ABSTRACT
The actin cytoskeleton is the engine that powers the inflammatory chemotaxis of immune cells to sites of tissue damage or infection. Here, we combine genetics with live in vivo imaging to investigate how cytoskeletal rearrangements drive macrophage recruitment to wounds in Drosophila. We find that the actin-regulatory protein Ena is a master regulator of lamellipodial dynamics in migrating macrophages, where it remodels the cytoskeleton to form linear filaments that can then be bundled together by the cross-linker Fascin (also known as Singed in flies). In contrast, the formin Dia generates rare, probing filopods for specialised functions that are not required for migration. The role of Ena in lamellipodial bundling is so fundamental that its overexpression increases bundling even in the absence of Fascin by marshalling the remaining cross-linking proteins to compensate. This reorganisation of the lamellipod generates cytoskeletal struts that push against the membrane to drive leading edge advancement and boost cell speed. Thus, Ena-mediated remodelling extracts the most from the cytoskeleton to power robust macrophage chemotaxis during their inflammatory recruitment to wounds.
KEY WORDS: Drosophila, Ena, Actin, Hemocyte, Macrophage, Migration
Summary: Macrophages must migrate to a variety of stimuli, including inflammatory wounds. We identify the actin-regulatory protein Ena as a master remodeller of the cytoskeleton within migrating macrophages in vivo.
INTRODUCTION
Cell migration is crucial to whole swathes of fundamental biology, including embryogenesis, cancer metastasis, wound healing and immunity. This is perhaps most evident in immune cells, such as macrophages, which are required to rapidly migrate to sites of damage and infection (Wood and Martin, 2017). Through chemotaxis, immune cells are drawn towards wounds by detecting and migrating towards signals released by damaged tissue. For example, both fly and fish leukocytes rapidly respond to the early damage signal, H2O2, detected via Src family kinases (Niethammer et al., 2009; Moreira et al., 2010; Yoo et al., 2011; Razzell et al., 2013).
The cellular protrusions that underlie motility are formed through rearrangements in the actin cytoskeleton enacted through the activity of highly conserved actin regulators. For example, the Arp2/3 complex creates dendritic networks of F-actin driving the extension of the lamellipod during cell motility (Mullins et al., 1998; Svitkina and Borisy, 1999). The spatial and temporal activity of the Arp2/3 complex is controlled by activators such as WASP, SCAR/WAVE and WASH (Machesky and Insall, 1998; Machesky et al., 1999; Linardopoulou et al., 2007). For example, SCAR recruits the Arp2/3 complex to promote the formation of lamellipods. In contrast, other nucleators such as Ena/VASP family proteins and formins generate unbranched, linear filaments that can be bundled together (Pruyne et al., 2002; Breitsprecher et al., 2008). These bundles of F-actin are found within the lamellipod or projecting out of the cell as filopods (Svitkina et al., 2003). Ultimately a subset of actin regulators are collectively deployed to drive the extension of a certain type of protrusion, which in turn promotes a specific cellular behaviour, such as chemotaxis.
Increasingly sophisticated biochemical approaches and chemotactic chambers are advancing our understanding of in vitro cell migration (Vignjevic et al., 2003; Lee et al., 2010; Muinonen-Martin et al., 2010; Reymann et al., 2010; Wu et al., 2012). However, our ability to apply these findings to an in vivo setting lags behind due to the difficulty of studying cells within the context of a living tissue.
Here, we have utilised Drosophila embryonic macrophages as a model of actin dynamics, combining the powerful genetics of the fly, with the excellent live in vivo imaging possible in the embryo (Evans et al., 2010). Like their mammalian counterparts, these macrophages chemotax towards a wide range of stimuli, including bacterial infection and tissue damage through the extension of actin-rich protrusions (Wood and Jacinto, 2007).
Here, we demonstrate that Ena rather than the formin Dia is operating to organise actin within the lamellipod into Fascin-decorated bundles (Fascin is also known as Singed in flies). Ena is such a potent remodeller within the lamellipod that its overexpression can even compensate for the loss of bundlers such as Fascin. Through these bundles, Ena acts to reinforce the lamellipod and drive the leading edge forward, and thus underlies robust macrophage motility during the inflammatory response. Our findings demonstrate that Ena is a master regulator of the actin cytoskeleton within chemotaxing macrophages in vivo, ensuring the swiftest possible response to tissue damage and infection.
RESULTS AND DISCUSSION
Ena, rather than Dia, organises F-actin into linear bundles within the lamellipod
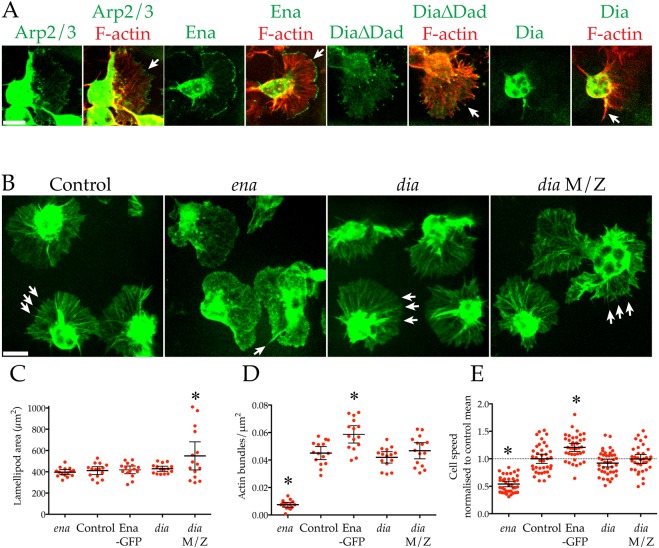
The actin within the lamellipods of Drosophila embryonic macrophages is highly organised and is arranged into linear bundles. We sought to understand how these lamellipodial bundles are formed and how they contribute to macrophage chemotaxis. Live, in vivo imaging revealed that both GFP-tagged Arp2/3 complex and Ena–GFP localise to the leading edge of the lamellipod where the latter interacts with the tips of the lamellipodial actin bundles (Fig. 1A; Tucker et al., 2011). Although not as smoothly localised to the lamellipod edge as Ena, DiaΔDad–GFP (a constitutively active, truncated Dia commonly used as a probe) also localises to the tips of actin bundles (Fig. 1A; Homem and Peifer, 2009, Bilancia et al., 2014). However, DiaΔDad–GFP severely disrupted the architecture of the lamellipod and significantly reduced lamellipodial bundle number compared to control cells (Fig. S1A,B). In contrast to DiaΔDad–GFP, full-length Dia–GFP is seldom utilised as a probe due to its poor localisation, and we likewise found it to be predominantly cytosolic (Homem and Peifer, 2008). However, in a rare few examples, full-length Dia–GFP localised to the entire length of an individual actin bundle (Fig. 1A, Movie 1; Davis et al., 2015). As a constitutively active fragment of Dia, the increased activity of DiaΔDad–GFP is unsurprising. However, the different localisations of Dia–GFP versus DiaΔDad–GFP was concerning.
Fig. 1.
Ena, but not Dia, is required for nearly all lamellipodial bundling and for efficient macrophage migration. (A) Live, in vivo imaging of F-actin (LifeAct–mCherry, red) and key, GFP-tagged actin regulators (green, arrows) within macrophage lamellipods. Scale bar: 10 µm. (B) Control, ena and dia (dia[2] and dia[2]/dia[5] M/Z) macrophages expressing LifeAct–GFP. Loss of ena (but not dia) results in loss of almost all lamellipodial bundles (arrows). Scale bar: 10 µm. (C–E) Quantification of motility in control, ena and dia (dia[2] and dia[2]/dia[5] M/Z) mutants and macrophages overexpressing Ena–GFP. (C) Lamellipodial area (ena=396.57±11.04, control=411.68±17.20, Ena=418.33±17.16, dia=428.79±11.41, dia M/Z=549.77±61.64 µm2, mean±s.e.m., n=15 cells/genotype), (D) actin bundle density (ena=0.008±0.001, control=0.045±0.002, Ena=0.059±0.003, dia=0.042±0.002, dia M/Z=0.047±0.003 bundles/µm2, mean±s.e.m., n=15 cells/genotype) and (E) basal cell speed normalised to control mean (dashed line, ena=0.54±0.025, control=1.0±0.042, Ena=1.21±0.037, dia=0.92±0.034, dia M/Z=1.0±0.041 mean±s.e.m., n≥35 cells/genotype). Error bars are 95% c.i. *P<0.05 vs control mean (ANOVA).
To distinguish the roles of Dia and Ena within the lamellipod, we visualised the actin cytoskeleton of ena and dia mutant macrophages (Fig. 1B; Movie 2). Ena is not required to extend lamellipods, in contrast to scar and arp3 (subunit of the Arp2/3 complex) mutants (Fig. 1C; Fig. S1C,D; Evans et al., 2013). However, as we have previously shown, ena mutants had a near total loss of lamellipodial bundles, which correlated with a decrease in basal motility (Fig. 1B,D,E; Movie 2; Tucker et al., 2011). Conversely, as previously shown, Ena–GFP expression increases lamellipodial bundling and basal cell speed (Fig. 1D,E; Tucker et al., 2011). In contrast, no significant difference in macrophage basal motility was detected in either of two dia mutants (Fig. 1E). In the more severe, maternally zygotic dia (dia[2]/dia[5] M/Z) mutant, many macrophages were significantly larger (Fig. 1B,C) and were likely multinucleate (Castrillon and Wasserman, 1994). Importantly, when normalised to lamellipod area, neither dia mutant exhibited any significant difference in bundle number compared to controls (Fig. 1D). Furthermore, Dia–GFP localised to the residual lamellipodial bundles found in ena mutant macrophages (Fig. S1E). These findings are consistent with the localisation of full-length Dia–GFP to only a rare subset of actin bundles involved in specialised roles such as contact-induced repulsion (Davis et al., 2015).
In summary, lamellipodial bundling is required for robust immune cell motility. However, exactly how Ena increases bundle formation and how these bundles contribute to cell migration remained an open question we next sought to answer.
Ena remodels actin within the lamellipod into Fascin cross-linked bundles
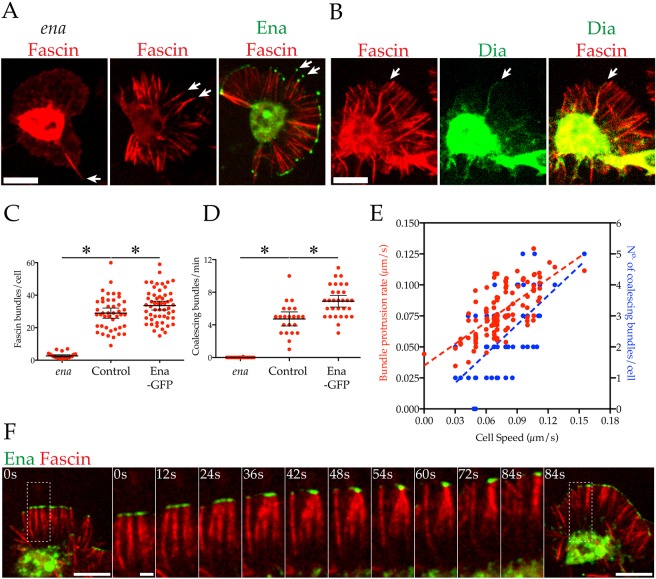
Ena remodels branched actin within the lamellipod into linear bundles. Purified Ena can bundle F-actin in vitro (Bachmann et al., 1999; Schirenbeck et al., 2006). However, within the lamellipod, Ena is confined to the leading edge and therefore cannot be directly responsible for bundling actin filaments (Fig. 1A, Rottner et al., 1999; Tucker et al., 2011). Instead, in vivo Ena co-operates with actin cross-linkers (Winkelman et al., 2014), and we found in Drosophila macrophages that Ena-capped lamellipodial bundles were indeed decorated with one such bundler, Fascin (Fig. 2A; Movie 3). Fascin also colocalised with Dia–GFP at the rare bundles that were positive for the latter (Fig. 2B).
Fig. 2.
Ena acts to generate and coalesce Fascin-decorated bundles within the lamellipod. (A,B) Fascin–mCherry (red) expressed (A) in ena/control macrophages with or without Ena–GFP or (B) with Dia–GFP (green). Arrows highlight distinct Fascin bundles. Scale bars: 10 µm. (C,D) Fascin-mediated bundling in ena/control macrophages with or without Ena–GFP. (C) Number of Fascin-decorated bundles/cell (ena=2.645±0.33, control=28.93±1.55, Ena=33.58± 1.35 bundes/cell, mean±s.e.m., n>25 cells/genotype). (D) Number of bundle coalescence events/min (ena=0.01±0.01, control=4.73±0.41, Ena=6.90±0.35 bundles/cell, mean±s.e.m., n≥20 cells/genotype). Error bars are 95% c.i. *P<0.05 vs control mean (ANOVA). (E) Control basal cell speed correlates positively with individual bundle protrusion rates (red, r2=0.49, n=109) and number of coalescing bundles/cell (blue, r2=0.39, n=88). Both slopes are significantly different from zero (P<0.0001). (F) Sequence showing coalescing Ena (green)-capped, Fascin (red)-decorated bundles. The area of the dashed box is expanded in intervening panels. Time: seconds. Scale bars: 10 µm (end panels), 2 µm intervening panels.
Loss of ena resulted in a near total loss of Fascin bundles (Fig. 2A,C). Conversely, overexpression of Ena significantly increased Fascin bundle number. By following individual Fascin-decorated and Ena-capped bundles, we observed bundle coalescence within advancing lamellipods (Fig. 2F; Movie 3). This process is initiated when Ena-capped Fascin-labelled bundles contact one another. Once joined via their Ena caps, the Fascin-decorated bundles proceed to coalesce from the Ena caps downwards in a zipper-like manner. Furthermore, overexpression of Ena significantly increased the number of coalescing events observed (Fig. 2D), including when normalised to mean Fascin bundle number (Fig. S1F).
Given their presence within the lamellipod and the suppressed migration of ena mutants, we next explored the relationship between Fascin bundle elongation/coalescence and motility. Tracking of the Ena–GFP cap on Fascin bundles revealed that the elongation rate of these bundles correlated positively with cell speed (Fig. 2E). The number of coalescing bundles/cell also increased with increasing speed (Fig. 2E).
In summary, we find Ena acts as a remodeller of lamellipodial actin, by firstly organising it into parallel aligned filaments cross-linked by Fascin, and secondly by mediating coalescence of these bundles into super-bundled structures. This remodelling of the lamellipod appears to be necessary for efficient cell migration.
Ena expression compensates for loss of Fascin by promoting bundling within the lamellipod
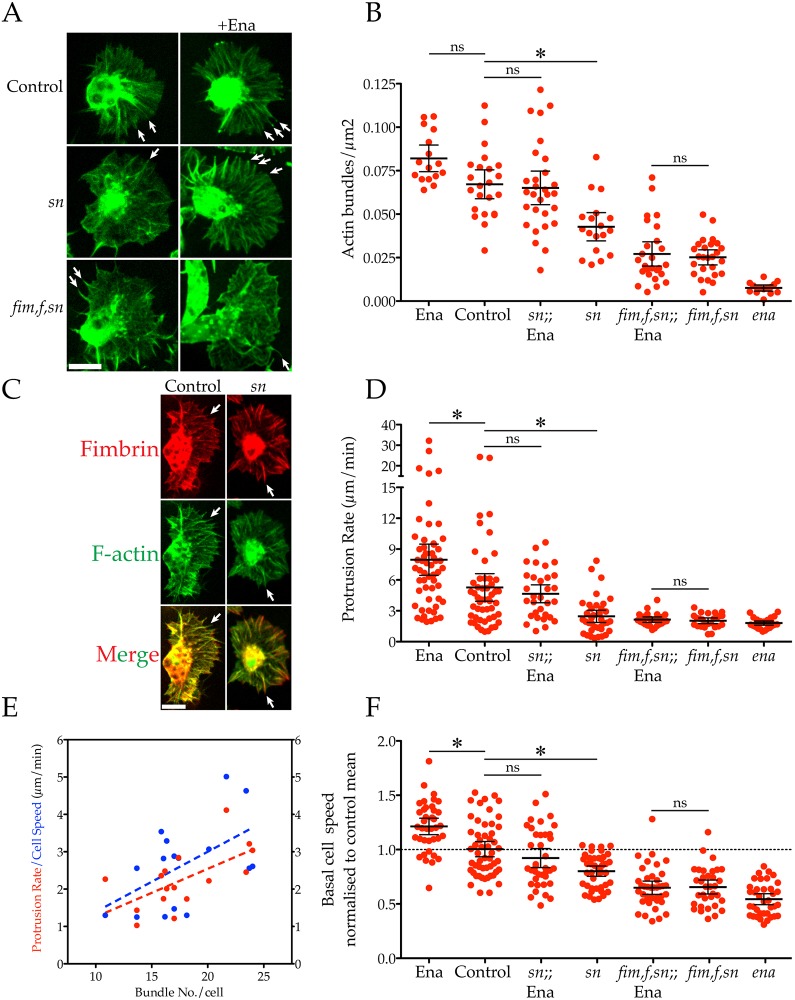
Given the dependence of Fascin on Ena for lamellipodial bundling, we next explored the activity of Ena in Fascin (sn) mutants. Although sn macrophages have a significantly reduced number of lamellipodial bundles compared to controls, this is not the severe loss observed in ena mutants (Fig. 3A,B; Movie 4; Zanet et al., 2009). Surprisingly, despite lacking Fascin and its bundling activity, overexpression of Ena in sn mutants significantly increased lamellipodial bundling, restoring bundle number to control levels (Fig. 3A,B; Movie 4). Like most organisms, flies possess additional parallel actin bundlers, including Fimbrin (Fim) and Forked (F). We reasoned that the other bundlers must be responsible for bundling in the absence of Fascin (Vignjevic et al., 2006). Consistent with this hypothesis, fimbrin–mCherry localised to the remaining lamellipodial bundles present in the sn mutant (Fig. 3C; Movie 5). Crucially, Ena overexpression failed to increase lamellipodial bundling in macrophages mutant for all three of these actin bundlers (fim, f, sn, Fig. 3A,B; Movie 4). From these data, we conclude that although the different actin bundlers can partially compensate for one another within the lamellipod, they all depend on Ena at the leading edge and at the tip of the nascent bundle.
Fig. 3.
Overexpression of Ena compensates for loss of Fascin by utilising the remaining actin bundlers to increase lamellipodial bundling, protrusion rate and basal speed. (A) Control, sn or fim, f, sn (triple bundler mutant) macrophages expressing LifeAct–GFP with or without Ena–GFP. Arrows highlight bundles. Scale bar: 10 µm. (B) Quantification of actin bundle density within the lamellipods of control, sn, fim, f, sn and ena macrophages with or without Ena. Ena overexpression fails to increase bundling in fim, f, sn mutants (Ena=0.082±0.004, control=0.067±0.004, sn;; Ena=0.065±0.005, sn=0.043±0.004, fim, f sn;; Ena=0.027±0.003, fim, f sn=0.025±0.002, ena=0.008±0.001 bundles/µm2, mean±s.e.m., n≥15 cells/genotype). Error bars are 95% c.i. *P<0.05 vs control mean; ns, not significant (P>0.05) (ANOVA). (C) Colocalisation of fimbrin–mCherry (red) with LifeAct–GFP (green) at lamellipodia bundles (arrows) in control/sn mutants. Scale bar: 10 µm. (D) Quantification of lamellipodia protrusion rate of control, sn, fim, f, sn and ena macrophages with or without Ena. Ena overexpression fails to increase protrusion rate in fim, f, sn mutants (Ena=7.967±0.758, control=5.273±0.667, sn;; Ena=4.653±0.425, sn=2.461±0.295, fim, f sn;; Ena=2.031±0.133, fim, f sn=2.149±0.127, ena=1.807±0.101 µm/min, mean±s.e.m., n≥25 cells/genotype). Error bars are 95% c.i. *P<0.05 vs control mean; ns, not significant (P>0.05) (ANOVA). (E) Control bundle number/cell correlates positively with lamellipodial protrusion rate (red, r2=0.39, n=16) and basal cell speed (blue, r2=0.29, n=16). Both slopes are significantly different from zero (P<0.05). (F) Quantification of basal cell speed of control, sn, fim, f, sn and ena macrophages with or without Ena normalised to control mean (dashed line). Ena overexpression fails to increase basal speed in fim, f, sn mutants (Ena=1.208±0.037, control=1.000±0.034, sn;; Ena=0.918±0.043, sn=0.798±0.023, fim, f sn;; Ena=0.654±0.030, fim, f sn=0.654±0.031, ena=0.541±0.025 µm/min, mean±s.e.m., n>35 cells/genotype). Error bars are 95% c.i. *P<0.05 vs control mean; ns, not significant (P>0.05) (ANOVA).
We next sought to understand how bundle number affected lamellipodial dynamics and how this related to cell speed. We used kymography to analyse leading edge extension and found that the decreasing bundle number found across the different genotypes was mirrored by incremental decreases in lamellipod protrusion rates (Fig. 3D). The overexpression of Ena in a control or sn background increased or rescued the lamellipod protrusion rate respectively. However, again this effect depended on cells retaining some bundling capacity, as Ena overexpression failed to increase the suppressed lamellipod protrusion rate of fim, f, sn triple mutants (Fig. 3D). Furthermore, these differences in lamellipodial dynamics translated into discrete differences in basal cell speed (Fig. 3F). Again, the overexpression of Ena restored the basal speed of sn, but not fim, f, sn macrophage migration.
In summary, increasing lamellipodial bundling promotes greater leading edge extension, which in turn drives the cell forward faster during migration. Consistent with these findings, increased bundle number correlated with both increased protrusion rate and basal speed in control macrophages (Fig. 3E). From these data, we conclude that although the different actin cross-linkers can partially compensate for one another, they all depend on Ena at the leading edge and at the tip of the nascent bundle to co-ordinate remodelling of the lamellipod and support robust migration.
Increased bundling induced by Ena overexpression improves sn basal motility and chemotaxis during inflammation
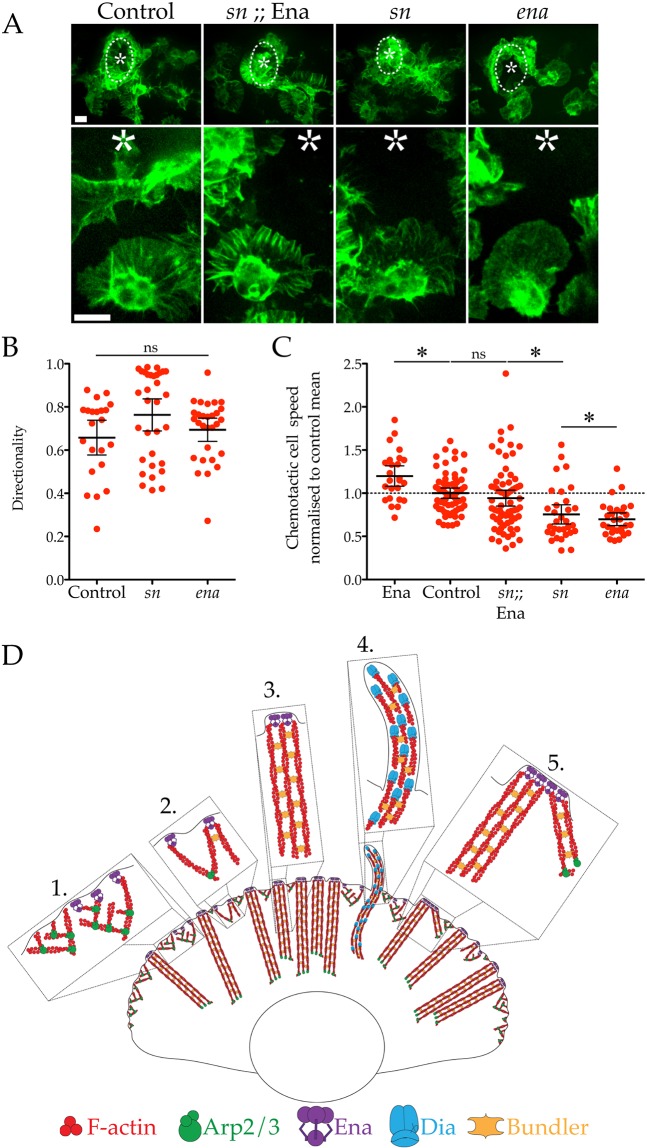
Given that Ena co-ordinates bundling within the lamellipod to promote efficient basal macrophage migration, we explored whether Ena also contributed to the inflammatory chemotaxis of macrophages towards wounds. We generated epithelial wounds through laser ablation and tracked macrophages during their recruitment (Fig. 4A). Neither sn or ena were required for macrophage recruitment to wounds and the directionality of these mutants during their inflammatory chemotaxis was indistinguishable from controls (Fig. 4A,B). However, loss of either sn or ena reduces macrophage speed towards such wounds (Fig. 4C; Zanet et al., 2009; Tucker et al., 2011). Again, we found that overexpression of Ena in sn mutants resulted in increased chemotactic speed and a more robust inflammatory response (Fig. 4C). Given the critical importance of rapid immune cell recruitment to sites of tissue injury, these data highlight the crucial role that Ena-generated actin bundles play in powering inflammatory chemotaxis in vivo.
Fig. 4.
Ena-mediated lamellipodial bundling drives robust recruitment during inflammation. (A) Inflammatory response of control and sn macrophages with or without Ena–GFP, and ena macrophages recruited to laser-induced wounds (dashed outlines and asterisks). Top panels show low magnification of wounds at 10 min post ablation. Lower panels show cropped images of individual macrophages during inflammatory chemotaxis to wounds. Scale bars: 10 µm. (B) Quantification of control, sn and ena macrophage directionality during inflammatory chemotaxis. No significant differences were detected between any of the genotypes. ns, not significant (P>0.05) (ANOVA). (C) Quantification of control, sn and ena with or without Ena–GFP cell speed during inflammatory chemotaxis to wounds (Ena=1.198±0.057, control=1.000±0.030, sn;; Ena=0.943± 0.046, sn=0.754±0.055, ena=0.698±0.036 µm/min, mean± s.e.m., n≥25 cells/genotype). All values normalised to control mean (dashed line). *P<0.05 vs control mean; ns, not significant (P>0.05) (ANOVA). (D) Diagram highlighting the role of Ena in remodelling actin within lamellipodia. (1) The Arp2/3 complex (green) generates dendritic actin (red) in order to extend a lamellipodia. Ena (purple) at the leading edge captures growing filaments and elongates them linearly. (2) The unbranched, linear actin filaments (elongated and brought into proximity of each other by Ena) can now be cross-linked by bundlers such as Fascin and/or Fimbrin (orange). (3) Lamellipodial actin bundles cross-linked with Fascin or Fimbrin and capped by Ena act as struts to reinforce dynamic leading edge extensions, aiding cell migration. (4) In contrast, Dia (blue) generates distinct, Fascin cross-linked filopods. (5) Through multimerisation, Ena can coalesce lamellipodial bundles into super bundles. Ultimately, Ena remodels lamellipodial actin to promote efficient cell migration.
Taking all these data together, we propose that Ena captures branched actin filaments generated by the Arp2/3 complex by binding their barbed-end and overseeing their continued elongation (Fig. 4D). Ena achieves this by both preventing barbed-end capping by capping protein and/or through the ability of Ena to processively elongate actin filaments (Bear et al., 2002; Winkelman et al., 2014). Once in control of filament elongation, Ena can bring together other similarly elongating filaments to be bundled by cross-linkers such as Fascin (Winkelman et al., 2014). Through the ability of constitutively tetrameric Ena to further multimerise, Ena can promote coalescing of bundles into higher-order bundled structures (Breitsprecher et al., 2008). Positioned perpendicular to the membrane, we propose that these lamellipodial bundles act as cytoskeletal struts, exerting maximum force on the leading edge and reinforcing it when it does protrude (Mueller et al., 2017).
We envision Ena acting at the leading edge to generate lamellipodial bundles via convergent elongation and ultimately remodelling dendritic actin into linear actin bundles (Svitkina et al., 2003). Furthermore, owing to its known physical interactions with other actin regulators such as the SCAR complex and Dia, it is perfectly placed to act as a master regulator of the cytoskeleton by co-ordinating nucleators, cross-linkers and F-actin itself (Chen et al., 2014; Bilancia et al., 2014; Schirenbeck et al., 2006).
Concluding remarks
Since both Ena and Dia generate actin bundles, disentangling their activities from one another is challenging, especially since mammals possess multiple homologs of both. As demonstrated here, Drosophila macrophages are skewed towards high Ena rather than Dia activity, possibly maintained through negative regulation of Dia mediated by Ena (Bilancia et al., 2014). This has yielded a unique opportunity to clarify their roles, even when compared to other motile cells within Drosophila that exhibit a blend of Ena and Dia activity (Homem and Peifer, 2009; Nowotarski et al., 2014).
Here, we demonstrated that Ena acts to remodel F-actin within the lamellipod into Fascin-decorated bundles. We do not mean to dismiss the well-established role of Ena as a nucleator. In different cells, Ena does generate filopods de novo (Rotty et al., 2014). However, in highly motile cells, which are migrating through the use of broad lamellipods, we propose Ena primarily functions as a remodeller of dendritic actin to promote formation and elongation of lamellipodial bundles. In this role, Ena acts to marshall actin and the other actin regulators within the lamellipod in order to co-ordinate the cytoskeleton during critical processes, such as the inflammatory recruitment of macrophages to wounds.
How macrophages couple the recognition of inflammatory stimuli to rearrangements in the actin cytoskeleton remains poorly understood. In Drosophila, the immunoreceptor tyrosine-based activation motif (ITAM)-containing MEGF10 homolog, Draper, has a central role in relaying the detection of H2O2 released upon wounding to the Syk-family kinase Shark (Evans et al., 2015). However, exactly how this signalling feeds down to the Rho-family GTPases and actin regulators such as Ena, which are driving chemotaxis, is not known.
Further studies are required to bridge the gap between the signals that guide macrophages and the cytoskeletal regulators that power their motility. However, from this study it is clear that Ena is a master remodeller within the lamellipod, allowing macrophages to harness the full force of the actin cytoskeleton during inflammatory chemotaxis where the rapidity of this response determines survival of the organism as a whole.
MATERIALS AND METHODS
Fly stocks
SingedGAL4 (sn-GAL4, Zanet et al., 2012) was combined with serpentHemoGAL4 (srp-GAL4, Brückner et al., 2004) and croquemortGAL4 (crq-GAL4, Stramer et al., 2005) to drive expression of UAS constructs specifically in hemocytes. The following UAS constructs were used in this study: UAS-GFP-Ena, UAS-FPPPPmito-GFP (Gates et al., 2007), UAS-mCherry-Fascin (Zanet et al., 2009), UAS-LifeAct-GFP (Hatan et al., 2011), UAS-DiaΔDAD-GFP (Homem and Peifer, 2009) and UAS-Dia-GFP (Homem and Peifer, 2008). UAS-fimbrin-mCherry was generated in-house. UAS-LifeAct-mCherry flies were generated by introducing sequence encoding LifeAct-mCherry into pATTB-UASt, which was then sent for commercial injection (Best Gene Inc). The amorphic mutant alleles used in this study were: arp3[EP3640] (Hudson and Cooley, 2002), dia[2], dia[5] (Castrillon and Wasserman, 1994), ena[GC1] (Gertler et al., 1995), sn[28] (Cant and Cooley, 1996) and scar[37] (Zallen et al., 2002). dia[2]/dia[5] maternally zygotic embryos were generated as in Homem and Peifer, 2008. A fimbrin, forked, singed triple mutant was generated by recombining Df(1)BSC584 (Bloomington) with sn[28] in-house.
Embryo genotypes
A minimum of three embryos/genotype were imaged in every case. For assessing the colocalisation of actin regulators: (1); sn-Gal4, UAS-arp3-gfp; sn-Gal4, UAS-lifeact-mcherry, (2); sn-Gal4, UAS-lifeact-mcherry; crq-Gal4, UAS-ena-gfp, (3); sn-Gal4, UAS-lifeact-mcherry; UAS-diaΔdad-gfp, (4); sn-Gal4, UAS-dia-gfp; sn-Gal4, UAS-lifeact-mcherry, (5); ena[GC1], sn-Gal4, UAS-dia-gfp; sn-Gal4, UAS-lifeact-mcherry, (6); srp-Gal4, UAS-fimbrin-mcherry; sn-Gal4, UAS-lifeact-gfp and (7) sn[28], sn-Gal4, UAS-lifeact-gfp; srp-Gal4, UAS- fimbrin-mcherry. For the imaging and quantification of actin bundles: (1); sn-Gal4, UAS-lifeact-gfp, (2); ena[GC1], sn-Gal4, UAS-lifeact-gfp, (3); dia[2], sn-Gal4, UAS-lifeact-gfp, (4) dia[2], sn-Gal4, UAS-lifeact-gfp/dia[5], (5); scar[37], sn-Gal4, UAS-lifeact-gfp, (6);; arp3[EP3640], sn-Gal4, UAS-lifeact-gfp, (7); sn-Gal4, UAS-lifeact-gfp; crq-Gal4, UAS-ena-gfp, (8) sn[28], sn-Gal4, UAS-lifeact-gfp, (9) sn[28], sn-Gal4, UAS-lifeact-gfp; crq-Gal4, UAS-ena-gfp, (10) df(1)BSC584, sn[28]; sn-Gal4, UAS-lifeact-gfp, (11) df(1)BSC584, sn[28]; sn-Gal4, UAS-lifeact-gfp; crq-Gal4, UAS-ena-gfp, (12); sn-Gal4, UAS-lifeact-gfp; UAS-diaΔdad-gfp, (13); ena[GC1], sn-Gal4, UAS-lifeact-gfp; UAS- diaΔdad-gfp and (14); ena[GC1], sn-Gal4, UAS-lifeact-gfp; crq-Gal4, UAS-ena- gfp were used. For the imaging and quantification of Fascin bundles: (1) sn[28], sn-Gal4, UAS-sn-mcherry; ena[GC1], (2) sn-Gal4, UAS-sn-mcherry, (3) sn[28], sn-Gal4, UAS-sn-mcherry;; crq-Gal4, UAS-ena-gfp and (4) sn[28], sn-Gal4, UAS-sn-mcherry; sn-Gal4, UAS-dia-gfp were used.
Live imaging
Developmental stage 15 embryos were collected in cell strainers (Falcon), dechorionated with bleach (Jangro), washed vigorously with water and mounted between a glass slide and a supported coverslip in droplets of VOLTALEF oil (VWR) as previously described (Evans et al., 2010). Ventral hemocytes were then imaged using a spinning disc confocal microscope (Perkin Elmer Ultraview) with a plan-apochromat 63× objective with a NA of 1.4 and a Hamamatsu C9100-14 camera. The acquisition software used was Volocity (Perkin Elmer). Epithelial wounds were generated using laser ablation (nitrogen-pumped micropoint ablation laser tuned to 435 nm, Andor Technologies) as previously described (Wood et al., 2002).
Image processing and statistical analysis
All acquired images were imported into ImageJ (NIH) and maximally projected. Lamellipods were outlined by hand to measure their area (excluding cell body). Actin bundles were defined as any linear concentration of LifeAct–GFP and counted manually. In both cases, cell means were derived from the analysis of all frames between cell–cell collisions. Cell speed was derived from tracks generated using the ImageJ manual tracking plugin (severely enlarged dia M/Z macrophages were excluded from cell speed analysis). Kymographs were constructed using the ImageJ reslice tool and used to calculate leading edge protrusion rates. Fascin bundles were defined as any linear concentration of Fascin–mCherry and were counted manually. Bundle tips were tracked during extension to derive bundle protrusion rates and capture coalescence events. Coalescence was defined as the sustained alignment (>2 frames, 12 s) of two or more bundles per minute. Unpaired, two-tailed t-tests and one-way ANOVA with a Tukey's multiple comparisons test were used to test statistical significance and generate P values.
Supplementary Material
Acknowledgements
We would like to thank Philippa Tucker and Kate Comber for preliminary data, Prof. Mark Peifer and the Bloomington Stock Centre (University of Indiana, USA) for Drosophila stocks, and Flybase.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.J.D., W.W.; Methodology: A.J.D.; Validation: A.J.D.; Investigation: A.J.D., I.R.E.; Resources: T.H.M.; Writing - original draft: A.J.D., W.W.; Writing - review & editing: A.J.D., W.W.; Supervision: W.W.; Project administration: W.W.; Funding acquisition: W.W.
Funding
This work is funded by a Wellcome Trust Senior Fellowship to W.W. (107940/Z/15/Z) and a Wellcome Trust Sir Henry Wellcome Postdoctoral Fellowship to A.J.D. (107355/Z/15/Z). Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.224618.supplemental
References
- Bachmann C., Fischer L., Walter U. and Reinhard M. (1999). The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 274, 23549-23557. 10.1074/jbc.274.33.23549 [DOI] [PubMed] [Google Scholar]
- Bear J. E., Svitkina T. M., Krause M., Schafer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chaga O. Y., Cooper J. A., Borisy G. G. et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509-521. 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Bilancia C. G., Winkelman J. D., Tsygankov D., Nowotarski S. H., Sees J. A., Comber K., Evans I., Lakhani V., Wood W., Elston T. C. et al. (2014). Enabled negatively regulates diaphanous-driven actin dynamics in vitro and in vivo. Dev. Cell 28, 394-408. 10.1016/j.devcel.2014.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D., Kiesewetter A. K., Linkner J., Urbanke C., Resch G. P., Small J. V. and Faix J. (2008). Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943-2954. 10.1038/emboj.2008.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K., Kockel L., Duchek P., Luque C. M., Rørth P. and Perrimon N. (2004). The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7, 73-84. 10.1016/j.devcel.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Cant K. and Cooley L. (1996). Single amino acid mutations in Drosophila fascin disrupt actin bundling function in vivo. Genetics 143, 249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D. H. and Wasserman S. A. (1994). Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 120, 3367-3377. [DOI] [PubMed] [Google Scholar]
- Chen X. J., Squarr A. J., Stephan R., Chen B., Higgins T. E., Barry D. J., Martin M. C., Rosen M. K., Bogdan S. and Way M. (2014). Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell 30, 569-584. 10.1016/j.devcel.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. R., Luchici A., Mosis F., Thackery J., Salazar J. A., Mao Y., Dunn G. A., Betz T., Miodownik M. and Stramer B. M. (2015). Inter-cellular forces orchestrate contact inhibition of locomotion. Cell 161, 361-373. 10.1016/j.cell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans I. R., Zanet J., Wood W. and Stramer B. M. (2010). Live imaging of Drosophila melanogaster embryonic hemocyte migrations. J. Vis. Exp. 36, 1696 10.3791/1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans I. R., Ghai P. A., Urbančič V., Tan K.-L. and Wood W. (2013). SCAR/WAVE-mediated processing of engulfed apoptotic corpses is essential for effective macrophage migration in Drosophila. Cell Death Differ. 20, 709-720. 10.1038/cdd.2012.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans I. R., Rodrigues F. S. L. M., Armitage E. L. and Wood W. (2015). Draper/CED-1 mediates an ancient damage response to control inflammatory blood cell migration in vivo. Curr. Biol. 25, 1606-1612. 10.1016/j.cub.2015.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J., Mahaffey J. P., Rogers S. L., Emerson M., Rogers E. M., Sottile S. L., Van Vactor D., Gertler F. B. and Peifer M. (2007). Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development 134, 2027-2039. 10.1242/dev.02849 [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Comer A. R., Juang J. L., Ahern S. M., Clark M. J., Liebl E. C. and Hoffmann F. M. (1995). Enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 9, 521-533. 10.1101/gad.9.5.521 [DOI] [PubMed] [Google Scholar]
- Hatan M., Shinder V., Israeli D., Schnorrer F. and Volk T. (2011). The Drosophila blood brain barrier is maintained by GPCR-dependent dynamic actin structures. J. Cell Biol. 192, 307-319. 10.1083/jcb.201007095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C. C. F. and Peifer M. (2008). Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development 135, 1005-1018. 10.1242/dev.016337 [DOI] [PubMed] [Google Scholar]
- Homem C. C. F. and Peifer M. (2009). Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol. Biol. Cell 20, 5138-5155. 10.1091/mbc.e09-02-0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M. and Cooley L. (2002). A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 156, 677-687. 10.1083/jcb.200109065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Gallop J. L., Rambani K. and Kirschner M. W. (2010). Self-assembly of filopodia-like structures on supported lipid bilayers. Science 329, 1341-1345. 10.1126/science.1191710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M. and Trask B. J. (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 3, e237 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M. and Insall R. H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347-1356. 10.1016/S0960-9822(98)00015-3 [DOI] [PubMed] [Google Scholar]
- Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., May R. C., Hall M. E. and Pollard T. D. (1999). Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 96, 3739-3744. 10.1073/pnas.96.7.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S., Stramer B., Evans I., Wood W. and Martin P. (2010). Prioritization of competing damage and developmental signals by migrating macrophages in the Drosophila embryo. Curr. Biol. 20, 464-470. 10.1016/j.cub.2010.01.047 [DOI] [PubMed] [Google Scholar]
- Mueller J., Szep G., Nemethova M., de Vries I., Lieber A. D., Winkler C., Kruse K., Small J. V., Schmeiser C., Keren K. et al. (2017). Load adaptation of lamellipodial actin networks. Cell 171, 188-200.e16. 10.1016/j.cell.2017.07.051 [DOI] [PubMed] [Google Scholar]
- Muinonen-Martin A. J., Veltman D. M., Kalna G. and Insall R. H. (2010). An improved chamber for direct visualisation of chemotaxis. PLoS ONE 5, e15309 10.1371/journal.pone.0015309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R. D., Heuser J. A. and Pollard T. D. (1998). The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95, 6181-6186. 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P., Grabher C., Look A. T. and Mitchison T. J. (2009). A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459, 996-999. 10.1038/nature08119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotarski S. H., McKeon N., Moser R. J. and Peifer M. (2014). The actin regulators Enabled and Diaphanous direct distinct protrusive behaviors in different tissues during Drosophila development. Mol. Biol. Cell 25, 3147-3165. 10.1091/mbc.e14-05-0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A. and Boone C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612-615. 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Razzell W., Evans I. R., Martin P. and Wood W. (2013). Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr. Biol. 23, 424-429. 10.1016/j.cub.2013.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A.-C., Martiel J.-L., Cambier T., Blanchoin L., Boujemaa-Paterski R. and Théry M. (2010). Nucleation geometry governs ordered actin networks structures. Nat. Mater. 9, 827-832. 10.1038/nmat2855 [DOI] [PubMed] [Google Scholar]
- Rottner K., Behrendt B., Small J. V. and Wehland J. (1999). VASP dynamics during lamellipodia protrusion. Nat. Cell Biol. 1, 321-322. 10.1038/13040 [DOI] [PubMed] [Google Scholar]
- Rotty J. D., Wu C., Haynes E. M., Suarez C., Winkelman J. D., Johnson H. E., Haugh J. M., Kovar D. R. and Bear J. E. (2014). Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 32, 54-67. 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A., Arasada R., Bretschneider T., Stradal T. E. B., Schleicher M. and Faix J. (2006). The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc. Natl. Acad. Sci. USA 103, 7694-7699. 10.1073/pnas.0511243103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Wood W., Galko M. J., Redd M. J., Jacinto A., Parkhurst S. M. and Martin P. (2005). Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168, 567-573. 10.1083/jcb.200405120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M. and Borisy G. G. (1999). Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009-1026. 10.1083/jcb.145.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M. and Borisy G. G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409-421. 10.1083/jcb.200210174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. K., Evans I. R. and Wood W. (2011). Ena drives invasive macrophage migration in Drosophila embryos. Dis. Model. Mech. 4, 126-134. 10.1242/dmm.005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T. and Borisy G. G. (2003). Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951-962. 10.1083/jcb.200208059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T. and Borisy G. G. (2006). Role of fascin in filopodial protrusion. J. Cell Biol. 174, 863-875. 10.1083/jcb.200603013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman J. D., Bilancia C. G., Peifer M. and Kovar D. R. (2014). Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc. Natl. Acad. Sci. USA 111, 4121-4126. 10.1073/pnas.1322093111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. and Jacinto A. (2007). Drosophila melanogaster embryonic haemocytes: masters of multitasking. Nat. Rev. Mol. Cell Biol. 8, 542-551. 10.1038/nrm2202 [DOI] [PubMed] [Google Scholar]
- Wood W. and Martin P. (2017). Macrophage functions in tissue patterning and disease: new insights from the fly. Dev. Cell 40, 221-233. 10.1016/j.devcel.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., Jacinto A., Grose R., Woolner S., Gale J., Wilson C. and Martin P. (2002). Wound healing recapitulates morphogenesis in Drosophila embryos. Nat. Cell Biol. 4, 907-912. 10.1038/ncb875 [DOI] [PubMed] [Google Scholar]
- Wu C., Asokan S. B., Berginski M. E., Haynes E. M., Sharpless N. E., Griffith J. D., Gomez S. M. and Bear J. E. (2012). Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973-987. 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Starnes T. W., Deng Q. and Huttenlocher A. (2011). Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature 480, 109-112. 10.1038/nature10632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Cohen Y., Hudson A. M., Cooley L., Wieschaus E. and Schejter E. D. (2002). SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156, 689-701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Stramer B., Millard T., Martin P., Payre F. and Plaza S. (2009). Fascin is required for blood cell migration during Drosophila embryogenesis. Development 136, 2557-2565. 10.1242/dev.036517 [DOI] [PubMed] [Google Scholar]
- Zanet J., Jayo A., Plaza S., Millard T., Parsons M. and Stramer B. (2012). Fascin promotes filopodia formation independent of its role in actin bundling. J. Cell Biol. 197, 477-486. 10.1083/jcb.201110135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.