ABSTRACT
Intratumor heterogeneity associates with cancer progression and may account for a substantial portion of therapeutic resistance. Although extensive studies have focused on the origin of the heterogeneity, biological interactions between heterogeneous malignant cells within a tumor are largely unexplored. Glioblastoma (GBM) is the most aggressive primary brain tumor. Here, we found that the expression of Yes-associated protein (YAP, also known as YAP1) is intratumorally heterogeneous in GBM. In a xenograft mouse model, differential YAP expression in glioma cells promotes tumorigenesis and leads to clonal dominance by cells expressing more YAP. Such clonal dominance also occurs in vitro when cells reach confluence in the two-dimensional culture condition or grow into tumor spheroids. During this process, growth of the dominant cell population is enhanced. In the tumor spheroid, such enhanced growth is accompanied by increased apoptosis in cells expressing less YAP. The cellular interaction during clonal dominance appears to be reminiscent of cell competition. RNA-seq analysis suggests that this interaction induces expression of tumorigenic genes, which may contribute to the enhanced tumor growth. These results suggest that tumorigenesis benefits from competitive interactions between heterogeneous tumor cells.
KEY WORDS: Intratumor heterogeneity, Glioma, YAP, Cell competition, Clonal dominance
Summary: This study reveals a functional consequence of intratumor heterogeneity in YAP expression, and suggests that tumorigenesis benefits from competitive interactions between heterogeneous tumor cells.
INTRODUCTION
Malignant cells within individual tumors display striking heterogeneity in their cellular morphology, proliferation rate, genetic lesion, epigenetic modification and therapeutic response. Such intratumor heterogeneity is a recurrent feature of most human tumors, and is associated with progression to malignancy (Andor et al., 2016; Heppner, 1984; Marusyk et al., 2012). Intratumor heterogeneity may account for a substantial portion of observed tumor relapses as well as accompanied resistance to initial therapies (Dexter and Leith, 1986). Currently, two major concepts that attempt to explain the origin of this heterogeneity are the cancer stem cell hypothesis and the clonal evolution model (Campbell and Polyak, 2007). The conceptual difference between these two views could have distinct clinical implications. Besides tumor origin, biological interactions between malignant cells within a tumor compose another key issue of intratumor heterogeneity, and these interactions have been implied to be able to significantly modulate tumor progression and therapeutic response (Calbo et al., 2011; Cleary et al., 2014; Costa et al., 2015; Merlo et al., 2006). The nature of these interactions, and how they affect tumor progression as well as therapeutic outcomes are largely unexplored experimentally.
Glioblastoma (GBM) is the most aggressive primary brain tumor. Gene expression analysis has allowed GBM to be classified into several subtypes differing in treatment responses and survival rates (Phillips et al., 2006; Verhaak et al., 2010). Among these subtypes, the mesenchymal group associates with the worst prognosis (Phillips et al., 2006; Wang et al., 2017). The genetic alterations leading to these differential gene expression and prognosis profiles are not fully understood. Transcriptional coactivator with PDZ-binding motif (TAZ, also known as WWTR1) was proposed to be one of the transcriptional regulators driving the gene expression program of mesenchymal differentiation (Bhat et al., 2011). TAZ and its paralog protein, Yes-associated protein (YAP, also known as YAP1), are the two paralogous nuclear effectors of the Hippo signaling pathway (Zanconato et al., 2016). A comprehensive analysis of brain tumor samples by immunohistochemistry found that YAP expression is increased in a variety of human brain tumors, especially in infiltrating astrocytomas, oligodendrogliomas and GBM (Orr et al., 2011). Remarkably, YAP expression is significantly higher in the mesenchymal subtype of GBM, and this higher expression is also found in more aggressive gliomas associated with poor prognosis (Orr et al., 2011). Considering that both TAZ and YAP have been linked to GBM aggressive progression, it is likely that the transcriptional program controlled by them is responsible for driving GBM mesenchymal transformation.
Like other tumors, intratumor heterogeneity commonly exists in GBM (Morokoff et al., 2015; Parker et al., 2015; Soeda et al., 2015). In addition to the heterogeneous properties that have been described above, the GBM subtypes distinct in gene expression also display heterogeneity (Sottoriva et al., 2013). This suggests that the transcriptional regulators controlling these gene expression profiles might have heterogeneous activities (Sottoriva et al., 2013). In this study, we examined the expression of YAP and TAZ in samples collected from different regions within the same individual human GBM tumors (Sottoriva et al., 2013), and found that the expression of YAP, but not TAZ, is heterogeneous. To functionally study the heterogeneity of YAP expression in GBM, we constructed lines of human GBM cells (LN229 glioma cells) expressing YAP at differential levels and used them to develop tumorigenic models. With these models, we studied the interaction of these heterogeneous cells and its impact on tumorigenesis. Our studies suggested that differential YAP expression induces a competitive interaction, which promotes tumorigenesis.
RESULTS
Differential YAP expression in GBM promotes tumorigenesis
To examine whether the expression of YAP and TAZ is heterogeneous in GBM, we analyzed the gene expression dataset of 27 samples collected from five GBM patient tumors (5 samples/tumor for 4 tumors, and 7 samples/tumor for 1 tumor; Fig. 1A and data not shown) (Sottoriva et al., 2013). Besides intertumorally heterogeneous expression (2.5-fold difference in maximum gene expression, tumors sp42 versus sp52; Fig. 1A), we found that the expression of YAP also displays intratumoral heterogeneity (1.8-, 1.4-, 2.1-, 2.3- and 1.6-fold increase from minimum to maximum expression in tumors sp42, sp49, sp52, sp54 and sp55, respectively, P≤0.01 for each tumor; Fig. 1A). In contrast, the intratumoral heterogeneity of TAZ expression is not significant (1.5-, 1.2-, 1.3-, 1.6- and 1.5-fold increase from minimum to maximum expression in tumors sp42, sp49, sp52, sp54 and sp55, respectively, P>0.05; data not shown). This result indicated that the expression of YAP in GBM is heterogeneous.
Fig. 1.
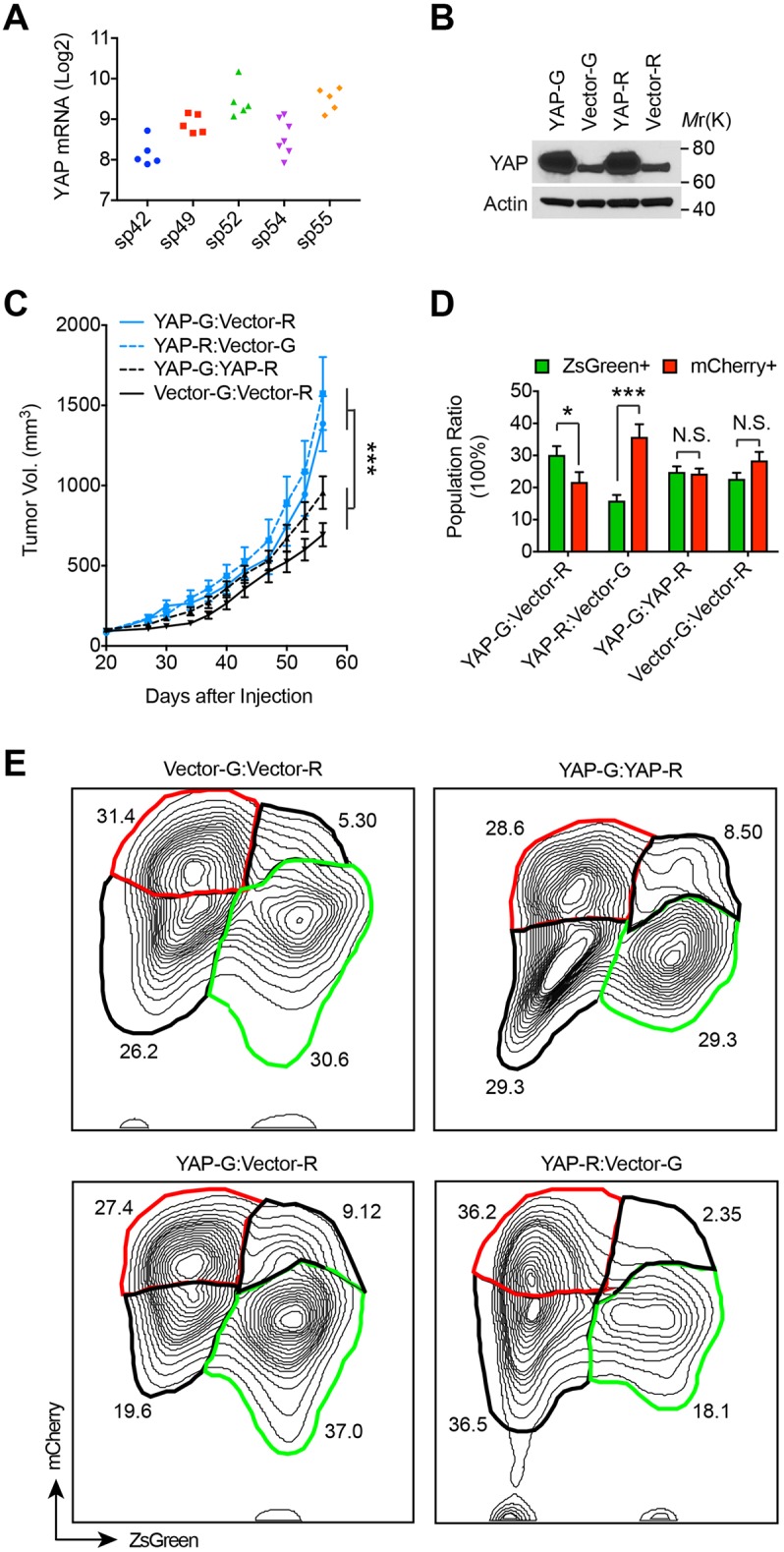
Differential YAP expression in GBM promotes tumorigenesis. (A) Analysis of YAP mRNA level from gene expression dataset in Sottoriva et al. (2013) reveals that human GBM samples from within the same tumors show heterogeneous expression of YAP. Sp42, sp49, sp52, sp54 and sp55 represent five individual tumors, and each dot represents an individual tumor sample. Sp42, P=0.0033; sp49, P=0.01; sp52, P=1×10−8; sp54, P=0.0013; sp55, P=0.0001. Statistical significance was calculated by χ2 variance test. (B) LN229 cells expressing ZsGreen (G) or mCherry (R) were stably transduced with a vector expressing YAP or empty vector, and subjected to western blotting. Representative blots from two independent experiments. (C) CYAP-G, CYAP-R, CVector-G, or CVector-R cells were mixed (1:1) as indicated and co-injected subcutaneously into nude mice. Formed tumors were measured at indicated days and mean±s.e.m. tumor size is shown. For YAP-G:Vector-R and YAP-R:Vector-G, n=10 tumors; YAP-G:YAP-R, n=16 tumors, and Vector-G:Vector-R, n=13 tumors. Statistical significance was calculated by two-way ANOVA. ***P<0.001 at day 56. (D,E) Tumors from mice as described in C were collected, dissociated and subjected to flow cytometry analysis for ZsGreen and mCherry expression. Mean±s.e.m. percentages of ZsGreen- or mCherry-positive cells out of the total scored cells (D) and typical flow cytometry results for each group of tumors (E) are shown. Statistical significance in D was calculated by Student’s t-test. N.S., no significance; *P<0.05; ***P<0.001.
To examine the impact of the heterogeneous expression of YAP on tumorigenesis, we stably expressed recombinant YAP in ZsGreen (G)- or mCherry (R)-expressing LN229 human glioma cells. These cells are denoted CYAP-G (YAP-G in figures) and CYAP-R (YAP-R in figures) hereafter, with results combined as CYAP. In parallel, the differentially labeled cells were transduced with vector alone and were denoted CVector-G (Vector-G in figures) and CVector-R (Vector-R in figures) hereafter, with results combined as CVector. By this method, CYAP cells express higher levels of YAP compared to CVector cells (Fig. 1B). Differentially labeled CYAP and CVector were mixed together (1:1) and co-injected subcutaneously into nude mice to form tumors. Two control experiments were conducted in parallel. In one control, CYAP-G and CYAP-R alone were premixed (1:1) and co-injected, while in the other control CVector-G and CVector-R alone were used. Growth of these tumors was assessed by measuring tumor sizes. Interestingly, although tumors containing CYAP only grow into tumors at a slightly increased speed comparing to those containing CVector only, tumors arising from the mixtures of these two populations grow much faster than either population alone (Fig. 1C). We collected these tumors and analyzed their cellular compositions using flow cytometry. In tumors arising from CYAP or CVector cells alone, differentially labeled cells appear to equally contribute the tumor mass. However, in tumors arising from the mixtures of CYAP and CVector, the CVector population is consistently smaller than the CYAP one (Fig. 1D,E). These results suggested that cells expressing YAP at a higher level become dominant over those expressing less YAP during tumorigenesis. Certain interaction during the clonal dominance may promote tumorigenesis.
Glioma cells expressing more YAP obtain enhanced growth during clonal dominance
To further investigate the interaction between glioma cells expressing YAP at different levels, we co-cultured the differentially labeled CVector and CYAP cells under regular two-dimensional (2D) conditions using the four indicated strategies (Fig. 2A), and compared the growth of each population. Before reaching confluence (day 1–6 after seeding), we found that CYAP cells proliferated at a similar speed to CVector when cultured separately (co-1 and co-2, Fig. 2B,C). After day 6, the growth of both CYAP and CVector cells slows down, presumably due to contact inhibition. Under these conditions, CYAP cells reached a slightly higher density than CVector cells before growth stopped (Fig. 2B,C). In contrast, when CYAP cells were grown together with CVector cells (co-3 and co-4), the confluence density of CYAP could reach to as much as twofold that of CVector (Fig. 2D,E). During co-culture of CYAP and CVector cells, the growth dynamics of CVector did not change compared to growth of these cells alone (comparing co-3, co-4 with co-2; Fig. 2F). However, CYAP cells in co-culture with CVector cells displayed enhanced growth compared to conditions when they were cultured alone (comparing co-3, co-4 with co-1; Fig. 2G). These results suggest that certain interactions between CYAP and CVector cells promote the growth of CYAP. This effect appears to be more pronounced when cells reach confluence.
Fig. 2.
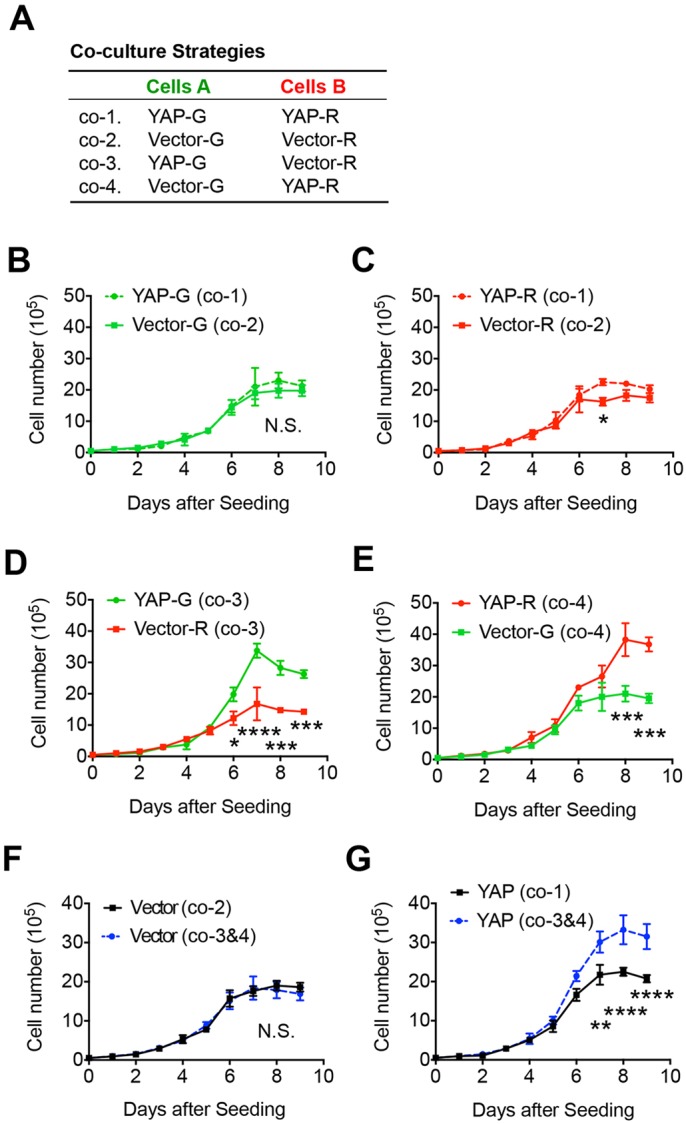
Glioma cells expressing more YAP display enhanced growth when co-cultured with cells expressing less YAP. (A) The co-culture strategies of differentially labeled CVector and CYAP LN229 cells under regular two-dimensional (2D) culture conditions. (B–G) Comparisons of cell population growth across a range of co-culture strategies. Mean±s.e.m. cell numbers of each indicated cell population from the indicated co-culture strategies were counted and plotted. n=2 cultures. N.S., no significance; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Statistical significance was calculated by two-way ANOVA.
Differential YAP expression induces clonal dominance in hybrid spheroids
Multicellular tumor spheroids possess many features mimicking tumors (Hirschhaeuser et al., 2010; Sutherland, 1988), and therefore have been suggested to be a valuable tool to model tumors in vitro. To further study the interaction between heterogeneous tumor cells, we developed a hybrid multicellular tumor spheroid model (Fig. 3A). The tumor spheroids from LN229 cells are able to grow from ∼70 to ∼600 μm in diameter within 15 days (Fig. 3B). To trace cell populations in the spheroids, clones of cells were pre-labeled through stable expression of ZsGreen or mCherry fluorescent protein (Fig. 3A,C). Because the spheroids are formed by cell aggregation, the initial population composition can be controlled by seeding certain numbers of cells. With this model, we could conduct spatial and temporal studies of the growth of each cell population.
Fig. 3.
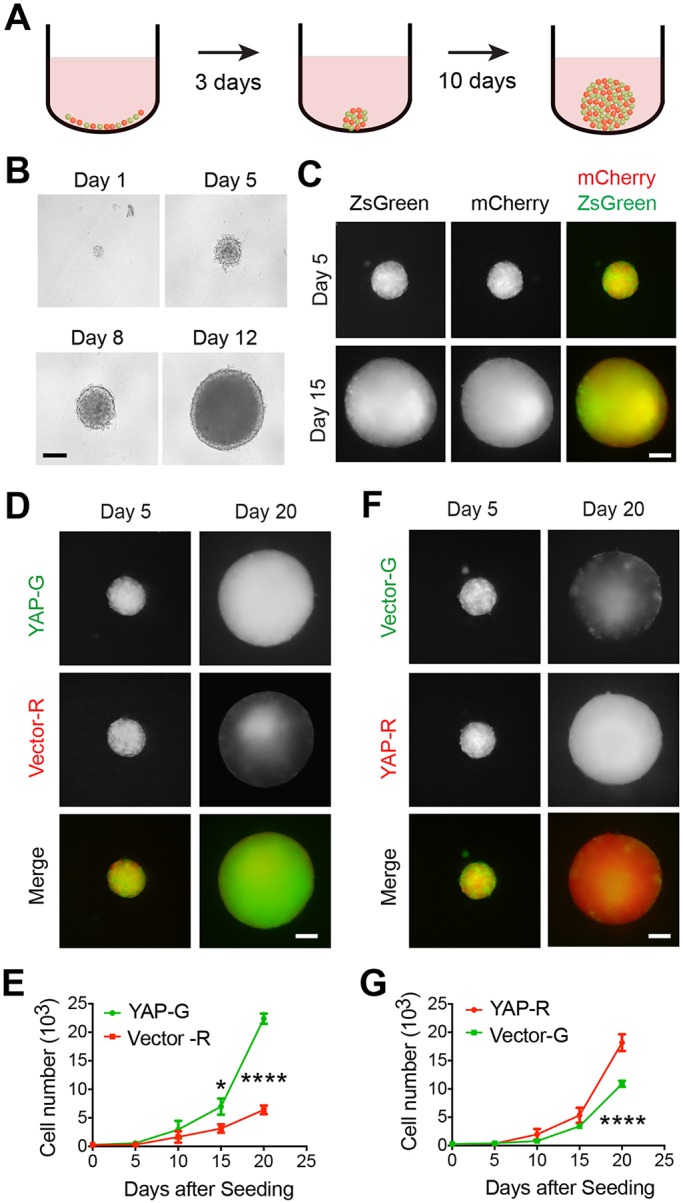
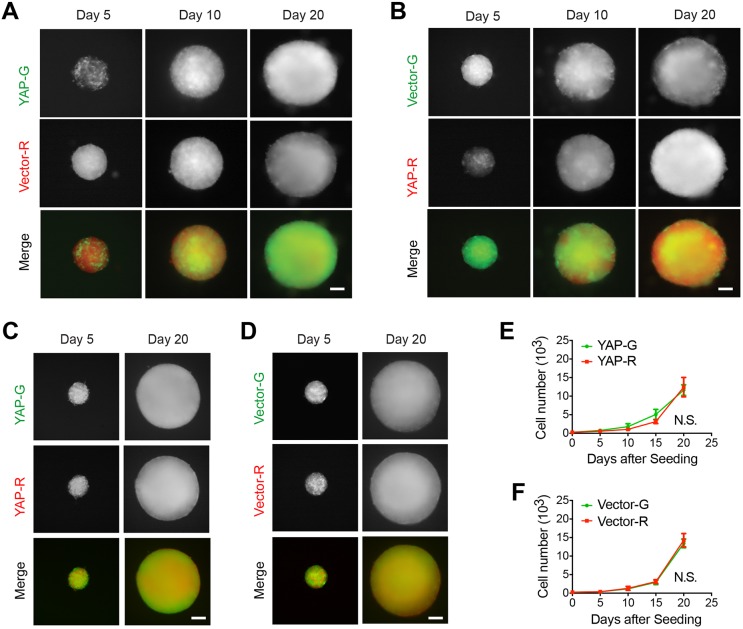
Differential YAP expression induces clonal dominance in hybrid spheroids. (A) Diagram showing the 3D hybrid tumor spheroid model. (B) Phase-contrast images of tumor spheroids from LN229 cells cultured for indicated number of days. Representative images from three independent experiments. (C) Fluorescence images of hybrid tumor spheroids containing ZsGreen- or mCherry-expressing LN229 cells after being cultured for indicated number of days. (D) Fluorescence images of spheroid containing ZsGreen (G)-expressing CYAP (YAP-G) and mCherry (R)-expressing CVector (Vector-R) cells after being cultured for indicated days. Representative images from three independent experiments. (E) Mean±s.e.m. cell numbers of each indicated populations as shown in D were counted and plotted. n=4 independent experiments. (F) Fluorescence images of spheroid containing mCherry (R)-expressing CYAP (YAP-R) and ZsGreen (G)-expressing CVector (Vector-G) after being cultured for indicated number of days. Representative images from three independent experiments. (G) Mean±s.e.m. cell numbers of each indicated populations as shown in F were counted and plotted. n=4 independent experiments. N.S., no significance; *P<0.05; ****P<0.0001. Statistical significance was calculated by two-way ANOVA. Scale bars: 200 μm.
We seeded a combination of CYAP-G and CVector-R cells (1:1) to form a multicellular spheroid. At day 5 after seeding, both CYAP-G and CVector-R populations were evenly distributed in the spheroid (Fig. 3D), suggesting that differential expression of YAP does not lead to distinct cellular adhesion properties. As the spheroid grew, the CYAP-G population expanded evenly across the whole spheroid, whereas the CVector-R population's expansion appeared to be limited to the internal part of the spheroid. At day 20 after seeding, most CVector-R cells were located at the internal regions, leaving the periphery largely occupied by CYAP-G (Fig. 3D). These results indicated that the proportion of the CYAP-G population becomes progressively higher than that of CVector-R in the spheroid. To confirm the imaging observation, we dissociated the spheroids and quantified the number of cells in each population. The results showed that both populations expand; however, the CYAP-G one displays a faster speed during the expansion (Fig. 3E). To rule out that the disproportionate expansion of the CYAP population compared to CVector was due to a difference between ZsGreen- and mCherry-expressing cells, we switched the labeling strategies. Consistently, we observed a similar clonal dominance by the CYAP-R population over CVector-G cells (Fig. 3F,G). These results suggest that CYAP cells possess a certain growth advantage over CVector cells during the growth of spheroids.
Clonal dominance induced by differential YAP expression is independent of initial population proportion
Our results above showed that CYAP cells grow into the dominant population when they are seeded with CVector cells at a 1:1 ratio. To examine whether such an initial proportion is required for this clonal dominance, we reduced the initial proportion of CYAP from 50% to 10% by seeding CYAP-G and CVector-R cells at a ratio of 1:9. Although CYAP-G was a minor population initially (Fig. 4A, day 5 after seeding), it progressively expanded within the spheroid and eventually grew into the major population (Fig. 4A, day 20 after seeding). Similarly, this clonal dominance also occurred when the labeling strategy was switched (Fig. 4B). Therefore, the dominancy is likely determined by a higher level of YAP expression, not the initial population proportion.
Fig. 4.
Clonal dominance induced by differential YAP expression is independent of initial population proportion. (A) CYAP cells marked by expression of ZsGreen (YAP-G) were seeded with CVector cells marked by expression of mCherry (Vector-R) at a ratio of 1:9 (total 1000 cells) and co-cultured for indicated number of days. (B) CYAP cells marked by expression of mCherry (YAP-R) were seeded with CVector cells marked by expression of ZsGreen (Vector-G) at a ratio of 1:9 (total 1000 cells) and co-cultured for indicated number of days. (C) CYAP cells marked by expression of ZsGreen (YAP-G) or mCherry (YAP-R) were seeded at a ratio of 1:1 (total 500 cells) and co-cultured for indicated number of days. (D) CVector cells marked by expression of ZsGreen (Vector-G) or mCherry (Vector-R) were seeded at a ratio of 1:1 (total 500 cells) and co-cultured for indicated number of days. Representative images from three independent experiments. Scale bars: 200 μm. (E) Mean±s.e.m. cell numbers of indicated populations as shown in C were counted and plotted. (F) Mean±s.e.m. cell numbers of indicated populations as shown in D were counted and plotted. n=4 independent experiments. N.S., no significance. Statistical significance was calculated by two-way ANOVA.
To further examine whether differential YAP expression is required for inducing clonal dominance, we seeded CYAP-G and CYAP-R cells together in a 1:1 ratio. In the developed spheroids, both populations were equally represented (Fig. 4C,E), indicating no clonal dominance occurs. Similarly, when differentially labeled CVector cells were seeded together (1:1), no clonal dominance was detected in the spheroids (Fig. 4D,F). Therefore, it is the differential, but not the intrinsic, expression levels of YAP that lead to clonal dominance.
A competitive interaction between CVector and CYAP cells leads to the clonal dominance
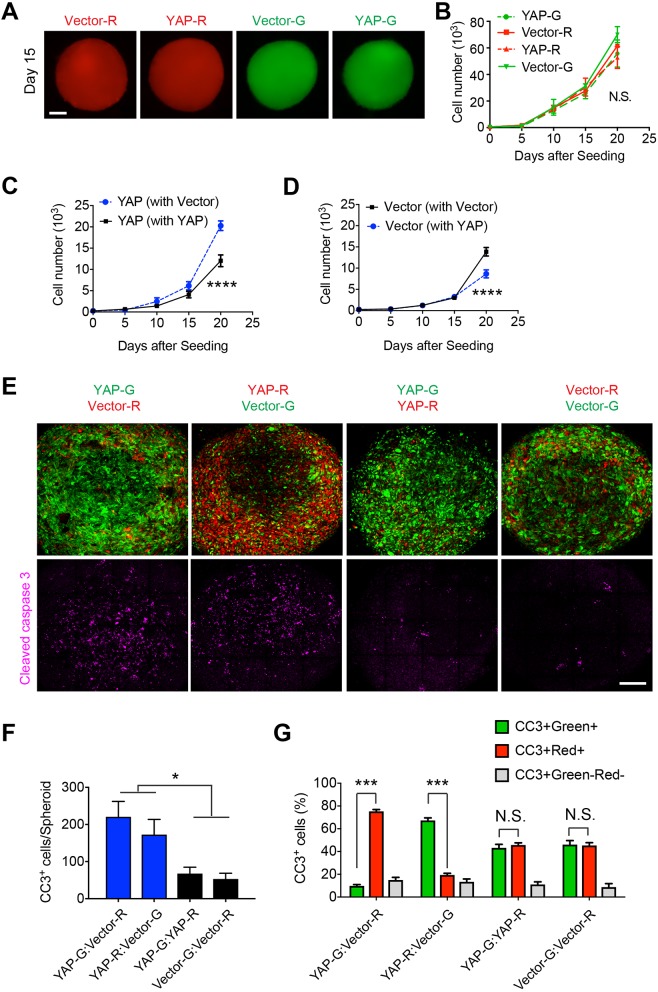
To examine how clonal dominance was achieved, we first tested whether CYAP cells autonomously possess a higher proliferation rate than CVector cells under 3D culture condition. Interestingly, spheroids containing CYAP or CVector cells alone are similar in size after they are cultured for the same number of days (Fig. 5A), suggesting no apparent difference between the growth speeds of these spheroids. We confirmed the imaging observation by dissociating the spheroids and quantifying cell numbers (Fig. 5B). Therefore, clonal dominance is not likely due to an autonomous difference in growth. We then compared the growth of CYAP cells when they are co-cultured with either another population of CYAP or with CVector cells. The CYAP population growing with CVector cells expanded at a faster speed than those cultured with additional CYAP cells (Fig. 5C). In contrast, when comparing the growth of CVector cells when they are cultured with different populations (CYAP versus CVector), we found that the CVector population growing with CYAP expands at a lower speed than that cultured with additional CVector (Fig. 5D). These results indicate that when growing together, the intrinsic growth properties of CYAP and CVector populations are altered, with this alteration leading to clonal dominance.
Fig. 5.
A competitive interaction between CVector and CYAP cells leads to clonal dominance. (A) Spheroids formed by differentially labeled CYAP cells (YAP-G or YAP-R) or by differentially labeled CVector cells (Vector-G or Vector-R) alone were separately cultured for indicated number of days. Scale bar: 200 μm. (B) Mean±s.e.m. cell numbers in indicated spheroids as shown in A were counted and plotted. n=2 independent experiments. (C) Mean±s.e.m. cell numbers of CYAP cells in spheroids also containing CVector or differentially labeled CYAP cells as indicated were counted and plotted. n=8 spheroids. (D) Mean±s.e.m. cell numbers of CVector cells in spheroids also containing CYAP or differentially labeled CVector cells as indicated were counted and plotted. n=8 spheroids. (E) Cleaved caspase 3 staining was used to detect apoptosis in hybrid spheroids containing indicated differentially labeled CYAP and CVector cells. Representative images from three independent experiments. Scale bar: 50 μm. (F) Mean±s.e.m. numbers of cleaved caspase 3-positive (CC3+) cells from each spheroid in each group as shown in E were quantified. n=10 spheroids. (G) Mean±s.e.m. percentages of CC3+ cells overlapped with CVector or CYAP cells (indicated by ZsGreen or mCherry signals) were quantified. For YAP-G:Vector-R, n=9 spheroids; YAP-R:Vector-G, n=8 spheroids; YAP-G:YAP-R and Vector-G:Vector-R, n=6 spheroids. Statistical significance in B–D was calculated by two-way ANOVA and in F,G by Student’s t-test. N.S., no significance; *P<0.05; ***P<0.001; ****P<0.0001.
The clonal dominance and growth dynamics changes that we observed in the hybrid spheroids are reminiscent of cell competition, in which the cell population with certain growth advantages outcompetes the other cell population. The competitive interaction induces changes in both cell populations. In this process, the ‘winner’ cells become dominant while the ‘loser’ cells are eliminated through various mechanisms such as apoptosis, senescence, extrusion, etc (Baker, 2017; Maruyama and Fujita, 2017). To examine whether apoptosis is involved in the reduced expansion of the CVector population, we conducted immunofluorescence staining of cleaved caspase 3 (denoted CC3), a typical marker for apoptotic cells. In spheroids containing CYAP and CVector cells, overall apoptotic signal is increased compared to spheroids containing CYAP or CVector cells alone (Fig. 5E,F). We then examined the identities of these apoptotic cells. In spheroids containing CYAP and CVector cells, a large proportion (65–75%) of CC3+ cells are CVector (Fig. 5G). Notably, quantification of CC3+ cells in the spheroids containing CVector cells alone revealed no increased cell death compared to spheroids containing CYAP cells alone (Fig. 5E,F). These results suggest that CVector cells undergo increased apoptosis during competitive interaction with CYAP cells. Taken together, the combination of increased cell death observed in CVector cells and enhanced growth observed in CYAP cells accounts for clonal dominance.
Competitive cell interaction is determined by relative expression levels of YAP
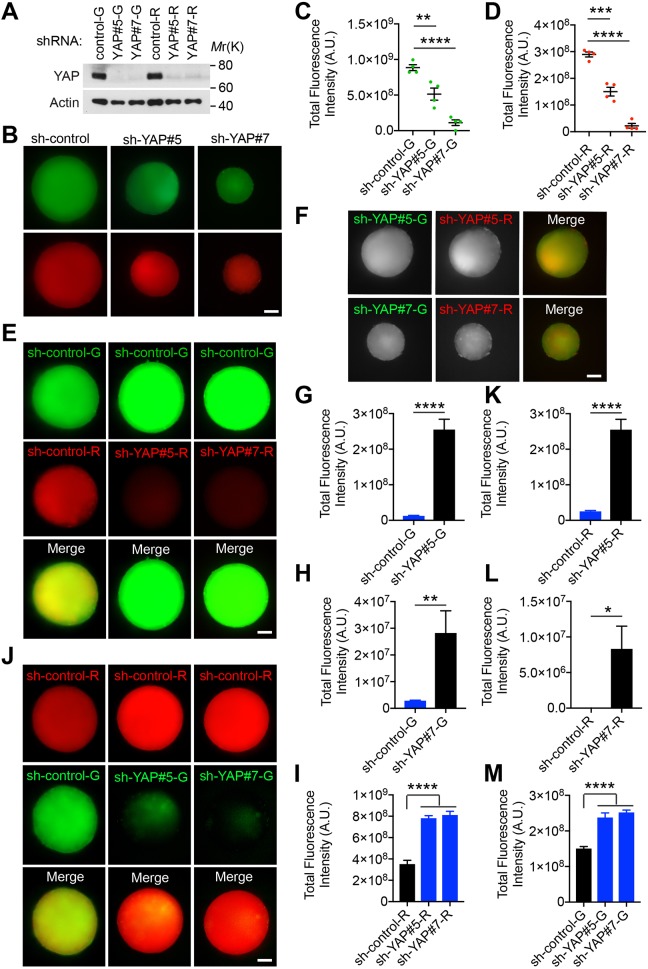
In our above studies, higher levels of YAP are expressed in CYAP cells compared to CVector cells, due to exogenous expression of YAP in the former (Fig. 1B). To examine if reducing endogenous YAP expression can also lead to similar competitive interaction, we knocked down YAP in LN229 cells stably expressing ZsGreen or mCherry, using two different shRNAs against YAP (Fig. 6A). These cells were denoted as Csh-YAP#5 or Csh-YAP#7 (sh-YAP#5-G/R and sh-YAP#7-G/R in figures). Scramble shRNA-transduced control cells were denoted as Csh-control (sh-control-G/R in figures). Spheroids of Csh-YAP#5-R or Csh-YAP#7-R cells grew more slowly than Csh-control-G cells (Fig. 6B–D), indicating that YAP is important for their growth. When cultured in hybrid spheroids with Csh-control-G cells (Fig. 6E), expansion of Csh-YAP#5-R and Csh-YAP#7-R populations was further markedly inhibited compared to when they grew alone (Fig. 6F,G,H). Similar inhibition also occured when the labeling strategy was switched (Fig. 6J,K,L). In addition, expansion of the Csh-control population is enhanced when they are grown with Csh-YAP#5 or Csh-YAP#7 cells, compared to when grown alone (Fig. 6E,J,I,M). These results suggest that the clonal dominance by Csh-control cells is a consequence not only of the intrinsic difference in their growth dynamic, but also of altered growth due to a potential competitive interaction. Furthermore, the results support the notion that competitive interaction is determined by relative, but not absolute, expression levels of YAP.
Fig. 6.
Differential YAP expression induces clonal dominance in hybrid spheroids. (A) LN229 cells expressing ZsGreen (G) or mCherry (R) were stably transduced with control shRNA or shRNA targeting YAP, and subjected to western blotting. Representative blots from one independent experiment. (B) Spheroids formed by differentially labeled cells transduced with control shRNA (sh-control) or by differentially labeled cells transduced with shRNA targeting YAP (sh-YAP) cells alone were separately cultured for 20 days. (C,D) Mean±s.e.m. total fluorescence intensity of each spheroid as shown in B were quantified. n=4 spheroids. (E,F,J) Spheroids cultured for 20 days contain differentially labeled cells transduced with sh-control or sh-YAP, co-cultured in combination as indicated. Representative images from 10 spheroids. (G) Mean±s.e.m. total fluorescence intensity of Csh-YAP#5-R cells when co-cultured with indicated ZsGreen-expressing cells transduced with sh-control or sh-YAP as shown in E (n=6 spheroids) and F (n=10 spheroids). (H) Mean±s.e.m. total fluorescence intensity of Csh-YAP#7-R cells when co-cultured with indicated ZsGreen-expressing cells transduced with sh-control or sh-YAP as shown in E and F. n=10 spheroids. (I) Mean±s.e.m. total fluorescence intensity of Csh-control-G cells when co-cultured with indicated mCherry-expressing cells transduced with sh-control or sh-YAP as shown in E. n=10 spheroids. (K) Mean±s.e.m. total fluorescence intensity of Csh-YAP#5-G cells when co-cultured with indicated mCherry-expressing cells transduced with sh-control or sh-YAP as shown in F and J. n=10 spheroids. (L) Mean±s.e.m. total fluorescence intensity of Csh-YAP#7-G cells when co-cultured with indicated mCherry-expressing cells transduced with sh-control or sh-YAP as shown in F and J. n=10 spheroids. (M) Mean±s.e.m. total fluorescence intensity of Csh-control-R cells when co-cultured with indicated ZsGreen-expressing cells transduced with sh-control or sh-YAP as shown in J. n=10 spheroids. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Scale bars: 200 μm.
Expression of tumorigenesis-related genes is enhanced during competitive interaction
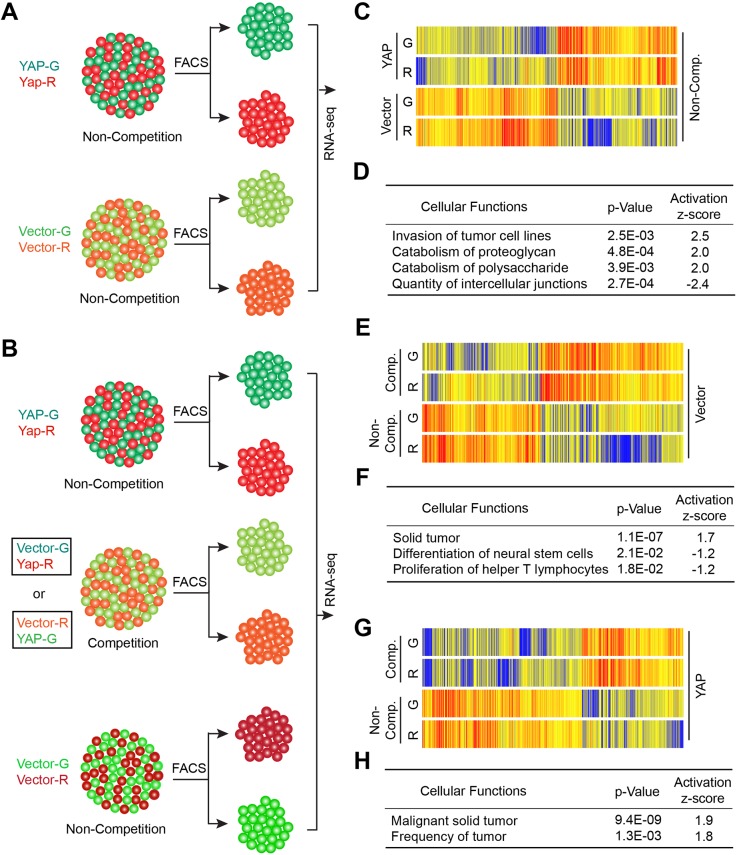
To understand the mechanism underlying the competitive interaction between CVector and CYAP cells, we examined the gene expression profiles of these cells using RNA sequencing (RNA-seq). Each cell population was isolated from the indicated hybrid spheroids through fluorescence-activated cell sorting (FACS) before RNA-seq analysis (Fig. 7A,B). First, we compared the gene expression profile of CYAP cells to that of CVector cells when each of them was grown with their differentially labeled counterparts (Fig. 7A,C). The expression levels of 319 genes are increased, whereas those of 375 genes are decreased, in CYAP cells compared to CVector cells (Table S1) (≥twofold increase, FDR<0.05). The upregulated genes include the well-known YAP target Cyr61. Ingenuity Pathway Analysis of these genes suggested that multiple cellular functions are significantly different between the two cell populations (Fig. 7D). In CYAP cells, cell invasion and catabolism-related genes are activated (z-score≥2, P<0.05), whereas intercellular junctions-related genes are inhibited (z-score<−2, P<0.05). These differences may contribute the growth advantage displayed by CYAP cells in the spheroids.
Fig. 7.
The expression of tumorigenesis-related genes is enhanced during competitive interaction. (A,B) Diagrams showing how indicated differentially labeled CYAP and CVector cell populations were isolated for RNA-seq analysis. (C) Comparison of gene expression in CYAP and CVector cells isolated from spheroids cultured without competition (Non-comp.) as indicated in A. (D) Cellular functions were predicted to be activated (z-score≥2) or inactivated (z-score≤−2) in CYAP cells by Ingenuity Pathway Analysis of genes as shown in C. (E) Comparison of gene expression in CVector cells isolated from spheroids with competition (Comp.) or without competition as indicated in A and B. (F) Cellular functions were predicted to be activated (z-score≥1) or inactivated (z-score≤−1) in CVector cells involved in competition by Ingenuity Pathway Analysis of genes as shown in E. (G) Comparison of gene expression in CYAP cells isolated from spheroids with competition or without competition as indicated in A and B. (H) Cellular functions were predicted to be activated (z-score≥1) in CVector cells involved in competition by Ingenuity Pathway Analysis of genes as shown in G.
We then compared the gene expression profiles of CVector or CYAP cells isolated from conditions without competition to those of their counterparts isolated from conditions with competition (Fig. 7E–H). Both cell types showed marked changes in gene expression when they are cultured under conditions of competition compared to non-competition conditions (Fig. 7E,G). For CYAP cells, the expression of 239 genes is upregulated, whereas that of 377 genes is downregulated (Table S2) (≥twofold increase, FDR<0.05). For CVector cells, the expression of 294 genes is upregulated, and of 245 genes downregulated (Table S3) (≥twofold increase, FDR<0.05). These changes in gene expression further support that competitive interaction has a marked impact on both CVector and CYAP cells when they grow together in spheroids. Notably, Ingenuity Pathway Analysis of these genes suggested that tumorigenesis-related genes are activated in both CVector and CYAP cells (Fig. 7F,H). In addition, genes related to neural stem cell differentiation and helper T lymphocyte proliferation are inhibited in CVector. The changes in gene expression during competitive interaction between CVector and CYAP cells may contribute the enhanced tumorigenesis observed in vivo (Fig. 1C).
DISCUSSION
The oncogenic capacity of YAP has been well demonstrated (Dong et al., 2007; Zanconato et al., 2016). Depending on the situation, YAP expression can promote cell proliferation, survival and/or stemness (Yu et al., 2015), and these cell-autonomous effects contribute to its tumor-promoting ability. Recent studies have found that YAP can also regulate the immune system, therefore suggesting that remodeling the tumor immune microenvironment is an additional way for YAP to promote tumorigenesis (Taha et al., 2018). In this study, we found that YAP can promote tumor growth by inducing intercellular interaction in the process of clonal dominance. The interaction occurs when tumor cells express YAP at differential levels and is reminiscent of cell competition. During the interaction, the growth of cells expressing YAP at a higher level is enhanced. In addition, the interaction induces the expression of cancer-related genes. Both of these changes may cause enhanced tumor growth. Considering intratumor heterogeneity is commonly found in tumors (Andor et al., 2016; Heppner et al., 1984; Marusyk et al., 2012), our study suggests that induction of the competitive interaction is another underlying mechanism of YAP-driven tumorigenesis.
By analyzing the gene expression dataset of samples collected from the same GBM tumors as detailed in Fig. 1A (Sottoriva et al., 2013), we found that expression of YAP is intratumorally heterogeneous (Fig. 1A). Previous studies have found intratumor heterogeneity of YAP expression in human colon cancers (Zhou et al., 2011) and meningiomas (Tanahashi et al., 2015). Therefore, heterogeneous YAP expression appears to exist in multiple human cancers.
To study the heterogeneity of YAP expression in GBM, we constructed human GBM cell lines (LN229 glioma cells) expressing YAP at differential levels. Interestingly, tumors arising from the co-culture of cell populations with differential YAP expression grow faster than either population alone (Fig. 1C). These results suggest that certain interactions between cells expressing YAP at differential levels might promote tumorigenesis. When analyzing the cellular composition of the resulting tumors, we found that CYAP cells form the dominant population compared to CVector cells (Fig. 1D). The autonomously stronger tumorigenicity of CYAP than CVector cells (Fig. 1C) may cause such clonal dominance. However, the dominance of CYAP cells in the heterogeneous tumors is unlikely to be solely caused by the autonomously faster growth of CYAP cells. Because the heterogeneous tumors showed even faster growth than either homogenous tumor, it is likely that enhanced growth was induced in CYAP cells in the heterogeneous tumors.
Increased expression of YAP in LN229 glioma cells (CYAP) per se does not promote their proliferation in either 2D or 3D culture conditions. However, these cells demonstrate stronger abilities to form tumors than CVector cells. One explanation is that a certain oncogenic property other than proliferation is responsible for promoting tumor growth. Alternatively, since CVector cells already have a strong ability to proliferate, a proliferation enhancement in vitro by YAP expression may be below the detectable threshold. Nevertheless, when growing in co-culture with CVector cells, both the growth of CYAP cells in vitro and the oncogenic ability of CYAP cells are enhanced (Fig 1C, Fig 2G, Fig 5C). These results suggest that CVector cells provide certain stimulations for the growth of CYAP cells. Previous studies in lung cancer and breast cancer have shown that cooperation between subclones of tumor cells could benefit tumor maintenance, growth and metastasis (Calbo et al., 2011; Cleary et al., 2014; Costa et al., 2015). Our results are in line with these previous findings, supporting the notion that heterogeneous cellular composition in tumors benefits tumor growth and progression. Interestingly, the interaction between CYAP and CVector cells is not a simple cooperation. On the contrary, the enhanced growth in CYAP cells and induced cell death in CVector cells observed in the spheroid model recapitulate classical cellular crosstalk in cell competition.
Competition has long been thought to be a biological interaction between malignant cells. Tumor cells might compete for limited resources under selective pressures. In this process, a clone of advantageous cells may thrive while other clones become extinct, thereby leading to clonal dominance (Kerbel et al., 1988; Miller et al., 1988). Although little is known about competition between tumor cells, studies of cell competition in other circumstances have provided important information about this interesting cellular crosstalk (Baker, 2017; Vincent et al., 2013). A definitive feature of cell competition is that the fitness of the cells involved is not determined by the cells themselves, but by their relative competitive status when confronting their neighbors. In addition, the outcome of competition is not a cell-autonomous process but occurs through dynamic interactions between the cells involved. The cells that ‘win’ the competition eventually take up the tissue territory. Compared to winner cells, the cells that ‘lose’ the competition may face diverse negative consequences, such as cell death, senescence, autophagy and extrusion (Maruyama and Fujita, 2017). The behaviors of CYAP and CVector cells in our in vitro models fit these canonical features of cell competition. In tumors containing CYAP and CVector cells, we saw dominance of CYAP over CVector cells. It appears that the competition between CYAP and CVector cells is milder in tumors than in spheroids (compare Fig. 1D,E with Fig. 3D–G). This is consistent with the thought that competitors can coexist in tumors, probably due to suppression of competition by various factors (Merlo et al., 2006). Accumulation of clonal diversity is a principal property of tumor progression (Maley et al., 2006). Therefore, uncontrolled clonal dominance resulting from competition may be deleterious to progressive tumor growth and be suppressed in cancer. Overall, our studies suggest that clonal oncogenic lesions could benefit tumors by recruiting surrounding tumor cells into a regulated competition process. Eliminating this competition may benefit tumor therapies.
MATERIALS AND METHODS
Cells
LN229 (CRL-2611) human glioblastoma cell lines were from ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM) (Corning, 10-013-CV) supplemented with 10% fetal bovine serum (Gibco, 10437028) and 1% antibiotic-antimycotic solution (Corning, 30-004-CI) at 37°C with 5% CO2. The cell lines were not authenticated in this study. The cell lines were examined to be mycoplasma-negative before experiments. Unless otherwise indicated, experiments were performed with cells grown to 50% confluency. To generate CYAP and CVector cells lines, LN229 cells were transduced in vitro with a lentivirus vector expressing ZsGreen or mCherry [plvx-ires-zsgreen1 (#632187) and plvx-ires-mCherry (#631237), Clontech]. ZsGreen- and mCherry-expressing LN229 cells were then further transduced with a retrovirus vector only or a vector expressing YAP (pBABEpuro-Flag-YAP2, Addgene 27472, deposited by Marius Sudol) (Oka et al., 2010). Retroviral generation and infection were as described previously (Li et al., 2014). For generation of YAP knockdown cell lines, lentiviral vectors encoding shRNAs targeting YAP (#5: TRCN0000107266; #7: TRCN00000107268) were from Sigma-Aldrich. Control shRNA, pLKO.1-TRC control, Addgene 10879, deposited by David Root (Moffat et al., 2006).
Mice
A total of 1×106 CYAP and/or CVector LN229 cells were injected subcutaneously into six- to eight-week-old female nude mice [Nu(NCr)-Foxn1nu, from Charles River, Strain Code 490]. Tumor size was measured by digital caliper twice per week. All experimental protocols were approved by the Penn State University Institutional Animal Care and Use Committee. All methods were performed in accordance with the relevant guidelines and regulations.
Hybrid spheroid culture
LN229 cells were trypsinized, resuspended and counted. Unless otherwise indicated, 500 cells total were seeded in neural sphere medium [DMEM/F12 (Corning, #15-090-CV), 2 mM L-glutamine (Invitrogen, #25030-081), 1× N-2 supplement (Invitrogen, #17502048), 1× B-27 supplement (Invitrogen, #17504044), 50 μg/ml BSA (Invitrogen, #15260037), 20 ng/ml each of EGF and bFGF (Invitrogen, #PHG0311 and #13256029), 1% antibiotic-antimycotic solution (Corning, #30-004-CI)] per well in 96-well ultra low cluster plates (Costar). After 24–48 h, medium was replaced with regular culture medium (10% FBS in DMEM). The spheroids were incubated at 37°C with 5% CO2 for 2–3 weeks. Spheroid growth was monitored daily based on detected ZsGreen and mCherry signals. To generate hybrid spheroids containing CYAP and CVector cells at different proportions (1:9), 100 CYAP and 900 CVector cells were seeded.
Cell number quantification
For cells cultured under 2D conditions, 1×105 (total) ZsGreen- or mCherry-expressing LN229 CYAP or CVector cells were seeded into a 6-well plate. The regular culture medium was replaced daily. After detaching the cells, cell number was manually counted using a hemocytometer based on ZsGreen and mCherry signals under a fluorescence microscope. For cells cultured under 3D conditions, hybrid spheroids were cultured according to the protocol described above. At days 5, 10, 15 and 20, ten spheroids were collected and dissociated with Accutase (Innovative Cell Technologies, Inc. #AT-104). Cell number was manually counted using a hemocytometer based on ZsGreen and mCherry signals under a fluorescence microscope. The total fluorescence intensity of each spheroid was quantified using IncuCyte Live-Cell Analysis System (Essen BioScience).
Immunoblotting
For western blotting, cells were seeded in complete medium on Petri dishes at a density of 4×104/cm2 24 h before collection. Immunoblotting procedure was described previously (Li et al., 2014). Briefly, cells were lysed in SDS-lysis buffer (10 mM Tris pH 7.5, 1% SDS, 50 mM NaF, 1 mM NaVO4) and subjected to SDS-PAGE on 4–12% Bis-Tris SDS-PAGE gels (Invitrogen) and transferred to Immobilon-P membranes (Millipore). Membranes were incubated in blocking buffer (5% skim milk, 0.1% Tween, 10 mM Tris at pH 7.6, 100 mM NaCl) for 1 h at room temperature and then with anti-YAP (1:1000, #12395, Cell Signaling Technology) and anti-β-actin (1:2000, #A5316, Sigma-Aldrich) primary antibodies diluted in blocking buffer overnight at 4°C. After three washes, the membranes were incubated with goat anti-rabbit HRP-conjugated antibody (1:5000, #7074, Cell Signaling Technology) or goat anti-mouse HRP-conjugated antibody (1:5000, #7076, Cell Signaling Technology) at room temperature for 2 h and subjected to chemiluminescence using ECL (Pierce #1856136).
Immunofluorescence staining for spheroid
Spheroids were fixed and permeabilized for 3 h at 4°C in PBS containing 4% paraformaldehyde and 1% Triton X-100. They were then dehydrated in an ascending series of methanol in PBS (25%, 50%, 75% and 95%, 15 min each) at 4°C, rehydrated in the opposite descending series and washed in PBS (3×10 min). Spheroids were then blocked with 5% BSA/PBS at 4°C for 1 h and followed by incubating overnight at 4°C with anti-cleaved caspase-3 (Asp175) (1:100, 9664, Cell Signaling Technology) primary antibody diluted in 2.5% BSA/0.05% Triton X-100/PBS. After washing with 0.1% Triton X-100/PBS, cells were incubated with donkey anti-rabbit Alexa Fluor 647 secondary antibody (1:200, #711-605-152, Jackson ImmunoResearch) diluted in 2.5% BSA/0.05% Triton X-100/PBS for 24 h at 4°C. Cells were washed with 0.1% Triton X-100/PBS, rinsed with PBS, and mounted in ProLong Gold Mountant (Invitrogen #P10144). When indicated, nuclei were stained with DAPI.
RNA-sequencing and data processing
A total of 96 tumor spheroids were collected and dissociated with Accutase. After cell sorting based on ZsGreen and mCherry signals using FACS, cells were lysed and total RNA was extracted using TRIzol (Thermo Fisher Scientific) following the manufacturer's instruction. RNA integration number (RIN) was measured using BioAnalyzer (Agilent) RNA 6000 Nano Kit to confirm RIN above 7. The cDNA libraries were prepared using the NEXTflex Illumina Rapid Directional RNA-Seq Library Prep Kit (Bioo Scientific) as per the manufacturer's instructions. Briefly, polyA RNA was purified from 100 ng of total RNA using oligo (dT) beads. The extracted mRNA fraction was subjected to fragmentation, reverse transcription, end repair, 3′-end adenylation, and adaptor ligation, followed by PCR amplification and SPRI bead purification (Beckman Coulter). The unique barcode sequences were incorporated in the adaptors for multiplexed high-throughput sequencing. The final product was assessed for its size distribution and concentration using BioAnalyzer High Sensitivity DNA Kit (Agilent Technologies). Pooled libraries were diluted to 2 nM in EB buffer (Qiagen) and then denatured using the Illumina protocol. The denatured libraries were diluted to 10 pM by pre-chilled hybridization buffer and loaded onto a TruSeq Rapid flow cell on an Illumina HiSeq 2500 and run for 50 cycles using a single-read recipe according to the manufacturer's instructions. Sequencing data were analyzed using Strand NGS. Briefly, reads were aligned to reference human genome and annotation file (GRCh38, build 38, RefSeq genes and transcripts, 2017_01_13). After filtering the reads by minimal 10 in at least one sample, Audic Claverie Test was performed when comparing each pair of samples. During the analysis, Benjamini Hochberg FDR correction was used for multiple testing corrections and the P-value cut-off was set as 0.05. After this processing, fold change was calculated and the threshold was set as ≥twofold. For hierarchical clustering analysis, genes showing changes above twofold were used. For Ingenuity Pathway Analysis, twofold change was used as the cut-off. Direct relationships were chosen. To investigate the expression of YAP and TAZ in human GBM cells, the gene expression dataset was downloaded from Sottoriva et al. (2013) and preprocessed with quantile normalization and log2 transformation. The significance of the variance was calculated by chi-square test.
Statistical methods
For statistical analyses, samples sizes were chosen based on whether the differences between groups were biologically meaningful and statistically significant. No data were excluded from the analyses. For cell experiments, all cells in one experiment were from the same pooled parental cells. All mice were from the same cohort. The mice were randomly picked to implant different types of cells. For data collection relying on objective instruments, such as FACS, microscopy software and western blotting, the investigators were not blinded to group allocation during data collection. For animal studies, the investigators were not blinded to group allocation during data collection. Statistical significance was determined by unpaired two-tailed Student's t-test unless indicated otherwise. All error bars shown are standard error of the mean (s.e.m.). All statistical calculations and plotting were performed using GraphPad Prism 7.
Supplementary Material
Acknowledgements
We thank Thomas Abraham and Wade Edris in the Microscopy Imaging Core Facility, Nate Sheaffer and Joe Bednarczyk in the Flow Cytometry & Cell Sorting Core Facility of Penn State College of Medicine for technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Zhijun Liu, P.P.Y., W.L.; Methodology: Zhijun Liu, P.P.Y., Y.I.K., W.L.; Validation: Zhijun Liu, P.P.Y., Y.W., W.L.; Formal analysis: Zhijun Liu, P.P.Y., Zhenqiu Liu, W.L.; Investigation: Zhijun Liu, P.P.Y., Y.W., Zhenqiu Liu, Y.I.K., W.L.; Resources: W.L.; Data curation: Zhenqiu Liu, Y.I.K.; Writing - original draft: Zhijun Liu, Zhenqiu Liu, W.L.; Writing - review & editing: Zhijun Liu, P.P.Y., W.L.; Supervision: W.L.; Project administration: W.L.; Funding acquisition: W.L.
Funding
This work was supported by the National Institutes of Health Grant K22 5K22CA190440 (to W.L.), and the Four Diamonds Fund for Pediatric Cancer Research (to W.L.). Deposited in PMC for release after 12 months.
Data availability
The datasets generated during the RNA-seq are available in the Gene Expression Omnibus under accession number GSE123755 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123755).
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.225714.supplemental
References
- Andor N., Graham T. A., Jansen M., Xia L. C., Aktipis C. A., Petritsch C., Ji H. P. and Maley C. C. (2016). Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105-113. 10.1038/nm.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E. (2017). Mechanisms of cell competition emerging from Drosophila studies. Curr. Opin. Cell Biol. 48, 40-46. 10.1016/j.ceb.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. P. L., Salazar K. L., Balasubramaniyan V., Wani K., Heathcock L., Hollingsworth F., James J. D., Gumin J., Diefes K. L., Kim S. H. et al. (2011). The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 25, 2594-2609. 10.1101/gad.176800.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calbo J., van Montfort E., Proost N., van Drunen E., Beverloo H. B., Meuwissen R. and Berns A. (2011). A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19, 244-256. 10.1016/j.ccr.2010.12.021 [DOI] [PubMed] [Google Scholar]
- Campbell L. L. and Polyak K. (2007). Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle 6, 2332-2338. 10.4161/cc.6.19.4914 [DOI] [PubMed] [Google Scholar]
- Cleary A. S., Leonard T. L., Gestl S. A. and Gunther E. J. (2014). Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508, 113-117. 10.1038/nature13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E. T., Barnabe G. F., Li M., Dias A. A. M., Machado T. R., Asprino P. F., Cavalher F. P., Ferreira E. N., del Mar Inda M., Nagai M. H. et al. (2015). Intratumoral heterogeneity of ADAM23 promotes tumor growth and metastasis through LGI4 and nitric oxide signals. Oncogene 34, 1270-1279. 10.1038/onc.2014.70 [DOI] [PubMed] [Google Scholar]
- Dexter D. L. and Leith J. T. (1986). Tumor heterogeneity and drug resistance. J. Clin. Oncol. 4, 244-257. 10.1200/JCO.1986.4.2.244 [DOI] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M., Anders R. A., Maitra A. and Pan D. (2007). Elucidation of a universal size-control mechanism in drosophila and mammals. Cell 130, 1120-1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner G. H. (1984). Tumor heterogeneity. Cancer Res. 44, 2259-2265. [PubMed] [Google Scholar]
- Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W. and Kunz-Schughart L. A. (2010). Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 148, 3-15. 10.1016/j.jbiotec.2010.01.012 [DOI] [PubMed] [Google Scholar]
- Kerbel R. S., Waghorne C., Korczak B., Lagarde A. and Breitman M. L. (1988). Clonal dominance of primary tumours by metastatic cells: genetic analysis and biological implications. Cancer Surv. 7, 597-629. [PubMed] [Google Scholar]
- Li W., Cooper J., Zhou L., Yang C., Erdjument-Bromage H., Zagzag D., Snuderl M., Ladanyi M., Hanemann C. O., Zhou P. et al. (2014). Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 26, 48-60. 10.1016/j.ccr.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley C. C., Galipeau P. C., Finley J. C., Wongsurawat V. J., Li X., Sanchez C. A., Paulson T. G., Blount P. L., Risques R.-A., Rabinovitch P. S. et al. (2006). Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 38, 468-473. 10.1038/ng1768 [DOI] [PubMed] [Google Scholar]
- Marusyk A., Almendro V. and Polyak K. (2012). Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323-334. 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- Maruyama T. and Fujita Y. (2017). Cell competition in mammals - novel homeostatic machinery for embryonic development and cancer prevention. Curr. Opin. Cell Biol. 48, 106-112. 10.1016/j.ceb.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Merlo L. M. F., Pepper J. W., Reid B. J. and Maley C. C. (2006). Cancer as an evolutionary and ecological process. Nat. Rev. Cancer 6, 924-935. 10.1038/nrc2013 [DOI] [PubMed] [Google Scholar]
- Miller B. E., Miller F. R., Wilburn D. and Heppner G. H. (1988). Dominance of a Tumor Subpopulation Line in Mixed Heterogeneous Mouse Mammary Tumors. Cancer Res. 48, 5747-5753. [PubMed] [Google Scholar]
- Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K. et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283-1298. 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- Morokoff A., Ng W., Gogos A. and Kaye A. H. (2015). Molecular subtypes, stem cells and heterogeneity: Implications for personalised therapy in glioma. J. Clin. Neurosci. 22, 1219-1226. 10.1016/j.jocn.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Oka T., Remue E., Meerschaert K., Vanloo B., Boucherie C., Gfeller D., Bader G., Sidhu S., Vandekerckhove J., Gettemans J. et al. (2010). Functional complex between YAP2 and ZO-2 is PDZ domain dependent, regulates YAP2 nuclear localization and signaling. Biochem. J. 432, 461-472. 10.1042/BJ20100870 [DOI] [PubMed] [Google Scholar]
- Orr B. A., Bai H., Odia Y., Jain D., Anders R. A. and Eberhart C. G. (2011). Yes-associated protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J. Neuropathol. Exp. Neurol. 70, 568-577. 10.1097/NEN.0b013e31821ff8d8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker N. R., Khong P., Parkinson J. F., Howell V. M. and Wheeler H. R. (2015). Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol 5, 55 10.3389/fonc.2015.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H. S., Kharbanda S., Chen R., Forrest W. F., Soriano R. H., Wu T. D., Misra A., Nigro J. M., Colman H., Soroceanu L. et al. (2006). Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157-173. 10.1016/j.ccr.2006.02.019 [DOI] [PubMed] [Google Scholar]
- Soeda A., Hara A., Kunisada T., Yoshimura S., Iwama T. and Park D. M. (2015). The evidence of glioblastoma heterogeneity. Sci. Rep. 5, 7979 10.1038/srep07979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A., Spiteri I., Piccirillo S. G. M., Touloumis A., Collins V. P., Marioni J. C., Curtis C., Watts C. and Tavare S. (2013). Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 110, 4009-4014. 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland R. M. (1988). Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240, 177-184. 10.1126/science.2451290 [DOI] [PubMed] [Google Scholar]
- Taha Z., Janse van Rensburg H. J. and Yang X. (2018). The Hippo pathway: immunity and cancer. Cancers (Basel) 10, E94 10.3390/cancers10040094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi K., Natsume A., Ohka F., Motomura K., Alim A., Tanaka I., Senga T., Harada I., Fukuyama R., Sumiyoshi N. et al. (2015). Activation of yes-associated protein in low-grade meningiomas is regulated by merlin, cell density, and extracellular matrix stiffness. J. Neuropathol. Exp. Neurol. 74, 704-709. 10.1097/NEN.0000000000000211 [DOI] [PubMed] [Google Scholar]
- Verhaak R. G., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P. et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98-110. 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. P., Fletcher A. G. and Baena-Lopez L. A. (2013). Mechanisms and mechanics of cell competition in epithelia. Nat. Rev. Mol. Cell Biol. 14, 581-591. 10.1038/nrm3639 [DOI] [PubMed] [Google Scholar]
- Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., deCarvalho A. C., Lyu S., Li P., Li Y. et al. (2017). Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42-56 e46. 10.1016/j.ccell.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. X., Zhao B. and Guan K. L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811-828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M. and Piccolo S. (2016). YAP/TAZ at the roots of cancer. Cancer Cell 29, 783-803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Zhang Y., Wu H., Barry E., Yin Y., Lawrence E., Dawson D., Willis J. E., Markowitz S. D., Camargo F. D. et al. (2011). Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. USA 108, E1312-E1320. 10.1073/pnas.1110428108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.