Abstract
Protein kinases achieve substrate selective phosphorylation through their conformational flexibility and dynamic interaction with the substrate. Designing substrate selective or kinase selective small molecule inhibitors remains a challenge because of a lack of understanding of the dynamic mechanism by which substrates are selected by the kinase. Using a combination of all-atom molecular dynamics simulations and FRET sensors, we have delineated an allosteric mechanism that results in interaction among the DFG motif, G-loop, and activation loop and structurally links the nucleotide and substrate binding interfaces in protein kinase Cα and three other Ser/Thr kinases. ATP-competitive staurosporine analogues engage this allosteric switch region located just outside the ATP binding site to displace substrate binding to varying degrees. These inhibitors function as bitopic ligands by occupying the ATP binding site and interacting with the allosteric switch region. The conserved mechanism identified in this study can be exploited to select and design bitopic inhibitors for kinases.
To date, most small molecule kinase inhibitors (SMKIs) are designed to outcompete ATP binding through high-affinity interactions with the kinase catalytic domain. These type I ATP-competitive inhibitors often lack kinase selectivity because they target the highly conserved ATP binding site.1 The off-target effects when using such inhibitors become undesirable for the treatment of diseases.2 In contrast, type II allosteric SMKIs bind to a site topographically distinct from the ATP binding pocket and show higher selectivity but typically have lower binding affinity, thereby reducing their efficacy in cells.3–5 The combined strengths of type I and II inhibitors can be realized by bitopic inhibitors that simultaneously target the orthosteric ATP binding site and proximal allosteric sites.4,5 However, the challenge in designing bitopic inhibitors is the identification of allosteric sites that are proximal to the ATP binding site. In this study, we have identified an allosteric site that is proximal to the ATP binding site and demonstrated that n-propyl amine derivatives of staurosporine, commonly known as bisindolylmaleimide (Bim) derivatives, occupy both the ATP binding site and the proximal allosteric site to function as bitopic ligands for protein kinase Cα (PKCα). The Bim derivatives inhibit binding of both ATP and peptide substrates. The combination of ATP displacement and triggering a conserved allosteric switch proximal to the ATP binding site that inhibits substrate binding makes them bitopic.
The kinase−substrate interaction is inherently dynamic, making it challenging to characterize using conventional structural biology techniques.6 Consequently, the structural dynamics of this interaction remain poorly understood, especially with regard to the allosteric mechanisms in the kinase catalytic domain that influence selective substrate binding and phosphorylation. We have recently demonstrated the combined strength of a novel FRET sensor technology and torsional molecular dynamics (MD) simulations7 to iteratively dissect the substrate binding interface of PKCα.8 In this study, we have used the combined approach to identify an allosteric switch region near the ATP binding site and shown that the allosteric switch region can be engaged to regulate inhibition of both ATP and substrate binding. This allosteric switch is conserved across many kinases, thus opening up the possibility of utilizing it to design bitopic inhibitors for kinases.
The staurosporine derivatives have been studied extensively for ATP-competitive inhibition of substrate phosphorylation. We recently demonstrated that BimI, a staurosporine derivative, inhibits binding of both ATP and substrate peptides to the PKCα catalytic domain.9 In this work, FRET measurements of a range of nucleotide and staurosporine analogues reveal a systematic correlation between inhibitor structure and substrate displacement. Combining FRET measurements with MD simulation analysis, we uncover an allosteric switch region located outside the ATP binding site. We demonstrate that BimI contacts this region to function as a bitopic inhibitor.
An Allosteric Switch Regulates the Kinase Conformation Compatible with Substrate Binding.
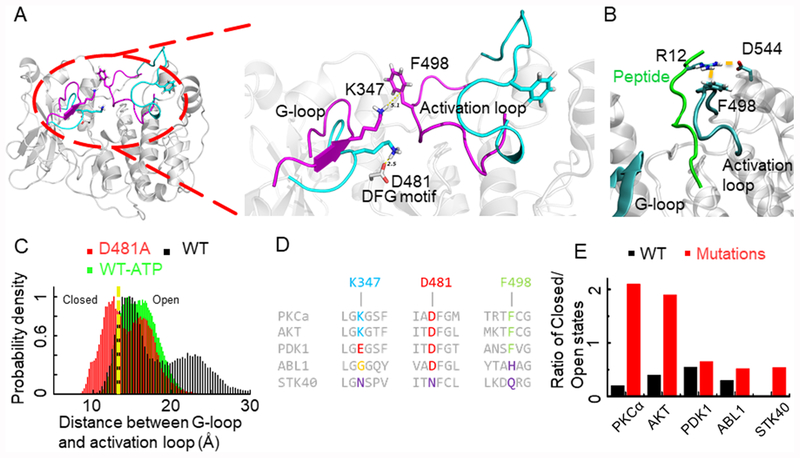
Using all-atom molecular dynamics (MD) simulations, we examined the conformational dynamics of the catalytic domain of PKCα in the apo form, in the ATP-bound state, and with several inhibitors bound (Supporting Information, Methods). The starting conformation for the MD simulations is the phosphorylated form with the DFG-in conformation. Two major conformational states were observed during the simulations, in the apo and ATP-bound simulations. The two conformational states were characterized on the basis of the relative positions of the glycine-rich G-loop, the activation loop, and the DFG motif (Figure 1A). We observed a “closed” conformation with an increased proximity of activation loop and the G-loop, as shown in Figure 1A in magenta. In this closed conformation, K347 in the G-loop comes close to F498 in the activation loop, forming a cation−π interaction (Figure 1A, inset). Previously, we have shown that the residues in the activation loop interact with the peptide substrate and form the floor of the substrate binding site in PKCα for 14 different peptides.8 A basic residue (K/R), three amino acids C-terminal to the phosphorylated Ser/Thr in the EGFR substrate, forms a strong electrostatic contact with D544 and a cation−π interaction with F498 in the activation loop (Figure 1B). However, if these residues in the activation loop interact with residues in the G-loop forming the closed state, they are no longer available for substrate binding. Thus, the closed state does not favor substrate binding (as described in section 1.4 of the Supporting Information). The other distinct conformational state of the kinase domain populated in our dynamics is the “open” state. In the open conformation, the activation loop is farther from the G-loop as shown in Figure 1A in cyan. The interaction between K347 and F498 is not formed because the K347 in the G-loop is engaged in an ionic lock with D481 of the DFG motif. This leaves the activation loop in an open conformation that enables substrate binding. Thus, interactions between K347 and D481 or K347 and F498 form the basis for the open and closed conformations observed in the kinase domain.
Figure 1.
(A) Representative structure of PKCα showing closed (purple) and open (cyan) conformations. The inset shows the ionic lock between K347 and D481. (B) Average binding conformation of the peptide substrate in which R12 in the C-terminus of the peptide substrate interacts with F498 in the activation loop and D544. (C) Distance distribution histogram for wild-type PKCα, bound with ATP, and the D481A mutant system. The yellow dotted line demarcates the closed (left) and open (right) states. (D) Sequence alignment of the G-loop, DFG motif, and part of the activation loop showing conservation in multiple kinases. (E) Ratio of closed to open states for PKCα, PDK1, AKT, STK40, and wild-type ABL1 protein (black) and the D481 equivalent residue mutated to A (red).
We quantified the relative population of open and closed conformations during the MD simulations using methods described in the section 1.3 of the Supporting Information. The population distribution of the PKCα apo form shows a bimodal distribution, with a small population in the closed conformation (black histogram in Figure 1C). The same distribution for PKCα with ATP bound shows a shift toward the open conformation (green histogram in Figure 1C) because the γ-PO4 group of ATP also engages K347 in an ionic lock, facilitating an open conformation for the activation loop. To confirm the significance of these interactions with respect to kinase conformation, simulations were next performed with D481 mutated to Ala. Disrupting the K347− D481 interaction through the D481A mutation caused the transition of the kinase to a closed conformation as evidenced by the enhanced proximity of the G-loop and activation loop (Figure 1C; black vs red histogram). However, the K347-D481-F498 triad is generally not conserved across a range of protein kinases (Figure 1D). Nonetheless, mutation of the D481A equivalent residue in Akt, Pdk1, Ab1, and Stk40 leads to a significant increase in the level of the closed state in MD simulations (Figure 1E). Hence, while the open and closed conformations are likely conserved in multiple kinases, the stabilization of these states is likely to involve mechanisms beyond the triad.
Role of K347 and D481 in the Design of Inhibitors with Bitopic Properties.
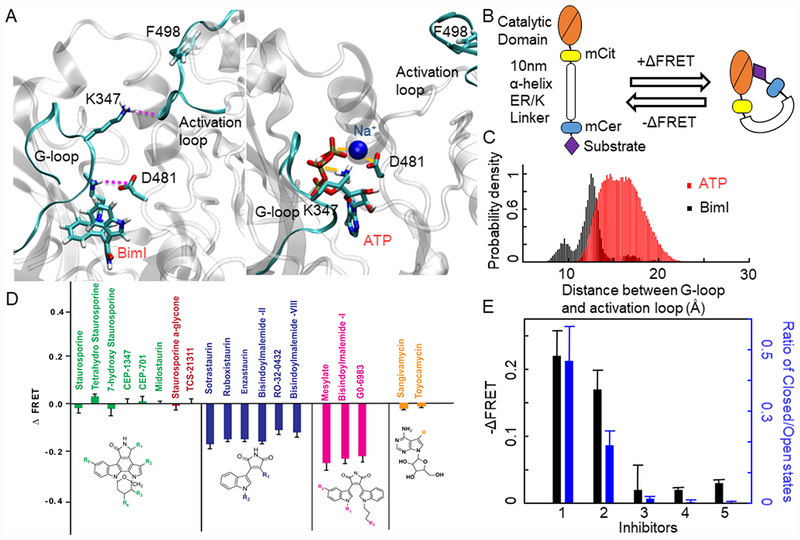
We used a FRET sensor technique termed systematic protein affinity strength modulation (SPASM) that can probe weak protein−protein interactions.10–12 The SPASM sensor construct we used in this study (shown in Figure 2B) involves the fusion of the substrate peptide at the N-terminus of the catalytic domain of PKCα.8,9 We have previously demonstrated that the potent ATP-competitive PKC inhibitor BimI displaces substrate binding to the catalytic domain of PKCα.9 To test a potential intersection between BimI and the allosteric switch mechanism involving K347−D481, we used MD simulations to examine the conformational dynamics of PKCα in the presence of BimI. The amine group at the end of the n-propyl chain of BimI makes a salt bridge with D481 to disrupt the D481−K347 interaction (Figure 2A). This leaves the K347 free to interact with the F498 of the activation loop. Consequently, the presence of BimI stabilizes the interaction between the G-loop and activation loop, resulting in a significant shift in population toward the closed state conformation (Figure 2C). Because BimI stabilizes the closed state, it inhibits peptide substrate binding as shown in Figure 2D. The allosteric effect of BimI, combined with its established ATP-competitive action, demonstrates that it functions as a bitopic ligand with dual effects on ATP and substrate binding.
Figure 2.
(A) Representative structure of BimI-PKCα. The amine group in BimI interacts with D481, thus breaking the ionic lock between D481 and K347. The inset shows γ-PO4 bridging D481 and the backbone of K347, leading to more open states. (B) FRET construct used. (C) Probability density of the distance between the G-loop and activation loop when BimI and ATP bound in PKCα. (D) ΔFRET measured for multiple inhibitors. These inhibitors can be categorized into several groups: pink for inhibitors with the most negative ΔFRET, blue for inhibitors with moderate negative ΔFRET, and green and yellow for inhibitors with no effect on ΔFRET. (E) Ratio of closed to open states when multiple inhibitors bound with PKCα and their corresponding −ΔFRET values. The inhibitors are (1) BimI, (2) sotrastaurin, (3) staurosporine, (4) sangivamycin, and (5) toyocamycin.
We have previously shown that BimI disrupts the interaction between the regulatory and catalytic domains of PKCα.11 The regulatory domain of PKCα has a pseudosubstrate domain that suppresses basal kinase activity by competitively inhibiting binding of the substrate to the catalytic domain.13 Given the substrate-competitive effects of BimI, we tested the effect of BimI on just the pseudosubstrate−catalytic domain interaction (Figure S1). In accordance with its effects on the substrate, BimI disrupts whereas ATP, ADP, and the nucleotide analogue inhibitor sangivamycin enhance the pseudosubstrate−catalytic domain interaction. The paradoxical release of the regulatory−catalytic domain interaction triggered by BimI is consistent with the enhanced translocation of PKCα to the plasma membrane of cells pretreated with this kinase inhibitor.14
BimI belongs to a broader class of staurosporine analogues that have found widespread use as effective kinase inhibitors.1,2,15 To understand the generality of the bitopic mechanism observed in BimI, we used SPASM sensors to profile the substrate-competitive behavior of 17 staurosporine analogues (Figure 2D). The SPASM sensors detect the strength of the PKCα−EGFR substrate peptide interaction using FRET.9 Displacement of substrate binding results in a decrease in the intensity of the FRET signal relative to that of the apo state (negative ΔFRET). We have previously established that changes in the FRET intensity ratio (ΔFRET) correlate linearly with the mole fraction of kinase bound to the substrate within the sensor.10 Hence, the larger the ΔFRET, the stronger the effect of the inhibitor on substrate binding. Interestingly, the magnitude of ΔFRET, which reflects on substrate binding to the kinase, correlates with the nature of the chemical groups that are attached to inhibitors. Mesylate, BimI, and Go6983, which have an extended n-propyl chain with a terminal amine group on the bisindole template, show similar, large negative ΔFRET.Sotrastaurin and related compounds shown in Figure 2B show medium ΔFRET and therefore moderate inhibition of substrate binding. On the other hand, staurosporine analogues have a negligible inhibitory effect on substrate binding as reflected by no discernible change in ΔFRET (Figure 2D). In addition, the nucleotide analogue inhibitors toyocamycin and sangivamycin have negligible effects on substrate binding, attesting to a unique bitopic mechanism for BimI analogues. The binding of these inhibitors to the PKCα catalytic domain at the concentrations used in the FRET assay is evident in their competitive inhibition of ATP consumption in a standard kinase assay (Figure S2). We next tested whether the differential effects of the staurosporine analogues stem from their competitive displacement of substrate binding by measuring the relative population of the catalytic domain in the closed versus open state in the presence of these inhibitors. We found a direct correlation between the stabilization of the closed conformation (ratio of closed to open states) and substrate displacement as measured by the change in ΔFRET (Figure 2E). The ratio of the population of the closed to the open conformation states directly reflects the relative free energy of these two states. The open state is more favored in all the systems studied here. However, Bim1 favors the closed state slightly less than the open state. Other inhibitors favor the open state much more than the closed state. We performed further analysis on how often the system with various inhibitors bound adopts the closed conformation compared to the open state. Figure S3 shows the time series plots of the open versus closed state for various inhibitor-bound PKCα forms. It is evident from this figure that Bim1-bound PKCα favors a longer residence time in the closed state during the MD simulations compared to other inhibitors. In summary, if the chemical group on the inhibitor is capable of forming a salt bridge with D481 as in BimI and derivatives, this leads to a higher ratio of the population of the closed to open states and thereby inhibits substrate binding. Taken together, these data demonstrate an inhibitor structure-dependent bitopic mechanism for PKCα.
This study advances the concept of bitopic inhibition of protein kinases through the dual displacement of ATP and substrate binding to the catalytic domain. ATP-competitive kinase inhibitors rely on their high potency (IC50) for a selected kinase or kinase family to achieve selective inhibition of a signaling pathway in cells. Given the high degree of conservation of the kinase catalytic domain, utilizing the allosteric switch region just outside the ATP binding site would advance drug discovery efforts toward bitopic kinase inhibitors.
Supplementary Material
Funding
This research was funded in part by the National Institute of Health (NIH) Director’s New Innovator Award (1DP2 CA186752–01) and NIH MIRA award (1R35GM126940–01) to S.S. The work performed in N.V.’s laboratory was funded by NIH Grant R01GM097261.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00729.
Experimental procedures, computational procedures, kinase activity assay results, and root-mean-square deviations for simulation trajectories (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Mackay HJ, and Twelves CJ (2007) Targeting the protein kinase C family: are we there yet? Nat. Rev. Cancer 7, 554–562. [DOI] [PubMed] [Google Scholar]
- (2).Mochly-Rosen D, Das K, and Grimes KV (2012) Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discovery 11, 937–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dar AC, and Shokat KM (2011) The Evolution of Protein Kinase Inhibitors from Antagonists to Agonists of Cellular Signaling. Annu. Rev. Biochem 80, 769–795. [DOI] [PubMed] [Google Scholar]
- (4).Fabbro D (2015) 25 Years of Small Molecular Weight Kinase Inhibitors: Potentials and Limitations. Mol. Pharmacol 87, 766–775. [DOI] [PubMed] [Google Scholar]
- (5).Keov P, López L, Devine SM, Valant C, Lane JR, Scammells PJ, Sexton PM, and Christopoulos A (2014) Molecular Mechanisms of Bitopic Ligand Engagement with the M 1 Muscarinic Acetylcholine Receptor. J. Biol. Chem 289, 23817–23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mayer BJ (2015) The discovery of modular binding domains: building blocks of cell signalling. Nat. Rev. Mol. Cell Biol 16, 691–698. [DOI] [PubMed] [Google Scholar]
- (7).Vaidehi N, and Jain A (2015) Internal coordinate molecular dynamics: a foundation for multiscale dynamics. J. Phys. Chem. B 119, 1233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lee S, Devamani T, Song HD, Sandhu M, Larsen A, Sommese R, Jain A, Vaidehi N, and Sivaramakrishnan S (2017) Distinct structural mechanisms determine substrate affinity and kinase activity of protein kinase Cα. J. Biol. Chem 292, 16300–16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Sommese RF, and Sivaramakrishnan S (2016) Substrate affinity differentially influences protein kinase C regulation and inhibitor potency. J. Biol. Chem 291, 21963–21970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sivaramakrishnan S, and Spudich JA (2011) Systematic control of protein interaction using a modular ER/K -helix linker. Proc. Natl. Acad. Sci. U. S. A 108, 20467–20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Swanson CJ, and Sivaramakrishnan S (2014) Harnessing the unique structural properties of isolated α-helices. J. Biol. Chem 289, 25460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ritt M, Guan JL, and Sivaramakrishnan S (2013) Visualizing and manipulating focal adhesion kinase regulation in live cells. J. Biol. Chem 288, 8875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Steinberg SF (2008) Structural Basis of Protein Kinase C Isoform Function. Physiol. Rev 88, 1341–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Swanson CJ, Ritt M, Wang W, Lang MJ, Narayan A, Tesmer JJ, Westfall M, and Sivaramakrishnan S (2014) Conserved modular domains team up to latch-open active protein kinase Cα. J. Biol. Chem 289, 17812–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wilkinson SE, Parker PJ, and Nixon JS (1993) Isoenzyme specificity of bisindolylmaleimides, selective inhibitors of protein kinase C. Biochem. J 294, 335–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.