued after drug washout and PI(3,5)P2/PI(5)P re-synthesis (Fig. 5I–K). These findings confirm that PIKfyve activity controls stage I melanosome size, number and dynamics by regulating fission processes for stage I melanosomes. Interference with PIKfyve activity by inhibiting the homotypic fission induces the accumulation of PMEL fragments in fewer but larger stage I melanosomes that would contribute to the impaired PMEL fibrillation.
PIKfyve activity controls endosomal branched actin required for tubule release
To refine the mechanisms involved in the inhibition of PMEL fibrillation upon PIKfyve activity we explored the downstream and upstream pathways regulated by this complex such as the activation of mucolipin channels by PI(3,5)P2 (Dong et al., 2010), PI(3)P levels (Zolov et al., 2012), pH (Jefferies et al., 2008) or endosomal actin branching (Hong et al., 2015). In MNT-1 cells, co-treatment of YM201636 with the mucolipin activator MLSA1 did not restore stage I melanosome size and membrane dynamics (Fig. S4A,B). These findings indicate that mucolipins may not be downstream effectors of PIKfyve activity in melanosome biogenesis, which is in line with our observations in HeLa cells (Bissig et al., 2017). Ratiometric fluorescence imaging with a short pulse-chase protocol excluded a pH defect in stage I melanosomes as a cause of impaired fibrillation of PMEL (Fig. S4C). Inhibition of PIKfyve activity has been shown to cause a slight increase in the level of the PI(3,5)P2 precursor PI(3)P in some, but not all, cell types (Jefferies et al., 2008; Zolov et al., 2012). Upon inhibition of PIKfyve activity, we did not observe any change in the localization or intensity of fluorescence associated with endogenous EEA1 nor with 2×FYVE–GFP, which both bind PI(3)P (Fig. S4D). Similarly, biochemical analysis showed no significant differences in membrane recruitment of endogenous EEA1 and expressed 2×FYVE–GFP between DMSO- and YM201636-treated cells (Fig. S4E,F). These results show that localization of PI(3)P-binding proteins is unaffected upon PIKfyve inhibition and indicate that, in melanocytes, PI(3)P is not likely to accumulate upon inhibition of PIKfyve activity.
Recently, PI(3,5)P2 has been shown to control dynamics of branched actin on endosomal membranes (Hong et al., 2015), which is itself involved in various membrane remodeling processes (Anitei and Hoflack, 2011). Thus, we asked whether PIKfyve activity promotes membrane remodeling through regulation of endosomal actin dynamics. As observed in human breast adenocarcinoma cells (Hong et al., 2015), we found that actin patches accumulated on EEA1-positive and enlarged stage I melanosomes upon PIKfyve inhibitor treatment. Actin accumulation was abolished upon inhibition of the Arp2/3 complex by treatment with CK666 (Fig. 6A). These data reveal that, in melanocytes, as in other cells, PI(3,5)P2/PI(5)P regulates branched endosomal actin.
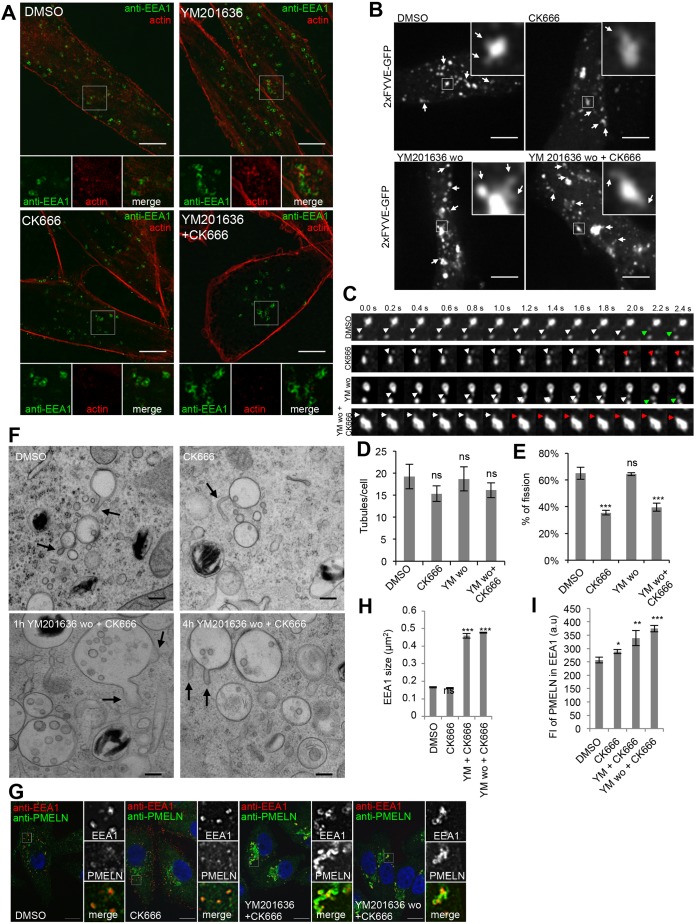
Fig. 6.
Endosomal actin dynamics is required for membrane tubule release. (A) MNT-1 cells treated for 2 h with DMSO or 1.6 µM YM201636 in the presence or absence of 200 µM CK-666 were fixed, permeabilized and immunolabeled using anti-EEA1 antibody (green) and phalloidin–TRITC (red) to label actin. Lower panels show magnifications of the boxed regions. (B) Snapshots of live 2×FYVE–GFP-expressing MNT-1 cells treated for 2 h with DMSO or 200 µM CK-666 and after washout (wo, for 1 h) of YM201636 in the absence and presence of 200 µM CK-666. Arrows highlight membrane tubules or buds. Magnifications of boxed regions illustrate membrane tubules. (C) Stills of compartments of cells in B illustrating membrane tubules and release. White arrows highlight membrane tubules or buds. Green and red arrows highlight membrane bud release and retraction, respectively. (D,E) Quantification of tubules per cell (D) and of the percentage of tubules undergoing release (E). (F) EM analysis of HPF/freeze substituted MNT-1 cells treated for 2 h 200 µM CK-666 or 1.6 µM YM201636, which was subsequently washed out (wo) for 1 h and 4 h in the presence of 200 µM CK-666. Arrows point at membrane tubules and deformations. Scale bars: 200 nm. (G) MNT-1 cells treated as indicated were fixed, permeabilized and immunolabeled using DAPI (blue) to stain nuclei and anti-PMEL-N (green) and anti-EEA1 (red) antibodies to label the PMEL/M-α and stage I melanosomes, respectively. Panels on the right show magnifications of the boxed regions. Scale bars: 10 µm. (H,I) Quantification of EEA1 size (H) and the mean fluorescence (FI) of PMELN in EEA1 compartment (I). Means±s.e.m. shown for n≥3 in all panels. *P<0.05; **P<0.01; ***P<0.001; ns, not significant (unpaired Student's t-test, unequal variance).
We performed live-cell imaging of 2×FYVE–GFP-transfected cells to study whether endosomal actin is involved in PI(3,5)P2/PI(5)P-mediated membrane remodeling. Stage I melanosomes of cells treated with the Arp2/3 inhibitor CK666 still displayed membrane tubules. Similarly, when YM201636 was washed out in the presence of CK666, membrane tubules were observed (Fig. 6B–E), showing that endosomal branched actin is not required for PI(3,5)P2/PI(5)P-mediated membrane remodeling. However, upon inhibition of branched actin dynamics by CK666 treatment, membrane buds and tubules were less readily released and often collapsed (Fig. 6C–E; Movies 12–15), suggesting that endosomal actin dynamics is involved in membrane tubule release.
EM analyses of high-pressure frozen MNT-1 cells confirmed that upon inhibition of Arp2/3 complex, tubules are formed on stage I melanosomes (Fig. 6F). Similarly, membrane tubules on enlarged stage I melanosomes were observed upon YM201636 washout in the presence of CK666 (Fig. 6F), but stage I melanosomes were still enlarged 4 h after YM201636 washout in the presence of CK666, indicating that although membrane tubules are formed the compartment size was not rescued (Fig. 6F), probably because the tubules were less readily released. Immunofluorescence microscopy showed that YM201636 induced accumulation of PMEL in EEA1-positive stage I melanosomes and their enlargement were rescued after drug washout and PI(3,5)P2/PI(5)P re-synthesis. However, both accumulation of PMEL and enlargement of EEA1-positive stage I melanosomes persisted when YM201636 was washed out in the presence of Arp2/3 complex inhibitor (Fig. 6G–I), showing that PI(3,5)P2/PI(5)P-mediated membrane remodeling and endosomal actin dynamics are involved in the same pathway. Thus, our findings indicate that PI(3,5)P2/PI(5)P spatially and temporally coordinates membrane remodeling and actin-dependent release of endosomal tubules and buds, which is required for stage I melanosome homeostasis and PMEL fibril formation.
DISCUSSION
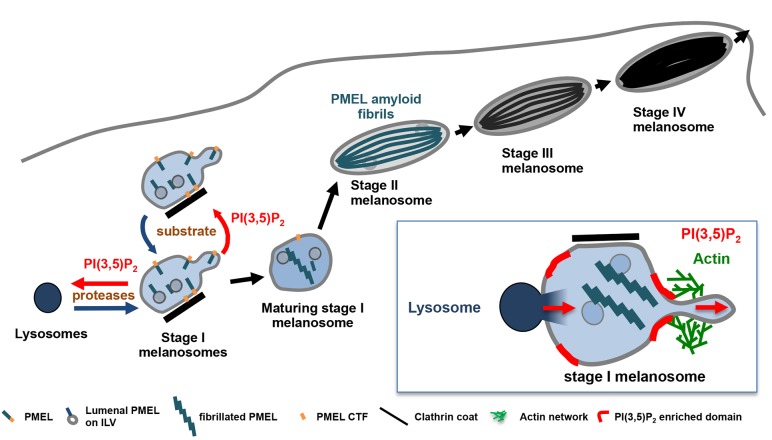
Our data show that the formation of PMEL amyloid fibrils and melanosome identity rely on tight regulation of the homeostasis of stage I melanosomes and their transient interaction with lysosomes. By controlling membrane remodeling at stage I melanosomes, PIKfyve activity balances the import of lysosomal proteases and the steady-state level of their substrate PMEL, and thereby regulates the formation of amyloid matrix and the preservation of melanosome identity (Fig. 7). Functionally, PIKfyve activity regulates the morphology, size and number of stage I melanosomes by promoting the formation of membrane tubules and buds that are released in a mechanism involving branched endosomal actin dynamics (Fig. 7, inset). Inhibition of PIKfyve activity abrogates membrane remodeling, causing an imbalance favoring stage I melanosomes homotypic fusion over fission, which reduces the number and increases the size of stage I melanosomes. The imbalance between homotypic fusion and fission causes an accumulation of PMEL, which saturates amyloidogenic processing and sorting machineries. These defects result in accumulation and aberrant aggregation of amyloidogenic PMEL fragments, impairing fibril formation.
Fig. 7.
Working model of PIKfyve function in melanosome biogenesis. Stage I melanosomes are highly dynamic organelles undergoing frequent homotypic fusion and fission. In these compartments, PMEL fibril formation is initiated by PMEL processing and sorting to ILVs. PMEL is processed by lysosomal proteases that are delivered to stage I melanosomes via transient interorganelle interactions. PI(3,5)P2 synthesized by PIKfyve regulates the morphology, size and number of stage I melanosomes by promoting formation of membrane tubules and buds that are released in a mechanism involving branched endosomal actin dynamics. Inhibition of PIKfyve activity abrogates membrane remodeling, reduces the number and increases the size of stage I melanosomes, leading to accumulation and unstructured aggregation of PMEL, and thus impairing fibril formation and melanosome maturation. PIKfyve inhibition also prolongs interactions between stage I melanosomes and lysosomes, probably because PIKfyve activity and PI(3,5)P2 are also involved in membrane remodeling processes that lead to segregation of the two compartments. Prolonged interorganelle interactions most likely cause mislocalization of endolysosomal proteins to melanosomes and a loss of melanosomal identity during the whole melanogenesis process. Inset: schematic representation of the role of PIKfyve activity on the regulation of import/export processes in stage I melanosomes.
Interorganelle interactions ensure PMEL amyloid formation
The data presented here confirm the requirement of lysosomal proteases for PMEL processing and amyloid formation, although the localization of these proteases is separated from PMEL (Ho et al., 2016; Kawaguchi et al., 2015; Leonhardt et al., 2013; Raposo et al., 2001). We describe a model in which transient kiss-and-run interactions and fusions between endolysosomes and PMEL-containing stage I melanosomes ensure PMEL processing by lysosomal proteases. These transient interactions provide a dynamic means to coordinate melanosome maturation with PMEL processing and fibril formation, and thus ensure proper amyloid assembly. The kiss-and-run interactions that we describe do not lead to full fusion of both compartments, and are thus consistent with previous observations showing that endolysosomes and melanosomes co-exist as distinct, but proximate organelles, in melanocytes (Raposo et al., 2001). By regulating interorganelle interaction, the PIKfyve activity appears to be a key element of the acquisition of melanosome identity.
Inhibition of PIKfyve activity results in longer interorganelle contacts, which correlates with mislocalization of endolysosomal proteins to melanosomes. Such impaired segregation of melanosomal and lysosomal cargo has been suggested to affect PMEL fibril formation and early melanogenesis (Giordano et al., 2009). One can imagine that the increased lysosomal character of maturing melanosomes may affect PMEL assembly rates.
PIKfyve activity promotes membrane remodeling
Our data demonstrate that PIKfyve activity promotes membrane remodeling and the tubule formation required to maintain stage I melanosome number, homeostasis and function in melanocytes. The molecular mechanism by which PIKfyve activity induces the formation of membrane tubules in mammalian cells still remains elusive. PI(3,5)P2 produced by PIKfyve activates the transient receptor potential mucolipin (TRPML) channel family, and mice carrying a mutation in TRPML3, which is highly expressed in melanocytes, show pigmentation abnormalities (Di Palma et al., 2002; Dong et al., 2010). Activation of the TRPML protein family by the small molecule activator MLSA1 has been shown to rescue the enlarged compartment phenotype of PI(3,5)P2-depleted FIG4- and VAC14-knockout cells (Dong et al., 2010; Zou et al., 2015). However, in melanocytes MLSA1 co-treatment with YM201636 neither counteracted stage I melanosome enlargement nor rescued membrane tubule formation (Fig. S4A,B). These findings indicate that during melanosome biogenesis, TRPMLs may not be downstream effectors of PIKfyve activity, which is in line with our observations in HeLa cells (Bissig et al., 2017). It is worth noting that mice mutant for TRPML3, VAC14 and FIG4 exhibit different fur pigmentation phenotypes (Atiba-Davies and Noben-Trauth, 2007; Chow et al., 2007; Jin et al., 2008; Zhang et al., 2007), which may reflect distinct molecular roles. PI(3,5)P2 has also been shown to activate the melanosomal two-pore sodium channel 2 (TPC2), which negatively regulates pigmentation (Bellono et al., 2016). However, TPC2 acts mainly on pigmented melanosomes, whereas PIKfyve activity is involved in homeostasis of unpigmented melanosomes, and inhibition of PIKfyve activity does not impair melanin production.
It is attractive to speculate that PI(3,5)P2 synthesis by PIKfyve triggers the recruitment of PI(3,5)P2-binding effector proteins, which may have membrane bending properties, as shown for the BAR domain of SNX1, which is involved in retrograde transport (Carlton et al., 2004). Recently, the PROPPIN domain of yeast Atg18, which binds to PI(3)P and PI(3,5)P2, has been shown to drive membrane tubule formation and fission via bilayer insertion of an amphipathic α-helix and oligomerization (Gopaldass et al., 2017). This membrane deformation and fission function may be shared by other PROPPINs, such as the mammalian WIPI proteins, which also contain potential amphipathic α-helical features. Interestingly, mammalian WIPI1 is involved in early melanogenesis, and is thus a promising PI(3,5)P2 effector in melanocytes (Ho et al., 2011).
Our data further provide evidence that membrane tubules generated by PIKfyve activity are released by a mechanism involving branched actin dynamics. As PI(3,5)P2 regulates endosomal branched actin, it is likely that PIKfyve activity also controls actin-dependent membrane release. A role of actin in membrane release is exemplified by the fission of Shiga-toxin carriers induced by actin-driven lipid-phase separation (Allain et al., 2004; Romer et al., 2010; Roux et al., 2005) and in endosomal fission associated with ER contact sites (Rowland et al., 2014). The actin network may also promote membrane reorganization and neck constriction by inducing mechanical forces, which may involve the action of actin-based motors (Derivery et al., 2009; Ripoll et al., 2018). In that respect, it is interesting to note that overexpression of myosin 1b affects early endosome morphology, slows PMEL processing and impairs PMEL fibril formation (Salas-Cortes et al., 2005) – phenotypes reminiscent of PIKfyve inhibition.
PIKfyve-mediated membrane remodeling is required for formation of PMEL amyloid matrix
Our study provides evidence that formation of early melanosomal membrane tubules initiated by PIKfyve activity is co-ordinated with PMEL amyloid assembly and melanosome maturation. Similarly, endosome maturation was described to be co-ordinated with endosome tubule formation (van Weering et al., 2012), indicating that melanocytes have fine-tuned a ubiquitous endosomal process for melanosome biogenesis. Membrane tubules formed during endosome maturation serve as cargo sorting to different cellular locations, which is ensured by the assembly of various sorting machineries. In melanocytes, tubules emerging in an actin-dependent manner from recycling endosomes transport melanogenic enzymes, transporters and accessory proteins to melanosomes for pigment synthesis (Delevoye et al., 2016; Dennis et al., 2016; Setty et al., 2007; Sitaram et al., 2012). These tubules are, however, distinct from the PIKfyve-driven tubules described here, as we do not observe any defect in pigment synthesis. Endosomal tubules formed by PIKfyve activity are involved in endosome-to-TGN retrograde transport (de Lartigue et al., 2009; Rutherford et al., 2006), a process that also involves the endosomal actin network (Seaman et al., 2013). However, such retrograde transport has not been described for PMEL. The membrane remodeling initiated by the PIKfyve activity described here instead contributes to homotypic fusion–fission processes that allow a homogenous repartition of PMEL within the small and highly dynamic population of stage I melanosomes. Impaired membrane remodeling then results in the accumulation of PMEL lumenal fragments in enlarged, immotile and fewer stage I melanosomes. This accumulation saturates the PMEL processing and sorting machinery and leads to aberrant aggregation of PMEL, and thus a defect in PMEL fibril formation and melanosome maturation. Interestingly, evidence suggests that terminal proteolytic maturation of lumenal PMEL fragments only occurs after aggregation into a pre-matrix (Leonhardt et al., 2013). Thus, impaired pre-matrix assembly may lead to accumulation of partially processed lumenal PMEL fragments that aberrantly aggregate due to their amyloidogenic nature.
Implications for pathological amyloid formation
The PMEL fibrils that assemble within melanosomes are amyloids (Fowler et al., 2006). In contrast to pathological amyloids that are associated with neurodegenerative diseases such as Alzheimer's, Parkinson's or Huntington's disease, PMEL amyloid fibrils are not pathogenic and serve beneficial functions. They are thus called functional or physiological amyloids. Nevertheless, the amyloidogenic nature of PMEL presents a potential danger for the cell, as aberrant aggregation may be toxic (Watt et al., 2011). It is possible that long-term inhibition of PIKfyve activity may damage melanosome integrity by triggering an accumulation of toxic PMEL aggregates and/or melanin intermediates, perhaps leading to melanocyte death. Interestingly, mice with a specific knockout for PIKfyve in pigment cells (Liggins et al., 2018) exhibit hair graying that is similar to what is seen in mouse models with impaired PMEL fibril formation (Rochin et al., 2013), and their melanocytes display vacuolar compartments that are reminiscent of our observations. Lower melanocyte survival rates may also explain why VAC14 and FIG4 mutant mice show hypopigmentation phenotypes, while we did not observe impaired pigment synthesis. Since PMEL fibrils sequester melanin and thus might facilitate melanin transfer to keratinocytes, fur hypopigmentation may also result from inefficient pigment transfer. These two models are not mutually exclusive.
In view of the interesting similarities between intracellular trafficking and processing of PMEL and amyloid precursor protein (APP), which is involved in Alzheimer's disease (Bissig et al., 2016; Sannerud et al., 2016), our studies may also have important implications for pathological amyloid formation during Alzheimer's disease. Intriguingly, PIKfyve depletion leads to massive neurodegeneration in individuals with mutations of FIG4 and VAC14 (Baulac et al., 2014; Campeau et al., 2013; Chow et al., 2007; Lenk et al., 2011; Lenk et al., 2016), and PIKfyve influences APP (Balklava et al., 2015; Currinn et al., 2016). It is therefore tempting to speculate that, in neurons, PIKfyve activity regulates the abundance of an amyloidogenic substrate and its proteases in a compartment known to generate the Aβ intracellular pool (Peric and Annaert, 2015; Rajendran and Annaert, 2012; Sannerud et al., 2016). Further work will determine whether PIKfyve activity plays analogous roles in physiological and pathological amyloid formation.
MATERIALS AND METHODS
Cell culture, transfections and drug treatment
Human melanocytic MNT-1 cells and primary melanocytes were maintained as previously described (Berson et al., 2001; Raposo et al., 2001; van Niel et al., 2015). Cells were transfected according to manufacturer's recommendation with DNA and siRNA using JetPrime (Polyplus Transfection) and Oligofectamine (Invitrogen), respectively. Experiments were performed 16 h after transfection with DNA. siRNA transfections were performed twice at 48 h intervals, and experiments were performed 96 h after the first siRNA transfection. If not otherwise indicated, cells were incubated for 2 h with 1.6 µM YM201636 (Abcam), 200 µM CK-666 (Tocris), 1 µM Vps34 inhibitor (IN1, Calbiochem) or DMSO (Sigma-Aldrich). YM201636 was washed out for 1 h to 4 h at 37°C after three washes with cold medium. If indicated, 200 µM CK-666 (Tocris) was present during the washout.
Reagents, antibodies, siRNAs and plasmids
Reagents were obtained from the following sources: 4′,6-diamidino-2-phenylindole (DAPI), Dextran 10,000 kDa conjugated to Alexa Fluor 488, 555 or 647 and DQ-BSA from Thermo Fisher Scientific; MagicRed probe from Immuno Chemistry Technologies; pepstatinA and E-64d from Calbiochem; leupeptin from Euromedex and other reagents were obtained from Sigma-Aldrich.
Antibodies were obtained from the following sources: anti-FIG4 [ab186269, 1:500 dilution for western blotting (WB)], anti-VAC14 (ab67369, 1:1000 dilution for WB), anti-melanoma gp100 (HMB45) [ab787, 1:200 dilution for WB and immunofluorescence (IF)], anti-PMEL NKI-beteb (ab34165, 1:200 dilution for IF), anti-β-TUB (ab6046, 1:10,000 dilution for WB), anti-PSEN2 (ab51249, 1:100 dilution for IF), anti-TYRP1 (ab3312, 1:200 dilution for IF) anti-EEA1 (ab70521, 1:200 dilution for IF) and horseradish peroxidase (HRP)-conjugated goat polyclonal antibodies to rabbit IgG (ab6721, 1:10,000 dilution for WB) and to mouse IgG (ab6789, 1:10,000 dilution for WB) antibodies from Abcam; anti-PIKfyve antibody (PCR-PIKFYVE-3C9, 1:100 dilution for WB) from the Developmental Studies Hybridoma Bank (DSHB); mouse anti-LAMP1 antibody (555798, 1:200 for IF) from BD Biosciences; and secondary goat anti-rabbit-IgG or anti-mouse IgG antibodies conjugated to Alexa Fluor 488, 555 or 647 from Thermo Fisher Scientific and used at 1:200. Protein A conjugated to 15 nm gold particles (PAG15) and BSA conjugated to 5 nm gold particles (BSA-gold 5 nm) were from Cell Microscopy Center (AZU, Utrecht University, Netherlands). Affinity-purified anti-peptide antibodies recognizing the PMEL N-terminus (anti-PMEL-N, 1:200 dilution for IF, 1:3000 dilution for WB and 1:40 for EM) (Berson et al., 2003) and C-terminus (anti-PMEL-C, 1:100 dilution for IF and 1:200 dilution for WB) (Raposo et al., 2001) were described previously.
The siRNAs targeting FIG4 (L019141), VAC14 (L015729), PIKfyve (L005058) and non-targeting control (D001810) ON-TARGET plus SMARTpool were from Thermo Fisher Scientific.
The 2×FYVE–GFP (Gillooly et al., 2000) was a kind gift from Harald Stenmark (Department of Molecular Cell Biology, Institute for Cancer Research, The Norwegian Radium Hospital, Oslo, Norway) and LAMP1–mRuby2 was Addgene plasmid #55902, deposited by Michael Davidson (Lam et al., 2012).
Dextran internalization assays
To internalize Dextran 10,000 AF555 into endolysosomes, MNT-1 cells were pulsed with 1 mg/ml DextranAF555 at 37°C for 4 h, washed three times with cold medium and chased with conjugate-free medium for 20 h at 37°C (Bright et al., 2005). For DextranAF647 cointernalization with DQ-BSA, MNT-1 cells were pretreated for 2 h with 1.6 µM YM201636 or DMSO. Then DextranAF647 and DQ-BSA were co-internalized at a concentration of 50 µg/ml each for 2 h at 37°C. Cells were washed three times with cold medium and incubated for 1 h with conjugate-free medium at 37°C. Snapshots of live cells were taken at 37°C in imaging buffer (20 mM Hepes-NaOH pH 7.4 140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 4.5 g/l D-glucose). YM201636 or DMSO was present throughout the experiment.
Electron microscopy
For conventional EM, MNT-1 were grown on coverslips, fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 24 h, post-fixed with 1% osmium tetroxide, dehydrated in ethanol and embedded in Epon as described previously (Raposo et al., 2001). For EM analysis of RPE sections, tissues from newborn mice were processed as previously (Rochin et al., 2013). All animal use was performed in compliance with guidelines of the University Committee on Use and Care of Animals of the University of Michigan and National Institutes of Health. For high-pressure freezing, MNT-1 cells were grown on carbonated sapphire discs (30 mm in diameter) and high-pressure frozen using a HPM 100 (Leica Microsystems) or HPM Light µ (CryoCapCell) in FBS serving as filler. High-pressure frozen samples were transferred to an AFS (Leica Microsystems) with precooled (−90°C) anhydrous acetone containing 2% osmium tetroxide and 1% H2O. Freeze substitution and peon embedding was performed as described previously (Hurbain et al., 2008). Ultrathin sections of cell monolayers or RPE were prepared with a Reichert UltracutS ultramicrotome (Leica Microsystems) and contrasted with uranyl acetate and lead citrate.
For ultrathin cryosectioning and immunogold labeling, cells were fixed in 2% paraformaldehyde (PFA) with 0.2% glutaraldehyde in 0.1 M phosphate buffer pH 7.4. Cells were processed for ultracryomicrotomy and immunogold labeled using PAG 15 as described previously (Raposo et al., 2001).
All samples were examined with a FEI Tecnai Spirit electron microscope (Thermo Fisher Scientific), and digital acquisitions were made with a numeric camera (Quemesa; EMSIS).
Live-cell microscopy and immunofluorescence analysis
MNT-1 cells grown on coverslips were fixed with 2% PFA, permeabilized with 0.05% saponin/1% BSA in PBS, quenched with 50 mM glycine and processed for indirect immunolabeling. Images were captured on a LSM 780 confocal microscope (Zeiss) using an oil-immersion plan Apo 63× A/1.40 NA objective lens.
Live-cell imaging experiments were performed in fluorodishes (World Precision Instruments) and viewed with an inverted spinning disc microscope (Nikon) using an oil-immersion plan Apo 100× A/1.40 NA objective lens. Live-cell imaging was performed at 37°C in imaging buffer (20 mM Hepes-NaOH pH 7.4, 140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2 and 4.5 g/l D-glucose) and movies were taken at 0.4 s frame rate for 2 min. Data were collected using Metamorph Software, and analyzed using ImageJ, Icy and CellProfiler software.
Image analysis and quantification
Melanosome stages were defined by morphology (Raposo et al., 2001; Seiji et al., 1963). The length and width of melanosomes, as well as the size were measured using ImageJ software. To measure contact times, organelles were segmented using Icy software and the time that two organelles partially overlap was assessed. The size of organelles was measured after segmentation using Icy software. Colocalization was quantified using CellProfiler software, calculating the correlation coefficient. Dextran transfer to 2×FYVE–GFP compartments [defined as the region of interest (ROI)] was quantified measuring the fluorescence intensity of the Dextran channel in the ROI using ImageJ. P-values were determined by performing an unpaired Student's t-test for unequal variance (*P<0.05, **P<0.01, ***P<0.001).
Other methods
Western blotting
For western blots, a Triton X-soluble lysate was prepared in 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% TX-100, 1 mM EDTA and protease inhibitors. The Triton X-insoluble fraction was resuspended in 1% SDS and 1% B-mercaptoethanol in PBS containing protease inhibitors, incubated for 10 min at room temperature and then heated for 10 min at 100°C.
Membrane cytosol fractionation
Cells are scraped and centrifuged at 900 rpm for 5 min at 4°C. The pellet is homogenized in homogenization buffer (0.25 M sucrose, 10 mM Hepes-NaOH pH 7.4) and then centrifuged at 500 g for 10 min at 4°C. The pellet is resuspended in the homogenization buffer and after centrifugation at 500 g for 10 min, the post nuclear supernatant (PNS) is collected and centrifuged at 100,000 g for 1 h at 4°C. The supernatant (cytosol fraction) is collected and the pellet (membrane fraction) is resuspended in 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5 mM EDTA solution containing protease inhibitors.
Melanin assay
Cells were disrupted by sonication in 50 mM Tris-HCl pH 7.4, 2 mM EDTA, 150 mM NaCl, 1 mM DTT, and protease inhibitors. The pigment was pelleted at 16,000 g for 15 min at 4°C, rinsed once in ethanol/ether (1:1), and dissolved in 2 M NaOH in 20% DMSO at 60°C. Melanin content was measured as the optical density (OD) at 492 nm.
Ratiometric pH measurement
The pH-sensitive fluorophore Oregon Green 488 (DextranOG) and pH-insensitive fluorophore Alexa Fluor 647 (DextranAF647) were internalized and imaged as described above in Dextran internalization assays. To convert fluorescence values to pH, the emission of the two dyes was recorded separately and the fluorescence ratio was converted to pH using an internal calibration curve. To acquire the calibration curve, cells were sequentially bathed for 5 min in 143 mM KCl, 5 mM glucose, 1 mM MgCl2, 1 mM CaCl2, 20 mM Hepes buffered to a pH ranging from 4.0 to 7.5 in solution containing 10 µM nigericin and 5 µM monensin.
Supplementary Material
Acknowledgements
We are grateful to Michael S. Marks and Cedric Delevoye for fruitful discussions and critical reading of the manuscript. We thank the PITC-IBiSA Imaging Facility, the Institut Curie (Paris), the Nikon Imaging Center and the members of the France-BioImaging national research infrastructure for assistance with microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.B., G.v.N.; Methodology: C.B., P.C., X.H., I.H., G.v.N.; Validation: G.v.N.; Formal analysis: C.B., P.C.; Investigation: C.B., P.C., X.H., I.H., R.S., G.v.H.; Resources: G.M.L., E.K., R.S., M.H.M., L.S.W.; Data curation: C.B.; Writing - original draft: C.B., G.v.N.; Writing - review & editing: C.B., W.A., M.H.M., L.S.W., G.R., G.v.N.; Supervision: G.v.N.; Project administration: G.v.N.; Funding acquisition: G.R., G.v.N.
Funding
This work was supported by Institut Curie, Centre National de la Recherche Scientifique (CNRS), by the Fondation ARC pour la Recherche sur le Cancer (grant SL220100601359) (to G.R.), by the Fondation pour la Recherche Médicale (AJE20160635884) (to G.v.N.), by a Research Grant from the Amyloidosis Foundation (to G.v.N.), from the French National Research Agency (Agence Nationale de la Recherche) through the “Investments for the Future” program (France-BioImaging, ANR-10-INSB-04), the CelTisPhyBio Labex (no. ANR-10-LBX-0038) part of the IDEX PSL (no. ANR-10-IDEX-0001-02 PSL) and the Swiss National Fund for Research for the Early Postdoc. Mobility fellowship P2GEP3-151589 and the Advanced Postdoc. Mobility fellowship P300PA_167618 (to C.B.), the Federation of European Biochemical Societies for the Long-Term Fellowship (to C.B.) and Labex CelTisPhyBio for the post-doctoral fellowship (C.B.). W.A. and R.S. are supported by Vlaams Instituut voor Biotechnologie (VIB) and grants of KU Leuven (C16/15/073), Fonds Wetenschappelijk Onderzoek (FWO) (G078117N and SBO-S006617N), Hercules Foundation (AKUL/09/037) and Foundation for Alzheimer Research (SAO-FRA) (S#16018; 2017/033). We also acknowledge support from the National Institutes of Health (NIH) grant R01 GM24872 (M.H.M.) and R01 NS NS064015 (L.S.W.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.229500.supplemental
References
- Allain J. M., Storm C., Roux A., Ben Amar M. and Joanny J. F. (2004). Fission of a multiphase membrane tube. Phys. Rev. Lett. 93, 158104 10.1103/PhysRevLett.93.158104 [DOI] [PubMed] [Google Scholar]
- Anitei M. and Hoflack B (2011). Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat. Cell Biol. 14, 11-19. 10.1038/ncb2409 [DOI] [PubMed] [Google Scholar]
- Atiba-Davies M. and Noben-Trauth K (2007). TRPML3 and hearing loss in the varitint-waddler mouse. Biochim. Biophys. Acta 1772, 1028-1031. 10.1016/j.bbadis.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Balklava Z., Niehage C., Currinn H., Mellor L., Guscott B., Poulin G., Hoflack B. and Wassmer T (2015). The amyloid precursor protein controls PIKfyve function. PLoS ONE 10, e0130485 10.1371/journal.pone.0130485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S., Lenk G. M., Dufresnois B., Ouled Amar Bencheikh B., Couarch P., Renard J., Larson P. A., Ferguson C. J., Noe E., Poirier K. et al. (2014). Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology 82, 1068-1075. 10.1212/WNL.0000000000000241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellono N. W., Escobar I. E. and Oancea E (2016). A melanosomal two-pore sodium channel regulates pigmentation. Sci. Rep. 6, 26570 10.1038/srep26570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson J. F., Harper D. C., Tenza D., Raposo G. and Marks M. S. (2001). Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol. Biol. Cell 12, 3451-3464. 10.1091/mbc.12.11.3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson J. F., Theos A. C., Harper D. C., Tenza D., Raposo G. and Marks M. S. (2003). Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 161, 521-533. 10.1083/jcb.200302072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C., Rochin L. and van Niel G. (2016). PMEL amyloid fibril formation: the bright steps of pigmentation. Int. J. Mol. Sci. 17 10.3390/ijms17091438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C., Hurbain I., Raposo G. and van Niel G. (2017). PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes. Traffic 18, 747-757. 10.1111/tra.12525 [DOI] [PubMed] [Google Scholar]
- Bonangelino C. J., Nau J. J., Duex J. E., Brinkman M., Wurmser A. E., Gary J. D., Emr S. D. and Weisman L. S. (2002). Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 156, 1015-1028. 10.1083/jcb.200201002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright N. A., Gratian M. J. and Luzio J. P. (2005). Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 15, 360-365. 10.1016/j.cub.2005.01.049 [DOI] [PubMed] [Google Scholar]
- Campeau P. M., Lenk G. M., Lu J. T., Bae Y., Burrage L., Turnpenny P., Roman Corona-Rivera J., Morandi L., Mora M., Reutter H. et al. (2013). Yunis-Varon syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am. J. Hum. Genet. 92, 781-791. 10.1016/j.ajhg.2013.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B. J., Oorschot V. M., Rutherford A., Mellor H., Klumperman J., McMahon H. T. and Cullen P. J. (2004). Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 14, 1791-1800. 10.1016/j.cub.2004.09.077 [DOI] [PubMed] [Google Scholar]
- Chow C. Y., Zhang Y., Dowling J. J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M. E., Li J., Zhang X. et al. (2007). Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448, 68-72. 10.1038/nature05876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currinn H., Guscott B., Balklava Z., Rothnie A. and Wassmer T (2016). APP controls the formation of PI(3,5)P-2 vesicles through its binding of the PIKfyve complex. Cell. Mol. Life Sci. 73, 393-408. 10.1007/s00018-015-1993-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S. A., Urbe S. and Clague M. J. (2009). PIKfyve regulation of endosome-linked pathways. Traffic 10, 883-893. 10.1111/j.1600-0854.2009.00915.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C., Heiligenstein X., Ripoll L., Gilles-Marsens F., Dennis M. K., Linares R. A., Derman L., Gokhale A., Morel E., Faundez V. et al. (2016). BLOC-1 brings together the actin and microtubule cytoskeletons to generate recycling endosomes. Curr. Biol. 26, 1-13. 10.1016/j.cub.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M. K., Delevoye C., Acosta-Ruiz A., Hurbain I., Romao M., Hesketh G. G., Goff P. S., Sviderskaya E. V., Bennett D. C., Luzio J. P. et al. (2016). BLOC-1 and BLOC-3 regulate VAMP7 cycling to and from melanosomes via distinct tubular transport carriers. J. Cell Biol. 214, 293-308. 10.1083/jcb.201605090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D. and Gautreau A (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712-723. 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Di Palma F., Belyantseva I. A., Kim H. J., Vogt T. F., Kachar B. and Noben-Trauth K (2002). Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA 99, 14994-14999. 10.1073/pnas.222425399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L. S., Delling M et al. et al. (2010). PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun. 1, 38 10.1038/ncomms1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S. K., McEwen R. K., Mayes A., Hughes D. C., Beggs J. D. and Michell R. H. (2002). Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12, 885-893. 10.1016/S0960-9822(02)00891-6 [DOI] [PubMed] [Google Scholar]
- Duex J. E., Nau J. J., Kauffman E. J. and Weisman L. S. (2006a). Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot. Cell 5, 723-731. 10.1128/EC.5.4.723-731.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex J. E., Tang F. and Weisman L. S. (2006b). The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J. Cell Biol. 172, 693-704. 10.1083/jcb.200512105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn L. C. and Thigpen L. W. (1930). The silver mouse: a recessive color variation. J. Heredity 495-498. 10.1093/oxfordjournals.jhered.a103275 [DOI] [Google Scholar]
- Ferguson C. J., Lenk G. M. and Meisler M. H. (2009). Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 18, 4868-4878. 10.1093/hmg/ddp460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E. and Kelly J. W. (2006). Functional amyloid formation within mammalian tissue. PLoS Biol. 4, e6 10.1371/journal.pbio.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G. and Stenmark H (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577-4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F., Bonetti C., Surace E. M., Marigo V. and Raposo G (2009). The ocular albinism type 1 (OA1) G-protein-coupled receptor functions with MART-1 at early stages of melanogenesis to control melanosome identity and composition. Hum. Mol. Genet. 18, 4530-4545. 10.1093/hmg/ddp415 [DOI] [PubMed] [Google Scholar]
- Gopaldass N., Fauvet B., Lashuel H., Roux A. and Mayer A (2017). Membrane scission driven by the PROPPIN Atg18. EMBO J. 36, 3274-3291. 10.15252/embj.201796859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom A. R., Watt B., Fard S. S., Tenza D., Mannstrom P., Narfström K., Ekesten B., Ito S., Wakamatsu K., Larsson J. et al. (2011). Inactivation of Pmel alters melanosome shape but has only a subtle effect on visible pigmentation. PLoS Genet. 7, e1002285 10.1371/journal.pgen.1002285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H., Kapadia R., Al-Tahan S., Ahmad S. and Ganesan A. K. (2011). WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J. Biol. Chem. 286, 12509-12523. 10.1074/jbc.M110.200543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. Y., Choy C. H., Wattson C. A., Johnson D. E. and Botelho R. J. (2015). The Fab1/PIKfyve phosphoinositide phosphate kinase is not necessary to maintain the pH of lysosomes and of the yeast vacuole. J. Biol. Chem. 290, 9919-9928. 10.1074/jbc.M114.613984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T., Watt B., Spruce L. A., Seeholzer S. H. and Marks M. S. (2016). The kringle-like domain facilitates post-endoplasmic reticulum changes to premelanosome protein (PMEL) oligomerization and disulfide bond configuration and promotes amyloid formation. J. Biol. Chem. 291, 3595-3612. 10.1074/jbc.M115.692442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong N. H., Qi A. and Weaver A. M. (2015). PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions. J. Cell Biol. 210, 753-769. 10.1083/jcb.201412127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurbain I., Geerts W. J., Boudier T., Marco S., Verkleij A. J., Marks M. S. and Raposo G (2008). Electron tomography of early melanosomes: implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc. Natl. Acad. Sci. USA 105, 19726-19731. 10.1073/pnas.0803488105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O. C., Sbrissa D., Fenner H. and Shisheva A (2009). PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J. Biol. Chem. 284, 35794-35806. 10.1074/jbc.M109.037515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incerti B., Cortese K., Pizzigoni A., Surace E. M., Varani S., Coppola M., Jeffery G., Seeliger M., Jaissle G., Bennett D. C. et al. (2000). Oa1 knock-out: new insights on the pathogenesis of ocular albinism type 1. Hum. Mol. Genet. 9, 2781-2788. 10.1093/hmg/9.19.2781 [DOI] [PubMed] [Google Scholar]
- Jefferies H. B., Cooke F. T., Jat P., Boucheron C., Koizumi T., Hayakawa M., Kaizawa H., Ohishi T., Workman P., Waterfield M. D. et al. (2008). A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 9, 164-170. 10.1038/sj.embor.7401155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N., Chow C. Y., Liu L., Zolov S. N., Bronson R., Davisson M., Petersen J. L., Zhang Y., Park S., Duex J. E. et al. (2008). VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 27, 3221-3234. 10.1038/emboj.2008.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M., Hozumi Y. and Suzuki T (2015). ADAM protease inhibitors reduce melanogenesis by regulating PMEL17 processing in human melanocytes. J. Dermatol. Sci. 78, 133-142. 10.1016/j.jdermsci.2015.02.020 [DOI] [PubMed] [Google Scholar]
- Lam A. J., St-Pierre F., Gong Y., Marshall J. D., Cranfill P. J., Baird M. A., McKeown M. R., Wiedenmann J., Davidson M. W., Schnitzer M. J. et al. (2012). Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005-1012. 10.1038/nmeth.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Z. H., Hou L., Moellmann G., Kuklinska E., Antol K., Fraser M., Halaban R. and Kwon B. S. (1996). Characterization and subcellular localization of human Pmel 17/silver, a 110-kDa (pre)melanosomal membrane protein associated with 5,6,-dihydroxyindole-2-carboxylic acid (DHICA) converting activity. J. Invest. Dermatol. 106, 605-610. 10.1111/1523-1747.ep12345163 [DOI] [PubMed] [Google Scholar]
- Lenk G. M., Ferguson C. J., Chow C. Y., Jin N., Jones J. M., Grant A. E., Zolov S. N., Winters J. J., Giger R. J., Dowling J. J. et al. (2011). Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 7, e1002104 10.1371/journal.pgen.1002104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk G. M., Szymanska K., Debska-Vielhaber G., Rydzanicz M., Walczak A., Bekiesinska-Figatowska M., Vielhaber S., Hallmann K., Stawinski P., Buehring S. et al. (2016). Biallelic mutations of VAC14 in pediatric-onset neurological disease. Am. J. Hum. Genet. 99, 188-194. 10.1016/j.ajhg.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt R. M., Vigneron N., Rahner C. and Cresswell P (2011). Proprotein convertases process Pmel17 during secretion. J. Biol. Chem. 286, 9321-9337. 10.1074/jbc.M110.168088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt R. M., Vigneron N., Hee J. S., Graham M. and Cresswell P (2013). Critical residues in the PMEL/Pmel17 N-terminus direct the hierarchical assembly of melanosomal fibrils. Mol. Biol. Cell 24, 964-981. 10.1091/mbc.e12-10-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy F., Muehlethaler K., Salvi S., Peitrequin A. L., Lindholm C. K., Cerottini J. C. and Rimoldi D (2005). Ubiquitylation of a melanosomal protein by HECT-E3 ligases serves as sorting signal for lysosomal degradation. Mol. Biol. Cell 16, 1777-1787. 10.1091/mbc.e04-09-0803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggins M. C., Flesher J. L., Jahid S., Vasudeva P., Eby V., Takasuga S., Sasaki J., Sasaki T., Boissy R. E. and Ganesan A. K. (2018). PIKfyve regulates melanosome biogenesis. PLoS Genet. 14, e1007290 10.1371/journal.pgen.1007290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes V. S., Wasmeier C., Seabra M. C. and Futter C. E. (2007). Melanosome maturation defect in rab38-deficient retinal pigment epithelium results in instability of immature Melanosomes during transient melanogenesis. Mol. Biol. Cell 18, 3914-3927. 10.1091/mbc.e07-03-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney A. J., Zhang Y. and Weisman L. S. (2014). Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays 36, 52-64. 10.1002/bies.201300012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric A. and Annaert W. (2015). Early etiology of Alzheimer's disease: tipping the balance toward autophagy or endosomal dysfunction? Acta Neuropathol. 129, 363-381. 10.1007/s00401-014-1379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L. and Annaert W (2012). Membrane trafficking pathways in Alzheimer's disease. Traffic 13, 759-770. 10.1111/j.1600-0854.2012.01332.x [DOI] [PubMed] [Google Scholar]
- Raposo G., Tenza D., Murphy D. M., Berson J. F. and Marks M. S. (2001). Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 152, 809-824. 10.1083/jcb.152.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll L., Heiligenstein X., Hurbain I., Domingues L., Figon F., Petersen K. J., Dennis M. K., Houdusse A., Marks M. S., Raposo G et al. et al. (2018). Myosin VI and branched actin filaments mediate membrane constriction and fission of melanosomal tubule carriers. J. Cell Biol. 217, 2709-2726. 10.1083/jcb.201709055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochin L., Hurbain I., Serneels L., Fort C., Watt B., Leblanc P., Marks M. S., De Strooper B., Raposo G. and van Niel G (2013). BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc. Natl. Acad. Sci. USA 110, 10658-10663. 10.1073/pnas.1220748110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer W., Pontani L. L., Sorre B., Rentero C., Berland L., Chambon V., Lamaze C., Bassereau P., Sykes C., Gaus K et al. et al. (2010). Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell 140, 540-553. 10.1016/j.cell.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Roux A., Cuvelier D., Nassoy P., Prost J., Bassereau P. and Goud B (2005). Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 24, 1537-1545. 10.1038/sj.emboj.7600631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A. A., Chitwood P. J., Phillips M. J. and Voeltz G. K. (2014). ER contact sites define the position and timing of endosome fission. Cell 159, 1027-1041. 10.1016/j.cell.2014.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge S. A., Anderson D. M. and Emr S. D. (2004). Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell 15, 24-36. 10.1091/mbc.e03-05-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford A. C., Traer C., Wassmer T., Pattni K., Bujny M. V., Carlton J. G., Stenmark H. and Cullen P. J. (2006). The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 119, 3944-3957. 10.1242/jcs.03153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Cortes L., Ye F., Tenza D., Wilhelm C., Theos A., Louvard D., Raposo G. and Coudrier E (2005). Myosin Ib modulates the morphology and the protein transport within multi-vesicular sorting endosomes. J. Cell Sci. 118, 4823-4832. 10.1242/jcs.02607 [DOI] [PubMed] [Google Scholar]
- Sannerud R., Esselens C., Ejsmont P., Mattera R., Rochin L., Tharkeshwar A. K., De Baets G., De Wever V., Habets R., Baert V. et al. (2016). Restricted Location of PSEN2/gamma-secretase determines substrate specificity and generates an intracellular abeta pool. Cell 166, 193-208. 10.1016/j.cell.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O. C., Filios C., Delvecchio K. and Shisheva A (2012). Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. American journal of physiology. Cell Physiol. 303, C436-C446. 10.1152/ajpcell.00105.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N. J., Gautreau A. and Billadeau D. D. (2013). Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 23, 522-528. 10.1016/j.tcb.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiji M., Fitzpatrick T. B., Simpson R. T. and Birbeck M. S. (1963). Chemical composition and terminology of specialized organelles (melanosomes and melanin granules) in mammalian melanocytes. Nature 197, 1082-1084. 10.1038/1971082a0 [DOI] [PubMed] [Google Scholar]
- Setty S. R., Tenza D., Truschel S. T., Chou E., Sviderskaya E. V., Theos A. C., Lamoreux M. L., Di Pietro S. M., Starcevic M., Bennett D. C. et al. (2007). BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell 18, 768-780. 10.1091/mbc.e06-12-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A. and Marks M. S. (2012). Mechanisms of protein delivery to melanosomes in pigment cells. Physiology 27, 85-99. 10.1152/physiol.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A., Dennis M. K., Chaudhuri R., De Jesus-Rojas W., Tenza D., Setty S. R., Wood C. S., Sviderskaya E. V., Bennett D. C., Raposo G. et al. (2012). Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol. Biol. Cell 23, 3178-3192. 10.1091/mbc.e11-06-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A. C., Berson J. F., Theos S. C., Herman K. E., Harper D. C., Tenza D., Sviderskaya E. V., Lamoreux M. L., Bennett D. C., Raposo G. et al. (2006a). Dual loss of ER export and endocytic signals with altered melanosome morphology in the silver mutation of Pmel17. Mol. Biol. Cell 17, 3598-3612. 10.1091/mbc.e06-01-0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., Thomas P. C., Raposo G. and Marks M. S. (2006b). A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev. Cell 10, 343-354. 10.1016/j.devcel.2006.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M. S., Rubinstein E. and Raposo G (2011). The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 21, 708-721. 10.1016/j.devcel.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., Bergam P., Di Cicco A., Hurbain I., Lo Cicero A., Dingli F., Palmulli R., Fort C., Potier M. C., Schurgers L. J. et al. (2015). Apolipoprotein E regulates amyloid formation within endosomes of pigment cells. Cell Rep 13, 43-51. 10.1016/j.celrep.2015.08.057 [DOI] [PubMed] [Google Scholar]
- van Weering J. R., Verkade P. and Cullen P. J. (2012). SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic 13, 94-107. 10.1111/j.1600-0854.2011.01297.x [DOI] [PubMed] [Google Scholar]
- Viaud J., Boal F., Tronchere H., Gaits-Iacovoni F. and Payrastre B (2014). Phosphatidylinositol 5-phosphate: a nuclear stress lipid and a tuner of membranes and cytoskeleton dynamics. BioEssays 36, 260-272. 10.1002/bies.201300132 [DOI] [PubMed] [Google Scholar]
- Watt B., van Niel G., Fowler D. M., Hurbain I., Luk K. C., Stayrook S. E., Lemmon M. A., Raposo G., Shorter J., Kelly J. W. et al. (2009). N-terminal domains elicit formation of functional Pmel17 amyloid fibrils. J. Biol. Chem. 284, 35543-35555. 10.1074/jbc.M109.047449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt B., Tenza D., Lemmon M. A., Kerje S., Raposo G., Andersson L. and Marks M. S. (2011). Mutations in or near the transmembrane domain alter PMEL amyloid formation from functional to pathogenic. PLoS Genet. 7, e1002286 10.1371/journal.pgen.1002286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zolov S. N., Chow C. Y., Slutsky S. G., Richardson S. C., Piper R. C., Yang B., Nau J. J., Westrick R. J., Morrison S. J. et al. (2007). Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. USA 104, 17518-17523. 10.1073/pnas.0702275104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S. N., Bridges D., Zhang Y., Lee W. W., Riehle E., Verma R., Lenk G. M., Converso-Baran K., Weide T., Albin R. L. et al. (2012). In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA 109, 17472-17477. 10.1073/pnas.1203106109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Hu B., Arpag S., Yan Q., Hamilton A., Zeng Y. S., Vanoye C. G. and Li J (2015). Reactivation of lysosomal Ca2+ efflux rescues abnormal lysosomal storage in FIG4-deficient cells. J. Neurosci. 35, 6801-6812. 10.1523/JNEUROSCI.4442-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.