Abstract
Human milk is a dynamic protein-protease system that delivers bioactive peptides to infants. The pH of milk changes from the mother’s mammary gland to the infant’s digestive tract. Although the release of human milk peptides has been studied during in vivo or in vitro digestion, these models did not explicitly vary nor observe the effect of pH. The objective of this research was to determine the effect of pH on the proteolysis of human milk. Using high-resolution accurate-mass Orbitrap mass spectrometry, profiles of endogenous human milk peptides before and after incubation at various pH levels have been mapped. Over 5000 peptides were identified. Comparative analyses classified 74 peptides that were consistently found independent of pH alterations, and 8 peptides that were released only at pH 4 or 5 (typical infant gastric pH). Results documented that the proteolysis of milk proteins, particularly β-casein, polymeric immunoglobulin receptor, and α-lactalbumin, is pH-dependent.
Keywords: Peptidomics, Milk peptides, Bioactive peptides, Human milk, Protein digestion, Proteolysis, Mass spectrometry
1. Introduction
Under selective pressure throughout mammalian evolution, milk has been instrumental as a nourishing diet in the survival of offspring. Milk proteins have diverse biological functions beyond serving as nutritional sources of amino acids. A subset of the milk proteins consists of substrates, proteases, protease activators, and protease inhibitors, forming a dynamic proteolytic system (Dallas, Murray, & Gan, 2015). With the application of modern analytical tools to biological research, the release of human milk peptides has been followed from the mother’s milk to the infant’s digestive tract in vivo (Dallas, Guerrero, Khaldi, Borghese, Bhandari, Underwood, et al., 2014). Bioinformatic analyses have shown that milk-derived peptides generated in the infant stomach correspond with the cleavage patterns of proteases in human milk rather than pepsin in the infant’s stomach (Holton, Vijayakumar, Dallas, Guerrero, Borghese, Lebrilla, et al., 2014). Scientific discoveries on the dynamic proteolytic system delivered to infants through mothers’ milk are providing new insights into the biological process of selective proteolysis.
Peptides are important in almost all biological systems (Boonen, Creemers, & Schoofs, 2009; Petsalaki & Russell, 2008; Zasloff, 2002). Antimicrobial peptides are evolutionarily ancient weapons for both plants and animals in their defense against a broad range of microbes (Zasloff, 2002). Signaling peptides bind to protein domains and mediate cellular processes, particularly biochemical pathways (Petsalaki & Russell, 2008). Furthermore, bioactive peptides in signaling systems often coordinately function as a network, and cells respond to the integrated peptidome instead of a single peptide (Boonen, Creemers, & Schoofs, 2009). Therefore, the consortium of peptides delivered through milk may function in unison to regulate processes such as the metabolism of different cells and tissues in infants, and to mediate the development of the entire body at multiple sites within the regulatory hierarchy.
The diverse biogeography of the digestive tract affects the landscape of proteins and peptides in each location. The cellular and tissue environment changes dramatically from oral cavity to stomach and intestine, and pH varies significantly along the digestive tract. During ingestion, the environment of milk changes from neutral to acidic when entering the infant’s stomach, and then the pH increases in the intestine (Bourlieu, Menard, Bouzerzour, Mandalari, Macierzanka, Mackie, et al., 2014; Gan, Bornhorst, Henrick, & German, 2018). The typical gastric pH in a newborn infant’s stomach is around 4, but the pH varies from 2 to 6 depending on individual conditions, such as term or preterm birth, developmental stage, and health status (Agunod, Yamaguch, Lopez, Luhby, & Glass, 1969; He, Chen, Li, & Deng, 2017; Maffei & Nobrega, 1975; Mason, 1962; Sondheimer, Clark, & Gervaise, 1985). Fluctuation in pH may affect the ionization state of charged molecules, the structural stability of proteins, the activity of milk enzymes, the microstructure of milk compartments, and the substrate-enzyme molecular interactions within human milk. Therefore, we hypothesized that pH changes the dynamics of the proteolytic system of human milk and mediates the selective proteolysis of milk proteins to release specific peptides. Although recent studies characterized human milk peptides during infant digestion in vivo or in vitro (Dallas, et al., 2014; Su, Broadhurst, Liu, Gathercole, Cheng, Qi, et al., 2017; Wada & Lonnerdal, 2015), these models involved many variables and did not explicitly vary nor observe the effects of pH on human milk proteolysis.
The objective of this study was to determine how pH affects the proteolysis of human milk. Fresh human milk samples were incubated at various levels of physiologically relevant pH, and peptides were mapped using high-resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Identifying how human milk proteins are changed and what peptides are released upon pH alteration is important for understanding the complex biological functions of human milk and guiding innovations to improve infant health.
2. Materials and methods
2.1. Human milk collection
This study was approved by the University of California Davis Institutional Review Board. Human milk samples were provided by four healthy mothers (at 5 days, 2 months, 10 months, and 12 months of lactation, respectively) after informed consent. Each sample was expressed by an electric breast pump, collected into a sterile container, and transported to the laboratory on ice. A fraction of human milk was used immediately for pH adjustment, incubation, and proteolytic bacteria assay without freeze-thaw cycles. The remainder of milk was stored at −80°C for compositional analysis.
2.2. Compositional analysis of human milk
Composition of the four human milk samples was analyzed by the Fourier transform mid-infrared spectroscopy (Delta Instruments LactoScope FTIR Advanced, Advanced Instruments, Norwood, MA, USA). Prior to sample analysis, the instrument was calibrated using previously validated methods (Smilowitz, Gho, Mirmiran, German, & Underwood, 2014). Human milk samples were thawed at 4°C overnight, warmed to 38°C in a water bath and vortexed for 20 seconds to ensure homogenization for the analysis. Three measurements were taken for each sample.
2.3. Proteolytic bacteria assay
To evaluate and control for the impact of proteolytic bacteria in the experiments described below, the presence of proteolytic bacteria in the milk was tested using skim milk agar (Criterion, Hardy Diagnostics, Santa Maria, CA, USA). 100 μL of human milk was spread over the surface of skim milk agar and incubated at 37°C for 24 to 48 hours. Proteolytic bacteria were indicated by clean zones around the colonies (Storrs & Hull, 1956).
2.4. pH adjustment and incubation
Each human milk sample was divided into five aliquots of 500 μL. The first aliquot was kept on ice to serve as a control without pH adjustment or 37°C incubation; 500 μL of 2X protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA) was added to prevent proteolysis in the control group. The four treatment groups were incubated for 2 hours at 37°C under different pH conditions. One treatment group remained at its natural pH of approximately 7. For the other three treatment groups, 1M HCl was used to adjust the pH to 5 (5.0 ± 0.1), 4 (4.0 ± 0.1) and 2 (2.0 ± 0.1), respectively. After incubation, 500 μL of 2X protease inhibitor cocktail was added to stop proteolysis. Mixtures of 1M NaOH and 1M HCl were then added to neutralize the samples and adjust for salt variations. The final volume and amount of materials in each tube were the same after neutralization (500 μL milk, 500 μL protease inhibitor cocktail, 20 μL of 1M HCl, and 20 μL of 1M NaOH) in order to minimize the effect of pH or ion strength on the subsequent peptide extraction and analysis. The experimental procedure is depicted in Fig. 1.
Fig. 1.
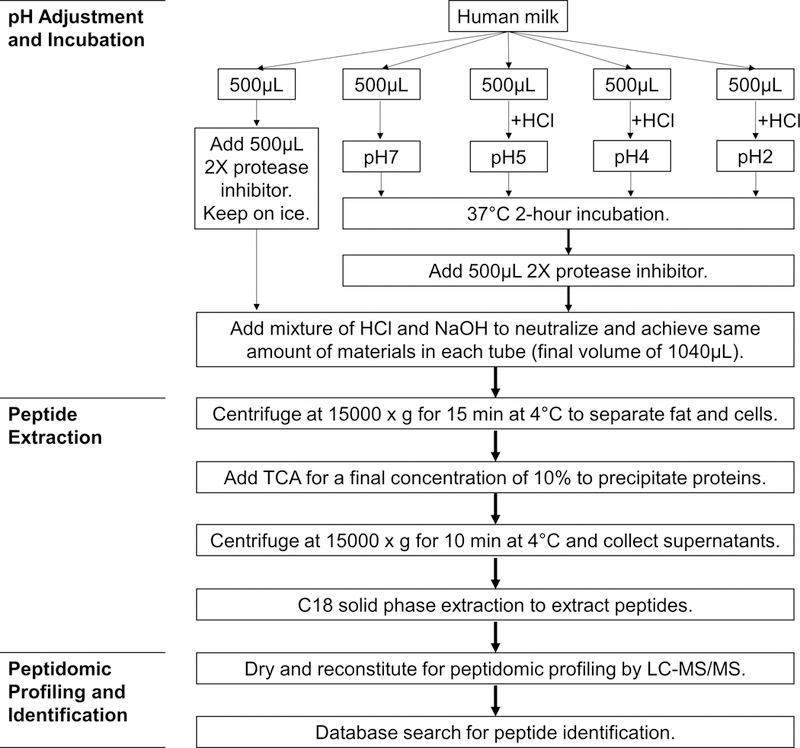
Flowchart depicting the experimental procedure of pH adjustment of human milk and peptide analysis by mass spectrometry. Human milk sample collected from each subject served as a biological replicate. The experimental procedure was applied to each biological replicate.
2.5. Peptide extraction
Samples were centrifuged at 15000 x g for 15 min at 4°C to separate the fat and precipitate the cells in human milk. The aqueous layer was transferred to a new tube without retrieving the cream layer or disturbing the cell pellet. An equal volume of 20% (w/v) trichloroacetic acid (TCA) was added to each sample, and the mixture was kept at 4°C for 10 min to precipitate proteins. After centrifugation at 15000 x g for 10 min at 4°C, the supernatant was purified by C18 solid-phase extraction. The C18 96-well plate (Glygen Corp, Columbia, MD, USA) was activated with 60% acetonitrile in 0.1% formic acid and equilibrated with 0.1% formic acid. Samples were loaded on the plate, and then washed with 0.1% formic acid to remove salts and carbohydrates. Peptides were eluted with 60% acetonitrile in 0.1% formic acid. Eluted peptides were dried in a centrifugal vacuum concentrator (Genevac MiVac Quattro concentrator, Genevac Ltd., Ipswitch, UK) for Orbitrap analysis.
2.6. Peptide quantification
Following extraction, peptide concentration of each sample was measured in duplicate with the Pierce Quantitative Fluorescent Peptide Assay (Thermo Scientific, Rockford, IL, USA). Peptides were labeled by an amine-reactive fluorescent detection reagent, and the fluorescence intensity was detected at Ex390nm/Em475nm with a microplate reader (SpectraMax M5, Molecular Devices, San Jose, CA, USA).
2.7. Peptidomic profiling by LC-MS/MS
LC-MS/MS analysis was performed based on previously published methods for milk samples with slight modifications (Beck, Weber, Phinney, Smilowitz, Hinde, Lonnerdal, et al., 2015). Specifically, samples were resuspended in nanopure water containing 2% acetonitrile and 0.1% formic acid. One microgram of peptides was loaded onto a 100 μm × 25 mm Magic C18 100Å 5U trap column before being separated on a 75 μm × 150 mm Magic C18 200Å 3U reverse phase column. Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in 80% acetonitrile. Peptides were separated over 120 min with a gradient from 0 to 100% solvent B at a flow rate of 3 μL/min. Eluted peptides were analyzed with an ESI source and a Q Exactive Plus Orbitrap mass spectrometer (Thermo Scientific). Mass spectra were collected in data-dependent mode with one precursor scan followed by 15 MS/MS scans. Full-scan MS spectra were acquired with a scan range of 350 to 1600 m/z, resolution of 70000, and target of 1 × 106 ions or maximum injection time of 30ms. MS/MS spectra were acquired with a scan range of 200 to 2000 m/z, resolution of 17500, and target of 5 × 104 ions or maximum injection time of 50ms. MS/MS fragmentation was performed using higher-energy collision-induced dissociation with normalized collision energy of 27. Precursors with unassigned, 1 or >4 charge states were excluded from fragmentation.
2.8. Peptide identification
The sequences of peptides were identified from MS/MS spectra using X!Tandem (Craig & Beavis, 2004) online advanced search against the SwissProt Homo sapiens (Human) proteome (May 2017, 20201 entries). A non-specific enzyme cleavage pattern was defined, and 50 missed cleavage sites were allowed. No complete modifications were set. Oxidation of methionine, deamidation of asparagine and glutamine, as well as phosphorylation of serine and threonine were selected as potential modifications. Mass error tolerance was 40 ppm for precursor ions and 20 ppm for fragment ions. For individual spectra, a peptide match was accepted if the e-value was below 0.01.
2.9. Data analysis
Outputs from X!Tandem were processed computationally using a custom Python script. The program extracts data from multiple X!Tandem xml files and exports as a csv file. Comparison of peptides in different experimental groups and statistical summarization of results were performed in R (version 3.3.3). Peptide concentration of 2-hour incubation samples was normalized by that of the corresponding 0-hour control sample; the normalized results were analyzed by ANOVA for Randomized Block Design, with pH as the treatment factor and milk as the blocking factor. Monoisotopic molecular weight of proteins was computed by ExPASy Compute pI/Mw tool (https://web.expasy.org/compute_pi/); the information was used in a bubble chart to visualize the pattern of proteolysis. Functional classification of identified proteins was achieved with the PANTHER (protein annotation through evolutionary relationship, version 12.0) database (Mi, Huang, Muruganujan, Tang, Mills, Kang, et al., 2017). Sequences of peptides were searched on bioactive peptide databases, including EROP-Moscow (Endogenous Regulatory OligoPeptide knowledgebase, release of Nov 2016, 14511 entries) (Zamyatnin, Borchikov, Vladimirov, & Voronina, 2006), BIOPEP (Minkiewicz, Dziuba, Iwaniak, Dziuba, & Darewicz, 2008), and MBPDB (Milk Bioactive Peptide Database) (Nielsen, Beverly, Qu, & Dallas, 2017).
3. Results and discussion
3.1. Compositional analysis and proteolytic bacteria assay of the human milk samples
The composition of human milk is well studied, and typical macronutrient concentrations are documented; thus, analyses were conducted to verify that the samples used in these experiments were within normal ranges. Results for the four human milk samples were 0.983 ± 0.426% (Mean ± SD) protein, 5.659 ± 5.118% fat, and 6.941 ± 0.674% lactose, which are similar to data from previous studies (Ballard & Morrow, 2013).
To demonstrate the effects of endogenous human milk proteolytic enzymes, contamination by exogenous bacteria that produce proteolytic enzymes needs to be controlled. In this study, milk samples were examined for proteolytic bacteria with skim milk agar. In contrast to raw cow’s milk with high bacterial counts (Storrs & Hull, 1956), freshly expressed human milk from healthy mothers has low viable bacterial counts on agar as detected on non-selective agar media (Jost, Lacroix, Braegger, & Chassard, 2013). No clear zones were observed surrounding the bacterial colonies on skim milk agar in this study, so the milk samples were suitable for addressing the research question.
Milk samples used for compositional analysis were stored at −80°C prior to analysis (Smilowitz, Gho, Mirmiran, German, & Underwood, 2014). However, freeze-thaw cycles may affect milk structure, bacterial survival, and enzyme activity; in addition, proteolysis may occur during thawing. Therefore, fresh milk samples without freeze-thaw cycles were used for the proteolytic bacteria assay and peptide analysis.
3.2. Effects of pH on the amount and profile of human milk peptides
To investigate the effects of pH on the human milk peptides, we measured the concentration of total peptides and mapped the profile of these peptides after incubation at various pH levels. Peptide concentrations in samples after 2-hour incubation were normalized by the corresponding 0-hour control before comparison (Fig. 2A). ANOVA results indicate that the total amounts of peptides between pH treatment groups were not significantly different (p>0.05).
Fig. 2.
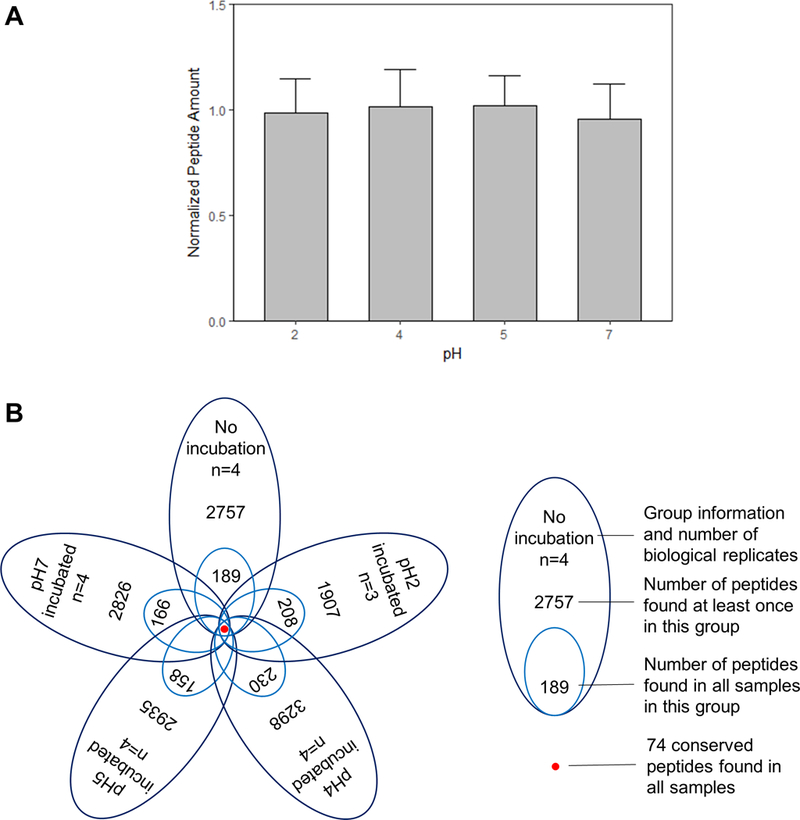
Effects of pH on the total amount (A) and profile (B) of human milk peptides.
Peptidomic profiling was employed with the goal of comprehensive identification of endogenous peptides in milk samples. Using high-resolution accurate-mass Orbitrap detection, we have identified a total of 5847 peptides among all 19 samples. There were 2757 peptides identified in the samples without incubation, and 5496 peptides in the samples after 2 hours of incubation. To reduce the influence of individual variation and focus on the effect of pH, peptides found from all biological replicates in each treatment group were analyzed (Table S1). The number of these common peptides ranged from 158 to 230, which was about 5 – 11% of the total peptides identified in the respective groups (Fig. 2B). One sample of the pH 2 group (collected at 5 days postpartum) had a pH higher than 2.1 after pH adjustment; thus, this sample was excluded from data analysis. Due to this missing data point, the number of peptides found at least once in the pH 2 group was lower than that of other groups.
MS/MS spectra were analyzed by database searching against the human proteome, and all the individual spectrum-to-sequence assignments were validated with statistical confidence (expectation value <0.01). The length of peptides was limited within the range of 6 to 50 amino acid residues for reliable sequence assignments. The current experimental design and acquisition settings of mass spectrometry focused on qualitative characterization of peptides with high confidence and sensitivity, facilitating the discovery of targeted peptides for further quantitation.
3.3. Conserved peptides found in all samples
In the analyzed samples, 74 peptides were found consistently in samples before and after incubation and from all biological replicates (Table 1). These conserved peptides are likely produced in the mammary gland, delivered to infants through milk, and maintaining their structure despite the pH change. To determine whether this number of conserved peptides was substantially lowered by a single influential sample, analyses were performed on the dataset with one sample removed each time, and the number of conserved peptides was calculated for the remaining samples. The average value for conserved peptides with one sample removed was 76 ± 2.49, indicating that the diversity in conserved peptides was similar across samples.
Table 1.
Conserved peptides found in all samples.
| Protein | UniPr ot AC |
Peptide | Pre a | Start b | End c | Post d |
|---|---|---|---|---|---|---|
| Polymeric immunoglobulin receptor | P01833 | AEEKAVADTRDQADGSR | PRLF | 606 | 622 | ASVD |
| AVADTRDQADGSRASVDSG | AEEK | 610 | 628 | SSEE | ||
| AVADTRDQADGSRASVDSGSSEEQGGSS | AEEK | 610 | 637 | RALV | ||
| AVADTRDQADGSRASVDSGSSEEQGGSSR | AEEK | 610 | 638 | ALVS | ||
| AVADTRDQADGSRASVDSGSSEEQGGSSRA | AEEK | 610 | 639 | LVST | ||
| AVADTRDQADGSRASVDSGSSEEQGGSSRALVST | AEEK | 610 | 643 | LVPL | ||
| DGSRASVDSGSSEEQGGSSRA | RDQA | 619 | 639 | LVST | ||
| DQADGSRASVDSGSSEEQGGSSR | ADTR | 616 | 638 | ALVS | ||
| DQADGSRASVDSGSSEEQGGSSRA | ADTR | 616 | 639 | LVST | ||
| DQADGSRASVDSGSSEEQGGSSRA [Q+Deamidated;] | ADTR | 616 | 639 | LVST | ||
| DQADGSRASVDSGSSEEQGGSSRALVST | ADTR | 616 | 643 | LVPL | ||
| DSGSSEEQGGSSRALVST | RASV | 626 | 643 | LVPL | ||
| DTRDQADGSRASVDSGSSEEQGGSSRA | KAVA | 613 | 639 | LVST | ||
| SGSSEEQGGSSRALVST | ASVD | 627 | 643 | LVPL | ||
| SRASVDSGSSEEQGGSSRA | QADG | 621 | 639 | LVST | ||
| SVDSGSSEEQGGSSRA | GSRA | 624 | 639 | LVST | ||
| TRDQADGSRASVDSGSSEEQGGSSRA | AVAD | 614 | 639 | LVST | ||
| VADTRDQADGSRASVDSGSSEEQGGSSR | EEKA | 611 | 638 | ALVS | ||
| VADTRDQADGSRASVDSGSSEEQGGSSRA | EEKA | 611 | 639 | LVST | ||
| VADTRDQADGSRASVDSGSSEEQGGSSRA [Q+Deamidated;] | EEKA | 611 | 639 | LVST | ||
| Fibrinogen alpha chain | P02671 | DSGEGDFLAEGGGVR | AWTA | 21 | 35 | GPRV |
| Beta-casein | P05814 | EKVKHEDQQQGEDEHQDK | KQKV | 37 | 54 | IYPS |
| ESLSSSEESITEYK | RETI | 20 | 33 | QKVE | ||
| ETIESLSSSEESITEYK | ALAR | 17 | 33 | QKVE | ||
| ETIESLSSSEESITEYK [S+Phospho;] | ALAR | 17 | 33 | QKVE | ||
| ETIESLSSSEESITEYK [S+Phospho;S+Phospho;] | ALAR | 17 | 33 | QKVE | ||
| ETIESLSSSEESITEYKQK | ALAR | 17 | 35 | VEKV | ||
| ETIESLSSSEESITEYKQKVEK [S+Phospho;] | ALAR | 17 | 38 | VKHE | ||
| EVPKAKDTVYTK | PEIM | 100 | 111 | GRVM | ||
| IESLSSSEESITEYK [S+Phospho;] | ARET | 19 | 33 | QKVE | ||
| KVEKVKHEDQQQGEDEHQDK | EYKQ | 35 | 54 | IYPS | ||
| LLNPTHQIYPVTQPLAPVHNPISV | NQEL | 203 | 226 | ] | ||
| LNPTHQIYPVTQPLAPVHNPISV | QELL | 204 | 226 | ] | ||
| LNQELLLNPTHQIYPVTQPLAPVHNPISV | QALL | 198 | 226 | ] | ||
| LSSSEESITEYK [S+Phospho;] | TIES | 22 | 33 | QKVE | ||
| LSSSEESITEYKQKVEK [S+Phospho;] | TIES | 22 | 38 | VKHE | ||
| NPTHQIYPVTQPLAPVHNPISV | ELLL | 205 | 226 | ] | ||
| NQELLLNPTHQIYPVTQPLAPVHNPISV | ALLL | 199 | 226 | ] | ||
| PTHQIYPVTQPLAPVHNPISV | LLLN | 206 | 226 | ] | ||
| QKVEKVKHEDQQQGEDEHQD [Q+Ammonia-loss;] | TEYK | 34 | 53 | KIYP | ||
| QKVEKVKHEDQQQGEDEHQDK [Q+Ammonia-loss;] | TEYK | 34 | 54 | IYPS | ||
| QKVEKVKHEDQQQGEDEHQDK [Q+Deamidated;Q+Ammonia-loss;] | TEYK | 34 | 54 | IYPS | ||
| QKVEKVKHEDQQQGEDEHQDKIYP [Q+Ammonia-loss;] | TEYK | 34 | 57 | SFQP | ||
| RETIESLSSSEESITEYK | LALA | 16 | 33 | QKVE | ||
| RETIESLSSSEESITEYK [S+Phospho;] | LALA | 16 | 33 | QKVE | ||
| RETIESLSSSEESITEYK [S+Phospho;S+Phospho;] | LALA | 16 | 33 | QKVE | ||
| RETIESLSSSEESITEYKQK [S+Phospho;] | LALA | 16 | 35 | VEKV | ||
| RETIESLSSSEESITEYKQKVE [S+Phospho;S+Phospho;] | LALA | 16 | 37 | KVKH | ||
| RETIESLSSSEESITEYKQKVEK | LALA | 16 | 38 | VKHE | ||
| RETIESLSSSEESITEYKQKVEK [S+Phospho;] | LALA | 16 | 38 | VKHE | ||
| RETIESLSSSEESITEYKQKVEK [S+Phospho;S+Phospho;] | LALA | 16 | 38 | VKHE | ||
| SEESITEYK | SLSS | 25 | 33 | QKVE | ||
| SLSSSEESITEYK [S+Phospho;] | ETIE | 21 | 33 | QKVE | ||
| SLSSSEESITEYKQKVEK | ETIE | 21 | 38 | VKHE | ||
| SLSSSEESITEYKQKVEK [S+Phospho;] | ETIE | 21 | 38 | VKHE | ||
| SSEESITEYKQK [S+Phospho;] | ESLS | 24 | 35 | VEKV | ||
| SSSEESITEYK | IESL | 23 | 33 | QKVE | ||
| SSSEESITEYKQKVEK [S+Phospho;] | IESL | 23 | 38 | VKHE | ||
| TIESLSSSEESITEYK [S+Phospho;] | LARE | 18 | 33 | QKVE | ||
| TIESLSSSEESITEYK [S+Phospho;S+Phospho;] | LARE | 18 | 33 | QKVE | ||
| VKHEDQQQGEDEHQDK | KVEK | 39 | 54 | IYPS | ||
| VKHEDQQQGEDEHQDKIYPSFQPQP | KVEK | 39 | 63 | LIYP | ||
| Osteopontin | P10451 | DIQYPDATDEDITSH | FRRP | 178 | 192 | MESE |
| DQSAETHSHKQSRLY [S+Phospho;] | SQLD | 232 | 246 | KRKA | ||
| SHELDSASSEVN [S+Phospho;] | KFRI | 303 | 314 | ] | ||
| Mucin-1 | P15941 | NPAVAATSANL | LSYT | 1245 | 1255 | ] |
| TNPAVAATSANL | SLSY | 1244 | 1255 | ] | ||
| Bile salt-activated lipase | P19835 | EGGFVEGVNKK | AVYT | 29 | 39 | LGLL |
| Alpha-S1-casein | P47710 | EKQTDEIKDTR | NILR | 58 | 68 | NEST |
| RLQNPSESSEPIPLESREEYMNGMN [M+Oxidation;M+Oxidation;] | RYPE | 26 | 50 | RQRN | ||
| YPERLQNPSESSEPIP | LPLR | 23 | 38 | LESR | ||
| Butyrophilin subfamily 1 member A1 | Q13410 | DADTLHSKLIPTQPSQGAP | SAPR | 508 | 526 | ] |
| DGREQEAEQMPEYR [M+Oxidation;] | LVHR | 79 | 92 | GRAT | ||
| DGREQEAEQMPEYRG [M+Oxidation;] | LVHR | 79 | 93 | RATL | ||
Amino acid residues before the starting position
Starting position of peptide in the complete protein sequence
Ending position of peptide in the complete protein sequence
Amino acid residues after the ending position
Among the 74 conserved peptides, sequences that matched entries in the bioactive peptide databases were mainly related to antimicrobial activities. The peptide YPERLQNPSESSEPIP from αS1-casein (residues 23–38) in this dataset is part of the antimicrobial peptide RPKLPLRYPERLQNPSESSEPIP (residues 16–38), which was produced by chymosin treatment of casein and active against Staphylococcus aureus (Lahov & Regelson, 1996). Peptides at the C-terminal end of human β-casein LLNPTHQIYPVTQPLAPVHNPISV (residues 203–226), LNPTHQIYPVTQPLAPVHNPISV (residues 204–226), NPTHQIYPVTQPLAPVHNPISV (residues 205–226), and PTHQIYPVTQPLAPVHNPISV (residues 206–226) are part of QELLLNPTHQIYPVTQPLAPVHNPISV (residues 200–226). This peptide was produced by hydrolysis of human sodium caseinate with a partially purified protease of Lactobacillus helveticus PR4, and has shown a broad spectrum of inhibition against Gram-positive and Gram-negative bacteria (Minervini, Algaron, Rizzello, Fox, Monnet, & Gobbetti, 2003). The unmatched peptides may exert biological functions that have not been explored at present.
3.4. Unique peptides released at infant gastric pH
Eight unique peptides with 12 to 29 amino acid residues were found in samples at pH 4 and 5 (typical infant gastric pH) that did not appear in the samples before incubation or after incubation at other pH levels (Table 2). The criteria to screen these unique peptides were strict in this study. First, identical modifications on the amino acid residues were required; second, the occurrence of peptides in samples at the corresponding pH condition was 100% across biological replicates; third, these peptides were not identified in any samples under other conditions. These conservative criteria were set to obtain a high certainty that the release of these peptides is related to pH, and to reduce the probability of false positives.
Table 2.
Unique peptides released at pH 4 and 5.
| Condition | Protein | UniProt AC | Peptide | Pre a | Start b | End c | Post d |
|---|---|---|---|---|---|---|---|
| pH4 | Alpha-lactalbumin | P00709 | HTSGYDTQAIVENNESTEYGL | CTMF | 51 | 71 | FQIS |
| IVENNESTEYGL | DTQA | 60 | 71 | FQIS | |||
| Beta-casein | P05814 | SVPQPKVLPIPQQVVPYPQRAVPVQALLL | QPLW | 170 | 198 | NQEL | |
| Osteopontin | P10451 | SYETSQLDDQSAETHSHKQSR [T+Phospho;] | RGKD | 224 | 244 | LYKR | |
| pH5 | Beta-casein | P05814 | LSSSEESITEYKQK [S+Phospho;] | TIES | 22 | 35 | VEKV |
| pH4&pH5 | Apolipoprotein A-I | P02647 | LSALEEYTKKLNTQ | KVSF | 254 | 267 | ] |
| SALEEYTKKLNTQ | VSFL | 255 | 267 | ] | |||
| Beta-casein | P05814 | PVTQPLAPVHNPIS | HQIY | 212 | 225 | V] | |
Amino acid residues before the starting position
Starting position of peptide in the complete protein sequence
Ending position of peptide in the complete protein sequence
Amino acid residues after the ending position
During ingestion, the environment of milk shifts from neutral to acidic. This alteration in pH may change the structure of milk and activate specific proteolytic enzymes, which could subsequently trigger proteolytic processing of other proteins, leading to changes in their structure and function. Development of gastric secretion takes years in early life; the pH in an infant’s stomach is generally higher than that of adults (Agunod, Yamaguch, Lopez, Luhby, & Glass, 1969; Klumpp & Neale, 1930). The unique peptides that are released only at infant-relevant pH have potential health implications. For example, the sequence PVTQPLAPVHNPIS (residues 212–225) from β-casein had an approximate match in all three bioactive peptide databases with QELLLNPTHQIYPVTQPLAPVHNPISV (residues 200–226), which is an antimicrobial peptide discussed above. No exact matches were found for these peptides among current entries in the bioactive peptide databases; further analyses are needed to explore their functions.
3.5. Source proteins of the identified peptides
The 5847 peptides identified in this study were derived from 516 proteins. The molecular functions of these proteins and their coding genes based on the PANTHER classification system are shown in Fig. 3. Common peptides found in all biological replicates of each group (Table S1) were derived from 29 proteins. The PANTHER analysis results of these 29 proteins are organized in Table S2. Among the 29 proteins, binding (GO:0005488) was annotated for 9 proteins, and transporter activity (GO:0005215) was annotated for 3 proteins.
Fig. 3.
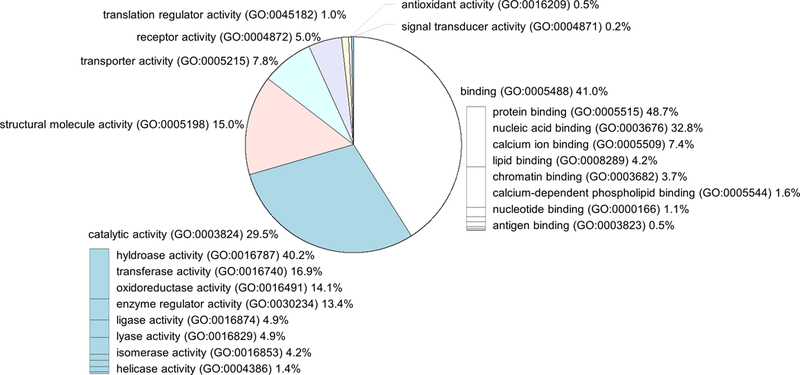
Molecular function of source proteins.
To elucidate the effect of pH on the proteolysis of individual human milk proteins, unique peptides from each protein were counted for every sample. The peptide counts from the 3 biological replicates of mature milk present in all treatment groups were averaged, and the top 10 proteins with highest averaged peptide count were plotted in a bubble chart (Fig. 4). Most of the identified peptides originated from β-casein, polymeric immunoglobulin receptor, and osteopontin. Our data show that the size of source proteins did not correlate with the occurrence of peptides. Instead, pH alteration changed the profile of peptides released from these proteins. Particularly, pH 4 incubation enhanced the diversity of peptides derived from β-casein and polymeric immunoglobulin receptor.
Fig. 4.
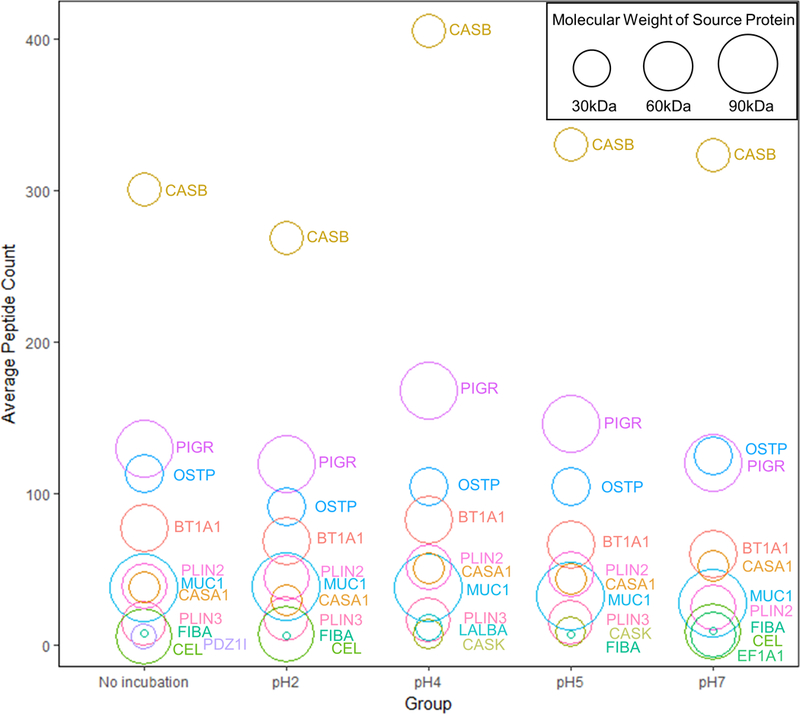
Bubble chart of average peptide count for source proteins. Each circle represents a protein (BT1A1: Butyrophilin subfamily 1 member A1; CASA1: Alpha-S1-casein; CASB: Beta-casein; CASK: Kappa-casein; CEL: Bile salt-activated lipase; EF1A1: Elongation factor 1-alpha 1; FIBA: Fibrinogen alpha chain; LALBA: Alpha-lactalbumin; MUC1: Mucin-1; OSTP: Osteopontin; PDZ1I: PDZK1-interacting protein 1; PIGR: Polymeric immunoglobulin receptor; PLIN2: Perilipin-2; PLIN3: Perilipin-3). The position of the circle indicates the average count of unique peptides released from this protein among biological replicates (n=3) in each group. The area of the circle is proportional to the size (molecular weight) of the protein.
The dominance of β-casein derived peptides has been reported in undigested human milk as well as after in vitro and in vivo digestion (Dallas, et al., 2014; Su, et al., 2017; Wada & Lonnerdal, 2015). One might speculate a correlation between the number of peptide sequences and the abundance of their source protein. However, our data show that the abundance of a source protein was not a major determinant of the count of its peptides. The most abundant proteins in human milk are β-casein (0.04 to 4.42 g/L), α-lactalbumin (2.75 to 3.72 g/L), lactoferrin (0.97 to 2.91 g/L), secretory IgA (0.22 to 0.61 g/L), and lysozyme (0.03 to 0.35 g/L) (Ballard & Morrow, 2013; Huang, Kailemia, Goonatilleke, Parker, Hong, Sabia, et al., 2017; Ke, Chen, Pan, Zhang, Mo, & Ren, 2016; Liao, Weber, Xu, Durbin-Johnson, Phinney, & Lonnerdal, 2017). Interestingly, peptides from the second most abundant human milk protein α-lactalbumin were not significantly released except for the pH 4 treatment group in this study. These results indicate that pH is a factor that regulates the dynamics of the proteolytic system in human milk and controls the release of peptides from α-lactalbumin.
Polymeric immunoglobulin receptor was the second major source of the released peptides in human milk. Following proteolytic processing, this protein yields a secretory component that mediates the epithelial transport of secretory IgA and the immune responses at mucosal surfaces (Kaetzel, 2005). The conserved peptides AVADTRDQADGSRASVDSGSSEEQGGSSRA (residues 610–639) and AVADTRDQADGSRASVDSGSSEEQGGSSR (residues 610–638) identified in our study were also found in human reflex tear fluid (Hayakawa, Landuyt, Baggerman, Cuyvers, Lavigne, Luyten, et al., 2013). It is likely that these peptides are important in mucosal immunity, interacting with the mucosal immune system in the oral cavity and the gastrointestinal tract.
Osteopontin was the third major source for human milk peptides. Christensen et al. have shown the milk proteases plasmin and cathepsin D are able to cleave osteopontin near its integrin-binding motifs (Christensen, Schack, Klaning, & Sorensen, 2010). Those cleavage sites account for many of the osteopontin-derived sequences identified in our study.
3.6. Discussion on the study model
Lactation is essential for the success of mammalian evolution, and the role of milk proteins in the regulation of growth and development of the infant is beyond simply providing nutrients (Lefèvre, Sharp, & Nicholas, 2010). Peptides released from milk proteins have been proposed to be intermediate products during protein digestion that further break down to serve as sources of amino acids; they can also be the products of proteolytic processing that are critical for physiological regulation (Neurath, 1989). In previous research studies, discoveries of bioactive peptides often involved the explicit hydrolysis of proteins using experimentally administered exogenous enzymes to yield peptides for subsequent screening. Here, this study provides a novel strategy to interrogate the release of peptides due to the protease-protein interactions within the milk system. Without the addition of exogenous enzymes, it becomes possible to discover the products of controlled proteolysis within milk and to reveal their natural functions in biology.
This study was designed to explicitly investigate the effect of pH variation on the proteolysis of the human milk system. A limitation of the study model is that the simplified incubation conditions could not represent all physiological conditions during in vivo digestion. The proteolysis of human milk during infant digestion is affected by many factors, such as dynamic changes of pH, amounts and activities of digestive enzymes, diverse biogeography along the digestive tract, as well as developmental stage and health status of the infant. Currently, this process is not well understood, and variables that are important for the regulation of proteolytic processing in human milk may have yet to be identified. Characterization of key controlling factors is needed to improve our understanding of the regulation process.
4. Conclusions
Peptides are revealing diverse functions in biology. Human milk is a dynamic system containing proteins and proteases, and is a vehicle that delivers numerous functional peptides to infants. During the ingestion of human milk, pH is a critical environmental change that affects the proteolysis of milk proteins. Our data showed that pH alteration did not significantly change the total concentration of peptides in milk, but did change the profiles of peptides released from specific proteins, such as β-casein, polymeric immunoglobulin receptor, osteopontin, and α-lactalbumin. Results illustrated that the effect of pH on the proteolytic system of human milk is related to selected proteolysis of targeted proteins to release specific peptides, rather than assisting protein breakdown in general for nutritional purposes. Comparative analyses of treatment groups classified unique collections of peptides with potential bioactivities. These findings have extended the scientific knowledge on the diversity of bioactive peptides and begun the difficult task of interrogating the regulation of proteolysis of human milk.
Supplementary Material
Common peptides found from all biological replicates in each treatment group.
PANTHER analysis results of source proteins in Table S1.
Highlights.
Human milk contains a dynamic proteolytic system.
Selective proteolysis of milk proteins into specific peptides was pH-dependent.
Seventy-four human milk peptides were consistently found despite pH alterations.
Eight peptides were released only at pH 4 or 5 (typical infant gastric pH).
Total concentration of peptides was not significantly changed by pH alterations.
Acknowledgements
The authors appreciate the mothers who donated milk for this research. We thank Tony Herren and Brett Phinney at the UC Davis Proteomics Core for mass spectrometer operation. We thank Tianxiao Gu and Hao Fu at the UC Davis Department of Computer Science for developing custom Python programs. This project was financially supported by the National Institutes of Health [grant numbers AT007079, AT008759], the UC Davis Jastro-Shields Research Scholarship (J. Gan) and Provost’s Undergraduate Fellowship (J. Wang, N. Krishnakumar).
Footnotes
Conflict of interest
None.
Appendix. Supplementary material
References
- Agunod M, Yamaguch N, Lopez R, Luhby AL, & Glass GBJ (1969). Correlative study of hydrochloric acid, pepsin, and intrinsic factor secretion in newborns and infants. American Journal of Digestive Diseases, 14(6), 400–414. [DOI] [PubMed] [Google Scholar]
- Ballard O, & Morrow AL (2013). Human milk composition: nutrients and bioactive factors. Pediatric Clinics of North America, 60(1), 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck KL, Weber D, Phinney BS, Smilowitz JT, Hinde K, Lonnerdal B, Korf I, & Lemay DG (2015). Comparative proteomics of human and macaque milk reveals species-specific nutrition during postnatal development. Journal of Proteome Research, 14(5), 2143–2157. [DOI] [PubMed] [Google Scholar]
- Boonen K, Creemers JW, & Schoofs L (2009). Bioactive peptides, networks and systems biology. Bioessays, 31(3), 300–314. [DOI] [PubMed] [Google Scholar]
- Bourlieu C, Menard O, Bouzerzour K, Mandalari G, Macierzanka A, Mackie AR, & Dupont D (2014). Specificity of infant digestive conditions: some clues for developing relevant in vitro models. Critical Reviews in Food Science and Nutrition, 54(11), 1427–1457. [DOI] [PubMed] [Google Scholar]
- Christensen B, Schack L, Klaning E, & Sorensen ES (2010). Osteopontin is cleaved at multiple sites close to its integrin-binding motifs in milk and is a novel substrate for plasmin and cathepsin D. Journal of Biological Chemistry, 285(11), 7929–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, & Beavis RC (2004). TANDEM: matching proteins with tandem mass spectra. Bioinformatics, 20(9), 1466–1467. [DOI] [PubMed] [Google Scholar]
- Dallas DC, Guerrero A, Khaldi N, Borghese R, Bhandari A, Underwood MA, Lebrilla CB, German JB, & Barile D (2014). A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. Journal of Nutrition, 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Murray NM, & Gan J (2015). Proteolytic systems in milk: perspectives on the evolutionary function within the mammary gland and the infant. Journal of Mammary Gland Biology and Neoplasia, 20(3), 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J, Bornhorst GM, Henrick BM, & German JB (2018). Protein digestion of baby foods: study approaches and implications for infant health. Molecular Nutrition & Food Research, 62(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa E, Landuyt B, Baggerman G, Cuyvers R, Lavigne R, Luyten W, & Schoofs L (2013). Peptidomic analysis of human reflex tear fluid. Peptides, 42, 63–69. [DOI] [PubMed] [Google Scholar]
- He L, Chen W, Li Y, & Deng M (2017). Gastric residual volume linked to gastric fluid pH in infants with very low birth weight. American Journal of Nursing Science, 6(4), 366. [Google Scholar]
- Holton TA, Vijayakumar V, Dallas DC, Guerrero A, Borghese RA, Lebrilla CB, German JB, Barile D, Underwood MA, Shields DC, & Khaldi N (2014). Following the digestion of milk proteins from mother to baby. Journal of Proteome Research, 13(12), 5777–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Kailemia MJ, Goonatilleke E, Parker EA, Hong QT, Sabia R, Smilowitz JT, German JB, & Lebrilla CB (2017). Quantitation of human milk proteins and their glycoforms using multiple reaction monitoring (MRM). Analytical and Bioanalytical Chemistry, 409(2), 589–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T, Lacroix C, Braegger C, & Chassard C (2013). Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. British Journal of Nutrition, 110(7), 1253–1262. [DOI] [PubMed] [Google Scholar]
- Kaetzel CS (2005). The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunological Reviews, 206, 83–99. [DOI] [PubMed] [Google Scholar]
- Ke X, Chen Q, Pan XD, Zhang JS, Mo WM, & Ren YP (2016). Quantification of lactoferrin in breast milk by ultra-high performance liquid chromatography-tandem mass spectrometry with isotopic dilution. RSC Advances, 6(15), 12280–12285. [Google Scholar]
- Klumpp TG, & Neale AV (1930). The gastric and duodenal contents of normal infants and children: the duodenal enzyme activity and the gastric and duodenal reactions (H-ion). American Journal of Diseases of Children, 40(6), 1215–1229. [Google Scholar]
- Lahov E, & Regelson W (1996). Antibacterial and immunostimulating casein-derived substances from milk: casecidin, isracidin peptides. Food and Chemical Toxicology, 34(1), 131–145. [DOI] [PubMed] [Google Scholar]
- Lefèvre CM, Sharp JA, & Nicholas KR (2010). Evolution of lactation: ancient origin and extreme adaptations of the lactation system. Annual Review of Genomics and Human Genetics, 11, 219–238. [DOI] [PubMed] [Google Scholar]
- Liao YL, Weber D, Xu W, Durbin-Johnson BP, Phinney BS, & Lonnerdal B (2017). Absolute quantification of human milk caseins and the whey/casein ratio during the first year of lactation. Journal of Proteome Research, 16(11), 4113–4121. [DOI] [PubMed] [Google Scholar]
- Maffei HVL, & Nobrega FJ (1975). Gastric pH and microflora of normal and diarrhoeic infants. Gut, 16(9), 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S (1962). Some aspects of gastric function in the newborn. Archives of Disease in Childhood, 37(194), 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi HY, Huang XS, Muruganujan A, Tang HM, Mills C, Kang D, & Thomas PD (2017). PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research, 45(D1), D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minervini F, Algaron F, Rizzello CG, Fox PF, Monnet V, & Gobbetti A (2003). Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Applied and Environmental Microbiology, 69(9), 5297–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkiewicz P, Dziuba J, Iwaniak A, Dziuba M, & Darewicz M (2008). BIOPEP database and other programs for processing bioactive peptide sequences. Journal of AOAC International, 91(4), 965–980. [PubMed] [Google Scholar]
- Neurath H (1989). Proteolytic processing and physiological regulation. Trends in Biochemical Sciences, 14(7), 268–271. [DOI] [PubMed] [Google Scholar]
- Nielsen SD, Beverly RL, Qu Y, & Dallas DC (2017). Milk bioactive peptide database: a comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chemistry, 232, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E, & Russell RB (2008). Peptide-mediated interactions in biological systems: new discoveries and applications. Current Opinion in Biotechnology, 19(4), 344–350. [DOI] [PubMed] [Google Scholar]
- Smilowitz JT, Gho DS, Mirmiran M, German JB, & Underwood MA (2014). Rapid measurement of human milk macronutrients in the neonatal intensive care unit: Accuracy and precision of Fourier transform mid-infrared spectroscopy. Journal of Human Lactation, 30(2), 180–189. [DOI] [PubMed] [Google Scholar]
- Sondheimer JM, Clark DA, & Gervaise EP (1985). Continuous gastric pH measurement in young and older healthy preterm infants receiving formula and clear liquid feedings. Journal of Pediatric Gastroenterology and Nutrition, 4(3), 352–355. [DOI] [PubMed] [Google Scholar]
- Storrs AB, & Hull ME (1956). Proteolytic enzymes in human and cow’s milk. Journal of Dairy Science, 39(8), 1097–1103. [Google Scholar]
- Su MY, Broadhurst M, Liu CP, Gathercole J, Cheng WL, Qi XY, Clerens S, Dyer JM, Day L, & Haigh B (2017). Comparative analysis of human milk and infant formula derived peptides following in vitro digestion. Food Chemistry, 221, 1895–1903. [DOI] [PubMed] [Google Scholar]
- Wada Y, & Lonnerdal B (2015). Bioactive peptides released from in vitro digestion of human milk with or without pasteurization. Pediatric Research, 77(4), 546–553. [DOI] [PubMed] [Google Scholar]
- Zamyatnin AA, Borchikov AS, Vladimirov MG, & Voronina OL (2006). The EROP-Moscow oligopeptide database. Nucleic Acids Research, 34, D261–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M (2002). Antimicrobial peptides of multicellular organisms. Nature, 415(6870), 389–395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Common peptides found from all biological replicates in each treatment group.
PANTHER analysis results of source proteins in Table S1.


