Abstract
Objective
The zinc finger protein 804A (ZNF804A) gene encodes the protein 804A containing the C2H2 zinc finger structure, which plays an important role in embryonic nerve development and repair. Previous studies have shown a significant association between the ZNF804A genetic variation polymorphism rs1344706 and the risk of schizophrenia and brain structure abnormalities. However, the findings are inconsistent.
Materials and methods
Seventeen studies on structural magnetic resonance imaging (sMRI), with 1,031 schizophrenia patients and 3,416 healthy controls, were included in the meta-analysis. These analyses were performed using Anisotropic Effect-Size Signed Differential Mapping (AES-SDM) software and Comprehensive Meta-Analysis (CMA) software.
Results
rs1344706 risk allele carriers of schizophrenia had increased gray matter in the brain regions including frontal lobe, temporal lobe, and other brain regions, but the carriers of healthy individuals had decreased gray matter and white matter integrity in the frontal lobe, central network, and other brain regions. The results of sensitivity analysis are stable, but publication bias exists in a few analyses of indexes.
Conclusion
Abnormalities of brain structure have a strong relationship with ZNF804A gene rs1344706 polymorphism, but the association may be different in healthy individuals and those with mental disorders.
Keywords: ZNF804A, genetic variation, magnetic resonance imaging, brain structure, meta-analysis
Introduction
Brain damage can directly lead to mental disorders. Abnormalities in gray matter of the frontal lobe are found in schizophrenia patients.1 The central nervous system is composed of neurons and glial cells; the former is mainly responsible for the transmission and processing of information, while the latter supports neurons and plays a role in protection, nutrition, and formation of myelin sheath and repair.2 Neurons send signals through synaptic connections, and there is approximately one teraflop of brain operation occurring in the synaptic connections, forming a number of neural circuits that underlie mental and neural activity. Once these brain regions are changed, the individual may show noticeable psychiatric symptoms.3–5
Schizophrenia has a major genetic component, and it is both etiologically and clinically complex. A genome-wide association study found that the single-nucleotide polymorphism rs1344706 of the zinc finger protein 804A (ZNF804A) gene is associated with schizophrenia. The alleles of rs1344706 result in differential prediction of the presence of binding sites for two brain-expressed transcription factors. The MYT1L zinc finger protein is expressed in neural progenitors along with the related family member MYT1 (which is known to modulate proliferation and terminal differentiation of oligodendrocyte progenitors).6 Of note, the MYT1L gene was included in a copy number variant observed in an affected individual in a recent study of structural variation in schizophrenia. The POU3F1/Oct-6 POU domain transcription factor is involved in oligodendrocyte differentiation and the transition of promyelinating to myelinating Schwann cells and is also normally expressed in adult cortex and hippocampus.6 Researchers use “imaging genetics” to explore the relationship between the risk gene and brain structural changes in patients with mental disorders. Lencz et al7 investigated the effect of ZNF804A, a risk gene for schizophrenia, on neuroanatomy and neurocognitive phenotypes. They found that healthy individuals with homozygous risk allele T have larger overall white matter and smaller gray matter compared with C allele carriers. This phenotype may directly reflect altered neural connection status. Wassink et al8 found that rs1344706 has a significant effect on total, frontal, and parietal white matter content in patients with schizophrenic spectrum diseases and their health.
Although there is a number of evidence to support the influence of the ZNF804A gene on the brain structure of healthy individuals and those with mental disorders, they are inconsistent. There is still a lack of meta-analysis to comprehensively evaluate these disparate findings. Therefore, the aim of this study was to explore the relationship between brain structure and the ZNF804A gene rs1344706 polymorphism based on a signed difference map. It also provides a reference for studying genetic-related mechanisms of mental disorders such as schizophrenia.
Materials and methods
Study design
The study was designed using voxel-based and general meta-analysis methods based on clinical data of statistical maps, peak coordinates, and statistical effect size collected from previous association studies of brain structural changes and ZNF804A gene polymorphisms. Two researchers (SW and YH) independently reviewed the literature and selected studies to use in the meta-analysis. Any disagreement was resolved through a group discussion.
Searching strategies
Literature searches were performed in databases including Medline, ScienceDirect, and Scopus before December 2017. The following search terms were combined and used: “ZNF804A/rs1344706” and “MRI/structural MRI/sMRI”. Publications from conferences, monographs, theses, or reference lists in identified studies were also regarded as potential sources to be included in the meta-analysis.
Inclusion and exclusion criteria
According to the PRISMA guidelines,9 the following criteria were used for inclusion in the meta-analysis: 1) original research published in peer-reviewed journals, conferences, or monographs; 2) scanning methods were structural (structural magnetic resonance imaging, sMRI); 3) studies comparing the brain structure in individuals with different genotypes of rs1344706 in the ZNF804A gene; 4) for Anisotropic Effect-Size Signed Differential Mapping (AES-SDM) analysis, the coordinates (either Talairach or Montreal Neurological Institute) of altered brain regions were detailed; 5) for Comprehensive Meta-Analysis (CMA), the effect size of different brain regions was listed as F, t, z, or P-values; 6) the publication was in English. Meanwhile, the following studies were excluded: 1) studies with region of interest (ROI) approaches and 2) studies not performed using sMRI. Two researchers (SW and ZC) examined the abstract or full text of all searched articles to identify studies that fit the abovementioned criteria. When multiple studies used the same individual cohort, the one with the largest sample size was selected.
Data extraction and quality scores
Two researchers (SW and YH) independently extracted the basic data, including 1) general characteristics such as first author and year of publication; 2) samples’ information such as individuals (schizophrenia or healthy controls), race, sample size, age, and gender; 3) genetic data such as distribution of genotypes and alleles of rs1344706; and 4) imaging data such as tesla of magnetic resonance imaging (MRI) and full width at half-maximum. We also obtained voxel-based data, which included the peak coordinates and effect size of statistically significant differences of brain structure. Missing data were acquired from the corresponding authors of the study by e-mail. If missing data could not be acquired, the studies were excluded. Any disagreement about the data was resolved through group discussions with consensus. All data were checked for internal consistency. The quality of the included studies was evaluated using a checklist that focused on both the clinical and demographic aspects of individual study samples and the imaging-specific methodology used in the studies. We also adopted recently established preprocessing and computational parameters.
AES-SDM analysis
The meta-analysis of sMRI studies was performed using AES-SDM software, which has been previously used in several neuropsychiatric disorders.10 AES-SDM software is a voxel-based meta-analytic approach that enables the use of reported peak coordinates of gray and white matter difference in whole brain studies.11 The AES-SDM method has been previously described.12
Two researchers (SW and YH) performed the meta-analysis. The main threshold was set at uncorrected P<0.001 (empirically equivalent to P<0.05, corrected) with z score >1 (peak height) and cluster extent ≥20 voxels. The default settings in the AES-SDM software were used for other parameters. A leave-one-out jackknife analysis was used to determine the sensitivity of the reported results to the inclusion of individual studies. Heterogeneity among studies was assessed through the Q statistic with a threshold of P<0.05.
CMA
Some studies failed to provide the differential peak coordinates for the AES-SDM analysis. To integrate more data, we further analyzed the differences of brain structure between ZNF804A gene rs1344706 carriers and noncarriers with CMA software. Statistical effect value was set as Z. The heterogeneity of the results was assessed with the I2 statistic using the chi-squared test at a=0.1. When I2≤50%, it was considered that there was no heterogeneity, and the fixed-effect model was used for the meta-analysis. Otherwise, the studies were considered to involve significant heterogeneity, and the random-effect model was used. Publication bias was tested using Egger’s test. Then, P<0.05 was used to denote statistical significance.
Results
Basic information of included studies
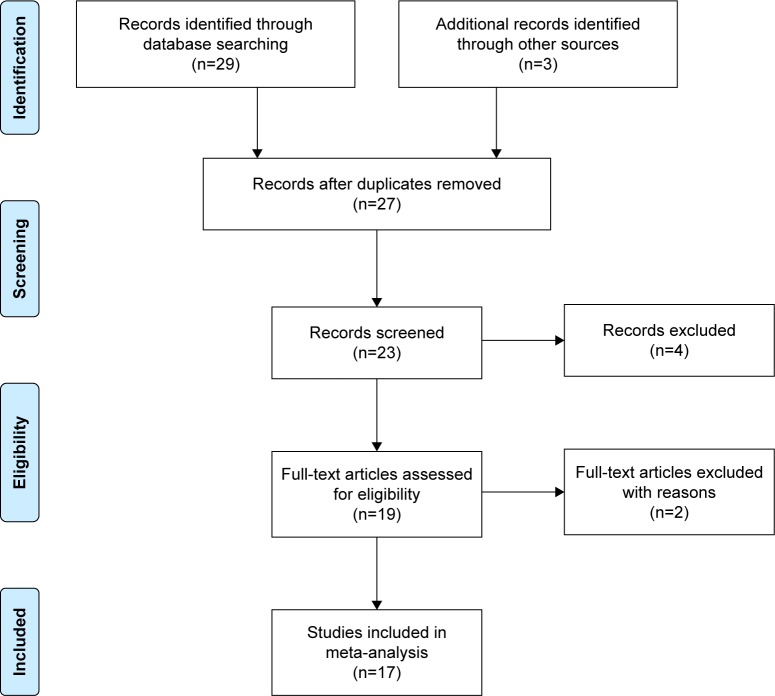
The entire search process is as described in the quorum-type flowchart (Figure 1). Seventeen studies7,8,13–27 on brain structure met the inclusion criteria, and one14 of which involved both gray matter and white matter. The overall sample was equivalent to a cohort of 1,031 schizophrenia patients and 3,416 healthy controls. The general, genotype, and scanning information and quality scores of each study are summarized in Tables 1 and 2.
Figure 1.
Flowchart of the selection process.
Table 1.
General characteristics of the included studies
| Study | Year of publication | Population (n) | Age (years) | Gender (male, n) | Ancestry | Medication | Genotype distribution | |||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | HWE | |||||||
| Gray matter | ||||||||||
| Lencz et al7 | 2010 | HC (39) | 34.4 | 19 | Caucasian | – | 18 | 14 | 7 | Yes |
| Donohoe et al13 | 2011 | SZ (70) | 40.4 | 46 | Caucasian | Yes | 27 | 30 | 13 | Yes |
| HC (38) | 32.5 | 20 | Caucasian | – | 19 | 14 | 5 | Yes | ||
| Voineskos et al14 | 2011 | HC (62) | 37.4 | 40 | Caucasian | – | 23 | 33 | 6 | Yes |
| Cousijn et al15 | 2012 | HC (892) | 22.8 | 369 | Caucasian | – | 297 | 434 | 160 | Yes |
| Wassink et al8 | 2012 | SZ (335) | 31.7 | 248 | Caucasian | Yes | 126 | 165 | 44 | NS |
| HC (198) | 30.7 | 103 | Caucasian | – | 69 | 105 | 24 | NS | ||
| Wei et al16 | 2012 | SZ (80) | 26.6 | 45 | Chinese | Yes | 15 | 46 | 19 | Yes |
| HC (69) | 25.4 | 39 | Chinese | – | 22 | 35 | 12 | Yes | ||
| Bergmann et al17 | 2013 | SZ (82) | 34.2 | 49 | Caucasian | Yes | NS | NS | NS | NS |
| HC (152) | 35.9 | 79 | Caucasian | – | NS | NS | NS | NS | ||
| Schultz et al18 | 2014 | SZ (55) | 29.3 | NS | Caucasian | Yes | 20 | 35 (AC + CC) | NS | |
| HC (40) | 27.3 | NS | Caucasian | – | 15 | 25 (AC + CC) | NS | |||
| Cousijn et al20 | 2015 | HC (1,300) | 22.9 | 43 | Caucasian | – | NS | NS | NS | NS |
| Nenadic et al19 | 2015 | SZ (62) | 31.6 | 44 | Caucasian | Yes | 23 | 33 | 6 | Yes |
| HC (54) | 29.5 | 29 | Caucasian | – | 19 | 28 | 7 | Yes | ||
| Wei et al21 | 2015 | SZ (59) | 26.3 | 31 | Chinese | No | 26 | 16 | 17 | Yes |
| HC (60) | 25.6 | 33 | Chinese | – | 38 | 12 | 10 | Yes | ||
| White matter | ||||||||||
| Voineskos et al14 | 2011 | HC (62) | 37.4 | 40 | Caucasian | – | 23 | 33 | 6 | Yes |
| Kuswanto et al22 | 2012 | SZ (125) | 34.3 | 90 | Chinese | Yes | 26 | 53 | 32 | Yes |
| HC (75) | 31.9 | 48 | Chinese | – | 11 | 37 | 19 | Yes | ||
| Sprooten et al23 | 2012 | HC (50) | 22.7 | 25 | Caucasian | – | 19 | 24 | 7 | Yes |
| Wei et al24 | 2013 | SZ (100) | 26.5 | 53 | Chinese | Yes | 18 | 58 | 24 | Yes |
| HC (69) | 25.4 | 39 | Chinese | – | 22 | 35 | 12 | Yes | ||
| Ikuta et al25 | 2014 | HC (107) | 31.8 | 56 | Caucasian | – | 43 | 42 | 22 | NS |
| Mallas et al26 | 2016 | SZ (63) | 33.8 | 50 | Mixed | Yes | 27 | 28 | 8 | Yes |
| HC (124) | 35.8 | 67 | Mixed | – | 59 | 51 | 14 | Yes | ||
| Zhang et al27 | 2016 | HC (87) | 27.6 | 68 | Chinese | – | 23 | 43 | 21 | Yes |
Abbreviations: HC, healthy control; SZ, schizophrenia.
Table 2.
Scanning characteristics and quality assessment of the included studies
| Study | Year of publication | fMRI information Tesla FWHM (mm) Task Indices
|
Quality | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Gray matter | ||||||
| Lencz et al7 | 2010 | 1.5 | NS | None | GM, WM, CSF | High |
| Donohoe et al13 | 2011 | 1.5 | 8 | None | GM | High |
| Voineskos et al14 | 2011 | 1.5 | NS | None | CT | High |
| Cousijn et al15 | 2012 | 1.5, 3.0 | 8 | None | GM, WM | High |
| Wassink et al8 | 2012 | 1.5 | NS | None | GM, WM, CSF | High |
| Wei et al16 | 2012 | 1.5 | 12 | None | WM | High |
| Bergmann et al17 | 2013 | 1.5 | 30 | None | CT | Medium |
| Schultz et al18 | 2014 | 1.5 | NS | None | CT, CF | Medium |
| Cousijn et al20 | 2015 | 1.5 | NS | None | GM, WM | Medium |
| Nenadic et al19 | 2015 | 1.5 | 12 | None | GM, WM | High |
| Wei et al21 | 2015 | 1.5 | 10 | None | CT, CSA, CV | High |
| White matter | ||||||
| Voineskos et al14 | 2011 | 1.5 | None | None | FA | High |
| Kuswanto et al22 | 2012 | 3 | None | None | FA | High |
| Sprooten et al23 | 2012 | 1.5 | None | None | FA, MD | High |
| Wei et al24 | 2013 | 1.5 | None | None | FA, MD | High |
| Ikuta et al25 | 2014 | 3 | None | None | FA | High |
| Mallas et al26 | 2016 | 1.5 | None | None | FA | High |
| Zhang et al27 | 2016 | 3 | None | None | FA | High |
Abbreviations: fMRI, functional magnetic resonance imaging; FWHM, full width at half-maximum; NS, not stated; GM, gray matter; WM, white matter; CSF, cerebrospinal fluid; CT, cortical thickness; CF, cortical fold; CSA, cortical surface area; CV, cortical volume; FA, fractional anisotropy; MD, mean diffusivity.
Significant genetic–neuroimaging associations in AES-SDM
Six studies on gray matter containing peak coordinates were included in this part of AES-SDM analysis. For healthy controls, rs1344706 risk allele carriers had decreased gray matter in the left superior temporal gyrus (z=−2.299, P=0.00027) and left median network (z=−1.949, P=0.00076) compared with noncarriers. However, for schizophrenia patients, the risk allele carriers had increased gray matter in the right inferior frontal gyrus (z=2.634, P=0.00019) and left superior temporal gyrus (z=2.479, P=0.00033) compared with noncarriers. The AES-SDM results of gray matter are summarized in Table 3 and Figure 2.
Table 3.
Influence of ZNF804A rs1344706 on gray matter (AES-SDM)
| Brain region | Peak coordinate | Cluster | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MNI (x, y, z) | SDM-z | P-value | Voxels | Breakdown (voxels) | |||
|
| |||||||
| Positive effects of rs1344706 risk alleles in HC | |||||||
| None | |||||||
| Negative effects of rs1344706 risk alleles in HC | |||||||
| Left superior temporal gyrus | −60 | 26 | 10 | −2.299 | 0.00027 | 252 | BA21 (93) |
| BA22 (89) | |||||||
| BA22 (25) | |||||||
| Corpus callosum (18) | |||||||
| Left median network, cingulum | −4 | −22 | 34 | −1.949 | 0.00076 | 52 | BA23 (29) |
| Positive effects of rs1344706 risk alleles in SZ | |||||||
| Right inferior frontal gyrus | 44 | 22 | −12 | 2.634 | 0.00019 | 248 | BA38 (102) |
| BA47 (52) | |||||||
| BA48 (29) | |||||||
| BA48 (13) | |||||||
| Left superior temporal gyrus | −54 | −6 | 6 | 2.479 | 0.00033 | 83 | BA48 (59) |
| BA48 (17) | |||||||
| Negative effects of rs1344706 risk alleles in SZ | |||||||
| None | |||||||
Abbreviations: AES-SDM, Anisotropic Effect-Size Signed Differential Mapping; MNI, Montreal Neurological Institute.
Figure 2.
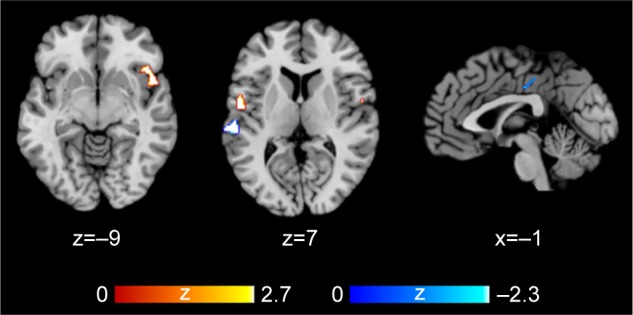
AES-SDM results of the effect of risk allele on gray matter.
Notes: Regions highlighted in red signify increased gray matter in schizophrenia patients carrying risk allele compared with noncarriers. Regions highlighted in blue signify decreased gray matter in healthy individuals carrying risk allele compared with noncarriers.
Abbreviation: AES-SDM, Anisotropic Effect-Size Signed Differential Mapping.
Two studies on white matter containing peak coordinates were included in this part of AES-SDM analysis. For healthy controls, rs1344706 risk allele carriers had decreased white matter integrity in the left median network (z=−1.100, P=0.00005) and corpus callosum (z=−1.099, P=0.00005) compared with noncarriers. However, no study involved schizophrenia patients. The AES-SDM results of white matter are summarized in Table 4 and Figure 3.
Table 4.
Influence of ZNF804A rs1344706 on white matter (AES-SDM)
| Brain region | Peak coordinate
|
Cluster size in voxels | ||||
|---|---|---|---|---|---|---|
| MNI (x, y, z) | SDM-z | P-value | ||||
|
| ||||||
| Positive effects of rs1344706 risk alleles | ||||||
| None | ||||||
| Negative effects of rs1344706 risk alleles | ||||||
| Left median network, cingulum | −16 | 42 | 8 | −1.100 | 0.00005 | 59 |
| Corpus callosum | 2 | 8 | 24 | −1.099 | 0.00005 | 52 |
| 20 | −42 | 36 | −1.101 | 0.00001 | 45 | |
Abbreviations: AES-SDM, Anisotropic Effect-Size Signed Differential Mapping; MNI, Montreal Neurological Institute.
Figure 3.
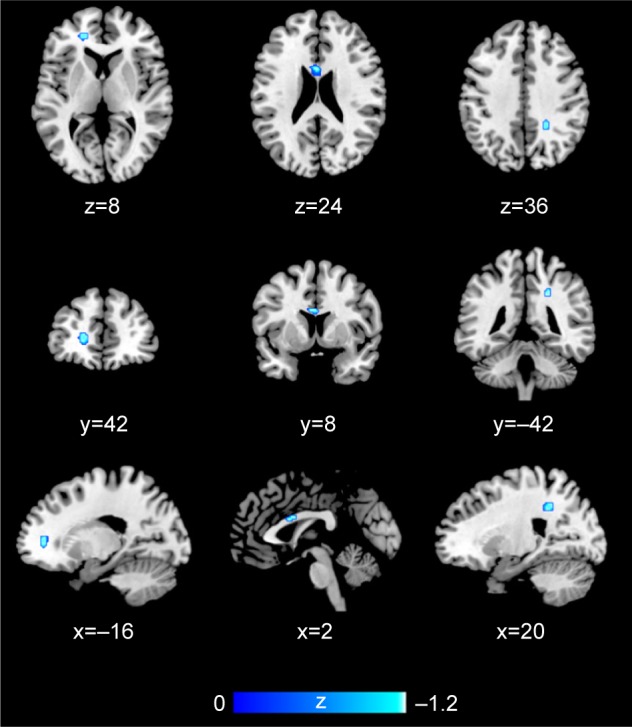
AES-SDM results of the effect of risk allele on white matter.
Note: Regions highlighted in blue signify decreased white matter integrity in healthy individuals carrying risk allele compared with noncarriers.
Abbreviation: AES-SDM, Anisotropic Effect-Size Signed Differential Mapping.
Significant genetic–neuroimaging associations in CMA
Seventeen studies containing effect sizes were included in the CMA analysis. For healthy controls, rs1344706 risk allele carriers had decreased gray matter in the frontal (z=−2.008, P=0.045), temporal (z=−6.099, P=0.000), and occipital (z=−3.106, P=0.002) areas compared with noncarriers. However, for schizophrenia patients, the risk allele carriers had increased gray matter in the frontal (z=3.445, P=0.001), temporal (z=2.140, P=0.032), parietal (z=5.346, P=0.000), occipital (z=3.605, P=0.000), limbic system (z=4.765, P=0.000), and basal nucleus (z=4.689, P=0.000) areas compared with noncarriers. In addition, there was no difference in white matter between the rs1344706 risk allele carriers and noncarriers in both patients and healthy individuals. The CMA results are summarized in Table 5.
Table 5.
Influence of ZNF804A rs1344706 on brain structure (CMA)
| Indices | Region | Population | Model | Effect size | Heterogeneity | Egger’s regression intercept | Publication bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z | P-value | D | 95% CI | I2 | P-value | T | P-value | |||||
| Gray matter | Frontal | HC | Random | −2.008 | 0.045 | −0.740 | −1.463~–0.018 | 89.022 | 0.000 | 15.644 | 0.004 | Yes |
| SZ | Fixed | 3.445 | 0.001 | 0.778 | 0.336~1.221 | 60.166 | 0.057 | 1.185 | 0.358 | No | ||
| Temporal | HC | Fixed | −6.099 | 0.000 | −0.958 | −1.266~–0.650 | 17.898 | 0.301 | 2.967 | 0.097 | No | |
| SZ | Random | 2.140 | 0.032 | 0.302 | 0.025~0.579 | 89.678 | 0.000 | 0.120 | 0.916 | No | ||
| Parietal | SZ | Fixed | 5.346 | 0.000 | 1.124 | 0.712~1.537 | 0.000 | 0.923 | – | – | – | |
| Occipital | HC | Fixed | −3.106 | 0.002 | −0.631 | −1.030~–0.233 | 33.581 | 0.220 | – | – | – | |
| SZ | Fixed | 3.605 | 0.000 | 0.748 | 0.342~1.155 | 54.182 | 0.140 | – | – | – | ||
| Limbic system | HC | Random | −1.485 | 0.138 | −0.900 | −2.087~0.288 | 93.838 | 0.000 | 11.921 | 0.053 | No | |
| SZ | Fixed | 4.765 | 0.000 | 0.939 | 0.553~1.325 | 0.000 | 0.631 | – | – | – | ||
| Basal nuclei | SZ | Fixed | 4.689 | 0.000 | 0.952 | 0.554~1.350 | 0.000 | 0.869 | – | – | – | |
| White matter | All | HC | Random | 0.081 | 0.936 | 0.018 | −0.430~0.467 | 80.609 | 0.000 | 1.325 | 0.256 | No |
| SZ | Random | −0.295 | 0.768 | −0.087 | −0.667~0.492 | 79.300 | 0.008 | 4.468 | 0.140 | No | ||
Abbreviations: CMA, Comprehensive Meta-Analysis; HC, healty control; SZ, schizophrenia.
Discussion
In this meta-analysis, AES-SDM software and CMA software were used to integrate the 17 studies and determine the association between ZNF804A gene rs1344706 polymorphism and brain structure. The present study not only analyzed mental disorders with schizophrenia but also included healthy individuals. In the AES-SDM analysis, only a part of the eligible studies were included due to the limitations of peak coordinates in the original literature. We found that schizophrenia patients carrying the risk allele of rs1344706 in ZNF804A had a larger gray matter in the right inferior frontal gyrus and the left superior temporal gyrus compared with noncarriers. Conversely, healthy controls with risk alleles had a smaller gray matter in the left superior temporal gyrus and left median network. It is important to note that the risk allele of rs1344706 has a completely opposite effect on the left superior temporal gyrus in different populations. In terms of white matter, the AES-SDM meta-analysis concluded that the white matter integrity was lower in healthy controls carrying the risk allele of rs1344706, including the left median network and corpus callosum compared with noncarriers. However, for schizophrenia patients, there are not enough data to conduct this meta-analysis.
To further verify the association, we then analyzed the included studies using the CMA method. This method allows more data (effect size) to be entered in the meta-analysis, but it cannot determine the precise brain area. The final result suggests that the risk allele of rs1344706 carriers in healthy controls had a smaller gray matter in the frontal and temporal lobes. Conversely, for schizophrenia patients, the risk allele carriers had larger gray matter in the frontal lobe, temporal lobe, parietal lobe, occipital lobe, limbic system, and basal nuclei. These results were basically consistent with those obtained using the AES-SDM method. However, the results of CMA did not suggest that the rs1344706 risk allele had any significant association with the white matter.
Compared with previous reports, the results of this meta-analysis are basically consistent with most of the studies. However, the results of the study by Wei et al21 showed that the risk allele had negative effects on brain structure (including cortical thickness, surface area, and volume) in schizophrenia patients. On the contrary, positive effects were found in healthy controls. Similarly, Voineskos et al14 believed that the risk allele of rs1344706 plays a positive role on the gray matter in healthy controls. In addition, Wassink et al8 found that the risk allele of rs1344706 had negative effects on brain structure in both healthy controls and schizophrenia patients. We speculated that the differences might come from the different baselines of the included samples. Furthermore, the ethnic difference of the subjects might contribute to the discriminate distribution frequency of rs1344706 allele and further affected the results of association studies.
The prevailing results of our meta-analysis and previous studies showed that the risk allele of rs1344706 in ZNF804A had opposite effects on the brain structure in mental disorders such as schizophrenia (positive) and healthy individuals (negative). It may be due to different genetic variations or genetic environment in subjects.28,29 For example, Guella et al30 found that the A allele of rs12476147 in exon 4 of ZNF804A is overexpressed in the dorsolateral prefrontal cortex of individuals heterozygous for rs1344706. Furthermore, this allele of rs12476147 could increase the risk of schizophrenia. In the same study, the risk allele of rs1344706 was found to be a cis-regulatory variant that could directly lead to an imbalance of this allele expression in adult cerebral cortex.30 Therefore, the different effects of rs1344706 on schizophrenia patients and healthy individuals may be related to the neuropathological mechanism of this risk variant. The increased gray matter in risk allele carriers may give the suggestion of the risk allele participation in schizophrenia and the brain changes of the patients.
Imaging genetic study is generally designed to assess the impact of specific risk genes (or risk scores) on brain structure. However, it is unclear how gene expression or gene products affect the brain structure and function. Several variants have been forthcoming in schizophrenia, in particular ZNF804A, located in the major histocompatibility complex. The strongest support is ZNF804A, encoding a zinc finger protein of unknown, but possibly regulatory function.31 An increasing number of evidence shows that the genetic effect on the brain begins at the early stages of neural development.32 The rs1344706 polymorphism affects the expression of ZNF804A in the brain during embryonic development, but this effect does not exist in the adult brain.32 However, autopsy studies found that ZNF804A expression is different in schizophrenia patients and healthy controls.33,34 These findings directly or indirectly demonstrate that there is a relationship between ZNF804A expression and brain development. However, the mechanisms of a given polymorphism impacting specific brain regions need further exploration.
Finally, the present meta-analysis has heterogeneity and publication bias in the discussion of some indicators. The greatest source of heterogeneity may come from the studies with opposite data. In addition, differences in the scan parameters and analytical indicators may also increase the heterogeneity. The trim and fill processes did not change the general result (although the strength of the association was slightly attenuated), suggesting that the association is not an artifact of unpublished negative studies. Nevertheless, that possibility is not fully excluded by this method. The unpublished articles in other languages have not been included in this meta-analysis and may result in publication bias. In general, the meta-analysis further confirmed the influence of the risk allele of ZNF804A gene rs1344706 on the brain structure and provided further insight into the genetic-related pathogenesis of psychiatric disorders such as schizophrenia.
Conclusion
Abnormalities on brain structure have a strong relationship with ZNF804A gene rs1344706 polymorphism, but the association may be different in healthy individuals and those with mental disorders. The risk allele of rs1344706 has a positive effect on brain structure in schizophrenia patients. However, this allele has a negative effect on brain structure in healthy individuals.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Chengdu Medical College (grant no. CYZ18-08).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shad MU, Keshavan MS. Neurobiology of insight deficits in schizophrenia: an fMRI study. Schizophr Res. 2015;165(2–3):220–226. doi: 10.1016/j.schres.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maria BL, Neufeld JA, Rosainz LC, et al. Central nervous system structure and function in Sturge-Weber syndrome: evidence of neurologic and radiologic progression. J Child Neurol. 1998;13(12):606–618. doi: 10.1177/088307389801301204. [DOI] [PubMed] [Google Scholar]
- 3.Brosch T, Neumann H. Computing with a canonical neural circuits model with pool normalization and modulating feedback. Neural Computation. 2014;26(12):2735–2789. doi: 10.1162/NECO_a_00675. [DOI] [PubMed] [Google Scholar]
- 4.Hopfield JJ, Tank DW. Computing with neural circuits: a model. Science. 1986;233(4764):625–633. doi: 10.1126/science.3755256. [DOI] [PubMed] [Google Scholar]
- 5.Liddle PF, Morris DL. Schizophrenic syndromes and frontal lobe performance. Br J Psychiatry. 1991;158(03):340–345. doi: 10.1192/bjp.158.3.340. [DOI] [PubMed] [Google Scholar]
- 6.Riley B, Thiselton D, Maher BS, et al. Replication of association between schizophrenia and ZNF804A in the Irish case-control study of schizophrenia sample. Mol Psychiatry. 2010;15(1):29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lencz T, Szeszko PR, Derosse P, et al. A schizophrenia risk gene, ZNF804A, influences neuroanatomical and neurocognitive phenotypes. Neuropsychopharmacology. 2010;35(11):2284–2291. doi: 10.1038/npp.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassink TH, Epping EA, Rudd D, et al. Influence of ZNF804A on brain structure volumes and symptom severity in individuals with schizophrenia. Arch Gen Psychiatry. 2012;69(9):885–892. doi: 10.1001/archgenpsychiatry.2011.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1):1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao B, Wang S, Liu J, Meng T, He Y, Luo X. Abnormalities of localized connectivity in schizophrenia patients and their unaffected relatives: a meta-analysis of resting-state functional magnetic resonance imaging studies. Neuropsychiatr Dis Treat. 2017;13:467–475. doi: 10.2147/NDT.S126678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radua J, Mataix-Cols D, Phillips ML, et al. A new meta-analytic method for neuroimaging studies that combines reported peak coordinates and statistical parametric maps. Eur Psychiatry. 2012;27(8):605–611. doi: 10.1016/j.eurpsy.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Radua J, Mataix-Cols D. Voxel-wise meta-analysis of grey matter changes in obsessive-compulsive disorder. Br J Psychiatry. 2009;195(5):393–402. doi: 10.1192/bjp.bp.108.055046. [DOI] [PubMed] [Google Scholar]
- 13.Donohoe G, Rose E, Frodl T, et al. ZNF804A risk allele is associated with relatively intact gray matter volume in patients with schizophrenia. Neuroimage. 2011;54(3):2132–2137. doi: 10.1016/j.neuroimage.2010.09.089. [DOI] [PubMed] [Google Scholar]
- 14.Voineskos AN, Lerch JP, Felsky D, et al. The ZNF804A gene: characterization of a novel neural risk mechanism for the major psychoses. Neuropsychopharmacology. 2011;36(9):1871–1878. doi: 10.1038/npp.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cousijn H, Rijpkema M, Harteveld A, et al. Schizophrenia risk gene ZNF804A does not influence macroscopic brain structure: an MRI study in 892 volunteers. Mol Psychiatry. 2012;17(12):1155–1157. doi: 10.1038/mp.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Q, Kang Z, Diao F, et al. Association of the ZNF804A gene polymorphism rs1344706 with white matter density changes in Chinese schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2012;36(1):122–127. doi: 10.1016/j.pnpbp.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Ørjan Bergmann, Haukvik UK, Brown AA. ZNF804A and cortical thickness in schizophrenia and bipolar disorder. Psychiatry Research: Neuroimaging. 2013;212(2):154–157. doi: 10.1016/j.pscychresns.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Schultz CC, Nenadic I, Riley B. ZNF804A and cortical structure in schizophrenia: in vivo and postmortem studies. Schizophrenia bull. 2014;40(3):532–541. doi: 10.1093/schbul/sbt123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nenadic I, Maitra R, Basmanav FB, et al. ZNF804A genetic variation (rs1344706) affects brain grey but not white matter in schizophrenia and healthy subjects. Psychol Med. 2015;45(1):143–152. doi: 10.1017/S0033291714001159. [DOI] [PubMed] [Google Scholar]
- 20.Cousijn H, Tunbridge EM, Rolinski M, et al. Modulation of hippocampal theta and hippocampal-prefrontal cortex function by a schizophrenia risk gene. Hum Brain Mapp. 2015;36(6):2387–2395. doi: 10.1002/hbm.22778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Q, Li M, Kang Z, et al. ZNF804A rs1344706 is associated with cortical thickness, surface area, and cortical volume of the unmedicated first episode schizophrenia and healthy controls. Am J Med Genet. 2015;168(4):265–273. doi: 10.1002/ajmg.b.32308. [DOI] [PubMed] [Google Scholar]
- 22.Kuswanto CN, Woon PS, Zheng XB, et al. Genome-wide supported psychosis risk variant in ZNF804A gene and impact on corticolimbic WM integrity in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(3):255–262. doi: 10.1002/ajmg.b.32032. [DOI] [PubMed] [Google Scholar]
- 23.Sprooten E, Mcintosh AM, Lawrie SM, et al. An investigation of a genomewide supported psychosis variant in ZNF804A and white matter integrity in the human brain. Magn Reson Imaging. 2012;30(10):1373–1380. doi: 10.1016/j.mri.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Q, Kang Z, Diao F, et al. No association of ZNF804A rs1344706 with white matter integrity in schizophrenia: a tract-based spatial statistics study. Neurosci Lett. 2013;532:64–69. doi: 10.1016/j.neulet.2012.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikuta T, Peters BD, Guha S, et al. A schizophrenia risk gene, ZNF804A, is associated with brain white matter microstructure. Schizophr Res. 2014;155(1–3):15–20. doi: 10.1016/j.schres.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallas E-J, Carletti F, Chaddock CA, et al. Genome-wide discovered psychosis-risk gene ZNF804A impacts on white matter microstructure in health, schizophrenia and bipolar disorder. PeerJ. 2016;4(10):e1570. doi: 10.7717/peerj.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Chen X, Yu P, et al. Effect of rs1344706 in the ZNF804A gene on the connectivity between the hippocampal formation and posterior cingulate cortex. Schizophr Res. 2016;170(1):48–54. doi: 10.1016/j.schres.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Prata DP, Mechelli A, Fu CHY, et al. Epistasis between the DAT 3′UTR VNTR and the COMT Val158Met SNP on cortical function in healthy subjects and patients with schizophrenia. Proc Natl Acad Sci U S A. 2009;106(32):13600–13605. doi: 10.1073/pnas.0903007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicodemus KK, Law AJ, Radulescu E, et al. Biological validation of increased schizophrenia risk with Nrg1, ErbB4, and Akt1 epistasis via functional neuroimaging in healthy controls. Arch Gen Psychiatry. 2010;67(10):991–1001. doi: 10.1001/archgenpsychiatry.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guella I, Sequeira A, Rollins B, et al. Evidence of allelic imbalance in the schizophrenia susceptibility gene ZNF804A in human dorsolateral prefrontal cortex. Schizophr Res. 2014;152(1):111–116. doi: 10.1016/j.schres.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer-Lindenberg A. Imaging genetics of schizophrenia. Dialogues Clin Neurosci. 2010;12(4):449–456. doi: 10.31887/DCNS.2010.12.4/amlindenberg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am J Psychiatry. 2012;169(12):1301–1308. doi: 10.1176/appi.ajp.2012.11121845. [DOI] [PubMed] [Google Scholar]
- 33.Okada T, Hashimoto R, Yamamori H, et al. Expression analysis of a novel mRNA variant of the schizophrenia risk gene ZNF804A. Schizophr Res. 2012;141(2–3):277–278. doi: 10.1016/j.schres.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Buonocore F, Hill MJ, Campbell CD, et al. Effects of cis-regulatory variation differ across regions of the adult human brain. Hum Mol Genet. 2010;19(22):4490–4496. doi: 10.1093/hmg/ddq380. [DOI] [PMC free article] [PubMed] [Google Scholar]