Abstract
Purpose:
Several case reports suggest sorafenib exposure and sorafenib-induced hyperbilirubinemia may be related to a (TA)5/6/7 repeat polymorphism in UGT1A1*28. We hypothesized that sorafenib inhibits UGT1A1 and individuals carrying UGT1A1*28 and/or UGT1A9 variants experience greater sorafenib exposure and greater increase in sorafenib-induced plasma bilirubin concentration.
Experimental Design:
Inhibition of UGT1A1-mediated bilirubin glucuronidation by sorafenib was assessed in vitro. UGT1A1*28 and UGT1A9*3 genotypes were ascertained using fragment analysis or direct sequencing in 120 cancer patients receiving sorafenib on five different clinical trials. Total bilirubin measurements were collected in prostate cancer patients prior to receiving sorafenib (n=41) and 19–30 days following treatment and were compared to UGT1A1*28 genotype.
Results:
Sorafenib exhibited mixed-mode inhibition of UGT1A1-mediated bilirubin glucuronidation (IC50=18μM; Ki=11.7μM) in vitro. Five patients carrying UGT1A1*28/*28 (n=4) or UGT1A9*3/*3 (n=1) genotypes had first-dose, dose-normalized sorafenib AUCs that were in the 93rd percentile, while three patients carrying UGT1A1*28/*28 had AUCs in the bottom quartile of all genotyped patients. DMET genotyping on six patients revealed the ABCC2–24C>T genotype cosegregated with sorafenib AUC phenotype. Sorafenib exposure was related to plasma bilirubin increases in patients carrying 1 or 2 copies of UGT1A1*28 alleles (n=12 and n=5; R2=0.38 and R2=0.77; P=0.032 and P=0.051, respectively). UGT1A1*28 carriers demonstrated two distinct phenotypes that could be explained by ABCC2–24C>T genotype and are more likely to experience plasma bilirubin increases following sorafenib if they had high sorafenib exposure.
Conclusions:
This pilot study indicates that genotype status of UGT1A1, UGT1A9, and ABCC2 and serum bilirubin concentration increases reflect abnormally high AUC in patients treated with sorafenib.
Keywords: Sorafenib, glucuronidation, hyperbilirubinemia, pharmacokinetics, pharmacogenetics
INTRODUCTION
Sorafenib tosylate inhibits VEGF receptors and several tyrosine kinases and thus is considered an antiangiogenic agent with dual multikinase activity (1). Although sorafenib is currently approved for the treatment of renal cell and unresectable hepatocellular carcinomas, it may also have activity in other solid tumors, which led to the initiation of several sorafenib-based clinical trials (2–5). There is wide variation in the response and toxicity between patients following sorafenib treatment that appears to be, at least in part, related to cumulative drug exposure (3). For this reason, studies investigating the sources of inter-individual variation in sorafenib exposure are needed.
In humans, the majority (77%) of the sorafenib dose is either not absorbed or is eliminated through the hepatobiliary route (50% unchanged), while 19% of the dose (mostly glucuronides) is excreted in urine (6). Both routes of elimination require glucuronidation catalyzed by UGT1A9 (7, 8), although it remains unclear if other UGTs are responsible for glucuronidation of oxidized sorafenib metabolites formed through CYP3A4/5 metabolism. Moreover, these enzymes demonstrate phenotypic variability based on multiple polymorphisms, (i.e., UGT1A9*3, UGT1A9 −118dT9/10, UGT1A9 IVS+1 399 C>T (9); CYP3A4*1b and CYP3A5*3C (10). Once in the gut, intestinal microflora deglucuronidate and reduce sorafenib resulting in enterohepatic circulation allowing systemic re-exposure (10, 11). However, renal elimination appears to be irreversible and individuals with low creatinine clearance (CrCl<60mL/min) require more sorafenib dose reductions than patients with normal renal function (12). Thus, sorafenib glucuronidation is a significant route of sorafenib metabolism (6, 8) and can potentially alter sorafenib exposure.
It is known that seven TA nucleotide repeats in the (TA)nTAA promoter region of UGT1A1 (UGT1A1*28) leads to decreased expression of UGT1A1, resulting in high plasma bilirubin levels and is often diagnosed as Gilbert’s syndrome (13). Previous reports suggested that bilirubin concentrations were elevated by sorafenib (14, 15). Interestingly, one report suggested that sorafenib induced jaundice in individuals carrying UGT1A1*28 alleles due to a proposed UGT1A1 inhibition (14). This is consistent with three additional reports that also suggested sorafenib might inhibit UGT1A1-mediated bilirubin glucuronidation resulting in elevated bilirubin concentration (14–17). Another study profiled a patient receiving sorafenib who had yellow skin coloration despite a normal serum concentration of bilirubin and determined that the outcome was likely, if not definitely, attributable to sorafenib treatment (18). Furthermore, a phase I dose-escalation trial (n=34) of sorafenib with irinotecan, a UGT1A1 and UGT1A9 substrate, resulted in elevated irinotecan and SN-38 exposure with the highest sorafenib dose (400 mg BID) (17). In that study, sorafenib was reported to have an in vitro inhibitor constant (Ki) of 2.7 μM in human liver microsomes. This suggested that the increased SN-38 exposure was due to sorafenib-induced inhibition of UGT1A1- and/or UGT1A9-mediated SN-38 glucuronidation. However, none of the above case reports evaluated sorafenib plasma concentration in these patients, thus further confirmation of these results in larger patient cohorts undergoing sorafenib treatment is needed.
Herein, we present a case report of a child with Gilbert’s syndrome who underwent sorafenib treatment and experienced abnormally high sorafenib exposure. Based on this observation and the aforementioned case studies, we hypothesized that sorafenib and/or CYP3A4/5-mediated sorafenib oxide may be glucuronidated by UGT1A1 and/or UGT1A9, and that sorafenib acts as an inhibitor of UGT1A1-mediated bilirubin glucuronidation. Additionally, sorafenib AUC and sorafenib-induced hyperbilirubinemia might be related to the UGT1A1*28 allele that is responsible for most cases of Gilbert’s syndrome. Since UGT1A9 and CYP3A4/5 are known to metabolize sorafenib (6, 8), we hypothesized that sorafenib exposure would also be related to allelic variation in the genes encoding these enzymes (19). To this end, we compared sorafenib AUC with genetic variation in CYP3A4/5 and UGT1A1/9 in patients with various solid tumors undergoing sorafenib therapy, as well as with sorafenib-related toxicities (hand-foot skin reaction; HFSR) and clinical outcome.
METHODS
Materials
The following chemicals were purchased from their respective suppliers: Sorafenib tosylate (CTEP, c/o Bayer Schering Pharma, Berlin, Germany), Human CYP3A4 supersomes containing CYP450 oxidoreductase and NADPH generating system, 0.5 M potassium phosphate, pH 7.4, 100 mM Tris buffer, pH 7.4, Human UGT1A1 and UGT1A9 Supersomes and UGT Reaction Mix (BD Biosciences, San Jose, CA), methanol and acetonitrile (Optima grade, Fisher Scientific, Pittsburgh, PA), formic acid and acetic acid (Sigma-Aldrich, St. Louis, MO). β-Glucuronidase was purchased from Roche (Roche Diagnostics, Indianapolis, IN). All water used was deionized and purified using a Millipore system.
In vitro studies
Sorafenib and cytochrome P450 (CYP)-mediated oxidized sorafenib (sorafenib-N-oxide; M-2) were subjected to uridine glucuronosyl transferase (UGT)-catalyzed glucuronidation by members of the UGT1A family, UGT1A1 and UGT1A9. Furthermore, the role of sorafenib as an inhibitor of UGT1A1-mediated bilirubin glucuronidation was also studied. Details are discussed in Supplemental Methods.
Patients and Treatment
Patients (n=120) from five clinical trials involving sorafenib treatment were used for subsequent pharmacogenetic analysis, consisting of two phase I trials and three phase II trials. The phase I trials were BAY-BEV (200 mg bid sorafenib with bevacizumab; n=27) (3), and BAY-KS (200 mg qd or 200–400 mg bid sorafenib with ritonavir in Kaposi’s sarcoma; n=8; data not yet published), AUC data from patients on the BAY-KS trial who received ritonavir were not available; thus potential AUC-influencing drug-drug interactions between ritonavir and sorafenib were not accounted for in future analyses. The phase II trials were BAY-CRPC (400 mg bid sorafenib in castration resistant prostate cancer; n=46) (2, 4), BAY-NSCLC (400 mg bid sorafenib in non small-cell lung cancer; n=22) (20, 21), and BAY-CRC (400 mg bid sorafenib with cetuximab in colorectal cancer; n=17; data not yet published). Written informed consent was obtained from all patients before enrollment on the trials and genotyping was approved by the Institutional Review Board of the National Cancer Institute. All inclusion/exclusion criteria and genotyping methods are detailed in the Supplemental Methods.
Sorafenib Exposure
Exposure (AUC) data were represented as day 1 dose-normalized AUC0–12h (ng·h/mL/mg), as previously reported (22). Steady-state exposures were not available for all 120 patients. Furthermore, exposure values were dose-normalized to compare AUC from the five different trials with patients administered different doses. Linear pharmacokinetics were not assumed from first-dose AUC values, rather individual patient exposures were correlated to physiological changes (i.e. HFSR, PFS, and plasma bilirubin concentration).
Statistical considerations
Hardy-Weinberg equilibrium was tested using the chi-squared test. Genetic linkage statistics were obtained using Haploview (Broad Institute, Cambridge, MA). Comparisons of genotype versus demographics, preclinical measures, and pharmacokinetics were conducted with nonparametric statistical tests including the Wilcoxon Rank-Sum test or Kruskal Wallis ANOVA. Kruskal Wallis ANOVA was also used to compare sorafenib AUC to the clinical grade of hand-foot skin reactions (HFSR). Fisher’s Exact Test was used to compare between different AUC percentiles and genotype. Linear regression was conducted to compare the creatinine clearance (CrCl) and serum glutamic oxaloacetic transaminase (SGOT) to sorafenib AUC and was also utilized in comparisons between AUC and the bilirubin change from baseline in the different genotype groupings. Cox model analysis was conducted to compare genotypes versus progression-free survival (PFS). Statistical significance was assigned if P<0.05 as this study was conducted in an exploratory mode for potential confirmation in independent data.
RESULTS
Case Report
The patient was a 12 year-old boy who enrolled on a phase I trial of sorafenib for children with neurofibromatosis type 1 and inoperable plexiform neurofibromas (23). Prior to treatment, he had a total elevated bilirubin of 2.0 mg/dL with a direct of 0.3 mg/dL, normal serum ALT and AST, and was clinically diagnosed with Gilbert’s syndrome. This was later confirmed with genetic testing as he was found to be homozygous for the A(TA)7TAA allele of the UGT1A1 gene (i.e., UGT1A1*28/*28). The protocol required a bilirubin concentration within normal limits for study entry, except for patients with Gilbert’s syndrome. He was treated with a dose of 115 mg/m2 twice daily (approximately 50% of the adult maximum tolerated dose (MTD) based on an average adult body surface area of 1.8 m2).
The patient had day 1 pharmacokinetics performed, and his AUC0–24h was noted to be 81 μg·h/mL, which is greater than the average AUC0–24h of 28 ± 17 μg·h/mL observed in children treated at the MTD of 200 mg/m2 on the refractory solid tumor phase I trial (24). The patient came off treatment after 9 days due to dose limiting grade 3 tumor pain. The protocol was subsequently amended to exclude patients with known Gilbert’s syndrome from trial participation.
Based on this case observation and the published case reports that sorafenib induced hyperbilirubinemia in a small number of patients who carry a UGT1A1*28 allele (14, 15), we hypothesized that genetic variation in UGT1A1 was a potential source of alterations in sorafenib exposure and sorafenib-induced hyperbilirubinemia. Moreover, since UGT1A9 is known to primarily glucuronidate sorafenib (8), we hypothesized that UGT1A9 alleles might also contribute to both endpoints; thus, we studied sorafenib and sorafenib-N-oxide glucuronidation by UGT1A1 and UGT1A9 in vitro (Supplemental Methods and Results), and ascertained UGT1A1 and UGT1A9 genotypes in patients treated with sorafenib for comparison with pharmacokinetics and sorafenib-induced hyperbilirubinemia endpoints. Patient characteristics are reported in Table 1.
Table 1.
Patient demographics and baseline characteristics
| Characteristic | Values n (%) or median (95%CI) |
|---|---|
| Total | 120 (100.0) |
| Male | 86(71.7) |
| Female | 34 (28.3) |
| Age | 63.2(60.9–65.4) |
| Race | |
| Caucasian | 99 (82.5) |
| African American | 11 (9.2) |
| Hispanic | 4 (3.3) |
| Asian | 5 (4.2) |
| Unknown | 1 (0.8) |
| BSA (m2) | 2.0(1.9–2.0) |
| Study/Disease | |
| BAY-BEV / Solid tumors | 27 (22.5) |
| BAY-KS / Kaposi’s sarcoma | 8 (6.7) |
| BAY-CRPC / Prostate | 46 (38.3) |
| BAY-NSCLC / Lung | 22(18.3) |
| BAY-CRC / Colorectal | 17(14.2) |
| Albumin (g/dL) | 3.6(3.5–3.7) |
| Alkaline Phosphatase (U/L) | 81.0(74.0–89.0) |
| Total Bilirubin (mg/dL) | 0.6 (0.6–0.7) |
| SGOT (U/L) | 26 (24–28) |
| CrCl (mL/min) | 90.9 (84.8–97.7) |
| CrCl <60mL/min | 17(14.2) |
In Vitro Sorafenib Glucuronidation
Sorafenib glucuronidation by UGT1A9 was confirmed via in vitro metabolism experiments with recombinant UGT1A9 via liquid chromatography-mass spectrometry (LC-MS), whereas a similar experiment using UGT1A1 did not metabolize sorafenib (see Supplemental Results). This is in agreement with literature (7, 8). Based on exploratory studies, neither UGT1A1 not UGT1A9 glucuronidated the CYP3A4-mediated sorafenib-N-oxide (M-2) (data not shown).
Based on previous reports (14, 15, 17), we hypothesized that UGT1A1 could bind sorafenib and that this binding event could inhibit bilirubin glucuronidation. Increasing amounts of sorafenib were added to an in vitro UGT1A1-catalyzed bilirubin glucuronidation enzyme activity assay to determine the extent and mechanism of inhibition by sorafenib. Bilirubin concentrations used were based on literature reports of its Km for UGT1A1-mediated glucuronidation, ranging between 0.2–26 μM (25–28). Graphical modeling suggested sorafenib best fits a mixed-mode (mixed-type) inhibitor of UGT1A1 (model correlation r=0.96), which demonstrates properties of both a competitive and noncompetitive inhibitor (Supplemental Results). Based on model correlations (noncompetitive r=0.93; competitive r=0.88), it was suggested that while sorafenib is a mixed-type inhibitor, it exhibits more noncompetitive-type inhibitor characteristics than competitive.
A bilirubin Km of 5.9 μM and an inhibitor constant (Ki) of 11.8 μM were determined empirically through the mixed-type model. The IC50 of sorafenib for UGT1A1-mediated bilirubin glucuronidation was determined to be 18 μM following a separate experiment (see Supplemental Methods). These values were slightly higher than the literature values obtained by studying the mixed-type sorafenib-mediated inhibition of SN38 glucuronidation by UGT1A1, where Ki was found to be 2.7 μM (17). The most likely reason for this discrepancy from literature is due to the different substrates used in the experiment (SN38 vs bilirubin). Since sorafenib demonstrates a competitive inhibition factor (model correlation 0.88), the substrate likely has an affect on inhibitor Ki.
Genotype versus sorafenib exposure
Based on the case report presented above, data indicating that sorafenib is glucuronidated by UGT1A9 (Supplemental Results), and the sorafenib-mediated inhibition of bilirubin glucuronidation through UGT1A1 (Supplemental Results), we next hypothesized that genetic variants in UGT1A1 and UGT1A9 would affect sorafenib pharmacokinetics and bilirubin metabolism in patients with solid tumors who received sorafenib.
We excluded patients with low CrCl (<60mL/min) and a single patient with abnormally high SGOT (90U/L) given the importance of hepatic and renal function in sorafenib PK (Supplemental Results). Carriers of UGT1A1*28/*28 tended to be younger and have higher median total bilirubin. Patients with wild-type CYP3A4 and CYP3A5 alleles had lower median SGOT and median CrCl than variant allele carriers, respectively. A more detailed summary can be found in the Supplemental Results section. None of the above associations are likely to have altered the results presented in later sections.
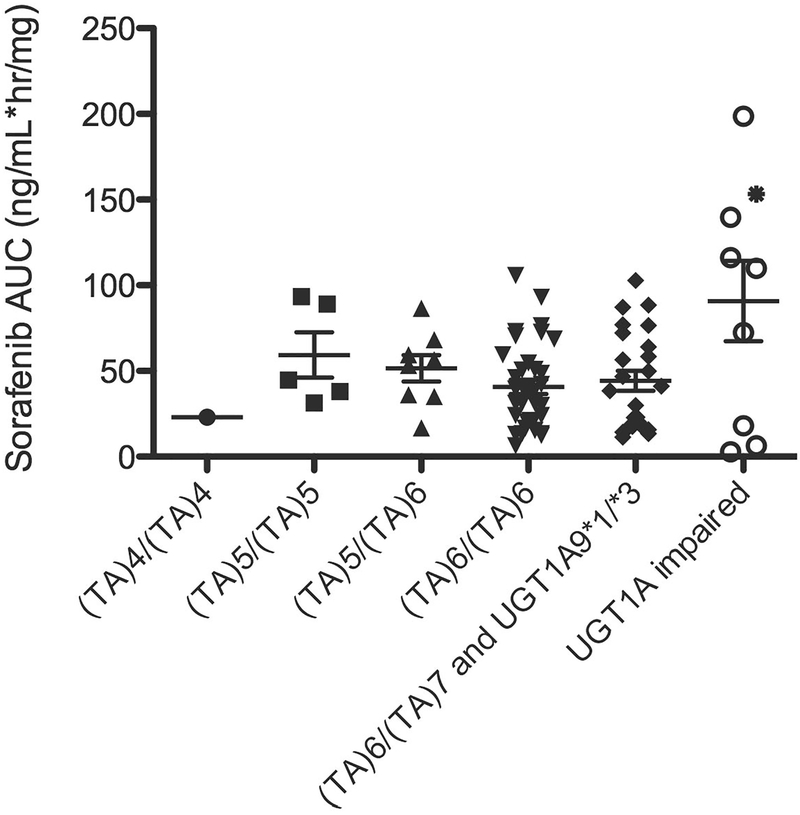
Initial analysis of UGT1A1 A(TA)nTAA and UGT1A9*3 genotypes versus day 1 dose-normalized sorafenib AUC0–12h revealed five patients carrying UGT1A1*28/*28 (n=4) and UGT1A9*3/*3 (n=1) have higher AUCs than those patients corresponding to any other genotypes with normal creatinine clearance (CrCl) and SGOT (i.e., all > 93rd percentile; range = 109.9 – 198.6 ng·h/mL/mg), and one patient carrying UGT1A1*28/*28 had an AUC in the 76th percentile (AUC = 72.5 ng·h/mL/mg). Interestingly, of the three remaining patients carrying UGT1A1*28/*28, two had the lowest AUCs (i.e. < 3rd percentile; range = 2.7 – 6.3 ng·h/mL/mg) while one had an AUC in the 21st percentile (AUC = 18.0 ng·h/mL/mg). For this reason, UGT1A1*28 status was considered to confer different phenotypes: those having an abnormally high exposure, those having exposures matching the rest of the cohort, and those having low exposure (Figure 1). Analysis of UGT1A1 and UGT1A9 genotypes versus sorafenib AUC in the different genotype groupings did not lead to a statistically significant result due to the wide variability in phenotype in the UGT1A1*28/*28 genotype grouping (P = 0.32; Kruskal-Wallis ANOVA; Figure 1); however, there were strongly significant differences in the odds of having AUCs >93rd percentile (i.e., ≥107 ng·h/mL/mg) and also carrying UGT1A1*28/*28 (n=4) or UGT1A9*3/*3 (n=1) (OR (95%CI) = 179.7 (8.5 – 3787); P<0.0001; Fisher’s Exact Test). In addition, there was a strongly significant difference in the odds of carrying UGT1A1*28/*28 and also having AUCs ≤ 3rd percentile (OR (95%CI) = 57.3 (2.5 – 1326); P = 0.0084; Fisher’s Exact Test), further justifying consideration of UGT1A1*28 alleles as conferring different phenotypes. Although UGT1A9 IVS+1 (399C>T) and −118dT9/10 were in linkage disequilibrium with UGT1A1 A(TA)nTAA (Supplementary Results), these polymorphisms were not associated with alterations in AUC (P≥0.39; data not shown). Neither CYP3A4*1B (P = 0.42) nor CYP3A5*3C (P = 0.52) status was associated with increased sorafenib exposure. Therefore, UGT1A1*28 and possibly the UGT1A9*3 SNP appeared to be the major predictive alleles associated with phenotype.
Figure 1.
Dose-normalized sorafenib AUC versus UGT1A genotype. UGT1A9*1/*3 carriers each also carried UGT1A1 (TA)6/(TA)7 (AUC = 17.3 and 20.2 ng/mL*hr/mg) while the single UGT1A9*3/*3 carrier also carried UGT1A1 (TA)6/(TA)6 (AUC = 153.3 ng/mL*hr/mg, as indicated by *). UGT1A-impaired implies patients with deficient UGT1A1 or UGT1A9 metabolism based on genetics. Excluded patients (n=38) are described in the supplementary results section and n=82 individuals were included in the present analysis. Of these, AUC data for n=8 patients participating on BAY-KS were not available; thus potential drug-drug interactions between ritonavir and sorafenib were not accounted for. There was no association between UGT1A1 and UGT1A9 genotype status when compared to sorafenib AUC (P = 0.20; Kruskal Wallis ANOVA).
To further study the apparently different phenotypes for patients carrying only UGT1A1*28 alleles (n=8), we genotyped 1,936 polymorphisms in 225 genes involved in clinical pharmacology using the DMET Plus panel (Coriell Institute, N.J., USA). DMET genotyping was only successful (≥90% call rate) in a total of 6 patients with the following AUCs (ng·h/mL/mg): 2.7, 6.3, 18.0, 109.9, 116.2, 139.6. DMET analysis revealed that only the ABCC2 −24C>T SNP cosegregated with sorafenib metabolism phenotype. After sequencing the ABCC2 −24C>T SNP in all patients carrying UGT1A1*28/*28, it was determined that the patient with the lowest observed AUC (i.e. 2.7 ng·h/mL/mg) was double variant, whereas patients with the next lowest AUCs (6.3, 18.0, and 72.5 ng·h/mL/mg) were heterozygous followed by those with the highest AUCs (109.9, 116.2, and 139.6) who carried homozygous wild-type alleles. A single patient with AUC = 198.6 ng·h/mL/mg was not ascertainable as ABCC2 genotyping by direct sequencing was not successful. Upon sequencing the whole population for ABCC2 −24C>T, it was determined that individuals carrying only variant alleles in this SNP tended to have lower median AUC (29.8 versus 40.5 ng·h/mL/mg) compared to individuals carrying one or two copies of wild-type allele, but this was not statistically significant (P=0.21). Therefore, the ABCC2 −24C>T SNP only appears to modify AUC phenotype in those carrying only UGT1A1*28 alleles.
Genotype versus bilirubin change following sorafenib
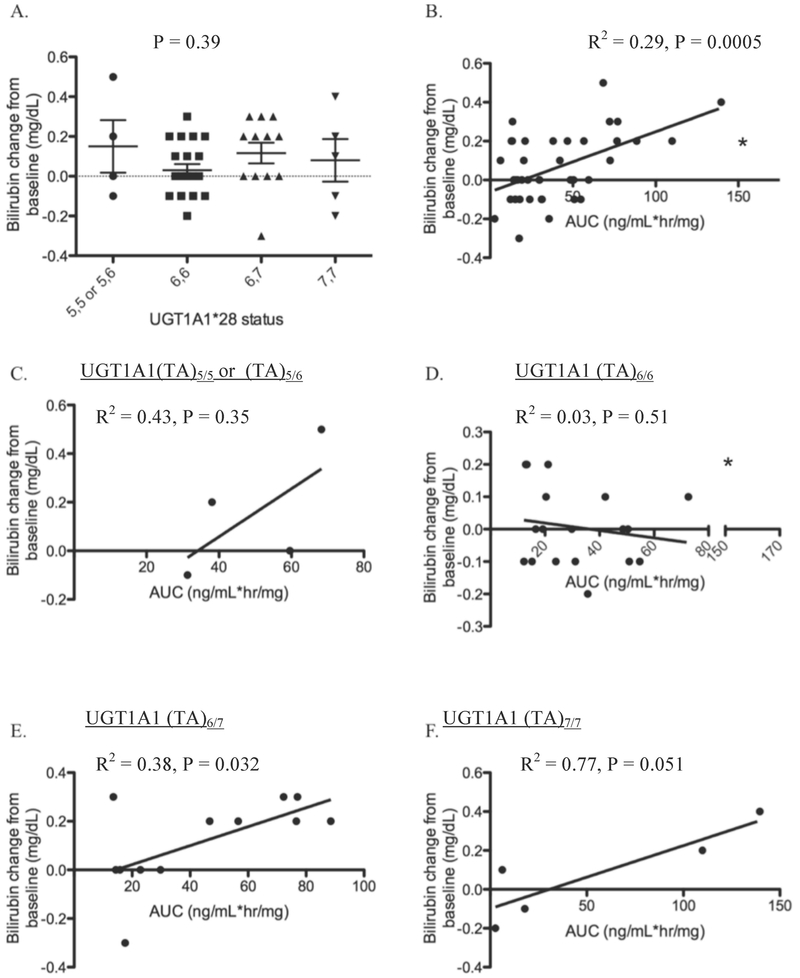
Since previous case-report data indicated that sorafenib might induce bilirubin changes in patients based on UGT1A1 allele status (14, 15), we hypothesized that sorafenib exposure would correspond to greater increases in post-sorafenib bilirubin concentration in those patients with low-functioning UGT1A1 alleles (i.e., UGT1A1*28). Analysis of bilirubin versus genotype was only conducted in men with prostate cancer receiving sorafenib as comprehensive bilirubin data were not obtained in other trials. The median change in bilirubin plasma concentration was 0 mg/dL (range = −0.3 to 0.5 mg/dL; n=45). A total of 3 patients with normal CrCL developed hyperbilirubinemia (i.e., bilirubin concentration ≥1.0mg/dL) following sorafenib (UGT1A1 (TA)5/(TA)6 n=1, (TA)6/(TA)7 n=2), and 2 patients that presented with hyperbilirubinemia prior to the sorafenib dose had a further rise in bilirubin concentration following sorafenib (UGT1A1 (TA)6/(TA)7 n=1, (TA)7/(TA)7 n=1; the latter patient had a 0.4mg/dL increase).
UGT1A1 A(TA)nTAA status was not related to change in bilirubin from baseline (P = 0.39; Figure 2A). However, regression analysis indicated that sorafenib exposure was related to bilirubin serum concentration in patients with normal CrCl (R2 = 0.29; P = 0.0005; Figure 2B). When regression analyses were stratified on the basis of UGT1A1 A(TA)nTAA genotype status, this analysis revealed that sorafenib exposure was not related to bilirubin increases in patients carrying either UGT1A1 (TA)5/5 or (TA)5/6, or UGT1A1 (TA)6/6 genotypes (R2 = 0.43 and R2 = 0.030 respectively; P = 0.35 and P = 0.51, respectively; Figure 2C–D). However, only 4 individuals carried a copy of UGT1A1 (TA)5, and there is an apparent (albeit non-significant) proportional increase in both AUC and sorafenib-induced bilirubin changes consistent with the rather high R2 for this genotype grouping. When the data were stratified by UGT1A1 (TA)6/7 and UGT1A1 (TA)7/7 genotypes, a significant (or marginally non-significant) relationship was observed in both cases with a relatively high correlation (R2 = 0.38 and R2 = 0.77 respectively; P = 0.032 and P = 0.051, respectively; Figure 2E–F). For patients with the UGT1A1 (TA)6/7 genotype, the data indicate that bilirubin increased by 0.1 mg/dL for every 25.7 (ng/mL*hr/mg) unit increase in sorafenib AUC. Carriers of UGT1A1 (TA)7/7 had a similar relationship between bilirubin and AUC (i.e., a 30.9 ng/mL*hr/mg unit increase in AUC corresponded to a 0.1 mg/dL increase in bilirubin). These data are consistent with previous case reports where UGT1A1 (TA)6/7 carriers developed jaundice following sorafenib treatment and are also consistent with our results that sorafenib is a mixed inhibitor of UGT1A1.
Figure 2.
UGT1A1 A(TA)nTAA status versus bilirubin change from baseline following sorafenib (mg/dL) (A); and sorafenib AUC (ng/mL*hr/mg) versus bilirubin change from baseline (mg/dL) (B) in patients with UGT1A1 (TA)5/5 or UGT1A1 (TA)5/6 (C), UGT1A1 (TA)6/6 (D), UGT1A1 (TA)6/7 (E), or UGT1A1 (TA)7/7 (F). * Indicates a patient who carried UGT1A1 (TA)6/6 that also carried two variants at UGT1A9*3 and was thus excluded from analysis in the UGT1A1 (TA)6/6 cohort.
Genotype versus progression-free survival and toxicity
The CYP3A4*1B allele was weakly associated with PFS according to a Cox model analysis accounting for the multiple clinical trials where the present patient cohort was ascertained (data not shown). Those patients carrying variant alleles at CYP3A4*1B (n=19) tended to have shorter PFS than those patients carrying homozygous wild-type alleles (n=99; P=0.034). None of the other alleles studied herein were related to PFS (P>0.05); however, the small numbers of variants within each trial led to wide confidence intervals for the individual hazard ratio estimates and the present results with respect to PFS should be interpreted with caution.
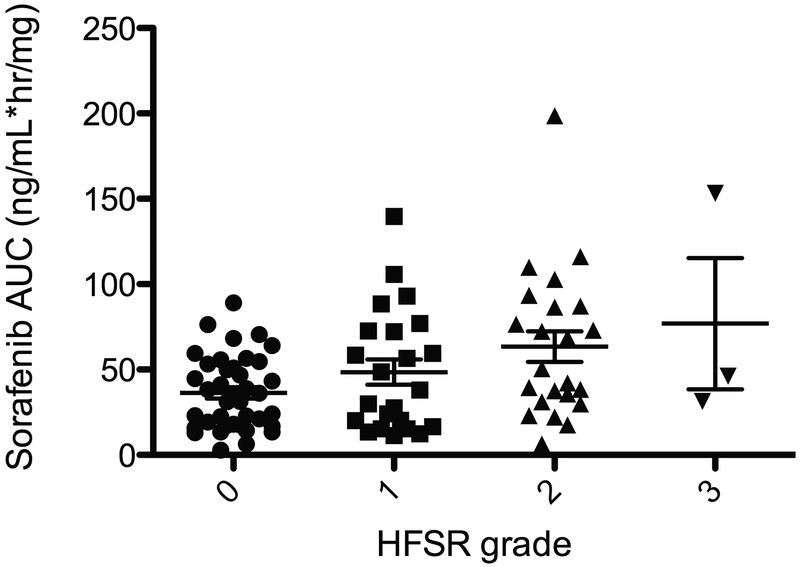
Consistent with previous literature (22), the incidence of HFSR was associated with increases in sorafenib AUC (P=0.0054; Figure 3). However, all patients who had AUC>100 ng·h/mL/mg (n=7) developed HFSR regardless of genotype (P=0.0085); thus, while UGT1A genotypes were not associated with HFSR in the present study (due to heterogeneity of phenotype and low allele frequency), it is likely that UGT1A1*28/*28 and UGT1A9*3 carriers are subject to increased incidence of HFSR as they likely have higher exposure to sorafenib, and this should be confirmed by future studies.
Figure 3.
Sorafenib exposure is related to hand-foot skin reaction. HFSR was associated with increases in sorafenib AUC (P=0.0054).
DISCUSSION
The case report presented here involved a child with Gilbert’s syndrome (UGT1A1*28/*28) who upon receiving sorafenib for treatment of a neurofibroma had higher than expected sorafenib exposure. Based on this and previously published observations (14–17), we hypothesized and assessed if UGT1A1 genotype was associated with sorafenib systemic exposure and serum bilirubin concentrations in patients treated with sorafenib. We confirmed previous hypotheses and findings (14–17) that sorafenib is an inhibitor of, but is not metabolized by, UGT1A1; rather, UGT1A9 is involved in sorafenib glucuronidation, which also confirms another report (8). It is unclear as to the exact reason for the approximately 4–10-fold difference in Ki values, however there is one plausible reason. Both this study and literature described sorafenib as a mixed-type inhibitor of UGT1A1, and although sorafenib demonstrates more noncompetitive characteristics (based on higher model correlation), there is a competitive inhibitor portion to the Ki calculation that is potentially altered based on the substrate used. The UGT1A1 substrate used in the literature was SN38, whereas bilirubin was used in this study, and the affinity of that substrate may affect the Ki of sorafenib. SN38 has a Km of 11 μM for UGT1A1 (29), whereas bilirubin has a reported Km range of 0.2–26 μM (25–28) due to the numerous stereoisomers present. Furthermore, the Ki of 11 μM and IC50 of 18 μM are clinically-relevant plasma concentrations following 400 mg BID dosing, which typically result in maximum plasma concentrations between 11–21 μM after a cycle lasting either 7 or 28 days (30). Although sorafenib is >99% plasma protein bound, there is a high enough intra-hepatic sorafenib concentration in patients to inhibit the UGT1A1-mediated bilirubin glucuronidation. From our data, patients carrying at least one allele of UGT1A1*28 and having sorafenib AUC >60 ng*h/mL, or 130 nM*h, were related to increases in serum bilirubin following sorafenib treatment (Figure 2E–F). This provides clinical evidence that intra-hepatic sorafenib concentrations can be achieved in high enough levels to inhibit UGT1A1.
The present results suggest that patients carrying only UGT1A1*28, and possibly UGT1A9*3, alleles are at an increased risk of elevated sorafenib concentrations, as well as a greater incidence of HFSR. However, there remained a group of patients carrying UGT1A1*28/*28 and ABCC2 −24C>T that had abnormally low sorafenib AUC, thereby complicating the present analysis. Nonetheless, sorafenib AUC was well correlated with change in bilirubin concentrations in patients carrying at least a single UGT1A1*28 allele; conversely, it was not correlated with AUC in patients who carried only wild-type genotypes at this site.
The mechanism underlying the observation that individuals carrying UGT1A1*28/*28 also have high AUC, a phenotype that appears to be modified by ABCC2 −24C>T, is currently unclear. We tested the hypothesis that UGT1A1*28 alleles were merely in linkage disequilibrium with UGT1A9 alleles (i.e., UGT1A9*3, UGT1A9 IVS+1 (399C>T) and −118dT9/10) that were truly responsible for the observed differences in AUC. While linkage was observed, the current UGT1A9 alleles did not explain the association between UGT1A1*28/*28 and AUC. We also tested whether or not ABCC2 −24C>T was itself related to AUC regardless of UGT1A1 genotype; this also revealed no apparent association outside of those patients carrying UGT1A1*28/*28. Only parent sorafenib pharmacokinetics were analyzed, however no bias was expected through analysis of metabolites, since it has previously been demonstrated that altering one metabolic pathway of sorafenib does not alter parent drug pharmacokinetics (6).
The present results demonstrate that UGT1A1 binds sorafenib; others demonstrated that sorafenib binds to ABCC2 and inhibits transport of other substrates but is not transported by ABCC2 (31). Elimination of sorafenib glucuronides through ABCC2 may also be involved, however it is unclear if sorafenib glucuronides are actually transported by ABCC2. One previous report has also shown that the liver contains binding sites for drugs that act as “sinks” to slowly dissociate bound drug that is subsequently metabolized by liver enzymes (32). It is therefore possible that given the extensive enterohepatic circulation of sorafenib (10, 11), individuals expressing relatively high levels of liver UGT1A1 (i.e., those not carrying UGT1A1*28) and possibly other proteins that bind sorafenib have a greater propensity to extrude sorafenib from the serum and hold sorafenib in the liver where it can be metabolized more extensively prior to hepatobiliary elimination. This may explain the significantly higher sorafenib AUCs of individuals who have reduced expression of UGT1A1 due to genetic polymorphisms. Still, our hypothesis does not explain why individuals carrying both UGT1A1*28 and ABCC2 −24C>T variants have some of the lowest AUCs. As both proteins are involved in bilirubin elimination (19), we expected that patients carrying both UGT1A1*28 and ABCC2 −24C>T would have less ability to glucuronidate and eliminate bilirubin due to decreased sorafenib-mediated UGT1A1 inhibition; however, this was not the case in the present patient cohort (data not shown). Therefore, the fact that both proteins regulate bilirubin concentrations that could in turn influence binding of sorafenib to UGT1A1 in the liver is not likely to be the cause of ABCC2 −24C>T modifying the sorafenib exposure phenotype of patients carrying UGT1A1*28/*28.
Although our data did not point to a specific mechanism underlying associations between UGT1A1*28/*28 and AUC, we observed a clear relationship between sorafenib exposure and bilirubin concentration. We demonstrated that sorafenib inhibits bilirubin glucuronidation as a mixed inhibitor in vitro. Based on this observation and the aforementioned case reports, we assessed whether or not sorafenib inhibited bilirubin glucuronidation in patients and whether or not this depended on UGT1A1 A(TA)nTAA status. The results indicated that sorafenib exposure was not correlated with total bilirubin concentration in patients that do not carry UGT1A1*28; however, there was an increasingly strong correlation between sorafenib AUC and total bilirubin in patients carrying a single copy or two copies of UGT1A1*28. Therefore, hyperbilirubinemia appears to be a marker of high sorafenib exposure in patients expressing low levels of UGT1A1 and care should be taken in monitoring patients carrying UGT1A1*28 that are known to have high sorafenib exposure. Nonetheless, none of the genes studied in this investigation were related to the PFS of the various sorafenib studies with the exception of the weak association with the CYP3A4*1B allele. The results of the present Cox model should be interpreted with caution.
To our knowledge, the present pilot study represents the first exploration of sorafenib AUC in patients based on UGT1A1 and UGT1A9 genotype status and suggests that future studies should focus on the UGT1A1 A(TA)nTAA and UGT1A9*3 alleles and bilirubin increases in relation to sorafenib exposure, and HFSR.
Supplementary Material
Translational Relevance.
We investigated UGT1A-mediated sorafenib glucuronidation in vitro, inhibition of UGT1A1 bilirubin conjugation by sorafenib, and ascertained whether patients carrying UGT1A1*28 treated with sorafenib had increased sorafenib exposure or increased risk of developing hyperbilirubinemia. We also investigated genetic variation in genes encoding sorafenib metabolizing enzymes (UGT1A9, CYP3A4/5) and the glucuronide transporter, ABCC2. In vitro data demonstrate that sorafenib inhibits UGT1A1. Patients carrying UGT1A1*28/*28 had abnormally low exposure to sorafenib if they also carried the −24C>T variant in ABCC2, but had abnormally high exposure to sorafenib if they were wild-type for ABCC2. Patients carrying UGT1A1*28/*28 also had greater increases in total bilirubin if sorafenib exposure was high. The clinical data suggest that sorafenib can cause hyperbilirubinemia in patients with Gilbert’s syndrome, which can lead to abnormally high or low sorafenib exposure. Therefore, patients with Gilbert’s syndrome must be carefully monitored when treated with sorafenib or alternative treatment options should be considered.
Acknowledgements
The authors thank Kathleen M. Wyvill and Thomas S. Uldrick for their contributions to the BAY-KS trial. The authors also thank all participating patients in each of the trials.
Abbreviations:
- UGT
UDP-glucuronysltransferease
- AUC
area under the plasma-concentration time curve
- VEGF
vascular endothelial growth factor
- CYP
cytochrome P450
- MTD
maximum tolerated dose
- CrCL
creatinine clearance
- SGOT
serum glutamic oxaloacetic transaminase
- CTEP
Cancer Therapy Evaluation Program
- NADPH
nicotinamide adenine dinucleotide phosphate
- DMSO
dimethyl sulfoxide
- LC/MS
liquid chromatography/mass spectrometry
Footnotes
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
REFERENCES
- 1.Jain L, Sissung TM, Danesi R, Kohn EC, Dahut WL, Kummar S, et al. Hypertension and hand-foot skin reactions related to VEGFR2 genotype and improved clinical outcome following bevacizumab and sorafenib. J Exp Clin Cancer Res 2010; 29:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragon-Ching JB, Jain L, Gulley JL, Arlen PM, Wright JJ, Steinberg SM, et al. Final analysis of a phase II trial using sorafenib for metastatic castration-resistant prostate cancer. BJU Int 2009; 103:1636–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad NS, Posadas EM, Kwitkowski VE, Steinberg SM, Jain L, Annunziata CM, et al. Combination targeted therapy with sorafenib and bevacizumab results in enhanced toxicity and antitumor activity. J Clin Oncol 2008; 26:3709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahut WL, Scripture C, Posadas E, Jain L, Gulley JL, Arlen PM, et al. A phase II clinical trial of sorafenib in androgen-independent prostate cancer. Clin Cancer Res 2008; 14:209–14. [DOI] [PubMed] [Google Scholar]
- 5.Jain L, Venitz J, Figg WD. Randomized discontinuation trial of sorafenib (BAY 43–9006). Cancer Biol Ther 2006; 5:1270–2. [DOI] [PubMed] [Google Scholar]
- 6.Lathia C, Lettieri J, Cihon F, Gallentine M, Radtke M, Sundaresan P. Lack of effect of ketoconazole-mediated CYP3A inhibition on sorafenib clinical pharmacokinetics. Cancer Chemother Pharmacol 2006; 57:685–92. [DOI] [PubMed] [Google Scholar]
- 7.Keating GM, Santoro A. Sorafenib: a review of its use in advanced hepatocellular carcinoma. Drugs 2009; 69:223–40. [DOI] [PubMed] [Google Scholar]
- 8.Nexavar (Sorafenib) Prescribing Information, Bayer Pharmaceuticals. 2009. [cited; Available from: http://www.nexavar.com/html/download/Nexavar_PI.pdf
- 9.Girard H, Villeneuve L, Court MH, Fortier LC, Caron P, Hao Q, et al. The novel UGT1A9 intronic I399 polymorphism appears as a predictor of 7-ethyl-10-hydroxycamptothecin glucuronidation levels in the liver. Drug Metab Dispos 2006; 34:1220–8. [DOI] [PubMed] [Google Scholar]
- 10.Jain L, Woo S, Gardner ER, Dahut WL, Kohn EC, Kummar S, et al. Population Pharmacokinetic Analysis of Sorafenib in Patients with Solid Tumors. Br J Clin Pharmacol 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Public Assessment Reports (EPAR)-Scientific Discussion. 2009. [cited; Available from: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Scientific_Discussion/human/000690/WC500027707.pdf
- 12.Parsa V, Heilbrun L, Smith D, Sethi A, Vaishampayan U. Safety and efficacy of sorafenib therapy in patients with metastatic kidney cancer with impaired renal function. Clin Genitourin Cancer 2009; 7:E10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strassburg CP. Hyperbilirubinemia syndromes (Gilbert-Meulengracht, Crigler-Najjar, Dubin-Johnson, and Rotor syndrome). Best Pract Res Clin Gastroenterol 2010; 24:555–71. [DOI] [PubMed] [Google Scholar]
- 14.Meza-Junco J, Chu QS, Christensen O, Rajagopalan P, Das S, Stefanyschyn R, et al. UGT1A1 polymorphism and hyperbilirubinemia in a patient who received sorafenib. Cancer Chemother Pharmacol 2009; 65:1–4. [DOI] [PubMed] [Google Scholar]
- 15.Miller AA, Murry DJ, Owzar K, Hollis DR, Kennedy EB, Abou-Alfa G, et al. Phase I and pharmacokinetic study of sorafenib in patients with hepatic or renal dysfunction: CALGB 60301. J Clin Oncol 2009; 27:1800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abou-Alfa GK, Amadori D, Santoro A, Figer A, De Greve J, Lathia C, et al. Safety and Efficacy of Sorafenib in Patients with Hepatocellular Carcinoma (HCC) and Child-Pugh A versus B Cirrhosis. Gastrointest Cancer Res 2011; 4:40–4. [PMC free article] [PubMed] [Google Scholar]
- 17.Mross K, Steinbild S, Baas F, Gmehling D, Radtke M, Voliotis D, et al. Results from an in vitro and a clinical/pharmacological phase I study with the combination irinotecan and sorafenib. Eur J Cancer 2007; 43:55–63. [DOI] [PubMed] [Google Scholar]
- 18.Dasanu CA, Alexandrescu DT, Dutcher J. Yellow skin discoloration associated with sorafenib use for treatment of metastatic renal cell carcinoma. South Med J 2007; 100:328–30. [DOI] [PubMed] [Google Scholar]
- 19.Jedlitschky G, Hoffmann U, Kroemer HK. Structure and function of the MRP2 (ABCC2) protein and its role in drug disposition. Expert Opin Drug Metab Toxicol 2006; 2:351–66. [DOI] [PubMed] [Google Scholar]
- 20.Kelly RJ, Rajan A, Force J, Keen C, Cao L, Yu Y, et al. Evaluation of KRAS mutations, angiogenic biomarkers and DCE-MRI in patients with advanced non-small cell lung cancer receiving sorafenib. Clin Cancer Res 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M Gutierrez SK AD, Turkbey B, Choyke P, Wright JJ, Kurkjian C, Giaccone G, Doroshow JH, Murgo AJ. A phase II study of multikinase inhibitor sorafenib in patients with relapsed non-small cell lun cancer (NSCLC). American Society of Clinical Oncology (ASCO) 2008. [Google Scholar]
- 22.Azad NS, Aragon-Ching JB, Dahut WL, Gutierrez M, Figg WD, Jain L, et al. Hand-foot skin reaction increases with cumulative sorafenib dose and with combination anti-vascular endothelial growth factor therapy. Clin Cancer Res 2009; 15:1411–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim A DE, Tepas K, Fox E, Balis FM, Korf B, and Widemann BC. Phase I trial of sorafenib in children with neurofibromatosis Type I and inoperable plexiform neurofibromas. Neuro-Oncology; 2010. [Google Scholar]
- 24.Widemann BC, Fox E, Adamson PC, Baruchel S, Kim A, Ingle AM, et al. Phase I study of sorafenib in children with refractory solid tumors: A Children’s Oncology Group Phase I Consortium Trial. Proc Am Soc Clin Oncology 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senafi SB, Clarke DJ, Burchell B. Investigation of the substrate specificity of a cloned expressed human bilirubin UDP-glucuronosyltransferase: UDP-sugar specificity and involvement in steroid and xenobiotic glucuronidation. Biochem J 1994; 303 ( Pt 1):233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seppen J, Bosma PJ, Goldhoorn BG, Bakker CT, Chowdhury JR, Chowdhury NR, et al. Discrimination between Crigler-Najjar type I and II by expression of mutant bilirubin uridine diphosphate-glucuronosyltransferase. J Clin Invest 1994; 94:2385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, Chando TJ, Everett DW, Patten CJ, Dehal SS, Humphreys WG. In vitro inhibition of UDP glucuronosyltransferases by atazanavir and other HIV protease inhibitors and the relationship of this property to in vivo bilirubin glucuronidation. Drug Metab Dispos 2005; 33:1729–39. [DOI] [PubMed] [Google Scholar]
- 28.Udomuksorn W, Elliot DJ, Lewis BC, Mackenzie PI, Yoovathaworn K, Miners JO. Influence of mutations associated with Gilbert and Crigler-Najjar type II syndromes on the glucuronidation kinetics of bilirubin and other UDP-glucuronosyltransferase 1A substrates. Pharmacogenet Genomics 2007; 17:1017–29. [DOI] [PubMed] [Google Scholar]
- 29.Jinno H, Tanaka-Kagawa T, Hanioka N, Saeki M, Ishida S, Nishimura T, et al. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38), an active metabolite of irinotecan (CPT-11), by human UGT1A1 variants, G71R, P229Q, and Y486D. Drug Metab Dispos 2003; 31:108–13. [DOI] [PubMed] [Google Scholar]
- 30.Strumberg D, Clark JW, Awada A, Moore MJ, Richly H, Hendlisz A, et al. Safety, pharmacokinetics, and preliminary antitumor activity of sorafenib: a review of four phase I trials in patients with advanced refractory solid tumors. Oncologist 2007; 12:426–37. [DOI] [PubMed] [Google Scholar]
- 31.Hu S, Chen Z, Franke R, Orwick S, Zhao M, Rudek MA, et al. Interaction of the multikinase inhibitors sorafenib and sunitinib with solute carriers and ATP-binding cassette transporters. Clin Cancer Res 2009; 15:6062–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin GM, Tozer TN. Hepatic binding and Michaelis-Menten metabolism of drugs. J Pharm Sci 1986; 75:660–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.