Abstract
Background
l-Histidine biosynthesis is embedded in an intertwined metabolic network which renders microbial overproduction of this amino acid challenging. This is reflected in the few available examples of histidine producers in literature. Since knowledge about the metabolic interplay is limited, we systematically perturbed the metabolism of Corynebacterium glutamicum to gain a holistic understanding in the metabolic limitations for l-histidine production. We, therefore, constructed C. glutamicum strains in a modularized metabolic engineering approach and analyzed them with LC/MS-QToF-based systems metabolic profiling (SMP) supported by flux balance analysis (FBA).
Results
The engineered strains produced l-histidine, equimolar amounts of glycine, and possessed heavily decreased intracellular adenylate concentrations, despite a stable adenylate energy charge. FBA identified regeneration of ATP from 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) as crucial step for l-histidine production and SMP identified strong intracellular accumulation of inosine monophosphate (IMP) in the engineered strains. Energy engineering readjusted the intracellular IMP and ATP levels to wild-type niveau and reinforced the intrinsic low ATP regeneration capacity to maintain a balanced energy state of the cell. SMP further indicated limitations in the C1 supply which was overcome by expression of the glycine cleavage system from C. jeikeium. Finally, we rerouted the carbon flux towards the oxidative pentose phosphate pathway thereby further increasing product yield to 0.093 ± 0.003 mol l-histidine per mol glucose.
Conclusion
By applying the modularized metabolic engineering approach combined with SMP and FBA, we identified an intrinsically low ATP regeneration capacity, which prevents to maintain a balanced energy state of the cell in an l-histidine overproduction scenario and an insufficient supply of C1 units. To overcome these limitations, we provide a metabolic engineering strategy which constitutes a general approach to improve the production of ATP and/or C1 intensive products.
Keywords: Modularized metabolic engineering, LC/MS-QToF-based systems metabolic profiling (SMP), Flux balance analysis (FBA), Energy engineering, l-Histidine production, Corynebacterium glutamicum
Background
l-Histidine (further referred to as histidine) was discovered in the late nineteenth century by Kossel and Hedin simultaneously [89] and the l-enantiomer is nowadays categorized as an essential amino acid for human infants and adults, belonging to the 20 standard proteinogenic amino acids [52]. Histidine has the ability to switch between the protonated and unprotonated states due to the pKa of about 6 of its imidazole group [64]. Thus, histidine is a common ligand of metalloproteins and part of the catalytic triad in several enzymes, underlining its physiologically prominent role [57, 70, 72]. Exceeding physiological levels of histidine in humans has shown to be connected with mutations in histidase and was named histidinemia, a benign inborn error of metabolism [6, 49]. Furthermore, histidine is a precursor for histamine, which is known to play an important role in regulating human immune response, and thus is linked to several allergic disorders [67, 69]. Beyond this, histidine is available as feed supplement and has been reported to have anti-inflammatory and antioxidant properties, which makes it attractive for applications in the medical industry [25, 33, 34, 87, 90, 91].
Corynebacterium glutamicum is a Gram-positive, facultatively anaerobic bacterium which can grow on a wide range of sugars, alcohols, and organic acids [58, 65] and is known as a workhorse for the production of l-glutamate and l-lysine [8, 23, 83]. Moreover, metabolic engineering approaches expanded the product portfolio to other amino acids such as l-methionine, l-valine, l-arginine, and l-tryptophan [8, 42, 66, 68, 71], organic acids [17, 53, 94, 95], alcohols [13, 43, 46, 80], vitamins [40], carotenoids [35, 36], fatty acids [82], polymers [59], terpenes [26, 48], and others. Most relevant, C. glutamicum possesses an intrinsic histidine synthesis pathway but, in contrast to other industrially relevant bacteria such as Pseudomonas and several Bacillus genera, lacks a histidine utilization system (reviewed in [11]). This renders C. glutamicum as an attractive platform for histidine production.
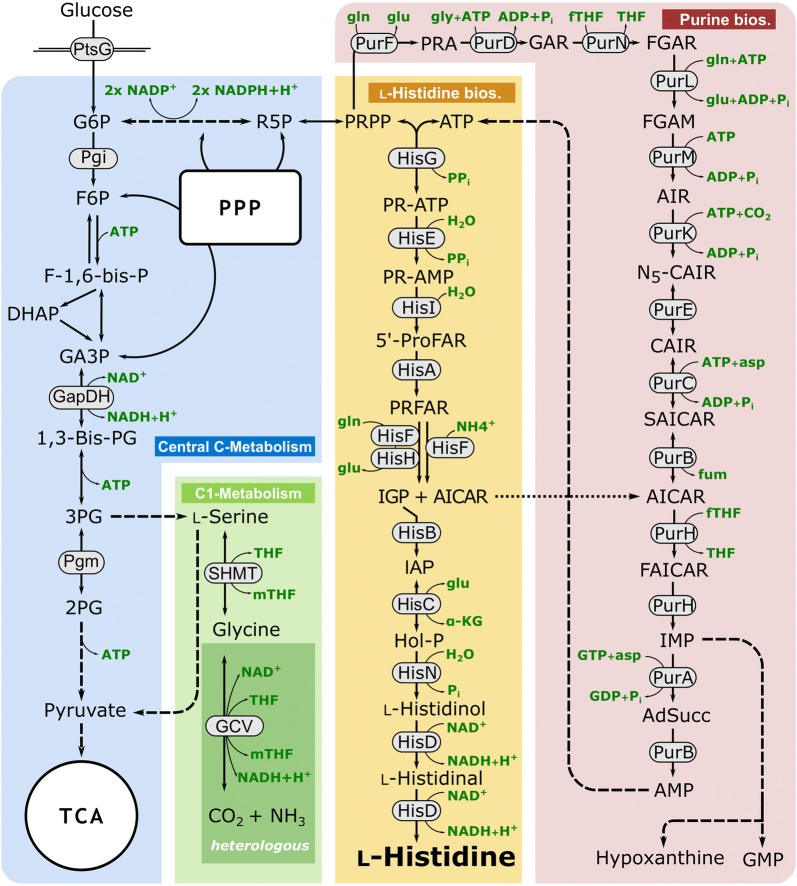
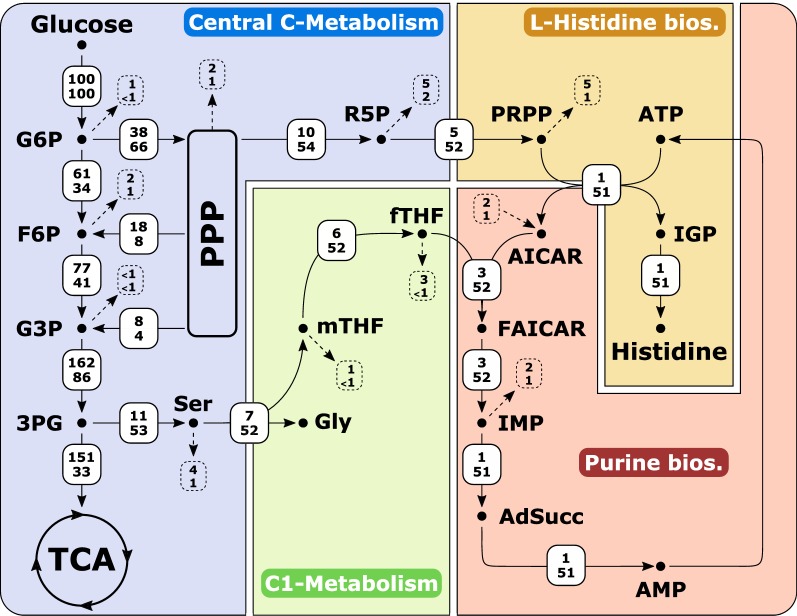
Biosynthesis of histidine has been extensively studied, mainly in Escherichia coli and Salmonella enterica serovar Typhimurium and several profound reviews are available [1, 28, 96]. Nowadays, histidine biosynthesis is considered fundamentally the same in all living organisms [1]. From a physiological point of view, the biosynthetic pathway of histidine is unique for an amino acid as it is closely entwined with other pathways, such as purine biosynthesis and C1 metabolism (Fig. 1). Both the purine and the histidine pathways have the same precursor, phosphoribosyl pyrophosphate (PRPP) (Fig. 1). The second precursor for histidine synthesis is ATP, which is commonly known as energy donor. However, in this unique reaction, the backbone of the ATP molecule is incorporated to give the first intermediate of the histidine pathway. In this reaction catalyzed by ATP phosphoribosyltransferase (HisG), PRPP and ATP are utilized to form phosphoribosyl-ATP, which is further converted in nine enzymatic reactions into histidine (Fig. 1). In the fifth step of the histidine pathway, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) is formed and rerouted to the purine biosynthesis, and thus is available to regenerate ATP (Fig. 1). Besides histidine biosynthesis, PRPP is also a precursor for the biosynthetic pathways of pyrimidines, tryptophan, and nicotinamide adenine dinucleotides [45]. The linkage with the C1 metabolism manifests in de novo synthesis of purines, where two molecules of 10-formyltetrahydrofolate (fTHF) are required as cofactors (Fig. 1). The major metabolic source for the generation of loaded THF molecules in C. glutamicum is the reaction of serine hydroxymethyltransferase (SHMT, encoded by glyA) that converts l-serine into glycine and simultaneously generates 5,10-methylene-THF (mTHF) from THF [60, 78, 79].
Fig. 1.
Overview of the modularized metabolism of C. glutamicum with focus on the l-histidine biosynthesis (yellow) and the related pathways central carbon metabolism (blue), purine biosynthesis (red), and one carbon metabolism (green). The glycine cleavage (GCV) system is not present in C. glutamicum ATCC 13032 and was heterologously produced (dark green). 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate; 5′-ProFAR, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4 carboxamide; α-KG, α-ketoglutarate; ADP, adenosine diphosphate; AdSucc, adenylosuccinate; AICAR, 1-(5′-phosphoribosyl)-5-amino-4-imidazolecarboxamide; AIR, 5-aminoimidazole ribotide; AMP, adenosine monophosphate; asp; l-aspartate; ATP, adenosine triphosphate; fTHF, 10-formyltetrahydrofolate; F-1,6-bis-P, fructose-1,6-bisphosphate; F6P, fructose 6-phosphate; FAICAR, 5-formamidoimidazole-4-carboxamide ribotide; fGAM, 5′phosphoribosylformylglycineamidine; fGAR, phosphoribosyl-N-formylglycineamide; fum, fumarate; G6P, glucose 6-phosphate; GA3P, glyceraldehyde 3-phosphate; GAR, glycineamide ribonucleotide; GCV, glycine cleavage system; gln, l-glutamine; glu, l-glutamate; GMP, guanosine monophosphate; HisA, 5′ProFAR isomerase; HisB, imidazoleglycerol phosphate dehydratase; HisC, histidinol phosphate aminotransferase; HisD, histidinol dehydrogenase; HisE, phosphoribosyl-ATP pyrophosphatase; HisF, synthase subunit of IGP synthase; HisG, ATP phosphoribosyltransferase; HisH, glutaminase subunit of IGP synthase; HisI, phosphoribosyl-AMP cyclohydrolase; HisN, histidinol phosphate phosphatase; Hol-P, l-histidinol phosphate; IAP, imidazole-acetole phosphate; IGP, imidazole-glycerol phosphate; IMP, inosine monophosphate; mTHF, 5,10-methylenetetrahydrofolate; N5-CAIR, 5′-phosphoribosyl-4-carboxy-5-aminoimidazole; NAD+/NADH, oxidized/reduced nicotine amide dinucleotide; NADP+/NADPH, oxidized/reduced nicotine amide dinucleotide phosphate; Pgi, phosphoglucoisomerase; Pi/PPi, inorganic phosphate/diphosphate; Pgm, phosphoglucomutase; PR-AMP, phosphoribosyl-AMP; PR-ATP, phosphoribosyl-ATP; PRA, phosphoribosylamine; PRFAR, 5-[(5-phospho-1-deoxyribulos-1-ylamino)methylideneamino]-1-(5-phosphoribosyl) imidazole-4-carboxamid; PRPP, phosphoribosyl pyrophosphate; PtsG, phosphoenolpyruvate-dependent phosphotransferase system for glucose; PurA, adenylosuccinate synthase; PurB, adenylosuccinate lyase; PurC, phosphoribosylaminoimidazolesuccinocarboxamide synthase; PurD, PRA-glycine ligase; PurE, phosphoribosylaminoimidazole mutase; PurF, amidophosphoribosyltransferase; PurH, bifunctional AICAR formyltransferase/IMP cyclohydrolase; PurK, phosphoribosylaminoimidazole carboxylase; PurL, phosphoribosylformylglycinamide synthase; PurM, phosphoribosylformylglycinamidine cycloligase; PurN, phosphoribosylglycinamide formyltransferase; R5P, ribose 5-phosphate; SAICAR, phosphoribosyl-aminoimidazolesuccinocarboxamide; SHMT, serine hydroxymethyltransferase; TCA, tricarboxylic acid cycle; THF, tetrahydrofolate
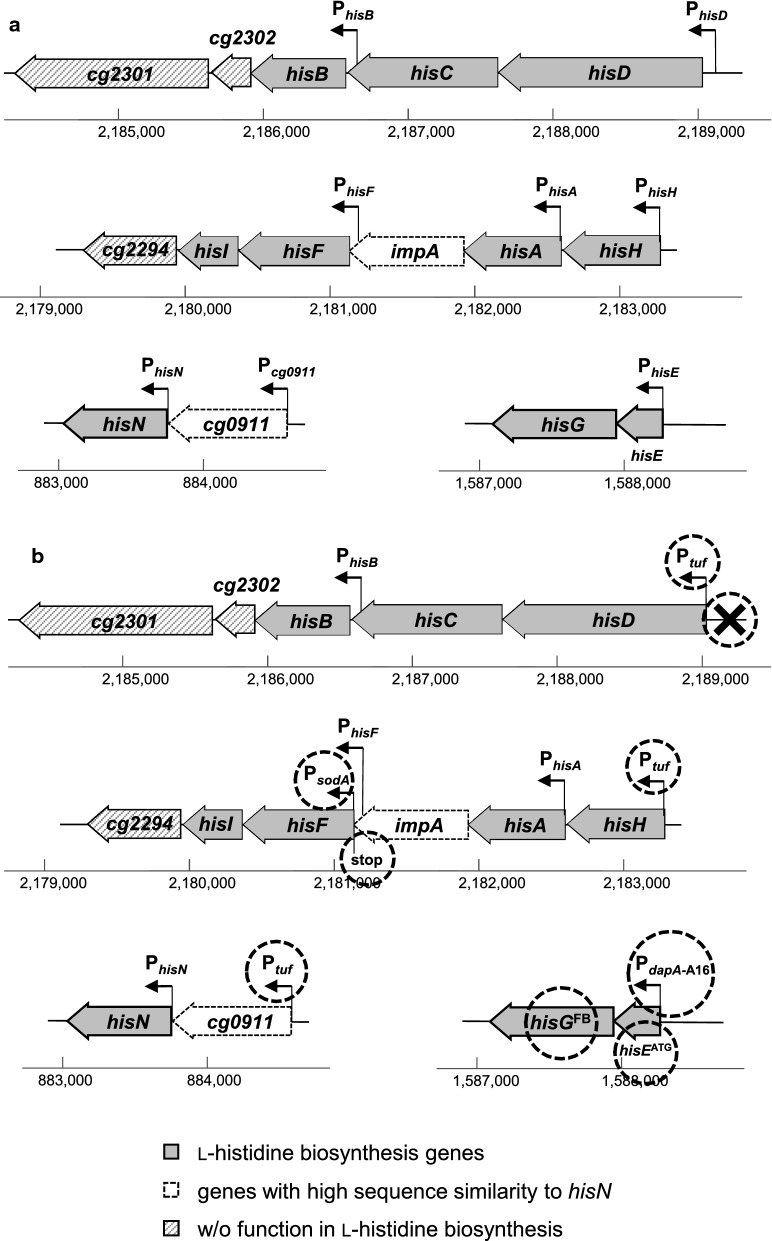
The biosynthesis of histidine in C. glutamicum consists of ten consecutive enzymatic reactions that are catalyzed by nine enzymes with histidinol dehydrogenase (HisD) being bifunctional [56]. The histidine genes are organized in four operons, comprising hisD–hisC–hisB–cg2302–cg2301, hisH–hisA–impA–hisF–hisI–cg2294, cg0911–hisN, and hisE–hisG (Fig. 7; [47, 56]) and eight of the corresponding histidine genes were described as essential [56]. Besides the feedback inhibition of HisG by histidine [2, 97], transcriptional control of histidine biosynthesis has been shown for the hisD operon of C. glutamicum AS019 to function via a T-box-mediated attenuation mechanism [47, 56]. Analysis of the 5′untranslated region (UTR) of hisD in C. glutamicum ATCC 13032, however, revealed a 103 base pair shorter 5′UTR region and it has been speculated that control of this operon occurs on translational rather than transcriptional level in C. glutamicum ATCC 13032 [56].
Fig. 7.
Schematic overview of the structure of the four operons containing l-histidine biosynthesis genes before (a) and after (b) genetic engineering applied in this study. Dashed circles show where modifications were introduced. The cross marks deletion of the promoter region. Modified after [56]
Concerning histidine production, the efforts that have been made with C. glutamicum are limited to a few examples and classically focused on mutagenesis approaches to increase resistance against histidine analogs and to free HisG from its feedback inhibition [2, 3, 63, 77, 97]. Rational approaches for strain engineering were done by promoter exchange of the hisD operon, overexpression of the hisEG genes [18], and elimination of feedback inhibition by deleting the C-terminal regulatory domain and mutating the catalytic domain of HisG, combined with hisEG overexpression [55]. In addition to modifications in the histidine biosynthesis, decreasing transketolase activity has been shown to improve precursor availability and histidine production [41]. However, a systems metabolic engineering approach to engineer histidine production strains has not yet been conducted. Due to the metabolic complexity of histidine synthesis, we combined rational strain engineering with systems metabolic profiling (SMP) and flux balance analysis (FBA) to identify bottlenecks in the intertwined pathways, and to finally engineer histidine producers with balanced metabolite pools for efficient production.
Results
Optimizing the histidine biosynthesis
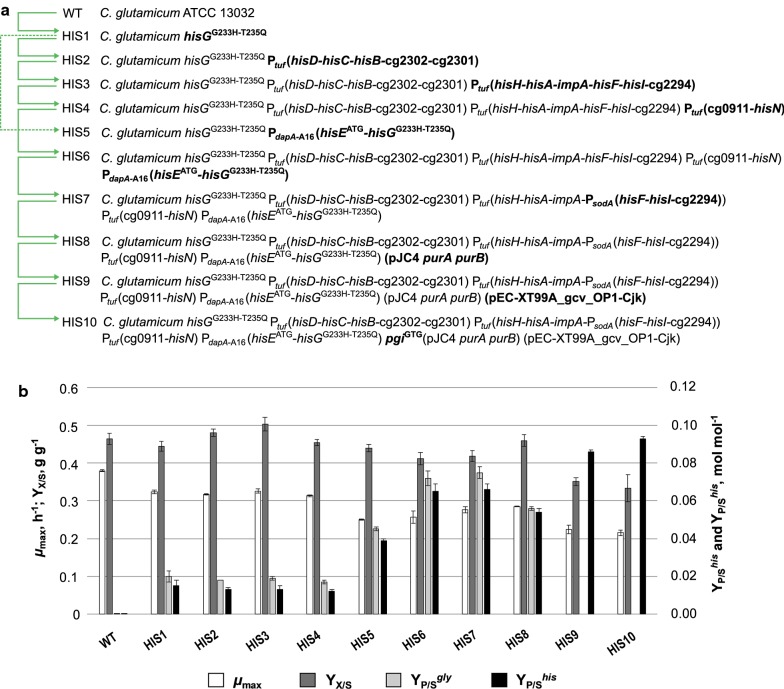
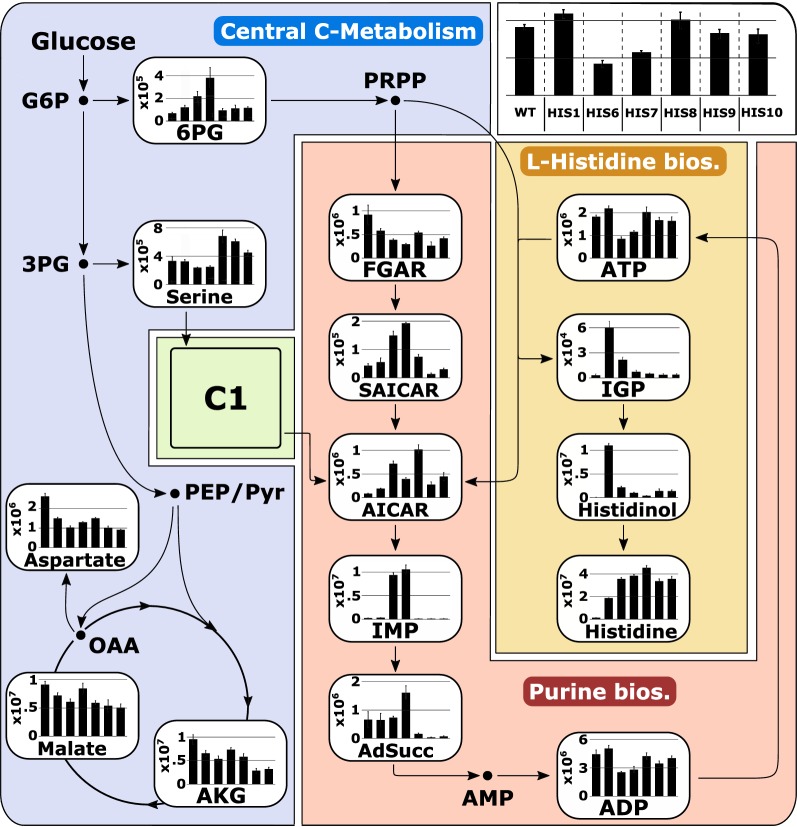
In the first step, we chromosomally introduced the nucleotide exchanges ggc to cat and acg to cag in hisG of C. glutamicum ATCC 13032 (Fig. 7) to relieve HisG from feedback inhibition, yielding variant HisGG233H−T235Q [77]. The resulting strain C. glutamicum HIS1 already secreted histidine into the culture supernatant with a product yield per unit substrate (YhisP/S) of 0.015 ± 0.003 mol histidine per mol glucose (Fig. 2). Compared to the WT strain, the maximum growth rate µmax of C. glutamicum HIS1 decreased from 0.38 ± 0.01 h−1 to 0.32 ± 0.01 h−1 and the biomass yield per unit substrate (YX/S) remained stable with 0.45 ± 0.01 and 0.46 ± 0.02 g biomass per g substrate, respectively. Beyond this, C. glutamicum HIS1 produced glycine as main byproduct besides histidine with a YglyP/S of 0.020 ± 0.003 mol glycine per mol glucose (Fig. 2). To gain a deeper insight into the metabolic state of C. glutamicum HIS1, systems metabolic profiling (SMP) was performed. Intracellular peak intensities of d-erythro-1-(imidazole-4-yl)glycerol 3-phosphate (IGP) and l-histidinol were 18 and 275 times higher compared to the WT (Fig. 3), respectively. To debottleneck histidine synthesis, we systematically exchanged the native promoters of all four canonical operons containing all histidine biosynthesis genes to stronger ones (Fig. 7), resulting in strains C. glutamicum HIS2–HIS6. Firstly, the native promoter of the hisD–hisC–hisB–cg2302–cg2301 operon in C. glutamicum HIS1 was replaced by the promoter of elongation factor TU (Ptuf) and in parallel the 5′UTR region of the hisD gene was deleted to eliminate suspected regulatory elements [56] (Fig. 7), which resulted in strain C. glutamicum HIS2. C. glutamicum HIS2 showed similar YhisP/S (0.013 ± 0.001 mol histidine per mol glucose), µmax (0.32 ± 0.01 h−1), YX/S (0.48 ± 0.01 g biomass per g glucose), and YglyP/S (0.018 mol glycine per mol glucose), compared to C. glutamicum HIS1. C. glutamicum HIS3 and HIS4 were generated, by additionally replacing the native promoters of the operons hisH–hisA–impA–hisF–hisI–cg2294 and cg0911–hisN in C. glutamicum HIS2 with Ptuf (Fig. 7), respectively. However, the performance of the resulting strains HIS3 and HIS4 remained constant, with YhisP/S of 0.013 ± 0.002 and 0.012 ± 0.001 mol histidine per mol glucose, µmax of 0.33 ± 0.01 and 0.31 ± 0.01 h−1, YX/S of 0.50 ± 0.02 and 0.45 ± 0.01 g biomass per g glucose, and YglyP/S of 0.019 ± 0.001 and 0.017 ± 0.001 mol glycine per mol glucose, respectively (Fig. 2). Since we did not succeed in replacing the native promoter of the hisEG operon with Ptuf or the promoter of manganese superoxide dismutase PsodA in C. glutamicum HIS4, we used PdapA–A16, a mutated variant of the promoter of dihydrodipicolinate synthase (encoded by dapA; [88]). Simultaneously, we replaced the native translational start codon GTG of hisE by ATG to improve translation efficiency (Fig. 7), which resulted in C. glutamicum HIS6. The introduced modifications significantly increased histidine production and strain C. glutamicum HIS6 showed a YhisP/S of 0.065 ± 0.004 mol histidine per mol glucose, which is about 5 times higher compared to the parental strain C. glutamicum HIS4. Accordingly, the YglyP/S increased to 0.072 ± 0.004 mol glycine per mol glucose and we observed reduction of 20% and 10% of µmax and YX/S, respectively (Fig. 2). To evaluate the cumulative effects of the applied modifications, we constructed strain C. glutamicum HIS5, which carries the HisGG233H−T235Q variant, the exchange of the translational start codon from GTG to ATG of hisE, and the replaced native promoter of hisE by PdapA–A16 (Fig. 7). Although, strain C. glutamicum HIS5 does not possess Ptuf in front of the remaining three histidine operons, it showed an intermediate YhisP/S of 0.039 ± 0.001 mol histidine per mol glucose which is 160% higher compared to C. glutamicum HIS1 and 40% lower compared to C. glutamicum HIS6 (Fig. 2). Thus, the applied modifications in C. glutamicum HIS6 are cumulatively beneficial for histidine production. Since the hisH–hisA–impA–hisF–hisI–cg2294 has a length of > 4000 bps, we finally replaced the internal promoter in front of hisF by PsodA downstream of an artificial stop codon (Fig. 7). The resulting strain C. glutamicum HIS7 showed similar characteristics like C. glutamicum HIS6 (Fig. 2), however the intracellular peak intensities of IGP and l-histidinol were reduced by a factor of 3 and 2, respectively (Fig. 3).
Fig. 2.
Strain genealogy (a) and characteristics (b) of the engineered l-histidine-producing strains developed in this work. Arrows indicate direct parentage; new modifications that differentiate the descendant from the progenitor are given in bold (a). Characteristics of these strains include maximum growth rate μmax (h−1) in white, biomass yield per unit substrate YX/S (g g−1) in dark gray, and (by)product yield per unit substrate for glycine in gray and l-histidine (mol mol−1) in black with the non-producing C. glutamicum ATCC 13032 (WT) as reference (b). Error bars give standard deviation of at least three independently performed experiments
Fig. 3.
Systems metabolic profiling (SMP) of l-histidine-producer strains C. glutamicum HIS1, HIS6–HIS10 and C. glutamicum ATCC 13032 (WT). Intracellular peak intensities of each metabolite are given as bar plots. Corresponding strains can be identified per legend in the top right corner. Error bars give standard deviation of three independently performed experiments. Abbreviations can be found in Fig. 1
In summary, using SMP, the applied genetic modifications were evaluated and allowed a stepwise increase to a YhisP/S of 0.065 ± 0.004 mol histidine per mol glucose, and readjusted the intracellular concentrations of IGP and l-histidinol to C. glutamicum WT-like levels (Fig. 3).
Overexpression of hisEG leads to diminished intracellular adenylate levels
Since histidine and purine biosynthesis are closely interlinked (Fig. 1), we determined the intracellular adenylate concentrations in strains C. glutamicum HIS1–HIS7 and calculated adenylate energy charges (ECs), as a measure for the energetic state of the strains. Interestingly, we found relatively stable ECs in all engineered strains with values between 0.88 and 0.92, which are comparable to the WT strain, with an EC of 0.89 ± 0.04 (Fig. 4). Despite the stable ECs, the absolute concentrations of ATP, ADP, and AMP were strongly affected (Fig. 4). The WT strain showed intracellular purine concentrations of 15.7 ± 0.4 µmol ATP g−1CDW, 6.0 ± 0.6 µmol ADP g−1CDW, and 2.1 ± 0.8 µmol AMP g−1CDW during exponential growth phase (AMP not shown). According to their intracellular ATP and ADP concentrations, strains C. glutamicum HIS1–HIS7 can be clustered into two groups. The first group (C. glutamicum HIS1–HIS4) exhibited solely diminished ADP levels, showing about half of the concentration of the WT strain; whereas ATP concentrations remained stable. The second group (C. glutamicum HIS5–HIS7) exhibited diminished ADP and also diminished ATP concentrations with values ranging from 6.6 to 9.7 µmol ATP g−1CDW and 1.8–2.5 µmol ADP g−1CDW. Strikingly, ATP concentrations were only affected in strains where the native promoter of the hisEG operon was exchanged by PdapA–A16. As described in the materials and methods part, intracellular AMP concentrations in the modified strains were below the detection limit of the applied method, and it, thus, can be concluded that AMP concentrations in strains C. glutamicum HIS1–HIS7 were significantly below the concentrations of the WT strain.
Fig. 4.
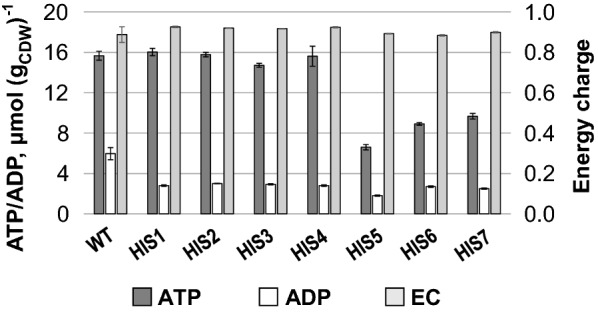
Intracellular ATP and ADP concentrations and the calculated energy charges for strains C. glutamicum HIS1–HIS7 with C. glutamicum ATCC 13032 (WT) as reference. Intracellular ATP and ADP contents from exponentially growing cells in μmol gCDW−1 are shown in dark gray and white, respectively. Energy charges are shown as light gray and have been calculated without considering AMP, since AMP concentrations in the histidine producing strains were below the detection limit of the applied HPLC system (see “Methods”). Experiments were performed in at least triplicates and standard deviations are given as error bars
Energy engineering to readjust ATP levels for histidine production
After optimizing the histidine biosynthesis and investigating intracellular adenylate concentrations, we performed flux balance analysis (FBA) to gain a holistic overview and further indications to improve histidine production with C. glutamicum (Fig. 5). Two different FBAs resembling WT-like growth and growth-coupled histidine production were performed, which showed that in addition to an increased flux through the histidine biosynthesis pathway, an equimolar supply of ATP is a prerequisite for efficient histidine production. Therefore, an unphysiologically high flux from AICAR to ATP has to be accomplished (Fig. 5). To identify bottlenecks in this ATP regeneration cascade, we performed further SMP, and found that in C. glutamicum, HIS7 strongly increased intracellular peak intensities of inosine monophosphate (IMP) and adenylosuccinate, which were 43- and twofold higher compared to the WT (Fig. 3). The strongly increased IMP level indicated a limitation on the level of adenylosuccinate synthetase (PurA) and/or adenylosuccinate lyase (PurB), which catalyze the subsequent reactions from IMP over adenylosuccinate to AMP, respectively. To improve the ATP regeneration capacity (designated as energy engineering), we overexpressed the native purA and purB genes from the plasmid pJC4 under control of Ptuf in C. glutamicum HIS7. Compared to the parental strain C. glutamicum HIS7, the plasmid-carrying derivative C. glutamicum HIS8 had a 17% lower YhisP/S of 0.054 ± 0.002 mol histidine per mol glucose. The growth rate remained stable with 0.29 ± 0.01 h−1 and the YX/S increased slightly to 0.46 ± 0.02 g biomass per g glucose. Concomitant with the reduced histidine yield, the glycine yield decreased to 0.056 ± 0.001 mol glycine per mol glucose (Fig. 2). However, SMP of C. glutamicum HIS8 showed that upon introduction of pJC4purApurB, the intracellular levels of IMP and adenylosuccinate were readjusted to WT-like levels. Additionally, intracellular peak intensities of ATP and ADP were restored from 64 and 63% (C. glutamicum HIS7) to 111% and 96% of the WT level in C. glutamicum HIS8 (Fig. 3). Taken together, the applied energy engineering approach identified the requirement of an unphysiologically high ATP regeneration capacity for histidine production which can be accounted for by overexpression of purA and purB. Although this modification readjusted the intracellular purine pool, the YhisP/S was not improved indicating further obstacles that must be overcome.
Fig. 5.
Flux distribution of WT-like growth (upper number) and growth coupled l-histidine production (lower number) in C. glutamicum. All fluxes are given in percent (%) of glucose uptake rate, which was set to 3.94 mmol gCDW−1 h−1 [12] for both simulations. Additional constraint for growth coupled l-histidine production: μ = 0.1 h−1. Dotted lines show respective flux to biomass. For abbreviations see Fig. 1
C1 supply is a further bottleneck for histidine production
For incorporation of the two carbonyl groups into the carbon skeleton of purines, the de novo biosynthesis requires two molecules of fTHF, which are provided as mTHF by the conversion of l-serine into glycine catalyzed by serine hydroxymethyltransferase (SHMT; Fig. 1). This is reflected in the conducted FBAs, yielding an equimolar flux into C1 metabolism and histidine synthesis (Fig. 5). Since the surplus of glycine cannot be further degraded or rerouted to the central metabolism of C. glutamicum, all strains of the so far engineered strain genealogy produced glycine as the main byproduct being inevitably present in equimolar amounts to histidine (Fig. 6). Although, overexpression of purA and purB reduced the intracellular peak intensities of IMP and adenylosuccinate to WT levels, we observed that the intracellular AICAR peak intensity was not reduced but even increased (2.6-fold) in C. glutamicum HIS8 compared to C. glutamicum HIS7 (Fig. 3). This result pointed to a fTHF limitation of the bifunctional AICAR formyltransferase/IMP cyclohydrolase (PurH, Fig. 1), which might negatively affect the flux in the upper part of histidine synthesis. Since the glycine cleavage (GCV) system converts glycine into CO2, ammonia, and simultaneously generates mTHF from THF (Fig. 1), we heterologously expressed the GCV system from C. jeikeium on plasmid pEC-XT99A_gcv_OP1-Cjk in strain C. glutamicum HIS8, resulting in C. glutamicum HIS9. This modification was expected to reduce glycine accumulation accompanied by an improvement of the supply with loaded THF molecules. C. glutamicum HIS9 showed a µmax of 0.22 ± 0.01 h−1, a YX/S of 0.35 ± 0.01 g biomass per g glucose and in contrast to C. glutamicum HIS8 did not secret glycine into the supernatant (Fig. 2). Furthermore, the YhisP/S in C. glutamicum HIS9 increased by 59% to 0.086 ± 0.001 mol histidine per mol glucose and the peak intensities of AICAR decreased by 73% compared to C. glutamicum HIS8 (Figs. 2, 3). In summary, the expression of a functional GCV system eliminated glycine as byproduct, increased the fTHF availability and, in combination with purA and purB overexpression, improved histidine production with C. glutamicum.
Fig. 6.
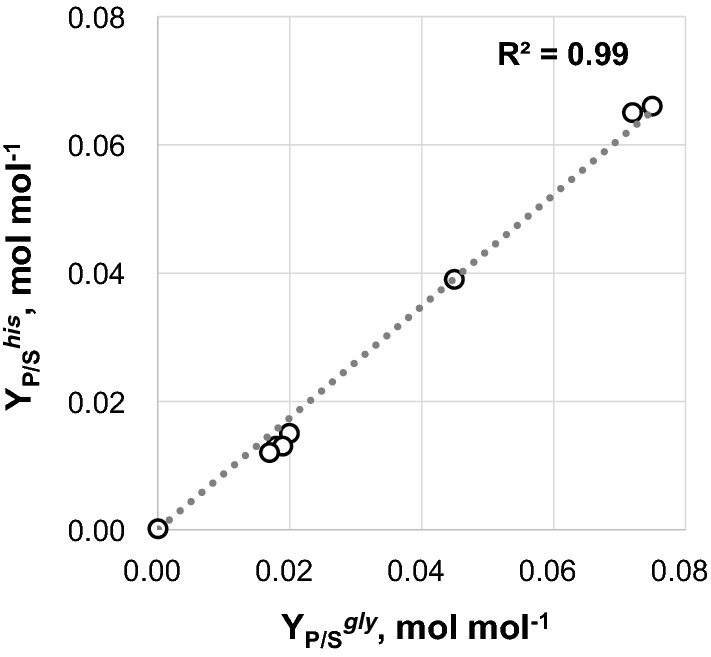
Correlation of the product substrate yields (mol mol−1) for histidine (YhisP/S) and glycine (YglyP/S) of strains C. glutamicum HIS1–HIS7 including values for C. glutamicum ATCC 13032. Given product substrate yields (open circles) are mean values calculated from at least three independent experiments
Engineering the glycolysis–PPP split ratio
To achieve an optimal flux distribution for histidine production with C. glutamicum, the FBA predicted to increase the carbon flux towards the pentose phosphate pathway (PPP) by 74% compared to the WT flux (Fig. 5). Thus, C. glutamicum HIS9 with optimized energy metabolism and enhanced C1 supply was further modified to reroute carbon from glycolysis to the PPP by changing the native translational start codon ATG of the pgi gene, encoding the glucose 6-phosphate isomerase (Pgi), to the weaker GTG. The constructed strain C. glutamicum HIS10 showed a similar µmax and YX/S, however, a YhisP/S of 0.093 ± 0.001 mol histidine per mol glucose which is 8% higher compared to C. glutamicum HIS9 (Fig. 2).
Discussion
Histidine is an attractive amino acid for various applications in the feed and medical sector [25, 87, 90, 93] and in 2003, the production by fermentation was estimated to be 400 t histidine per year [41]. Most efficient producer strains described in literature have been obtained by classical mutagenesis and show maximal YhisP/S values of about 0.15–0.20 g histidine per g substrate [9, 63], which is about 2.5-fold lower than the maximum theoretical product yield of 0.44 g per g achieved at μmax = 0.1 h−1 in growth-coupled manner (Fig. 5). Associated with the applied modifications in the histidine biosynthesis and connected pathways, µmax strongly decreased to a minimum of 0.22 ± 0.01 h−1 in C. glutamicum HIS9 and HIS10 (equals about 58% of WT µmax) which mostly can be attributed to the overexpression of the modified hisEG genes (Fig. 2). Both µ and YhisP/S, which is about 18% of the theoretical maximum, are crucial factors for industrial-scale application and therefore have to be optimized in further studies. So far, few studies provided knowledge for the targeted optimization of C. glutamicum as a histidine overproducer. As such, the deregulation of the biosynthesis and the improved precursor availability [41, 55, 63, 97] have been investigated. The moderate success to develop efficient production strains might be attributed to the demanding biosynthesis of histidine reflected by its tight connection to energy metabolism (Fig. 1). Therefore, to gain a more holistic understanding of the metabolic limitations for histidine production, we performed a modularized metabolic engineering approach, including flux balance analysis and LC/MS QToF-based systems metabolic profiling (SMP). Especially, the applied untargeted metabolomics workflow proofed an effective tool to monitor intracellular peak intensities of key metabolites in the engineered strain genealogy. The introduced modifications in the histidine biosynthesis enabled histidine overproduction (Fig. 2) and were shown for C. glutamicum HIS7 to maintain WT-like levels of the intermediates IGP and l-histidinol (Fig. 3). In contrast, the intracellular peak intensities of histidine increased stepwise with C. glutamicum HIS8 showing a 33 times higher level compared to the WT, indicating an export limitation. However, to our knowledge, no export system for histidine has been identified so far in C. glutamicum, whereas the gene product of cg1305 was proposed to be involved in histidine import [56]. If the prevention of re-import might be beneficial for histidine production has to be verified in future experiments.
The quantification of intracellular ATP and ADP concentrations in strains C. glutamicum HIS1–HIS7 (Fig. 4) showed that particularly the overexpression of hisEG (presumably the mutated hisG, since it encodes the first enzyme in the biosynthesis pathway, which catalyzes the covalent binding of ATP to PRPP) in strain C. glutamicum HIS5 drained ATP efficiently into histidine biosynthesis and led to strongly diminished purine concentrations. Interestingly, the perturbation of the energy metabolism did not manifest in an altered energy charge itself but was disclosed by consistent reduction of the ATP and ADP pools (Fig. 4), which underpins the relevance of a balanced energy state in the regulatory hierarchy of the cell [4, 5]. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry [4, 92]. The applied FBA already pointed to the requirement of a high ATP regeneration capacity of the cell for efficient histidine production (Fig. 5) and SMP finally hinted to PurA and/or PurB as the limiting step(s) by the observed strong increase of the IMP and adenylosuccinate levels in C. glutamicum HIS 6 and HIS7, compared to the WT (Fig. 3). Indeed, overexpression of purA and purB not only reduced the intracellular peak intensities of IMP and adenylosuccinate but also readjusted the ADP and ATP levels to WT level demonstrating that the natural capacity of the cell is not suited to regenerate ATP on top of the growth demands.
Recently, E. coli has also been engineered for histidine production and the observed intracellular accumulation of AICAR was overcome by introduction of an additional copy of purA into the chromosome [62]. Interestingly, although overexpression of purA and purB in our strains almost readjusted the levels of IMP, adenylosuccinate, ADP, and ATP, SMP revealed still increased peak intensities for AICAR in all histidine producing strains compared to the WT (Fig. 3) indicating a different regulatory pattern in C. glutamicum compared to E. coli. Since overexpression of purA and purB positively impacted the energy state of the cell but did not improve the histidine yield, we speculated that the increased intracellular AICAR levels feedback on the upper part of the histidine synthesis pathway and indicate another bottleneck in the metabolism of C. glutamicum. In accordance, Malykh et al. [62] recently suggested in E. coli a competitive inhibitory influence of AICAR on HisG. Furthermore, it has been shown for E. coli that upon folate limitation, AICAR accumulates and binds to a specific riboswitch, which negatively controls expression of purine genes [50]. Likewise, in C. glutamicum, the conversion of AICAR by PurH is fTHF dependent (Fig. 1), and consequently we speculated about a C1 limitation for histidine production, which was supported by the results of FBA, proposing a required high flux into the C1 metabolism (Fig. 5). Furthermore, strains C. glutamicum HIS1–HIS8 secreted glycine as inevitable equimolar byproduct to histidine, which has also been observed for other histidine producing mutants of C. glutamicum and Brevibacterium flavum [19, 44].
The required fTHF for purine biosynthesis is supplied by the reaction of the SHMT, converting l-serine into glycine, thereby generating mTHF, which might be further converted into fTHF [29, 39, 78, 79]. Unfortunately, the various THF species of the C1 metabolism are not accessible with the applied analytical system, due to low pool sizes caused by interconversion, polyglutamylation, and degradation [61]. However, the C1 cycle is a complex network of several oxidized/reduced forms of C1 units with THF as carrier molecule and has been investigated before for l-methionine- and l-serine-overproducing C. glutamicum strains [15, 32, 54, 81]. To overcome the proposed C1 limitation, we expressed the GCV system from C. jeikeium in C. glutamicum HIS8, which already overexpresses the purA and purB genes, and in fact observed the disappearance of glycine as byproduct (Fig. 2), a significant reduction of the AICAR pool (Fig. 3), and a significantly increased YhisP/S (Fig. 2). In a recent approach, a GCV system from E. coli has been heterologously produced in C. glutamicum, where it enabled increased l-serine accumulation in a glyA attenuated strain, by generating improved amounts of C1 units for incorporation in the purine biosynthesis [99]. Consistent with these data, the GCV system from C. jeikeium seems to be able to (partly) satisfy the need for loaded THF molecules in histidine-producing C. glutamicum. However, the remaining elevated AICAR levels in C. glutamicum HIS9 and HIS10 (Fig. 3) either indicate an even higher demand for fTHF or point to limiting AICAR formyltransferase/IMP cyclohydrolase activity, which might be overcome by overexpression of purH.
Conclusions
Taken together, the applied interplay of strain engineering, systems metabolic profiling, and flux balance analysis yielded a comprehensive view on the complex metabolic network of histidine biosynthesis. Energy engineering identified and reinforced the intrinsically low ATP regeneration capacity to maintain the balanced energy state of the cell. However, to utilize the readjusted ATP levels for histidine production, it is essential to provide sufficient C1 units avoiding the accumulation of AICAR, which seems to be a potent effector molecule to control the entry flux into histidine biosynthesis.
Methods
Strains and plasmids
All strains and plasmids used in this study are listed in Table 1. Oligonucleotides are given in Table 2.
Table 1.
Strains and plasmids that were used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or references |
|---|---|---|
| Strains | ||
| E. coli DH5α | F-ϕ80lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (r−K m+K) supE44 thi-1 gyrA96 relA1 phoA | Hanahan [31] |
| E. coli K-12 MG1655 | Wild-type strain DSM 18039; F-, λ-, ilvG-, rfb-50, rph-1 | German collection of microorganisms and cell cultures |
| C. glutamicum WT | Wild-type strain ATCC 13032 | American type culture collection |
| C. jeikeum K411 | Wild-type strain | Tauch et al. [84] |
| C. glutamicum HIS1 | C. glutamicum WT with the feedback inhibition released variant HisGG233H−T235Q (corresponding nucleotide exchanges: ggc742cat, acg748cag) | This work |
| C. glutamicum HIS2 | C. glutamicum HIS1 with additional implementation of Ptuf in front of the operon hisD–hisC–hisB–cg2302–cg2301 | This work |
| C. glutamicum HIS3 | C. glutamicum HIS2 with additional implementation of Ptuf in front of the operon hisH–hisA–impA–hisF–hisI–cg2294 | This work |
| C. glutamicum HIS4 | C. glutamicum HIS3 with additional implementation of Ptuf in front of the operon cg0911–hisN | This work |
| C. glutamicum HIS5 | C. glutamicum HIS1 with additional implementation of PdapA–A16 in front of the operon hisE–hisG, additional exchange of the translational start codon from the native GTG to ATG for hisE | This work |
| C. glutamicum HIS6 | C. glutamicum HIS4 with additional implementation of PdapA–A16 in front of the operon hisE–hisG, additional exchange of the translational start codon from the native GTG to ATG for hisE | This work |
| C. glutamicum HIS7 | C. glutamicum HIS6 with additional implementation of PsodA in front of gene hisF | This work |
| C. glutamicum HIS8 | C. glutamicum HIS7 containing pJC4 purA purB | This work |
| C. glutamicum HIS9 | C. glutamicum HIS8 containing pEC-XT99A_gcv_OP1-Cjk | This work |
| C. glutamicum HIS10 | C. glutamicum HIS9 with additional exchange of the translational start codon from ATG to GTG | This work |
| Plasmids | ||
| pK19mobsacB | Kmr, mobilizable (oriT), oriV | Schäfer et al. [75] |
| pK18mobsacB PaceE dapA-A16 | pK18mobsacB carrying the dapA-A16 promoter | Buchholz et al. [16] |
| pK19mobsacB hisGFB | pK19mobsacB carrying the nucleotide sequence of a modified hisG variant that encodes HisG with amino acid exchanges G233H and T235Q (corresponding nucleotide exchanges: ggc742cat, acg748cag) | This work |
| pK19mobsacB hisD-Ptuf | pK19mobsacB carrying promoter exchange to Ptuf for operon hisD–hisC–hisB–cg2302–cg2301 | This work |
| pK19mobsacB hisH-Ptuf | pK19mobsacB carrying promoter exchange to Ptuf for operon hisH–hisA–impA–hisF–hisI– | This work |
| pK19mobsacB hisN-Ptuf | pK19mobsacB carrying promoter exchange to Ptuf for operon cg0911–hisN | This work |
| pK19mobsacB hisE-PdapA–A16 | pK19mobsacB carrying promoter exchange to PdapA–A16 for operon hisE–hisG and an exchange of the translational start codon from the native GTG to ATG for hisE | This work |
| pJC4 | Kmr | Cordes et al. [20] |
| pJC4 purA purB | pJC4 carrying genes purA and purB from C. glutamicum ATCC 13032 under control of Ptuf and TrrnB | This work |
| pEC-XT99A | IPTG-inducible overexpression plasmid | Kirchner and Tauch [51] |
| pEC-XT99A_gcv_OP1-Cjk | IPTG-inducible overexpression plasmid for genes gcvP, gcvT, gcvH, lipA, and lipB from C. jeikeium xxx | (A. Hüser, Evonik Nutrition & Care GmbH) |
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Purpose |
|---|---|---|
| hisG1 | 5′-CGCGGATCCCAGCTCGATTTGGGTATCACCG-3′ | Outer primer #1 for introduction of hisGG233H−T235Q, BamHI site underlined |
| hisG2 | 5′-CCAGTTGTCGCGTGCCAGTGGGGATACCTGTGGATGGGATAAGCCTGGGGTTACTGC-3′ | Inner primer #1 for introduction of hisGG233H−T235Q, affected triplets underlined |
| hisG3 | 5′-GCAGTAACCCCAGGCTTATCCCATCCACAGGTATCCCCACTGGCACGCGACAACTGG-3′ | Inner primer #2 for introduction of hisGG233H−T235Q, affected triplets underlined |
| hisG4 | 5′-CGCGGATCCGTTGATGGTGGTTCGTGAGATTTGG-3′ | Outer primer #2 for introduction of hisGG233H−T235Q, BamHI site underlined |
| hisGseq | 5′-GGTATCCATCAAGCTTGG-3′ | Sequencing primer for hisGG233H−T235Q |
| hisD1 | 5′-CCCAAGCTTCGGTGTCGCTGAAGTTAAGTTCTG-3′ | Outer primer #1 for exchange of native promoter with Ptuf in operon hisD–hisC–hisB–cg2302–cg2301, HindIII site underlined |
| hisD2 | 5′-CGCAGGTCAGTGACATTCAACATATGTATTATGCATCTCGACCACCCCAGATTTACCTG-3′ | Inner primer #1 for exchange of native promoter with Ptuf in operon hisD–hisC–hisB–cg2302–cg2301, NdeI and NsiI sites underlined |
| hisD3 | 5′-CAGGTAAATCTGGGGTGGTCGAGATGCATAATACATATGTTGAATGTCACTGACCTGCG-3′ | Inner primer #2 for exchange of native promoter with Ptuf in operon hisD–hisC–hisB–cg2302–cg2301, NdeI and NsiI sites underlined |
| hisD4 | 5′-CGCGGATCCCTCATCAACACCCAAGATGGAAC-3′ | Outer primer #2 for exchange of native promoter with Ptuf in operon hisD–hisC–hisB–cg2302–cg2301, BamHI site underlined |
| tuf1 | 5′-AACTGCAGAACCAATGCATCCACAGGGTAGCTGGTAGTTTG-3′ | Primer #1 for amplification of Ptuf, NsiI site underlined |
| tuf2 | 5′-GGGAATTCCATATGATGTCCTCCTGGACTTCGTGGTGGC-3′ | Primer #2 for amplification of Ptuf, NdeI site underlined |
| hisH1 | 5′-CCCAAGCTTCGGAGTGAAATGAGGTCCTTGGTC-3′ | Outer primer #1 for exchange of native promoter with Ptuf in operon hisH–hisA–impA–hisF–hisI–cg2294, HindIII site underlined |
| hisH2 | 5′-GAGAAGGGCGACAGTTTTGGTCATATGTATTATGCATCTCCCTTCCTGAGAATGACGGCTAGTCG-3′ | Inner primer #1 for exchange of native promoter with Ptuf in operon hisH–hisA–impA–hisF–hisI–cg2294, NdeI and NsiI sites underlined |
| hisH3 | 5′-CGACTAGCCGTCATTCTCAGGAAGGGAGATGCATAATACATATGACCAAAACTGTCGCCCTTCTC-3′ | Inner primer #1 for exchange of native promoter with Ptuf in operon hisH–hisA–impA–hisF–hisI–cg2294, NdeI and NsiI sites underlined |
| hisH4 | 5′-CGCGGATCCGCACGAAGTAGAAACGCTCATCAGG-3′ | Inner primer #2 for exchange of native promoter with Ptuf in operon hisH–hisA–impA–hisF–hisI–cg2294, BamHI site underlined |
| hisN1 | 5′-CCCAAGCTTCGCATGAGCATTTCAGCCCAGTCC-3′ | Outer primer #1 for exchange of native promoter with Ptuf in operon cg0911–hisN, HindIII site underlined |
| hisN2 | 5′-GCTCTGGATTAGTCATGCCTTCCATATGTATTATGCATCTCGGTGGCGTTCTAAGAATCACAATCG-3′ | Inner primer #1 for exchange of native promoter with Ptuf in operon cg0911–hisN, NdeI and NsiI sites underlined |
| hisN3 | 5′-CGATTGTGATTCTTAGAACGCCACCGAGATGCATAATACATATGGAAGGCATGACTAATCCAGAGC-3′ | Inner primer #2 for exchange of native promoter with Ptuf in operon cg0911–hisN, NdeI and NsiI sites underlined |
| hisN4 | 5′-CGCGGATCCCACCGAACCACGTATAACCCATGG-3′ | Outer primer #2 for exchange of native promoter with Ptuf in operon cg0911–hisN, BamHI site underlined |
| hisE1 | 5′-CCCAAGCTTCTGACAGAGTTGAAGGCCCTCGAG-3′ | Outer primer #1 for exchange of native promoter with PdapA–A16 and change of the translational start codon from GTG to ATG in operon hisE–hisG, HindIII site underlined |
| hisE2 | 5′-CGTACAGCGAGTCAAATGTCTTCATATGTATTATGCATCTCGGAGGATATTAGTCGAATAATTTC-3′ | Inner primer #1 for exchange of native promoter with PdapA–A16 and change of the translational start codon from GTG to ATG in operon hisE–hisG, NdeI and NsiI sites underlined, base exchange bold |
| hisE3 | 5′-GAAATTATTCGACTAATATCCTCCGAGATGCATAATACATATGAAGACATTTGACTCGCTGTACG-3′ | Inner primer #2 for exchange of native promoter with PdapA–A16 and change of the translational start codon from GTG to ATG in operon hisE–hisG, NdeI and NsiI sites underlined, base exchange bold |
| hisE4 | 5′-CGCGGATCCGCAACGTAGATGGCGATATCTTTAGG-3′ | Outer primer #1 for exchange of native promoter with PdapA–A16 and change of the translational start codon from GTG to ATG in operon hisE–hisG, BamHI site underlined |
| dapA1 | 5′-AACTGCAGAACCAATGCATTGGTTCTGCAGTTATCACACCC-3′ | Primer #1 for amplification of PdapA–A16, NsiI site underlined |
| dapA2 | 5′-GGGAATTCCATATGAGGCTCCTTTTAAATCGAGCGGCTCCGGTCTTAGCTGTTAAACC-3′ | Primer #2 for amplification of PdapA–A16, NdeI site underlined |
| hisF1 | 5′-CGCAAGCTTCTCCATGCCCATGCTGGGTAAACGC-3′ | Primer #1 for integration of stop codon and PsodA in front of hisF, HindIII site underlined |
| hisF2 | 5′-GTCACAAGCCCGGAATAATTGGCAGTTATTTACTTGTACTCCTCATTTAACG-3′ | Primer #2 for integration of stop codon and PsodA in front of hisF, stop codon bold |
| sodA1 | 5′-CGTTAAATGAGGAGTACAAGTAAATAACTGCCAATTATTCCGGGCTTGTGAC-3′ | Primer #3 for integration of stop codon and PsodA in front of hisF, stop codon bold |
| sodA2 | 5′-GAATAACTCGAATTGCCACGCCCATGGGTAAAAAATCCTTTCGTAGGTTTC-3′ | Primer #4 for integration of stop codon and PsodA in front of hisF |
| hisF3 | 5′-GAAACCTACGAAAGGATTTTTTACCCATGGGCGTGGCAATTCGAGTTATTC-3′ | Primer #5 for integration of stop codon and PsodA in front of hisF |
| hisF4 | 5′-CGCGGATCCCGCCGCGCTTTGCCCACTCGATTGC-3′ | Primer #6 for integration of stop codon and PsodA in front of hisF, BamHI site underlined |
| hisFseq | 5′-CTGACACCGAAGGCCATC-3′ | |
| tuf2_1 | 5′-GATCAGCGACGCCGCAGGGTCTAGACCACAGGGTAGCTGGTAGTTTGAAAATC-3′ | Primer #1 for amplification of Ptuf, XbaI site underlined |
| tuf2_2 | 5′-CATTGAGCGCCGACAATAACGATTGCAGCCATTGTATGTCCTCCTGGACTTCG-3′ | Primer #2 for amplification of Ptuf |
| purA1 | 5′-CGAAGTCCAGGAGGACATACAATGGCTGCAATCGTTATTGTCGGCGCTCAATG-3′ | Primer #1 for amplification of purA (cg3063) |
| purA2 | 5′-GATCTTCTTTTTATCAGCCACTGTATGTCCTCCTGGACTTCCTAGTTGTCAGCTAGTACG-3′ | Primer #2 for amplification of purA (cg3063) |
| purB1 | 5′-CGTACTAGCTGACAACTAGGAAGTCCAGGAGGACATACAGTGGCTGATAAAAAGAAGATC-3′ | Primer #1 for amplification of purB (cg2876) |
| purB2 | 5′-CTTCTCTCATCCGCCAAAACAGTTAAAGAATCTCACCTGGTCGGTAGTC-3′ | Primer #2 for amplification of purB (cg2876) |
| rrnB1 | 5′-GACTACCGACCAGGTGAGATTCTTTAACTGTTTTGGCGGATGAGAGAAG-3′ | Primer #1 for amplification of terminator region of rrnB (E. coli) |
| rrnB2 | 5′-CTGCAGATATCCATCACACTGGCGGCCGCAGGAGAGCGTTCACCGACAAACAAC-3′ | Primer #2 for amplification of terminator region of rrnB (E. coli), NotI site underlined |
| ABseq1 | 5′-GCGATTGAAGACCGTC-3′ | Sequencing primer for pJC4 purA purB |
| ABseq2 | 5′-GCAATCGGCACCACCGGC-3′ | Sequencing primer for pJC4 purA purB |
| ABseq3 | 5′-GCCTGCATGGGACGAAG-3′ | Sequencing primer for pJC4 purA purB |
| ABseq4 | 5′-GTCACCGAACTGCTCAAC-3′ | Sequencing primer for pJC4 purA purB |
| ABseq5 | 5′-GATGGAAGCAGGATCGCG-3′ | Sequencing primer for pJC4 purA purB |
| pgi1 | 5′-CCCAAGCTTCAGCGTTGCGTGACGCACTCATTG-3′ | Outer primer #1 for introduction of pgiGTG, HindIII site underlined |
| pgi2 | 5′-CCAAACCTGGGTGGTCGAAATGTCCGCCACGAAAACTCCTTTATTGTCG-3′ | Inner primer #1 for introduction of pgiGTG, nucleotide exchange bold |
| pgi3 | 5′-CGACAATAAAGGAGTTTTCGTGGCGGACATTTCGACCACCCAGGTTTGG-3′ | Inner primer #2 for introduction of pgiGTG, nucleotide exchange bold |
| pgi4 | 5′-CGCGGATCCCCAATGTTGACGATCTTCTTGATCG-3′ | Outer primer #2 for introduction of pgiGTG, BamHI site underlined |
| pgiseq | 5′-CAAGCGTTGGGTTAAGGAGGA-3′ | Sequencing primer for pgiGTG |
| gcv_Cjk_start_EcoRV | 5′-ATCGATATCCGAGAGGAGACACAACATGTCTTCTGCAGCTACTCGC-3′ | Primer #1 for amplification of gene cluster gcvPTH from C. jeikeium genome |
| gcv_Cjk_MluI_XbaI | 5′-CAGTCTAGAACGCGTGGAACCGACCATAGGGTCTTG-3′ | Primer #2 for amplification of gene cluster gcvPTH from C. jeikeium genome |
| lipB-Cjk_start-EcoRV | 5′-GCGGATATCATGGGATTCCAGCAAGGC-3′ | Primer #1 for amplification of gene cluster lipAB from C. jeikeium genome |
| lipA-Cjk_stop-XbaI | 5′-GCGTCTAGATCCTTCGCCATGGATTCAAC-3′ | Primer #2 for amplification of gene cluster lipAB from C. jeikeium genome |
| pK19-fw | 5′-TAATGCAGCTGGCACGAC-3′ | Primer #1 for sequencing the pK19mobsacB inserts |
| pK19-rev | 5′-GTAGCTGACATTCATCCG-3′ | Primer #2 for sequencing the pK19mobsacB inserts |
Media and cultivation conditions
Escherichia coli DH5α was used as cloning host and was grown aerobically in 2 × YT complex medium [74] in a 5-mL glass test tube culture at 37 °C on a rotary shaker at 120 rpm. Precultures of C. glutamicum strains were prepared by thawing a glycerol stock (30% w v−1 glycerol) and streaking cell solution on a 2 × YT agar plate which was incubated at 30 °C for 2 days. A single colony of the respective strain was then used to inoculate 5 mL of 2 × YT complex medium in a glass test tube, which was incubated at 30 °C on a rotary shaker at 120 rpm for 6–8 h. The complete suspension of the glass test tube was transferred into 50 mL of 2 × YT medium in a 500-mL baffled shaking flask, which was incubated at 30 °C on a rotary shaker at 120 rpm overnight. To inoculate the main culture, cells were harvested by centrifugation (4500×g, 10 min, 4 °C), the pellet was resuspended in 0.9% w v−1 NaCl solution and used to inoculate CGXII minimal medium to an optical density at 600 nm (OD600) of about 2.5. The CGXII minimal medium [24] is composed of 20 g (NH4)2SO4 L−1, 5 g urea L−1, 21 g 3-morpholinopropanesulfonic acid (MOPS) L−1, 1 g K2HPO4 L−1, 1 g KH2PO4 L−1, 0.25 g MgSO4 L−1, 0.01 g CaCl2 L−1. The pH value of the medium was adjusted to 7.4 with 5 M KOH before autoclaving. Then, 16.4 mg FeSO4 × 7 H2O L−1, 10 mg MnSO4 × H2O L−1, 0.2 mg CuSO4 L−1, 1 mg ZnSO4 × 7 H2O L−1, 0.02 mg NiCl2 × 6 H2O L−1, 0.2 mg biotin L−1 were added sterilely. Standard cultivations in shaking flasks contained 10 g glucose L−1 as carbon source. For cultivations of strains bearing plasmid pJC4, 50-µg kanamycin mL−1 was added. For strains harboring plasmid pJC4 and pEC-XT99A, the kanamycin concentration was decreased to 12.5 µg mL−1 and 2.5-µg tetracycline mL−1 was added. The expression from Ptrc in pEC-XT99A_gcv_OP1-Cjk was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at the cultivation start. OD600 was measured with a photometer (Ultrospec 10 Cell Density Meter, GE Healthcare Company, Little Chalfont, UK). The cell dry weight (CDW in g L−1) was calculated using the correlation CDW = OD600 × 0.21 g L−1.
Determination of µmax, YX/S, and YP/S
The maximal growth rate µmax was determined by linear regression of ln(OD600), which was plotted against the cultivation time in h during the exponential growth phase of the respective strain. The biomass yield per unit substrate YX/S in g g−1 was determined by linear regression of the biomass concentration in g L−1, which was plotted against the corresponding glucose concentration in g L−1. The product yields per unit substrate for histidine (YhisP/S) and glycine (YglyP/S) in mol mol−1 were determined by dividing the product concentration (histidine) in mol L−1 and byproduct concentration (glycine) in mol L−1 after 24 h by the corresponding initial substrate concentration in mol L−1 at 0 h, respectively.
Genetic manipulation
Molecular cloning methods, such as PCR and DNA restriction, were carried out according to [74]. Plasmids were isolated with E.Z.N.A. Plasmid Mini Kit I (Omega Bio-tek Inc., Norcross, USA) and PCR fragments were purified with NucleoSpin Gel and PCR Clean-up Kit (Macherey–Nagel GmbH & Co. KG, Düren, Germany) according to the manufacturer’s instructions. Electrocompetent cells of E. coli and C. glutamicum were prepared as described before [22, 85]. Constructed plasmids were transformed into E. coli according to [22], and into C. glutamicum with a subsequent heat shock after transformation for 6 min at 46 °C according to Rest et al. [73]. Plasmids were transformed into electrocompetent E. coli and C. glutamicum strains with an Eporator (Eppendorf AG, Hamburg, Germany) at 2.5 kV with 600 Ω resistance. Enzymes for recombinant DNA work were obtained from Thermo Scientific Inc. (Darmstadt, Germany) and oligonucleotides were synthesized by biomers.net GmbH (Ulm, Germany, listed in Table 2).
Promoter exchanges and nucleotide substitutions were performed via a two-step homologous recombination by applying the respective pK19mobsacB derivative [75]. The plasmid to exchange the native HisG variant with the feedback-released HisGG233H–T235Q [77] was implemented into C. glutamicum ATCC 13032 by amplifying the flanking genomic regions of hisG up- and downstream of the mutations with primer pairs hisG1/hisG2 and hisG3/hisG4 (hisG2 and hisG3 harbor the exchanges). Both polymerase chain reaction (PCR) products were used as templates in a SOEing PCR [38] with primer pair hisG1/hisG4. The SOEing product and pK19mobsacB were digested with BamHI and fused together in a ligation reaction to give pK19mobsacB hisGFB. This plasmid was then transformed into E. coli DH5α, isolated, and its sequence integrity was verified by DNA sequencing with primers pK19-fw and pK19-rev (GATC Biotech AG, Constance, Germany). The verified plasmid was then transformed into C. glutamicum ATCC 13032. Applying the method described by Schäfer et al. [75], the native hisG sequence was replaced via homologous recombination (double crossover) by the mutated hisG sequence leading to amino acid exchanges G233H and T235Q. The screening of the C. glutamicum HIS1 mutants was done on 2 × YT agar plates containing 10% (w v−1) sucrose. For verification of the nucleotide exchanges, a PCR with primer pair hisG1/hisG4 was performed and sent for sequencing with primer hisGseq. To construct plasmids for the promoter exchanges in front of the operons containing histidine biosynthesis genes (C. glutamicum HIS2, HIS3, HIS4, and HIS6), the flanking regions of the respective promoter were amplified. For the exchange of the native promoter of the operon hisD–hisC–hisB–cg2302–cg2301 with the strong promoter of the gene tuf, encoding the elongation factor TU, the flanking regions were amplified with primer pairs hisD1/hisD2 and hisD3/hisD4. The products of both PCRs were used as templates in a SOEing PCR with primer pair hisD1/hisD4, and the SOEing product and the plasmid pK19mobsacB were digested with BamHI and HindIII and ligated together to give an intermediate plasmid. This plasmid was transformed into E. coli DH5α, isolated and sent for sequencing with primers pK19-fw and pK19-rev. In the next step, Ptuf was amplified with primer pair tuf1/tuf2, and the product and the intermediate plasmid were digested with NdeI and NsiI. Both were ligated to give plasmid pK19mobsacB hisD-Ptuf, which was transformed into E. coli DH5α, isolated and sent for sequencing with primers pK19-fw and pK19-rev. The verified pK19mobsacB hisD-Ptuf was transformed into C. glutamicum HIS1 and exchange of the native promoter region with the Ptuf promoter was performed as has been described above yielding C. glutamicum HIS2. The respective region was amplified with primer pair hisD1/Ptuf2 and sequenced with primer hisD1. The plasmids for exchanging the native promoter with Ptuf for operons hisH–hisA–impA–hisF–hisI–cg2294 and cg0911–hisN were constructed accordingly. Primer pairs hisH1/hisH2 and hisH3/hisH4 and hisN1/hisN2 and hisN3/hisN4 were used to amplify the flanking regions, respectively. After SOEing PCR, digestion, and ligation, plasmids were transformed into E. coli DH5α and prepared. In further steps, the mentioned plasmids were digested with NdeI and NsiI and fused with the Ptuf region. After sequencing, pK19mobsacB hisH–Ptuf was implemented in C. glutamicum HIS2 to yield C. glutamicum HIS3. The sequence was verified with primers hisH1, hisH4, tuf1, and tuf2. C. glutamicum HIS3 served as basis for implementing Ptuf in front of cg0911–hisN using pK19mobsacB hisN–Ptuf to yield C. glutamicum HIS4. This strain was verified with primers hisN1, hisN4, tuf1, and tuf2. Since we were not able to implement Ptuf in front of the hisE–hisG operon in C. glutamicum HIS4, we instead used PdapA–A16 [88], a modified version of the promoter of dihydrodipicolinate synthase, which was amplified with primer pair dapA1/dapA2 from pK18mobsacB PaceE dapA-A16 [16]. The flanking regions of the hisE–hisG promoter were amplified with primer pairs hisE1/hisE2 and hisE3/hisE4, a SOEing PCR was prepared with primer pair hisE1/hisE4. This product and pK19mobsacB were digested with BamHI and HindIII and ligated. PdapA-A16 and this plasmid were digested with NdeI and NsiI and ligated. Hence, on the basis of C. glutamicum HIS4, C. glutamicum HIS6 was created and verified with primers hisE1, hisE4, dapA1, and dapA2. C. glutamicum HIS5 was created by implementing PdapA–A16 in C. glutamicum HIS1. To exchange the internal promoter of hisF in the operon hisH–hisA–impA–hisF–hisI–cg2294 with the promoter of manganese superoxide dismutase (encoded by sodA), flanking regions and the promoter were amplified with primer pairs hisF1/hisF2, sodA1/sodA2, and hisF3/hisF4 and an additional stop codon (TAA) was integrated upstream of hisF. The SOEing PCR (with all three products as template and primer pair hisF1/hisF4) and pK19mobsacB were cut with HindIII and BamHI and ligated together. Integration of PsodA in front of hisF in strain C. glutamicum HIS6 yielded C. glutamicum HIS7, which was verified with primers hisF4 and hisFseq.
On the basis of plasmid pJC4 [20], we constructed pJC4 purA purB by amplifying Ptuf, purA, and purB with primer pairs tuf2_1/tuf2_2, purA1/purA2, and purB1/purB2 from the C. glutamicum genome. Furthermore, primer pair rrnB1/rrnB2 was used to amplify the TrrnB terminator region of the rrnB operon from the E. coli K-12 MG1655 genome. Isothermal plasmid assembly [27] was prepared with these four DNA fragments and pJC4, which had been digested with XbaI and NotI before. The sequence integrity was verified by sequencing with primers ABseq 1–5.
Plasmid pEC-XT99A [51] served as basis for the GCV system overproduction plasmid and was digested with Ecl136II and XbaI. The gene cluster gcvPTH was amplified from the C. jeikeium K411 genome [84] with primer pair gcv_Cjk_start_EcoRV/gcv_Cjk_MluI_XbaI and the resulting PCR product was digested with EcoRV and XbaI and ligated into the cut pEC-XT99A. After verification by sequencing, this intermediate plasmid served as basis for the second cloning step. The gene cluster lipAB was amplified from the C. jeikeium K411 genome with primer pair lipB-Cjk_start-EcoRV/lipA-Cjk_stop-XbaI and the PCR product was digested with SspI und EcoRV, and ligated into the XmnI cut intermediate plasmid to give pEC-XT99A_gcv_OP1-Cjk, which was verified by sequencing.
Plasmid pEC-XT99A_gcv_OP1-Cjk was transformed into strain C. glutamicum HIS8 resulting in C. glutamicum HIS9. The exchange of the translational start codon ATG of gene pgi to GTG [7] in C. glutamicum HIS9 was done with pK19mobsacB pgiGTG. For the construction, the flanking regions were amplified with primer pairs pgi1/pgi2 and pgi3/pgi4, containing the nucleotide exchange. The PCR products were used in a SOEing PCR with primer pair pgi1/pgi4. Then, the SOEing PCR and the vector were digested with HindIII and BamHI and ligated together. After sequence verification, the plasmid was introduced into C. glutamicum HIS9; the base exchange was done as described above, and verified by sequencing with primer pgiseq.
Intracellular adenylate measurements and energy charges
The determination of intracellular adenylate concentrations was done with a modified protocol as has been described before for E. coli [21]. 1 mL of culture suspension was collected during exponential growth phase (OD600 of 7.5) and sampled directly in − 20 °C cold 0.25 mL 35% (v v−1) perchloric acid containing 80 µM ethylenediaminetetraacetic acid (EDTA). The suspension was incubated for 15 min at 4 °C on a shaker, 0.25 mL 1 M K2HPO4 was added, and the suspension was neutralized with 5 M KOH to pH 7.0. After centrifugation (5 min, 4 °C, 7000×g), the supernatant was analyzed via HPLC (1200 series, Agilent Technologies, Santa Clara, CA, USA) equipped with a RP-C18 (octadecyl) phase column (Supelcosil LC-18-T, 3 µm, 150 cm × 4.6 mm) and a diode array detector (DAD). Buffer A (0.1 M KH2PO4, 0.1 M K2HPO4, 4 mM tetrabutylammonium bisulfate [TBAS], pH 6.0) and buffer B (0.1 M KH2PO4, 0.1 M K2HPO4, 4 mM TBAS, pH 7.2 with 30% (v v−1) methanol) were used to generate a gradient (3.5 min, 0% B; 20 min, 30% B, 22 min 35% B; 40 min, 60% B; 48 min, 100% B; 55 min, 100% B, 60 min 0% B) with a flow rate of 1 mL min−1. The energy charge of intracellular adenylates was calculated as has been defined before [4, 5]. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry [4, 5], with the difference that AMP was omitted from calculations, since AMP concentrations were below the detection limit of the applied analytical system. As such, current EC values are intrinsically higher by estimated 10% compared to the original definition:
Quantification of substrate and product concentrations
Substrates and products were quantified by harvesting 1 mL of cell suspension via centrifugation (12,100×g, 5 min, RT) at given time points. The supernatants were used for further analysis. The glucose concentration was determined with a test kit from Roche (Roche Diagnostics, Mannheim, Germany). Quantification of amino acids was performed with an Agilent 1200 series apparatus (Agilent Technologies, Santa Clara, CA, USA) equipped with an Agilent Zorbax Eclipse Plus C18 column (250 × 4.6 mm, 5 µm) protected by an Agilent Zorbax Eclipse Plus C18 guard column (12.5 × 4.6 mm, 5 µm). Automatic precolumn derivatization with ortho-phthaldialdehyde was followed by fluorometric detection (excitation at 230 nm and emission at 450 nm). The elution buffer consisted of a polar phase (10 mM Na2HPO4, 10 mM Na2B4O7, 0.5 mM NaN3, pH 8.2) and a nonpolar phase (45% [v v−1] methanol, 45% [v v−1] acetonitrile). Protocol details were described earlier [16]. Analytes were quantified using 200 µM l-ornithine as the internal standard to correct variabilities in analytes and a seven-point calibration curve for each component as an external reference standard.
Flux balance analysis
Metabolic fluxes of C. glutamicum ATCC 13032 were investigated by flux balance analysis (FBA), applying different objective functions and constraints [10]. All computations were carried out with MATLAB 2015b (The MathWorks, Natick, MA, USA) and the COBRA Toolbox v3.0 with glpk solvers [76], using the genome-scale metabolic model (GEM) of C. glutamicum ATCC 13032, iCW773 [98]. The glucose uptake rate was set to 3.94 mmol g−1CDW h−1 for all simulations [12]; however, objective functions and constraints were changed as follows: (a) Maximizing growth rate with no further constraints results in μ = 0.36 h−1 and (b) maximizing l-histidine yield with a fixed μ of 0.1 h−1 resulted in a maximum yield of 0.51 mol l-histidine mol−1 glucose.
Systems metabolic profiling (SMP)
Cultivation and extraction of metabolites
Corynebacterium glutamicum strains HIS1, HIS6, HIS7, HIS8, HIS9, and HIS10 were cultivated as described above. Sampling was performed at a CDW of approximately 1.8 g L−1 during the exponential growth phase. 2 mL of cell suspension was sampled by centrifugation (12,100×g, 20 s, 30 °C) and washed with 1.5 mL of 0.9% (w v−1) NaCl solution followed by centrifugation. Cells were quenched immediately with liquid nitrogen and temporarily stored at − 70 °C. Defined amounts of 250 μM l-norvaline solution (internal standard) were added to the cell pellets to obtain an extraction concentration of 20 gCDW L−1. Immediately after addition, suspensions were pre-incubated for 30 s at 100 °C in a water bath and homogenized by vortexing (20 s). Subsequently, samples were incubated for 3 min at 100 °C, chilled on ice and centrifuged (20,800×g, 10 min, 4 °C). Supernatants were stored at − 70 °C [14].
LC-QTOF analysis of intracellular metabolites
Differential metabolite analysis was performed on an Agilent 1260 bio-inert HPLC coupled to an Agilent 6540 Accurate-Mass LC–MS/MS Q-TOF system with ESI Jet Stream Technology (Agilent Technologies, Santa Clara, CA, USA). Two different hydrophilic interaction chromatography (HILIC) systems were used to get high metabolite coverage. The first method was ammonium acetate based (10 mM, pH 9.2) utilizing a Sequant ZIC-pHILIC column (150 × 2.1 mm, 5 μm) with guard column (Sequant ZIC-pHILIC, 20 × 2.1 mm, 5 μm) at 40 °C, 0.2 mL min−1, and 5 μL injection volume. For details see [86]. Additionally, an acidic HILIC method was established using ammonium formate buffer (10 mM, pH 2.8) and a Waters XBridge BEH Amide column (150 × 2.1 mm, 3.5 μm) coupled to a Waters XBridge BEH Amide VanGuard Cartridge (5 × 2.1 mm, 3.5 μm) at 35 °C, 0.2 mL min−1, and 5 μL injection volume. Mobile phases were composed as follows: Mobile phase A: 90% acetonitrile/10% water, 10 mM ammonium formate and mobile phase B: 10% acetonitrile/90% water, 10 mM ammonium formate. Both adjusted to pH 2.8 with formic acid. Gradient elution was carried out by the following program: Isocratic hold 0% B for 1 min, linear gradient to 62.5% B for 15 min, linear gradient to 100% B for 4 min, column wash at 100% B for 5 min, linear gradient to 0% B for 5 min and column equilibration at 0% B for 15 min. Samples were prepared in 60% (v v−1) acetonitrile and 10 mM ammonium acetate (pH 9.2) or ammonium formate (pH 2.8). All metabolite samples were separated with both HILIC methods in positive and negative MS mode (tuned in extended dynamic range) with following conditions: drying gas flow rate of 8 L min−1 with a gas temperature of 325 °C, nebulizer with 40 lb per square inch gauge, sheath gas flow rate of 12 L min−1 and sheath gas temperature of 350 °C, capillary voltage of 4000 V and fragmentor voltage of 100 V. Additionally, fragmentation experiments in the targeted MS/MS mode were carried out to investigate and verify structure integrity of IGP, adenylosuccinate, SAICAR, and FGAR. For this, precursor ions [M+H] or [M−H], verified by accurate mass, were selected and fragmented at their characteristic retention times via collision-induced dissociation (CID) at 10, 20, and 30 V. Since analytical standards of those compounds were not commercially available or only by custom synthesis, fragmentation patterns were computationally evaluated with MassHunter Molecular Structure Correlator (B05.00, Agilent Technologies, Santa Clara, CA, USA). By combining accurate mass and plausible fragmentation patterns IGP, adenylosuccinate, SAICAR, and FGAR could be identified.
Data analysis
System control and acquisition were performed using MassHunter Data Acquisition (B06.01, Agilent Technologies, Santa Clara, CA, USA). As first step, an untargeted differential analysis was carried out to generate hypothesis free data. Peak picking and integration were done in MassHunter ProFinder (B08.00, Agilent Technologies, Santa Clara, CA, USA) using “batch recursive feature extraction”. Subsequently, statistical analysis was performed in Mass Profiler Professional (13.1.1, Agilent Technologies, Santa Clara, CA, USA). Significance testing was done by one-way ANOVA and p values were filtered (p < 0.05). Peaks were identified by accurate mass and with a personal compound data library, containing retention times of authentic standards. Unidentified significant features were searched against the METLIN [30] and MassBank [37] database. After identification, peak integration was manually curated via “batch targeted feature extraction”. The following metabolites of the de novo purine and l-histidine biosynthesis could be identified and analyzed with the applied method: fGAR, phosphoribosyl-N-formylglycineamide; SAICAR, phosphoribosyl-aminoimidazolesuccinocarboxamide; AICAR, 1-(5-phosphoribosyl)-5-amino-4-imidazolecarboxamide; IMP, inosine monophosphate; AdSucc, adenylosuccinate; ADP, adenosine diphosphate; ATP, adenosine triphosphate; IGP, imidazole-glycerol phosphate; l-histidinol and l-histidine.
Authors’ contributions
AS, AF, RT, and BB conceived and designed the experiments; AS, AF, AH and EM performed the experiments and were supported by JS, IL, LF, and CM; AS, AF, and BB analyzed the data and were supported by JS, IL, and LF; AS, AF, JÖ, BG, RT, and BB wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the German Research Foundation (DFG) and the Technical University of Munich (TUM) in the framework of the Open Access Publishing Program.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was funded by the Fachagentur Nachwachsende Rohstoffe e.V. with the Grant Number 22008014.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
References
- 1.Alifano P, Fani R, Liò P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki K, Nakayama K. Feedback-resistant phosphoribosyl-ATP pyrophosphorylase in l-histidine-producing mutants of Corynebacterium glutamicum. Agric Biol Chem. 1974;38:2209–2218. doi: 10.1271/bbb1961.38.2209. [DOI] [Google Scholar]
- 3.Araki K, Shimojo S, Nakayama K. Histidine production by Corynebacterium glutamicum mutants, multiresistant to analogs of histidine, tryptophan, purine and pyrimidine. Agric Biol Chem. 1974;38:837–846. doi: 10.1080/00021369.1974.10861244. [DOI] [Google Scholar]
- 4.Atkinson DE. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry (Moscow, Russ. Fed.) 1986;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson DE, Walton GM. Adenosine triphosphate conservation in metabolic regulation. Rat liver citrate cleavage enzyme. J Biol Chem. 1967;242:3239–3241. [PubMed] [Google Scholar]
- 6.Auerbach VH, DiGeorge AM, Baldridge RC, Tourtellotte CD, Brigham MP. Histidinemia: a deficiency in histidase resulting in the urinary excretion of histidine and of imidazolepyruvic acid. J Pediatr. 1962;60:487–497. doi: 10.1016/S0022-3476(62)80109-7. [DOI] [PubMed] [Google Scholar]
- 7.Becker J, Buschke N, Bücker R, Wittmann C. Systems level engineering of Corynebacterium glutamicum—reprogramming translational efficiency for superior production. Eng Life Sci. 2010;10:430–438. doi: 10.1002/elsc.201000008. [DOI] [Google Scholar]
- 8.Becker J, Wittmann C. Advanced biotechnology: metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew Chem Int Ed Engl. 2015;54:3328–3350. doi: 10.1002/anie.201409033. [DOI] [PubMed] [Google Scholar]
- 9.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 11.Bender RA. Regulation of the histidine utilization (Hut) system in bacteria. Microbiol Mol Biol Rev. 2012;76:565–584. doi: 10.1128/MMBR.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blombach B, Buchholz J, Busche T, Kalinowski J, Takors R. Impact of different CO2/HCO3− levels on metabolism and regulation in Corynebacterium glutamicum. J Biotechnol. 2013;168:331–340. doi: 10.1016/j.jbiotec.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolten CJ, Kiefer P, Letisse F, Portais J-C, Wittmann C. Sampling for metabolome analysis of microorganisms. Anal Chem. 2007;79:3843–3849. doi: 10.1021/ac0623888. [DOI] [PubMed] [Google Scholar]
- 15.Bolten CJ, Schröder H, Dickschat J, Wittmann C. Towards methionine overproduction in Corynebacterium glutamicum—methanethiol and dimethyldisulfide as reduced sulfur sources. J Microbiol Biotechnol. 2010;20:1196–1203. doi: 10.4014/jmb.1002.02018. [DOI] [PubMed] [Google Scholar]
- 16.Buchholz J, Schwentner A, Brunnenkan B, Gabris C, Grimm S, Gerstmeir R, Takors R, Eikmanns BJ, Blombach B. Platform engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of l-lysine, l-valine, and 2-ketoisovalerate. Appl Environ Microbiol. 2013;79:5566–5575. doi: 10.1128/AEM.01741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bückle-Vallant V, Krause FS, Messerschmidt S, Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisocaproate production. Appl Microbiol Biotechnol. 2013;98:297–311. doi: 10.1007/s00253-013-5310-2. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Zhou Y, Yang L, Zhang C, Xu Q, Xie X, Chen N. Modification of histidine biosynthesis pathway genes and the impact on production of l-histidine in Corynebacterium glutamicum. Biotechnol Lett. 2013;35:735–741. doi: 10.1007/s10529-013-1138-1. [DOI] [PubMed] [Google Scholar]
- 19.Chim-Anage P, Shioya S, Suga K-I. Optimum conditions for histidine production by fed-batch culture of Brevibacterium flavum. J Ferment Bioeng. 1990;70:386–391. doi: 10.1016/0922-338X(90)90119-H. [DOI] [Google Scholar]
- 20.Cordes C, Möckel B, Eggeling L, Sahm H. Cloning, organization and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene. 1992;112:113–116. doi: 10.1016/0378-1119(92)90311-C. [DOI] [PubMed] [Google Scholar]
- 21.Cserjan-Puschmann M, Kramer W, Duerrschmid E, Striedner G, Bayer K. Metabolic approaches for the optimisation of recombinant fermentation processes. Appl Microbiol Biotechnol. 1999;53:43–50. doi: 10.1007/s002530051612. [DOI] [PubMed] [Google Scholar]
- 22.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggeling L, Bott M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2015;99:3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- 24.Eikmanns BJ, Metzger M, Reinscheid D, Kircher M, Sahm H. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl Microbiol Biotechnol. 1991;34:617–622. doi: 10.1007/BF00167910. [DOI] [PubMed] [Google Scholar]
- 25.Feng RN, Niu YC, Sun XW, Li Q, Zhao C, Wang C, Guo FC, Sun CH, Li Y. Histidine supplementation improves insulin resistance through suppressed inflammation in obese women with the metabolic syndrome: a randomised controlled trial. Diabetologia. 2013;56:985–994. doi: 10.1007/s00125-013-2839-7. [DOI] [PubMed] [Google Scholar]
- 26.Frohwitter J, Heider SAE, Peters-Wendisch P, Beekwilder J, Wendisch VF. Production of the sesquiterpene (+)-valencene by metabolically engineered Corynebacterium glutamicum. J Biotechnol. 2014;191:205–213. doi: 10.1016/j.jbiotec.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 27.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 28.Goldberger RF, Kovach JS. Regulation of histidine biosynthesis in Salmonella typhimurium. In: Horecker BL, Stadtman ER, editors. Current topics in cellular regulation. New York: Academic Press; 1972. pp. 285–308. [DOI] [PubMed] [Google Scholar]
- 29.Green JM, Matthews RG. Folate biosynthesis, reduction, and polyglutamylation and the interconversion of folate derivatives. EcoSal Plus 2. 2007 doi: 10.1128/ecosalplus.3.6.3.6. [DOI] [PubMed] [Google Scholar]
- 30.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, Wolan DW, Spilker ME, Benton HP, Siuzdak G. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Harvey RJ, Dev IK. Regulation in the folate pathway of Escherichia coli. Adv Enzyme Regul. 1975;13:97–124. doi: 10.1016/0065-2571(75)90010-2. [DOI] [PubMed] [Google Scholar]
- 33.Hasegawa S, Ichiyama T, Sonaka I, Ohsaki A, Hirano R, Haneda Y, Fukano R, Hara M, Furukawa S. Amino acids exhibit anti-inflammatory effects in human monocytic leukemia cell line, THP-1 cells. Inflamm Res. 2011;60:1013. doi: 10.1007/s00011-011-0362-1. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa S, Ichiyama T, Sonaka I, Ohsaki A, Okada S, Wakiguchi H, Kudo K, Kittaka S, Hara M, Furukawa S. Cysteine, histidine and glycine exhibit anti-inflammatory effects in human coronary arterial endothelial cells. Clin Exp Immunol. 2012;167:269–274. doi: 10.1111/j.1365-2249.2011.04519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heider SAE, Peters-Wendisch P, Netzer R, Stafnes M, Brautaset T, Wendisch VF. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2014;98:1223–1235. doi: 10.1007/s00253-013-5359-y. [DOI] [PubMed] [Google Scholar]
- 36.Heider SAE, Wolf N, Hofemeier A, Peters-Wendisch P, Wendisch VF. Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front Bioeng Biotechnol. 2014;2:28. doi: 10.3389/fbioe.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 38.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 39.Huennekens FM. Folic acid coenzymes in the biosynthesis of purines and pyrimidines. In: Harris RS, Wool IG, Loraine JA, Thimann KV, editors. Vitamins & hormones. New York: Academic Press; 1969. pp. 375–394. [DOI] [PubMed] [Google Scholar]
- 40.Hüser AT, Chassagnole C, Lindley ND, Merkamm M, Guyonvarch A, Elišáková V, Pátek M, Kalinowski J, Brune I, Pühler A, Tauch A. Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and genome-wide transcriptional profiling. Appl Environ Microbiol. 2005;71:3255–3268. doi: 10.1128/AEM.71.6.3255-3268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda M. Amino acid production processes, in microbial production of l-amino acids. Adv Biochem Eng Biotechnol. 2003;79:1–35. doi: 10.1007/3-540-45989-8_1. [DOI] [PubMed] [Google Scholar]
- 42.Ikeda M, Katsumata R. Hyperproduction of tryptophan by Corynebacterium glutamicum with the modified pentose phosphate pathway. Appl Environ Microbiol. 1999;65:2497–2502. doi: 10.1128/aem.65.6.2497-2502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol. 2004;8:243–254. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- 44.Ishino S, Kuga T, Yamaguchi K, Shirahata K, Araki K. 13C NMR studies of histidine fermentation with a Corynebacterium glutamicum mutant. Agric Biol Chem. 1986;50:307–310. doi: 10.1080/00021369.1986.10867392. [DOI] [Google Scholar]
- 45.Jensen KF, Dandanell G, Hove-Jensen B, WillemoËs M. Nucleotides, nucleosides, and nucleobases. EcoSal Plus 3. 2008 doi: 10.1128/ecosalplus.3.6.2. [DOI] [PubMed] [Google Scholar]
- 46.Jojima T, Noburyu R, Sasaki M, Tajima T, Suda M, Yukawa H, Inui M. Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2014;99:1165–1172. doi: 10.1007/s00253-014-6223-4. [DOI] [PubMed] [Google Scholar]
- 47.Jung S, Chun J-Y, Yim S-H, Lee S-S, Cheon C-I, Song E, Lee M-S. Transcriptional regulation of histidine biosynthesis genes in Corynebacterium glutamicum. Can J Microbiol. 2010;56:178–187. doi: 10.1139/w09-115. [DOI] [PubMed] [Google Scholar]
- 48.Kang M-K, Eom J-H, Kim Y, Um Y, Woo HM. Biosynthesis of pinene from glucose using metabolically-engineered Corynebacterium glutamicum. Biotechnol Lett. 2014;36:2069–2077. doi: 10.1007/s10529-014-1578-2. [DOI] [PubMed] [Google Scholar]
- 49.Kawai Y, Moriyama A, Asai K, Coleman-Campbell CM, Sumi S, Morishita H, Suchi M. Molecular characterization of histidinemia: identification of four missense mutations in the histidase gene. Hum Genet. 2005;116:340–346. doi: 10.1007/s00439-004-1232-5. [DOI] [PubMed] [Google Scholar]
- 50.Kim PB, Nelson JW, Breaker RR. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell. 2015;57:317–328. doi: 10.1016/j.molcel.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirchner O, Tauch A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J Biotechnol. 2003;104:287–299. doi: 10.1016/S0168-1656(03)00148-2. [DOI] [PubMed] [Google Scholar]
- 52.Kopple JD, Swendseid ME. Evidence that histidine is an essential amino acid in normal and chronically uremic man. J Clin Invest. 1975;55:881–891. doi: 10.1172/JCI108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krause FS, Blombach B, Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krömer JO, Wittmann C, Schröder H, Heinzle E. Metabolic pathway analysis for rational design of l-methionine production by Escherichia coli and Corynebacterium glutamicum. Metab Eng. 2006;8:353–369. doi: 10.1016/j.ymben.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Kulis-Horn RK, Persicke M, Kalinowski J. Corynebacterium glutamicum ATP-phosphoribosyl transferases suitable for l-histidine production—strategies for the elimination of feedback inhibition. J Biotechnol. 2015;206:26–37. doi: 10.1016/j.jbiotec.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Kulis-Horn RK, Persicke M, Kalinowski J. Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum. Microb Biotechnol. 2014;7:5–25. doi: 10.1111/1751-7915.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao S-M, Du Q-S, Meng J-Z, Pang Z-W, Huang R-B. The multiple roles of histidine in protein interactions. Chem Cent J. 2013;7:44. doi: 10.1186/1752-153X-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liebl W, Klamer R, Schleifer K-H. Requirement of chelating compounds for the growth of Corynebacterium glutamicum in synthetic media. Appl Microbiol Biotechnol. 1989;32:205–210. doi: 10.1007/BF00165889. [DOI] [Google Scholar]
- 59.Liu Q, Ouyang S, Kim J, Chen G-Q. The impact of PHB accumulation on l-glutamate production by recombinant Corynebacterium glutamicum. J Biotechnol. 2007;132:273–279. doi: 10.1016/j.jbiotec.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 60.Lorenz E, Stauffer GV. Characterization of the MetR binding sites for the glyA gene of Escherichia coli. J Bacteriol. 1995;177:4113–4120. doi: 10.1128/jb.177.14.4113-4120.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu W, Kwon YK, Rabinowitz JD. Isotope ratio-based profiling of microbial folates. J Am Soc Mass Spectrom. 2007;18:898–909. doi: 10.1016/j.jasms.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malykh EA, Butov IA, Ravcheeva AB, Krylov AA, Mashko SV, Stoynova NV. Specific features of l-histidine production by Escherichia coli concerned with feedback control of AICAR formation and inorganic phosphate/metal transport. Microb Cell Fact. 2018;17:42. doi: 10.1186/s12934-018-0890-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizukami T, Hamu A, Ikeda M, Oka T, Katsumata R. Cloning of the ATP phosphoribosyl transferase gene of Corynebacterium glutamicum and application of the gene to l-histidine production. Biosci Biotechnol Biochem. 1994;58:635–638. doi: 10.1271/bbb.58.635. [DOI] [PubMed] [Google Scholar]
- 64.Nelson D, Cox M. Lehninger biochemie. 4. Berlin: Springer; 2009. [Google Scholar]
- 65.Nishimura T, Vertès AA, Shinoda Y, Inui M, Yukawa H. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl Microbiol Biotechnol. 2007;75:889–897. doi: 10.1007/s00253-007-0879-y. [DOI] [PubMed] [Google Scholar]
- 66.Oldiges M, Eikmanns BJ, Blombach B. Application of metabolic engineering for the biotechnological production of l-valine. Appl Microbiol Biotechnol. 2014;98:5859–5870. doi: 10.1007/s00253-014-5782-8. [DOI] [PubMed] [Google Scholar]
- 67.O’Mahony L, Akdis M, Akdis CA. Regulation of the immune response and inflammation by histamine and histamine receptors. J Allergy Clin Immunol. 2011;128:1153–1162. doi: 10.1016/j.jaci.2011.06.051. [DOI] [PubMed] [Google Scholar]
- 68.Park SH, Kim HU, Kim TY, Park JS, Kim S-S, Lee SY. Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun. 2014;5(5):4618. doi: 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- 69.Parsons ME, Ganellin CR. Histamine and its receptors. Br J Pharmacol. 2006;147:127–135. doi: 10.1038/sj.bjp.0706440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polgár L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin T, Hu X, Hu J, Wang X. Metabolic engineering of Corynebacterium glutamicum strain ATCC13032 to produce l-methionine. Biotechnol Appl Biochem. 2014;62:563–573. doi: 10.1002/bab.1290. [DOI] [PubMed] [Google Scholar]
- 72.Rebek J. On the structure of histidine and its role in enzyme active sites. Struct Chem. 1990;1:129–131. doi: 10.1007/BF00675792. [DOI] [Google Scholar]
- 73.van der Rest ME, Lange C, Molenaar D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol. 1999;52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 74.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 75.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 76.Schellenberger J, Que R, Fleming RMT, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BØ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schendzielorz G, Dippong M, Grünberger A, Kohlheyer D, Yoshida A, Binder S, Nishiyama C, Nishiyama M, Bott M, Eggeling L. Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth Biol. 2014;3:21–29. doi: 10.1021/sb400059y. [DOI] [PubMed] [Google Scholar]
- 78.Schweitzer J-E, Stolz M, Diesveld R, Etterich H, Eggeling L. The serine hydroxymethyltransferase gene glyA in Corynebacterium glutamicum is controlled by GlyR. J Biotechnol. 2009;139:214–221. doi: 10.1016/j.jbiotec.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Simic P, Willuhn J, Sahm H, Eggeling L. Identification of glyA (encoding serine hydroxymethyltransferase) and its use together with the exporter ThrE to increase l-threonine accumulation by Corynebacterium glutamicum. Appl Environ Microbiol. 2002;68:3321–3327. doi: 10.1128/AEM.68.7.3321-3327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith KM, Cho K-M, Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stolz M, Peters-Wendisch P, Etterich H, Gerharz T, Faurie R, Sahm H, Fersterra H, Eggeling L. Reduced folate supply as a key to enhanced l-serine production by Corynebacterium glutamicum. Appl Environ Microbiol. 2007;73:750–755. doi: 10.1128/AEM.02208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takeno S, Takasaki M, Urabayashi A, Mimura A, Muramatsu T, Mitsuhashi S, Ikeda M. Development of fatty acid-producing Corynebacterium glutamicum strains. Appl Environ Microbiol. 2013;79:6776–6783. doi: 10.1128/AEM.02003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takors R, Bathe B, Rieping M, Hans S, Kelle R, Huthmacher K. Systems biology for industrial strains and fermentation processes—example: amino acids. J Biotechnol. 2007;129:181–190. doi: 10.1016/j.jbiotec.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 84.Tauch A, Kaiser O, Hain T, Goesmann A, Weisshaar B, Albersmeier A, Beke T, Bischoff N, Brune I, Chakraborty T, Kalinowski J, Meyer F, Rupp O, Schneiker S, Viehoever P, Pühler A. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J Bacteriol. 2005;187:4671–4682. doi: 10.1128/JB.187.13.4671-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tauch A, Kirchner O, Löffler B, Götker S, Pühler A, Kalinowski J. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol. 2002;45:362–367. doi: 10.1007/s00284-002-3728-3. [DOI] [PubMed] [Google Scholar]
- 86.Teleki A, Sánchez-Kopper A, Takors R. Alkaline conditions in hydrophilic interaction liquid chromatography for intracellular metabolite quantification using tandem mass spectrometry. Anal Biochem. 2015;475:4–13. doi: 10.1016/j.ab.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 87.Tuttle KR, Milton JE, Packard DP, Shuler LA, Short RA. Dietary amino acids and blood pressure: a cohort study of patients with cardiovascular disease. Am J Kidney Dis. 2012;59:803–809. doi: 10.1053/j.ajkd.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 88.Vašicová P, Pátek M, Nešvera J, Sahm H, Eikmanns B. Analysis of the Corynebacterium glutamicum dapA promoter. J Bacteriol. 1999;181:6188–6191. doi: 10.1128/jb.181.19.6188-6191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vickery HB, Leavenworth CS. On the separation of histidine and arginine IV. The preparation of histidine. J Biol Chem. 1928;78:627–635. [Google Scholar]
- 90.Waagbø R, Tröße C, Koppe W, Fontanillas R, Breck O. Dietary histidine supplementation prevents cataract development in adult atlantic salmon, Salmo salar L., in seawater. Br J Nutr. 2010;104:1460–1470. doi: 10.1017/s0007114510002485. [DOI] [PubMed] [Google Scholar]
- 91.Wade AM, Tucker HN. Antioxidant characteristics of l-histidine. Nutr Biochem. 1998;9:308–315. doi: 10.1016/S0955-2863(98)00022-9. [DOI] [Google Scholar]
- 92.Walker-Simmons M, Atkinson DE. Functional capacities and the adenylate energy charge in Escherichia coli under conditions of nutritional stress. J Bacteriol. 1977;130:676–683. doi: 10.1128/jb.130.2.676-683.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watanabe M, Suliman ME, Qureshi AR, Garcia-Lopez E, Bárány P, Heimbürger O, Stenvinkel P, Lindholm B. Consequences of low plasma histidine in chronic kidney disease patients: associations with inflammation, oxidative stress, and mortality. Am J Clin Nutr. 2008;87:1860–1866. doi: 10.1093/ajcn/87.6.1860. [DOI] [PubMed] [Google Scholar]
- 94.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol. 2013;6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wieschalka S, Blombach B, Eikmanns BJ. Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol. 2012;94:449–459. doi: 10.1007/s00253-011-3843-9. [DOI] [PubMed] [Google Scholar]
- 96.Winkler ME, Ramos-Montañez S. Biosynthesis of histidine. EcoSal Plus. 2009 doi: 10.1128/ecosalplus.3.6.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, Shang X, Deng A, Chai X, Lai S, Zhang G, Wen T. Genetic and biochemical characterization of Corynebacterium glutamicum ATP phosphoribosyltransferase and its three mutants resistant to feedback inhibition by histidine. Biochimie. 2012;94:829–838. doi: 10.1016/j.biochi.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Y, Cai J, Shang X, Wang B, Liu S, Chai X, Tan T, Zhang Y, Wen T. A new genome-scale metabolic model of Corynebacterium glutamicum and its application. Biotechnol Biofuels. 2017;10:169. doi: 10.1186/s13068-017-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y, Shang X, Lai S, Zhang Y, Hu Q, Chai X, Wang B, Liu S, Wen T. Reprogramming one-carbon metabolic pathways to decouple l-serine catabolism from cell growth in Corynebacterium glutamicum. ACS Synth Biol. 2018;7:635–646. doi: 10.1021/acssynbio.7b00373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.