ABSTRACT
Classically, canonical autophagy has been considered a survival mechanism initiated in response to nutrient insufficiency. We now understand that autophagy functions in multiple scenarios where it is necessary to maintain homeostasis. Recent evidence has established that a variety of non-canonical functions for autophagy proteins are mechanistically and functionally distinct from autophagy. LC3-associated phagocytosis (LAP) is one such novel function for autophagy proteins and is a contributor to immune regulation and inflammatory responses across various cell and tissue types. Characterized by the conjugation of LC3 family proteins to phagosome membranes, LAP uses a portion of the canonical autophagy machinery, following ligation of surface receptors that recognize a variety of cargos including pathogens, dying cells, soluble ligands and protein aggregates. However, instead of affecting canonical autophagy, manipulation of the LAP pathway in vivo alters immune activation and inflammatory responses. In this Cell Science at a Glance article and the accompanying poster, we detail the divergence of this distinctive mechanism from that of canonical autophagy by comparing and contrasting shared and unique components of each pathway.
KEY WORDS: LC3-associated phagocytosis, Autophagy, Phagocytosis, Autophagosome, LAPosome
Summary: This Cell Science at a Glance article and accompanying poster provide a broad overview of the LC3-associated phagocytic pathway and its role in a variety of physiological and pathological settings.
Introduction
Autophagy, or ‘self-eating’, has long been recognized as a vital mechanism for the regulation of cellular homeostasis through energy regulation, intracellular signaling and protection from damaged or malfunctioning organelles. The mechanistic insights obtained from the use of Saccharomyces cerevisiae have revealed a detailed pathway of key autophagy-related genes (ATGs) that function in a well-choreographed performance for the activation, engulfment and degradation of a variety of intracellular cargos (Ohsumi, 2014). We generally classify canonical autophagy into three groups, including macro-, micro- and chaperone-mediated autophagy. Here, we are concerned only with macro-autophagy, and will use the convention of referring to this simply as ‘autophagy’. Increasing evidence supports the presence of a variety of ‘autophagy-like’ pathways that are characterized by the shared usage of the autophagy machinery and distinct components that serve unique cellular locations and/or settings (Codogno et al., 2011; Dupont et al., 2017). Over the past decade, these distinctive functions for autophagy proteins have been referred to as non-canonical autophagy, although technically these do not involve ‘self-eating’ (hence our preference for referring to such processes as ‘non-canonical functions of autophagy proteins’). New roles for such non-canonical functions, including modulation of the host–pathogen interaction, regulating neuronal signaling and contribution to anti-cancer immunity, are being elucidated.
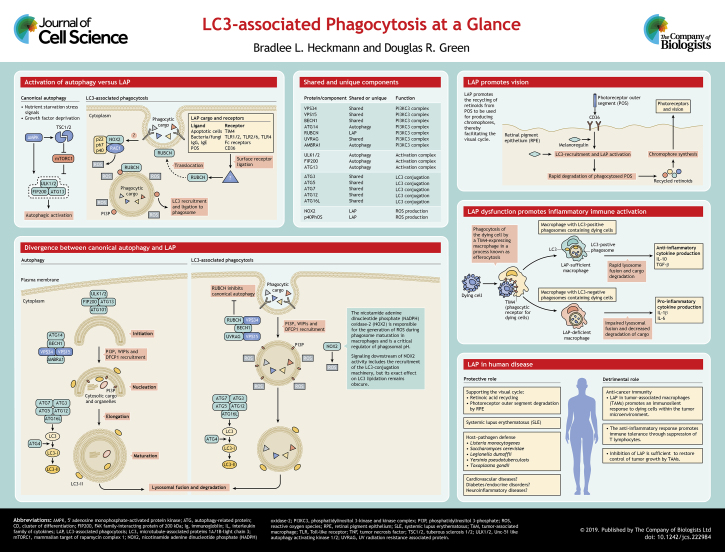
The pathway of LC3-associated phagocytosis (LAP) is one such non-canonical function for the autophagy proteins. This pathway utilizes components of the canonical autophagy machinery, including a subset of the ATGs to conjugate the family of microtubule-associated proteins 1A/1B light chain 3 (MAP1LC3A, MAP1LC3B, MAP1LC3C, referred to collectively here as LC3) to phagosome membranes (Martinez et al., 2011; Martinez et al., 2015; Sanjuan et al., 2007). As illustrated in this Cell Science at a Glance article and poster, the LAP pathway and that of canonical autophagy share several key molecular regulators and machinery; nonetheless, they are distinct pathways that have critical functions in normal physiology and disease pathology.
Activation of LAP versus canonical autophagy
It has long been recognized that basal levels of canonical autophagy are present in most cell types (Mizushima, 2005). This constitutive activation and maintenance of low levels of autophagic degradation is most closely associated with the regulation of cellular homeostasis and organelle and/or protein integrity (Jin, 2006). The most well-characterized stimulus for autophagy activation over the basal state is starvation or nutrient deprivation. Nutrient sensing is largely governed by the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) (Efeyan et al., 2015). mTORC1 is downstream of a multitude of nutrient signaling modalities responsive to growth factors, amino acids, hypoxia, hormones and ATP, to name a few, and inhibits autophagy by phosphorylation of ATG13 in the ULK1–FIP200–ATG13–ATG101 complex. Therefore, inhibition of mTOR activity (such as by amino acid deprivation) results in activation of autophagy (see poster). Similarly, the activation of adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) by low levels of ATP can also induce autophagy, both by inhibition of mTOR, and also by phosphorylation of ULK1 and ATG13 (at a site distinct from that of mTORC1), thereby directly promoting autophagy (Egan et al., 2011; Mao and Klionsky, 2011).
In contrast to canonical autophagy, LAP is not dependent on the AMPK–mTORC1–ULK1 axis and does not appear to be responsive to nutrient status or intracellular stress sensing (Heckmann et al., 2017; Kim et al., 2013; Martinez et al., 2011; Martinez et al., 2015). Since conjugation of LC3 to phagosomal membranes is the hallmark of LAP, it is not surprising that the activating stimulus stems from the exterior of the cell, upon induction of phagocytosis (see poster). A variety of ligands, including pathogen moieties, dying cells and immune complexes, have been shown to promote the recruitment of LC3 to the phagosome. Ligation of surface receptors, including pattern recognition receptors (PRR) such as toll-like receptors (TLRs; in particular, TLR1–TLR2 heterodimer, TLR2–TLR6 heterodimer, and TLR4), immunoglobulin (Ig) receptors that recognize opsonized foreign particles, and receptors mediating the clearance of cell corpses such as TIM4, are events that participate in cargo recognition (Henault et al., 2012; Kyrmizi et al., 2013; Martinez et al., 2011; Martinez et al., 2016; Sanjuan et al., 2007; Segawa and Nagata, 2015; Tam et al., 2014). Although a variety of receptors and ligands that activate LAP have been identified, it remains unclear how these ligation events lead to the recruitment of LAP regulators to the phagosome.
A divergence in machinery
While it remains unclear how the ligation of receptors stimulates the recruitment of the LAP effectors to the phagosome, LAP shares a variety of key regulators with canonical autophagy across various steps of the process culminating with lysosomal degradation. Following an activating stimulus, autophagy and LAP can essentially be delineated into three common stages, followed by lysosomal fusion, with both shared and unique components (see poster).
The first stage is phagophore formation, also referred to as nucleation (in the case of canonical autophagy) or phagosome cup development (for LAP), both of which serve as a scaffold for the assembly of the upstream regulatory kinase complexes in each pathway. Interestingly, the particular events that promote phagophore formation in canonical autophagy and the mechanism by which this is accomplished remains a fundamentally elusive question in cell biology. What is evident is that signaling from nutrient sensing leads to the formation of the initiation kinase complex that is composed of ULK1, FIP200 (also known as RB1CC1), ATG13 and ATG101, which is required for autophagy, but dispensable for activation of LAP (Martinez et al., 2011). In comparison to canonical autophagy, which is characterized by a double-membrane autophagosome, LAP is characterized by a single-membrane phagosome. Phagosome biogenesis that precedes LAP is governed by the same mechanisms that regulate canonical phagocytosis, and phagosome formation occurs prior to the recruitment of downstream regulators and LC3 conjugation as reviewed elsewhere (Heckmann et al., 2017; Mehta et al., 2014; Münz, 2017; Romao and Münz, 2014; Wong et al., 2018).
The second stage is characterized by the assembly of the multimeric phosphotidylinositol 3-kinase complex (PI3KC3) (see poster). In LAP, this is the first multi-protein complex involved in the regulation of the process. While the PI3KC3 complexes between autophagy and LAP are similar, they vary in certain components. The core components of the PI3KC3 complex are BECN1, VPS15 and VPS34, and these are shared between both autophagy and LAP (Backer, 2016). BECN1 is a coiled-coil BH3 domain-containing protein that regulates the lipid kinase function of VPS34 and is involved in autophagic regulation from yeast to mammals (Kametaka et al., 1998; Liang et al., 1999). VPS15 (also known as PIK3R4 in mammals) is a pseudokinase that regulates VPS34 activity, and VPS34 (also known as PI3KC3 in mammals) is the active catalytic subunit of the complex (Petiot et al., 2000). VPS34 functions by phosphorylating the inositol ring of the phosphotidylinositides (PtdIns) to produce phosphatidylinositol 3-phosphate (PI3P, also known as PtdIns3P), which is required for both autophagy and LAP (Heckmann et al., 2017; Martinez et al., 2015). Accordingly, inhibitors of PI3KC3 function or genetic deficiency in the core components BECN1 and VPS34 abrogate both autophagy and LAP (Martinez et al., 2011; Martinez et al., 2015; Sanjuan et al., 2007).
In addition to the shared core components described above, the PI3KC3 complex that participates in LAP includes UVRAG and RUBCN, whereas in canonical autophagy, it contains the subunits ATG14L and AMBRA1 that are dispensable for LAP, supporting the idea that a unique RUBCN-containing PI3KC3 complex exists and is required for LAP (Martinez et al., 2015). RUBCN is a RUN-domain-containing protein that has been best characterized as an inhibitor of canonical autophagy by abrogating PI3P production by the PI3KC3 complex (Matsunaga et al., 2009; Zhong et al., 2009). In contrast, upon induction of LAP, RUBCN is required for the generation of PI3P by VPS34 on phagosome membranes, resulting in downstream events, including reactive oxygen species (ROS) production by nicotamide adenine dinucleotide phosphate (NADPH) oxidase-2 (NOX2) and LC3 lipidation (Martinez et al., 2015).
One intermediate stage that is exclusive to the LAP pathway is the necessity to produce ROS at the phagosome membrane (see poster). ROS are not required for the formation of the autophagosome or lipidation of LC3 in canonical autophagy. ROS production in LAP occurs prior to LC3 lipidation, but subsequent to PI3KC3 assembly (Heckmann et al., 2017). NOX2 is a multiprotein complex that associates with the phagosome membrane and consists of multiple subunits, including RAC1, p22, p67 and p40, the last of which can bind membrane PI3P at the phagosome as reviewed in detail elsewhere (Bedard and Krause, 2007; Heckmann et al., 2017; Tian et al., 2008). NOX2 is responsible for the generation of ROS during phagosome maturation in macrophages and is a critical regulator of phagosomal pH (Bedard and Krause, 2007; Lambeth and Neish, 2014). Furthermore, activity of NOX2 is required for the recruitment of the LC3 conjugation machinery (the third stage, discussed below); however, this recruitment also requires PI3P generation. Thus, the precise role of NOX2 in LAP remains obscure. Oxidation of lipids at the phagosomal membrane occurs at the early stages of LAP, but it is not clear whether this or some other function of ROS is required for LAP (Martinez et al., 2015). Alternatively, ROS produced in the phagosomal lumen can diffuse to the cytosol where they might activate cytosolic enzymes that are required for LAP. Although NOX2 is indispensable for LAP in macrophages, NOX2 could be compensated for by other NOX family members, or possibly other forms of ROS in other cell types.
The third stage is assembly of the conjugation systems that participate in the processing and ligation of LC3 to either the phagophore (autophagy) or phagosome (LAP) (see poster). Components of these ubiquitin-like systems include ATG3, ATG5, ATG7, ATG10, ATG12 and ATG16L. ATG7 and ATG10 function as activating E1 and conjugating E2 enzymes, respectively, to produce isopeptide bonding between ATG12 and ATG5, which form a stable heterodimer. ATG5–ATG12 is then able to bind to membrane-associated ATG16L (recruited as described below), thereby forming the ATG16L complex, which functions as an E3 ligase in a second ubiquitin-like conjugation system, composed of ATG7, ATG3 and the ATG16L complex. Again, ATG7 functions as an E1 ligase for LC3 family members, whereas ATG3 serves as the E2 enzyme. LC3 is transferred from the ATG3–LC3 heterodimer to the lipid by the ATG16L complex (Nakatogawa, 2013). This second conjugating system is recruited to either the phagophore (autophagy) or phagosome (LAP) via membrane-localized PI3P that has been generated by the PI3KC3 complex described above. In canonical autophagy, PI3P acts as a platform and binds to the WIPI family protein, WIPI2, a WD β-propeller PROPPIN that utilizes an alternative lipid-binding domain from the common PX or FYVE domains (Proikas-Cezanne et al., 2015). WIPI2 is then able to associate with ATG16L, effectively labeling the membrane site where LC3 lipidation will occur. Interestingly, the WIPI proteins are dispensable for LAP, leaving the bridging mechanism between PI3KC3 and the lipidation machinery in LAP an open question. In addition, the WD repeat-containing C-terminal domain of ATG16L is essential for LC3 recruitment to endolysosomal membranes during non-canonical forms of autophagy including LAP, but dispensable for canonical autophagy (Fletcher et al., 2018). It is clear, however, that prior to LC3 conjugation, ATG4 has an important role in both pathways as it acts as a cysteine protease that cleaves LC3 (generating LC3-I) and reveals the glycine residue required for lipidation (generating LC3-II) (Fernandez and Lopez-Otin, 2015).
Another difference between canonical autophagy and LAP is the rate at which LC3 lipidation occurs. Recent evidence has demonstrated that LC3 is rapidly lipidated once the phagosome membrane has fully sealed and the lipidation machinery likely remains attached to the phagosome (Sanjuan et al., 2007; Martinez et al., 2015). Conversely, in canonical autophagy, LC3 conjugation occurs during membrane elongation of the phagophore (Kabeya et al., 2000; Kabeya et al., 2004). Moreover, in canonical autophagy, the ATG16L complex is likely released simultaneously to autophagosome formation and does not linger (Mizushima et al., 2003). LC3 likely serves various functions unique to each pathway. In canonical autophagy, LC3 is thought to have three primary roles; these include participating in cargo selection, promoting autophagosome closure and facilitating membrane fusion (Lee and Lee, 2016). Owing to the differences with regard to where cargo is selected in LAP, i.e. intra- versus extracellularly, it is unlikely that LC3 functions here in mediating cargo selection. Likewise, because LC3-conjugation occurs after phagosomal sealing in LAP, it does not function in promoting membrane closure. LC3 does function in downstream events of LAP, including phagosomal–lysosomal fusion (Martinez et al., 2015) and possibly phagosome–endosome fusion events (Henault et al., 2012).
Roles of LAP in immune regulation and inflammation
Following lysosome fusion in both canonical autophagy and LAP, luminal acidic hydrolases degrade the engulfed material and transmembrane pumps present in the lysosomal membrane facilitate the recycling of a cohort of nutrients, including sugars, lipids, amino acids and nucleotides to replenish intracellular stores. In LAP, this surplus in molecules derived from extracellular material is evocative of amoeboid feeding and can plausibly be utilized as energetic substrates within the phagocytic cell, suggesting that LAP can play a role in cellular metabolism (although this has not been explored).
The primary goal of phagocytosis is the degradation of extracellular cargo, including infectious agents, and the role of LAP in such degradation can therefore help to control infection. For example, LAP-deficient mice fail to efficiently clear Aspergillus fumigatus infection (Martinez et al., 2015; Sprenkeler et al., 2016), demonstrating that defects in the LAP pathway can indeed have immunologic consequences. Depending on the cargo, phagocytosis can also have inhibitory effects on the immune system. For example, engulfment of outer membrane vesicles from Bacteroides fragilis by dendritic cells induces regulatory T cells that control inflammatory bowel disease, and this process appears to be dependent on LAP (Chu et al., 2016).
The connection between LAP and the immune response is involved in the elimination of apoptotic cells by macrophages in a process known as efferocytosis (see poster). This homeostatic process does not normally elicit an immune response; it is therefore regarded as immunologically silent and is characterized by the inhibition of pro-inflammatory cytokines, including IL-6, IL-1β and CXCL-10 (Fadok et al., 1998; Kim et al., 2004; Martin et al., 2014; Mukundan et al., 2009; Xiao et al., 2008). LAP mediates this immunoregulation in response to dying cells, and is obligatory for the immune-silent, anti-inflammatory response of efferocytosis. For instance, mice that are LAP-deficient but can undergo canonical autophagy accumulate apoptotic bodies within their tissues and within phagocytic cells (Muñoz et al., 2010). These LAP-deficient mice spontaneously develop an auto-inflammatory, lupus-like syndrome upon aging that can be accelerated by chronic exposure to apoptotic thymocytes (Martinez et al., 2016).
Another example of how LAP modulates immunity is with regard to antigen presentation and pathogen clearance, and a number of studies have elucidated the importance of LAP in host–pathogen defense (Gluschko et al., 2018; Hubber et al., 2017; Lam et al., 2013; Mitchell et al., 2018; Oikonomou et al., 2018; Sprenkeler et al., 2016). LAP activation is important for promoting TLR signaling through IRF7, leading to a type-I interferon response, a process that promotes discrimination of self versus pathogenic DNA and is important for host–pathogen defense (Acharya et al., 2016; Hayashi et al., 2018; Henault et al., 2012; Raso et al., 2018). LAP-mediated activation of the C-type lectin receptor Dectin-1 (also known as CLEC7A) promotes the recruitment of LC3 to phagosomal membranes (Ma et al., 2012; Münz, 2015; Romao et al., 2013). Dectin-1 activation occurs in response to fungal infections and upon exposure to fungal-derived material. LAP activation in this setting promotes MHC class II recruitment to the phagosome, leading to sustained antigen presentation (Ma et al., 2012). These findings are consistent with and may in part explain why LAP-deficient animals show a reduced capacity in clearing fungal infections. Mice that are deficient in LAP also have difficulties in clearing other types of pathogens, including Listeria monocytogenes, Legionella dumoffii, Toxoplasma gondi and Helicobacter pylori (Deen et al., 2015; Florey et al., 2015; Oikonomou et al., 2018).
Differential roles for LAP in disease
As discussed above, LAP both contributes to immune regulation and is protective against autoimmunity in mice. The roles for LAP in disease may not be restricted to its functions in immune regulation, however. Studies investigating the convergence of autophagy and phagocytosis in retinal pigment epithelium (RPE) identified LAP as a protective mechanism that supports vision (Ferguson and Green, 2014) (see Box 1 and the poster for details of the role of LAP in the vision cycle).
Box 1. The role of LAP in the vision cycle.
The shedding and phagocytic clearance of photoreceptor outer segments (POS) is a vital daily process that allows for renewal of the photoreceptor disks and thus helps to maintain vision. The clearance of POS remnants by retinal pigment epithelium (RPE) cells thus is a homeostatic process that also functions to replenish necessary components of the visual cycle including retinoic acid as described below (Kim et al., 2013). In cultured RPE cells, LC3 is recruited to phagosomes that contain POS. LC3 recruitment is abrogated when there is a deficiency in either ATG5 or BECN1, but not ULK1, FIP200 or ATG13, indicating that LAP is activated in these conditions and not canonical autophagy (Kim et al., 2013).
Melanoregulin is an intracellular cargo-sorting protein that is required for the activation of LAP in order to phagocytose POS (Frost et al., 2015). Melanoregulin-mediated activation of LAP promotes the maturation of POS-containing phagosomes, their degradation and nutrient recycling (Frost et al., 2015). These findings are further corroborated in mice that lack ATG5 in RPE cells, which have reduced visual capacity owing to the inadequate recovery of retinoids (Zhang et al., 2017). In the visual cycle, all-trans retinal (ROL) is used to synthesize the chromophore 11-cis retinal (RAL). 11-cis RAL is converted to all-trans RAL following exposure to light and is released from opsin and reduced to ROL. ROL is then recycled to the RPE where it is used for de novo RAL synthesis. In the absence of ATG5, recovery of ROL by the RPE was diminished, resulting in suppressed RAL synthesis (Kim et al., 2013). This phenotype is not present in ULK1-deficient animals, which have normal vision. POS phagocytosis has been implicated in contributing to the recovery of ROL and thus indicates a correlation with LAP (Kim et al., 2013). Furthermore, POS-induced LAP is dependent on RUBCN, confirming a bona fide role for LAP as a protective mechanism in the visual cycle (Muniz-Feliciano et al., 2017).
In contrast to the beneficial effects of LAP, it is possible that this process opposes effective immune responses to cancers. The activation of LAP in tumor-associated macrophages (TAMs) following engulfment of dying cells promotes the production of anti-inflammatory cytokines in the tumor microenvironment, which leads to the establishment of immune tolerance through the suppression of T lymphocytes. Abrogation of LAP in TAMs imparts control of tumor growth upon phagocytosis of dying tumor cells (Cunha et al., 2018). In addition, the impairment of LAP in TAMs promotes the expression of pro-inflammatory genes and a STING-mediated type I interferon response that promote activity of tumor-infiltrating T lymphocytes, required for the control of tumors in LAP-deficient animals. These studies demonstrate that LAP is a dynamic mechanism that regulates immune system function and can be either beneficial or detrimental in a cell- and tissue-specific manner.
Conclusions and future directions
The importance of canonical autophagy to cellular homeostasis and the pathological consequences of autophagic perturbation have been recognized for decades. Congruent with the importance of canonical autophagy, the roles for LAP in inflammation, autoimmunity, pathogen clearance and host defense, reinforce the broad significance of this non-canonical use of autophagy proteins to not only immune, but cellular and organismal homeostasis and health as a whole.
The mechanistic divergence from canonical autophagy is an ideal starting point for specifically exploiting LAP as a putative treatment modality. While activation or inhibition of LAP would be dependent on the specific setting, manipulation in either direction can likely be achieved. Another aspect that should receive intense focus is the lack of mechanistic insight into the regulation of receptor ligation and the recruitment of RUBCN and the PI3KC3 complex. Elucidation of this dynamic signaling cascade will not only improve our understanding of how and why LAP diverged from canonical autophagy, but will further our ability to target this fascinating and important biological mechanism, one that is instrumental in shaping the immune response to cancer, autoimmunity and beyond.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the National Institutes of Health grant AI040646 to D.R.G. and grants AI138492 and CA231423 to B.L.H., a Distinguished Innovator award from the Lupus Research Alliance, and American Lebanese Syrian Associated Charities to D.R.G., and the John H. Sununu Endowed Fellowship to B.L.H. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.222984.supplemental.
References
- Acharya M., Sokolovska A., Tam J. M., Conway K. L., Stefani C., Raso F., Mukhopadhyay S., Feliu M., Paul E., Savill J. et al. (2016). alphav Integrins combine with LC3 and atg5 to regulate Toll-like receptor signalling in B cells. Nat. Commun. 7, 10917 10.1038/ncomms10917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J. M. (2016). The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 473, 2251-2271. 10.1042/BCJ20160170 [DOI] [PubMed] [Google Scholar]
- Bedard K. and Krause K.-H. (2007). The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87, 245-313. 10.1152/physrev.00044.2005 [DOI] [PubMed] [Google Scholar]
- Chu H., Khosravi A., Kusumawardhani I. P., Kwon A. H. K., Vasconcelos A. C., Cunha L. D., Mayer A. E., Shen Y., Wu W.-L., Kambal A. et al. (2016). Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116-1120. 10.1126/science.aad9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codogno P., Mehrpour M. and Proikas-Cezanne T. (2011). Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat. Rev. Mol. Cell Biol. 13, 7-12. 10.1038/nrm3249 [DOI] [PubMed] [Google Scholar]
- Cunha L. D., Yang M., Carter R., Guy C., Harris L., Crawford J., Quarato G., Boada-Romero E., Kalkavan H., Johnson M. D. et al. (2018). Regulation of myeloid function by LC3-associated phagocytosis promotes tumor immune tolerance. Cell 175, 429-441.e16. 10.1016/j.cell.2018.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen N. S., Gong L., Naderer T., Devenish R. J. and Kwok T. (2015). Analysis of the relative contribution of phagocytosis, LC3-associated phagocytosis, and canonical autophagy during helicobacter pylori infection of macrophages. Helicobacter 20, 449-459. 10.1111/hel.12223 [DOI] [PubMed] [Google Scholar]
- Dupont N., Nascimbeni A. C., Morel E. and Codogno P. (2017). Molecular mechanisms of noncanonical autophagy. Int. Rev. Cell Mol. Biol. 328, 1-23. 10.1016/bs.ircmb.2016.08.001 [DOI] [PubMed] [Google Scholar]
- Efeyan A., Comb W. C. and Sabatini D. M. (2015). Nutrient-sensing mechanisms and pathways. Nature 517, 302-310. 10.1038/nature14190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan D., Kim J., Shaw R. J. and Guan K.-L. (2011). The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 7, 643-644. 10.4161/auto.7.6.15123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok V. A., Bratton D. L., Konowal A., Freed P. W., Westcott J. Y. and Henson P. M. (1998). Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Invest. 101, 890-898. 10.1172/JCI1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson T. A. and Green D. R. (2014). Autophagy and phagocytosis converge for better vision. Autophagy 10, 165-167. 10.4161/auto.26735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. F. and López-Otín C. (2015). The functional and pathologic relevance of autophagy proteases. J. Clin. Invest. 125, 33-41. 10.1172/JCI73940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher K., Ulferts R., Jacquin E., Veith T., Gammoh N., Arasteh J. M., Mayer U., Carding S. R., Wileman T., Beale R. et al. (2018). The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 37, e97840 10.15252/embj.201797840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O., Gammoh N., Kim S. E., Jiang X. and Overholtzer M. (2015). V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy 11, 88-99. 10.4161/15548627.2014.984277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Lopes V. S., Bragin A., Reyes-Reveles J., Brancato J., Cohen A., Mitchell C. H., Williams D. S. and Boesze-Battaglia K. (2015). The contribution of melanoregulin to microtubule-associated protein 1 light chain 3 (LC3) associated phagocytosis in retinal pigment epithelium. Mol. Neurobiol. 52, 1135-1151. 10.1007/s12035-014-8920-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluschko A., Herb M., Wiegmann K., Krut O., Neiss W. F., Utermohlen O., Kronke M. and Schramm M. (2018). The beta2 Integrin Mac-1 Induces Protective LC3-Associated Phagocytosis of Listeria monocytogenes. Cell Host Microbe 23, 324-337.e5. 10.1016/j.chom.2018.01.018 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Taura M. and Iwasaki A. (2018). The interaction between IKKalpha and LC3 promotes type I interferon production through the TLR9-containing LAPosome. Sci. Signal. 11, eaan4144 10.1126/scisignal.aan4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann B. L., Boada-Romero E., Cunha L. D., Magne J. and Green D. R. (2017). LC3-associated phagocytosis and inflammation. J. Mol. Biol. 429, 3561-3576. 10.1016/j.jmb.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J., Martinez J., Riggs J. M., Tian J., Mehta P., Clarke L., Sasai M., Latz E., Brinkmann M. M., Iwasaki A. et al. (2012). Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37, 986-997. 10.1016/j.immuni.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A., Kubori T., Coban C., Matsuzawa T., Ogawa M., Kawabata T., Yoshimori T. and Nagai H. (2017). Bacterial secretion system skews the fate of Legionella-containing vacuoles towards LC3-associated phagocytosis. Sci. Rep. 7, 44795 10.1038/srep44795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. (2006). Autophagy, mitochondrial quality control, and oncogenesis. Autophagy 2, 80-84. 10.4161/auto.2.2.2460 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y. and Yoshimori T. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720-5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y. and Yoshimori T. (2004). LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805-2812. 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- Kametaka S., Okano T., Ohsumi M. and Ohsumi Y. (1998). Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284-22291. 10.1074/jbc.273.35.22284 [DOI] [PubMed] [Google Scholar]
- Kim S., Elkon K. B. and Ma X. (2004). Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity 21, 643-653. 10.1016/j.immuni.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Kim J.-Y., Zhao H., Martinez J., Doggett T. A., Kolesnikov A. V., Tang P. H., Ablonczy Z., Chan C. C., Zhou Z., Green D. R. et al. (2013). Noncanonical autophagy promotes the visual cycle. Cell 154, 365-376. 10.1016/j.cell.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrmizi I., Gresnigt M. S., Akoumianaki T., Samonis G., Sidiropoulos P., Boumpas D., Netea M. G., van de Veerdonk F. L., Kontoyiannis D. P. and Chamilos G. (2013). Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J. Immunol. 191, 1287-1299. 10.4049/jimmunol.1300132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam G. Y., Cemma M., Muise A. M., Higgins D. E. and Brumell J. H. (2013). Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy 9, 985-995. 10.4161/auto.24406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth J. D. and Neish A. S. (2014). Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu. Rev. Pathol. 9, 119-145. 10.1146/annurev-pathol-012513-104651 [DOI] [PubMed] [Google Scholar]
- Lee Y.-K. and Lee J.-A. (2016). Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 49, 424-430. 10.5483/BMBRep.2016.49.8.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H. and Levine B. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672-676. 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Ma J., Becker C., Lowell C. A. and Underhill D. M. (2012). Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J. Biol. Chem. 287, 34149-34156. 10.1074/jbc.M112.382812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K. and Klionsky D. J. (2011). AMPK activates autophagy by phosphorylating ULK1. Circ. Res. 108, 787-788. 10.1161/RES.0b013e3182194c29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. J., Peters K. N. and Behar S. M. (2014). Macrophages clean up: efferocytosis and microbial control. Curr. Opin. Microbiol. 17, 17-23. 10.1016/j.mib.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Almendinger J., Oberst A., Ness R., Dillon C. P., Fitzgerald P., Hengartner M. O. and Green D. R. (2011). Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA 108, 17396-17401. 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Malireddi R. K., Lu Q., Cunha L. D., Pelletier S., Gingras S., Orchard R., Guan J.-L., Tan H., Peng J. et al. (2015). Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17, 893-906. 10.1038/ncb3192 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez J., Cunha L. D., Park S., Yang M., Lu Q., Orchard R., Li Q.-Z., Yan M., Janke L., Guy C. et al. (2016). Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115-119. 10.1038/nature17950 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T. et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385-396. 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Mehta P., Henault J., Kolbeck R. and Sanjuan M. A. (2014). Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr. Opin. Immunol. 26, 69-75. 10.1016/j.coi.2013.10.012 [DOI] [PubMed] [Google Scholar]
- Mitchell G., Cheng M. I., Chen C., Nguyen B. N., Whiteley A. T., Kianian S., Cox J. S., Green D. R., McDonald K. L. and Portnoy D. A. (2018). Listeria monocytogenes triggers noncanonical autophagy upon phagocytosis, but avoids subsequent growth-restricting xenophagy. Proc. Natl. Acad. Sci. USA 115, E210-E217. 10.1073/pnas.1716055115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. (2005). The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12 Suppl. 2, 1535-1541. 10.1038/sj.cdd.4401728 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y. and Yoshimori T. (2003). Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116, 1679-1688. 10.1242/jcs.00381 [DOI] [PubMed] [Google Scholar]
- Mukundan L., Odegaard J. I., Morel C. R., Heredia J. E., Mwangi J. W., Ricardo-Gonzalez R. R., Goh Y. P., Eagle A. R., Dunn S. E., Awakuni J. U. et al. (2009). PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 15, 1266-1272. 10.1038/nm.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz-Feliciano L., Doggett T. A., Zhou Z. and Ferguson T. A. (2017). RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy 13, 2072-2085. 10.1080/15548627.2017.1380124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz L. E., Lauber K., Schiller M., Manfredi A. A. and Herrmann M. (2010). The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat. Rev. Rheumatol. 6, 280-289. 10.1038/nrrheum.2010.46 [DOI] [PubMed] [Google Scholar]
- Münz C. (2015). Of LAP, CUPS, and DRibbles - unconventional use of autophagy proteins for mhc restricted antigen presentation. Front Immunol. 6, 200 10.3389/fimmu.2015.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münz C. (2017). Autophagy Proteins in Phagocyte Endocytosis and Exocytosis. Front Immunol. 8, 1183 10.3389/fimmu.2017.01183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H. (2013). Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 55, 39-50. 10.1042/bse0550039 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. (2014). Historical landmarks of autophagy research. Cell Res. 24, 9-23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou V., Renga G., De Luca A., Borghi M., Pariano M., Puccetti M., Paolicelli G., Stincardini C., Costantini C., Bartoli A. et al. (2018). Autophagy and LAP in the fight against fungal infections: regulation and therapeutics. Mediators Inflamm. 2018, 6195958 10.1155/2018/6195958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E. F. C., Meijer A. J. and Codogno P. (2000). Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992-998. 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T., Takacs Z., Donnes P. and Kohlbacher O. (2015). WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 128, 207-217. 10.1242/jcs.146258 [DOI] [PubMed] [Google Scholar]
- Raso F., Sagadiev S., Du S., Gage E., Arkatkar T., Metzler G., Stuart L. M., Orr M. T., Rawlings D. J., Jackson S. W. et al. (2018). alphav Integrins regulate germinal center B cell responses through noncanonical autophagy. J. Clin. Invest. 128, 4163-4178. 10.1172/JCI99597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao S. and Münz C. (2014). LC3-associated phagocytosis. Autophagy 10, 526-528. 10.4161/auto.27606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao S., Gasser N., Becker A. C., Guhl B., Bajagic M., Vanoaica D., Ziegler U., Roesler J., Dengjel J., Reichenbach J. et al. (2013). Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J. Cell Biol. 203, 757-766. 10.1083/jcb.201308173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M. A., Dillon C. P., Tait S. W. G., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S. et al. (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253-1257. 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Segawa K. and Nagata S. (2015). An apoptotic ‘Eat Me’ signal: phosphatidylserine exposure. Trends Cell Biol. 25, 639-650. 10.1016/j.tcb.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Sprenkeler E. G., Gresnigt M. S. and van de Veerdonk F. L. (2016). LC3-associated phagocytosis: a crucial mechanism for antifungal host defence against Aspergillus fumigatus. Cell. Microbiol. 18, 1208-1216. 10.1111/cmi.12616 [DOI] [PubMed] [Google Scholar]
- Tam J. M., Mansour M. K., Khan N. S., Seward M., Puranam S., Tanne A., Sokolovska A., Becker C. E., Acharya M., Baird M. A. et al. (2014). Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J. Infect. Dis. 210, 1844-1854. 10.1093/infdis/jiu290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Li X. J., Stull N. D., Ming W., Suh C.-I., Bissonnette S. A., Yaffe M. B., Grinstein S., Atkinson S. J. and Dinauer M. C. (2008). Fc gamma R-stimulated activation of the NADPH oxidase: phosphoinositide-binding protein p40phox regulates NADPH oxidase activity after enzyme assembly on the phagosome. Blood 112, 3867-3877. 10.1182/blood-2007-11-126029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.-W., Sil P. and Martinez J. (2018). Rubicon: LC3-associated phagocytosis and beyond. FEBS J. 285, 1379-1388. 10.1111/febs.14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y. Q., Freire-de-Lima C. G., Schiemann W. P., Bratton D. L., Vandivier R. W. and Henson P. M. (2008). Transcriptional and translational regulation of TGF-beta production in response to apoptotic cells. J. Immunol. 181, 3575-3585. 10.4049/jimmunol.181.5.3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cross S. D., Stanton J. B., Marmorstein A. D., Le Y. Z. and Marmorstein L. Y. (2017). Early AMD-like defects in the RPE and retinal degeneration in aged mice with RPE-specific deletion of Atg5 or Atg7. Mol. Vis. 23, 228-241. 10.3390/molecules23020228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N. and Yue Z. (2009). Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468-476. 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]