Abstract
Whole-genome sequencing (WGS) has facilitated the first genome-wide evaluations of the contribution of de novo noncoding mutations to complex disorders. Using WGS, we assess genetic variation from 7,608 samples in 1,902 autism spectrum disorder (ASD) families, identifying 255,106 de novo mutations. In contrast to coding mutations, no noncoding functional annotation category, analyzed in isolation, is significantly associated with ASD. Casting noncoding variation in the context of a de novo risk score across multiple annotation categories, however, does demonstrate association with mutations localized to promoter regions. The strongest driver of this promoter signal emanates from evolutionarily conserved transcription factor binding sites distal to the transcription start site. These data suggest that de novo mutations in promoter regions, characterized by evolutionary and functional signatures, contribute to ASD.
De novo mutations play an important role in human disorders that impair reproductive fitness, including autism spectrum disorder (ASD) (1), severe developmental delay (2), epileptic encephalopathy (3), and a spectrum of congenital anomalies (4, 5). Analysis of de novo mutations in the 1.5% of the genome that encodes proteins has identified numerous genes associated with ASD (1), and these findings have provided a foundation from which to interrogate ASD etiology (6–9). The contribution of de novo variation in the 98.5% of sequence comprising the noncoding genome remains largely unknown (10, 11). Identifying noncoding variants that regulate gene function could provide important insights into when, where, and in which cell type ASD pathology occurs, which could have broad implications for targeted therapeutics (10).
Targeted sequencing of highly evolutionarily conserved loci in 7,930 families with a child affected by severe developmental delay identified a modest contribution from de novo mutations at loci that are active in the fetal brain (12). Whole-genome sequencing (WGS) represents the next critical step in such explorations, enabling the contribution of noncoding de novo mutations to be evaluated systematically across the genome; however, the multiplicity of hypotheses that can be tested in an unbiased screen requires careful consideration of statistical interpretation. WGS analyses to date of up to 519 families with a child affected by ASD have yet to identify a significant noncoding contribution from de novo mutations, after appropriate correction for the multiple comparisons necessary in genome-wide analyses (13–16).
WGS analyses are complicated by the sheer scale of the noncoding genome and limited methods to predict functional regions and disruptive variants. The category-wide association study (CWAS) framework applies multiple annotation methods to define thousands of annotation categories, each of which are tested for association with ASD. This CWAS approach is similar to that used in a genome-wide association study, with single nucleotide polymorphisms (SNPs) substituted for annotation categories, and uses similar correction for multiple comparisons (15, 17). The CWAS-defined categories can also be used to build a de novo risk score, akin to a polygenic risk score, by selecting multiple annotation categories in a training cohort for assessment in a testing cohort (15). This model is generated once, so it does not incur a multiple testing penalty. Here, we apply these methods to 1,902 families; our results demonstrate an association between de novo noncoding mutations and ASD that is driven by mutations in conserved promoter regions.
Identification of de novo mutations in 1,902 families
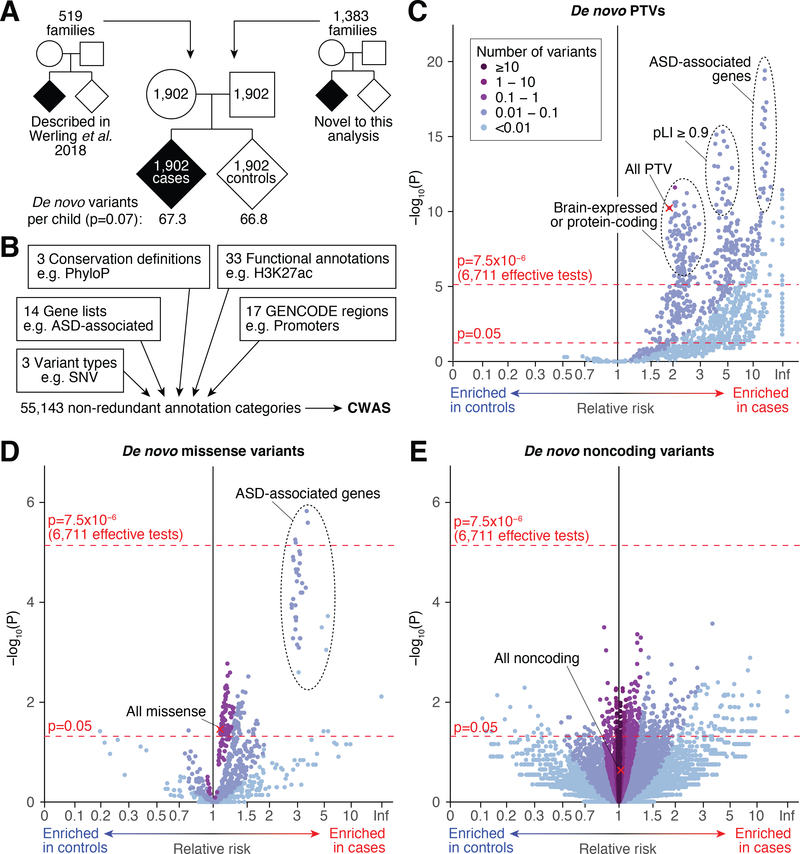
We present an analysis of WGS in 7,608 samples from 1,902 quartet families from the Simons Simplex Collection (18), composed of a mother and father, a child affected by ASD, and an unaffected sibling (Table S1). This family-based design enables the detection of newly arising de novo mutations that are rare, but can have dramatic effects, and a direct comparison between ASD cases and their unaffected siblings as controls. By comparing each affected and unaffected child to their parents, 255,106 de novo mutations were identified in 1,902 families (Fig. 1A; Table S2), with 61.5 de novo single nucleotide variants (SNVs) and 5.6 de novo insertion/deletions (indels, ≤50bp) per child, using a high-quality variant filter defined in our previous study (15). These mutation rates are similar to those reported previously (Fig. S1). Independent experimental validation confirmed 97.1% of SNVs (238/245) and 82.7% of indels (148/179) (19). No difference in noncoding de novo rate was observed between cases and controls after correcting for the established correlation between parental age and de novo frequency (20) (corrected relative risk (cRR)=1.005; p=0.15 by permutation of case-control labels; Table S3; Fig. S2). Ancestry was not a significant predictor of de novo mutation rate, thus it was not included in this correction (Fig. S3 and S4).
Figure 1. Category-wide association study on 1,902 ASD families.
A) De novo mutations were identified in 7,608 samples from 1,902 quartet families, each including an ASD case and an unaffected sibling control. The mean genome-wide mutation rate, corrected for paternal age, is shown for cases and controls. B) Each mutation was annotated against 70 annotation terms in five groups, combinations of which defined 55,143 annotation categories (Table S3, Fig. S5). C) A category-wide association study (CWAS) shows the degree to which de novo protein-truncating variants (PTVs) in each category (points) are enriched in cases (right x-axis) or controls (left x-axis) against the statistical evidence for this enrichment (y-axis). Red lines show the threshold for nominal significance (p=0.05) and significance after correction for 6,711 effective tests (19). The red ‘X’ shows the category of all PTVs without other annotations. The equivalent CWAS is shown for: D) de novo missense; and E) de novo noncoding variants. Statistical tests: B-D) Binomial exact test, two-tailed.
Only protein-coding categories show genome-wide enrichment in cases
In coding regions, ASD-associated mutations are found at a small number of critical loci, e.g. protein truncating variants (PTVs) in ~5% of genes (21). In the absence of an equivalent definition for critical noncoding loci, we annotated the mutations against gene definitions, ASD-associated gene lists, species conservation, types of mutation, and functional annotations (e.g. ChIP-Seq, ATAC-Seq, DNase-Seq), to define 55,143 annotation categories (Fig. 1B, Fig. S5, and Table S3). Considering each category separately in a category-wide association study (CWAS), 579 categories reached our correction threshold of 7.5×10−6, generated by Eigen decomposition of 20,000 simulated datasets (15). All 579 categories were enriched in cases rather than controls; 575 of these included de novo PTV mutations (cRR=1.92; p=2.9×10−11, binomial; Fig. 1C), and the remaining four categories are subsets of missense mutations in genes previously associated with ASD (cRR=2.90; p=5.7×10−6; Fig. 1D and Fig. S6). No noncoding categories reached the correction threshold (Fig. 1E). We note that many of the ASD-associated genes were identified by de novo PTVs, and to a lesser extent de novo missense mutations, in these same cases (1). To focus on classes of variation with more subtle impacts on ASD risk, all annotation categories that included PTVs were excluded from further analysis.
Previous analyses have used WGS data to screen the genome, but described analyses restricted to “candidate” noncoding categories, selected based on assumptions about functional impact as opposed to unbiased genome-wide analyses, in cohorts ranging from 39 to 516 ASD families (13, 14, 22). While these candidate categories were enriched at nominal significance in ASD cases in those initial discovery cohorts, no candidate categories reach nominal significance in this larger cohort, despite similar mutations rates (Table S4). Similarly, we did not observe enrichment of mutations in ASD cases in the conserved noncoding elements described with targeted sequencing of 6,239 families with severe developmental delay (12), though we note that our replication cohort is both substantially smaller than, and a different phenotype to, the discovery cohort.
Analysis across multiple noncoding categories highlights the role of promoters
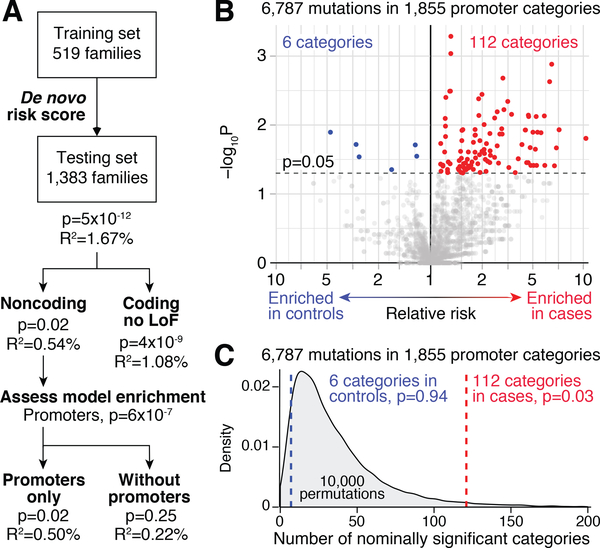
While no single noncoding annotation category passed our threshold of significance (Fig. 1E), we further explored the data by building a de novo risk score (15) to identify groups of categories in an unsupervised genome-wide analysis. To generate the score, we first restricted to annotation categories with a relatively small number of de novo mutations (19). This thresholding step is critical because the presence of numerous de novo mutations in an annotation category could represent false negatives in parents (i.e. apparent de novo mutations were actually inherited variants), highly mutable regions, regions with limited impact on natural selection, or categories covering large swathes of the genome; none of these possibilities are likely to enrich for ASD risk at a small number of critical loci. Next, to select annotations likely to be important for risk from the remaining annotations, a risk score was generated using a Lasso regression from 519 families, described previously (15), to identify annotation categories with rates of mutations that distinguish cases from controls. The resulting risk score was composed of 238 annotations categories, each with a coefficient reflecting the contribution of the category to the score (Table S5). Applying the risk score to 1,383 new families revealed it to be a significant predictor of case status (R2=1.67%; p=5×10−12; Fig. 2A). Of the 238 annotation categories, 75 were in coding regions (R2=1.08%; p=4×10−9; Table S5), while 163 were noncoding (R2=0.54%; p=0.02; Table S5), demonstrating a noncoding contribution of de novo mutations to ASD risk.
Figure 2. Enrichment of conserved promoters in cases.
A) After excluding categories with PTVs, which are known to have a strong contribution to ASD, a de novo risk score was generated using Lasso regression to distinguish cases and controls in the first 519 families and tested on 1,383 new families. The same risk score was tested considering 163 noncoding categories only and, based on the enrichment of promoter categories in the risk score, for 45 promoter categories and 118 noncoding categories without promoters (Table S5). B) Considering 1,855 promoter annotation categories with ≥7 mutations, 118 reached nominal significance, 112 of which had an excess of mutations in cases. C) The observation of 112 nominally significant case-enriched categories (red line) and 6 control-enriched categories (blue line) in B is compared to permuted expectation (grey distribution). Statistical tests: A) Lasso regression with permutation testing. B) Binomial, two-sided. C) Permutation testing.
To understand the nature of this noncoding contribution, we assessed the relative frequencies of the individual annotation terms from which the 163 noncoding categories are composed. The three annotation terms most frequently selected were PhastCons-defined (23) evolutionary conserved regions (68/163 categories), PhyloP-defined (24) evolutionary conserved nucleotides (49/163 categories), and promoter regions, defined as 2 kilobases (kb) upstream of the transcription start site (TSS) (45/163 categories). The inclusion of 45 promoter categories in the model is enriched 2.45-fold over expectation (p=6×10−7 after correcting for 62 noncoding annotation terms; Fig. 2A; Table S5). The risk score remained a significant predictor of case status with only these promoter categories included and accounted for the majority of the noncoding signal (R2=0.50%; p=0.01; Fig. 2A; Table S5). In contrast, the remaining 118 noncoding categories, without promoters, were not significant predictors of case status (R2=0.22%; p=0.25; Fig. 2A). The 45 promoter categories selected in the risk score encompassed 150 independent mutations, 112 in cases and 38 in controls (Table S6).
To examine whether this promoter signal was detectable beyond these 150 mutations, we considered the pattern of de novo mutation enrichment across all 1,855 promoter-defined annotation categories with ≥7 mutations. Of these, 112 are enriched in cases at nominal significance, which is more than expected (cross-category burden p=0.03; Fig. 2B, 2C), unlike the 6 categories enriched at nominal significance in controls (cross-category burden p=0.94; Fig. 2B, 2C). Ten of the 112 case-enriched categories were also selected for inclusion in the de novo risk score, compared to no control-enriched categories.
Promoter association is driven by evolutionary conservation
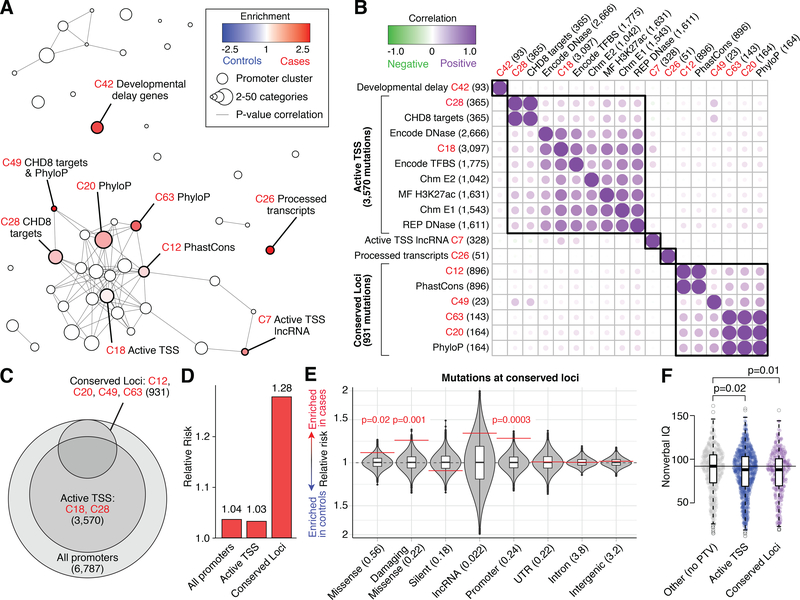
To understand the types of variants and genes that account for this association between promoter mutations and ASD, we performed an exploratory analysis of the 6,787 promoter region mutations and the 1,310 promoter annotation categories with at least 20 mutations. Considering the correlation of p-values across annotation categories, on the basis of 10,000 simulations (19), we identified 47 clusters, each composed of multiple highly-correlated categories (Fig. 3A and Table S7). Using the DAWN hidden Markov random field model (25) to refine the evidence for association based on the strength of association in neighboring clusters, nine of the 47 clusters were identified at a Bayesian false discovery rate of 0.01 (Fig. 3A; Table 1).
Figure 3. Mapping ASD association within promoter regions by annotation terms.
A) DAWN uses p-value correlations between 1,310 promoter categories with ≥20 mutations to define 47 clusters (nodes, with size representing the number of categories in the cluster). Evidence for ASD association is evaluated in the context of the local p-value correlation network (edges) to estimate false discovery rate (FDR). Enrichment is shown by color for the nine clusters with FDR≤0.01 (Table 1). B) The number of de novo mutations shared between these nine clusters and the annotation terms enriched in these clusters, is shown as a correlation with hierarchical clustering. The black boxes show the first five divisions based on hierarchical clustering with two large groups: Active Transcription Start Site (TSS) and Conserved Loci. The numbers of de novo mutations in each group are shown in parentheses. C) The size and relationship of the groups of promoter mutations identified in ‘A’ and ‘B’, based on de novo mutation counts. The number of mutations in each group is shown in parentheses. D) Estimates of relative risk based on the number of de novo mutations in cases and controls within each group. E) Considering mutations at Conserved Loci, the degree of enrichment of mutations in cases vs. controls (red line) is shown in relation to permuted expectation (grey distributions). The mean number of mutations per child is shown in parentheses on the left. Nominally significant uncorrected p-values are shown in red. F) Distribution of nonverbal IQ in cases with mutations at Active TSS (blue) and Conserved Loci (purple) promoters vs. cases with neither (grey). Cases with de novo PTVs were excluded from all groups. Statistical tests: A) DAWN. E) Permutation testing. F) Wilcoxon signed rank, two-sided. Boxplot in E and F shows the median (black line), interquartile range (white box), and a further 1.5 times the interquartile range (whiskers). DD: developmental delay; MF: Midfetal; REP: Roadmap Epigenome; TSS: Transcription Start Site; UTR: Untranslated Region.
Table 1.
Groups and clusters of categories within promoter regions.
| Cluster | Description | Active TSS | Conserved Loci | CHD8 binding targets | Total mutations (Case/Control) | Absolute RR | Binomial p-value | DAWN RR | DAWN p-value |
|---|---|---|---|---|---|---|---|---|---|
| C7 | Active TSS lncRNAs | 98% | 18% | 0% | 328 (178/150) | 1.19 | 0.14 | 1.66 | 0.03 |
| C12 | PhastCons | 59% | 100% | 8% | 896 (495/401) | 1.23 | 0.002 | 1.22 | 0.003 |
| C18 | Active TSS | 100% | 16% | 10% | 3,097 (1,600/1,497) | 1.07 | 0.07 | 1.1 | 0.03 |
| C20 | PhyloP | 82% | 100% | 14% | 164 (100/64) | 1.56 | 0.006 | 1.48 | 0.03 |
| C26 | Processed transcripts | 57% | 20% | 0% | 51 (34/17) | 2 | 0.02 | 2.39 | 0.009 |
| C28 | CHD8 targets | 100% | 21% | 100% | 365 (183/182) | 1.01 | 1 | 1.34 | 0.03 |
| C42 | Developmental delay genes | 77% | 11% | 10% | 93 (56/37) | 1.51 | 0.06 | 2.06 | 0.02 |
| C49 | CHD8 targets & PhyloP | 100% | 100% | 100% | 23 (16/7) | 2.29 | 0.09 | 2.43 | 0.01 |
| C63 | PhyloP | 79% | 100% | 12% | 143 (91/52) | 1.75 | 0.001 | 1.87 | 0.03 |
| NA | All Promoters | 53% | 14% | 5% | 6,787 (3,458/3,329) | 1.04 | 0.12 | NA | NA |
| NA | Active TSS Group | 100% | 16% | 10% | 3,570 (1,815/1,755) | 1.03 | 0.32 | NA | NA |
| NA | Conserved Loci Group | 60% | 100% | 8% | 931 (522/409) | 1.28 | 0.0002 | NA | NA |
Assessing the overlap of mutations between clusters and annotation terms identified two large groups of promoter mutations (Fig. 3B and 3C): an “Active Transcription Start Site (TSS)” group (RR=1.03; p=0.32, binomial test; Fig. 3D), distinguished by correlated epigenetic markers (C18 and C28; Fig. 3B) and a “Conserved Loci” group (RR=1.28; p=0.0002, binomial test; Fig. 3D), distinguished by PhastCons and/or PhyloP scores (C12, C20, C49, C63; Fig. 3B). Of the 931 de novo mutations in the Conserved Loci group, 557 (60%) are also in the Active TSS group (Fig. 3C) and removing these conserved loci from the Active TSS group removes almost all of the signal (RR=1.00).
The three remaining small clusters show limited overlap with the Active TSS and Conserved Loci groups (Fig. 3B and Table 1): C7 defined by lncRNAs at active TSSs (RR=1.19), C42 defined by developmental delay genes (2) (RR=1.51), and C26 defined by processed transcripts (RR=2.00).
When we consider all mutations in promoters as a single category, we see a non-significant trend towards weak enrichment in cases (3,458 in cases vs. 3,329 in controls; cRR=1.03; p=0.16 permutation test). Since the cluster analysis highlighted the role of evolutionary conservation (Fig. 3D), we assessed case-control burden for all 30,891 conserved mutations, split by GENCODE-defined (26) genic regions (Fig. 3E). We observed an excess of mutations in cases at conserved loci in promoters (522 vs. 409; cRR=1.26; p=0.0003 permutation test), but not for mutations in other noncoding regions (Fig. 3E and Fig. S7). In coding regions, de novo mutations that are not observed in the general population based on the Genome Aggregation Database (27) (gnomAD) are more likely to be associated with ASD (28). Similarly, we observe stronger ASD association at promoter regions if mutations seen in gnomAD are excluded (470 vs. 350; cRR=1.34; p=3×10−5 permutation test). Given the rarity and high effect sizes of protein disrupting de novo mutations, we might expect a marginally higher rate of risk-mediating mutations in the 1,759 ASD cases without previously identified ASD-associated mutations (1) compared to the 143 families with prior findings (Table S1). However, no such difference was observed between these two groups in conserved promoters (p=0.61 permutation test; Fig. S8) or for conserved missense mutations (p=0.20 permutation test; Fig. S8).
Gene set enrichment and phenotype in the Conserved Loci group
The Conserved Loci group includes the promoters of 886 unique genes, of which 53% are protein coding, 15% are processed pseudogenes, and 14% are lncRNAs (Table S6) with similar distributions in cases and controls except for processed transcripts (17 in cases, 0 in controls). In cases, genes with promoter mutations in the Conserved Loci group are enriched for “regulation of cell differentiation” (GO:0045595, FDR=0.02), “transcription, DNA-templated” (GO:0006351, FDR=0.04), and “regulation of transcription by RNA polymerase II” (GO:0006357, FDR=0.04), while no biological processes are enriched in controls (Table S8). Comparing cases to controls, there are non-significant trends towards enrichment in cases for ASD-associated genes (5 in cases, 2 in controls) and several ASD-related gene lists: brain-expressed (29), constrained (27), or CHD8 targets (8, 9, 30) (Fig. S9 and Table S8).
In coding regions, ASD-associated genes can be identified by the presence of multiple independent PTVs in different cases disrupting the same gene (1). In the WGS data, this approach did not yield specific promoters, since similar numbers of promoters had multiple Conserved Loci mutations in cases and controls (11 promoters in cases vs. 7 in controls; p=0.81, Fisher’s exact test). An equivalent analysis of damaging missense mutations split into 2,000bp blocks to simulate promoters, suggests we lack the power to detect specific promoters in a cohort of this size (22 in cases, 17 in controls; p=1.00).
Prior analyses of coding mutations have found large comorbid effects on nonverbal IQ, with ASD cases that carry ASD-associated mutations having a lower nonverbal IQ, on average (1). Excluding cases with de novo PTVs, we observed a 4-point reduction in median nonverbal IQ for cases with mutations in either the Active TSS (p=0.02, Wilcoxon signed rank test (WRST)) and/or Conserved Loci (p=0.01, WSRT) groups, compared to cases without such mutations (Fig. 3F). Furthermore, individuals with Conserved Loci promoter mutations show a trend towards a higher rate of mutations in female ASD cases (OR=1.13; 95%CI=0.74–1.73; p=0.31 Fisher’s Exact Test) and increased incidence of non-febrile seizures (OR=1.46; 95%CI=0.90–2.36; p=0.07 Fisher’s Exact Test); both trends are consistent with results seen in coding mutations.
The distal promoter shows the strongest evidence of association, especially at transcription factor binding sites
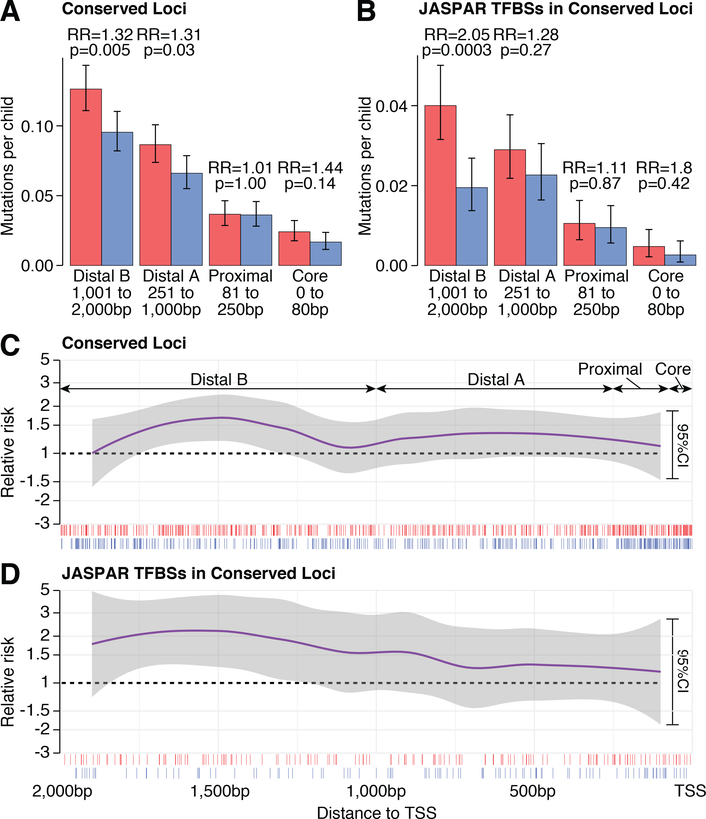
Since promoters are defined by their relationship to the TSS (31), we considered how ASD-association varied by TSS distance, with the expectation that association would diminish with distance from the TSS. We first examined four bins: the core promoter (≤80bp), which we would expect to contain the TATA box, initiator element (INR), and/or downstream promoter element (DPE), the proximal promoter (81–250bp), and two divisions of distal promoters (251–1,000bp, and 1,001bp-2,000bp). In contrast to this expectation, mutations in the Conserved Loci group are most strongly enriched in the distal region (RR=1.32; p=0.005, binomial test; Fig. 4A). This distal association prompted us to consider only mutations at experimentally-defined transcription factor binding sites (JASPAR CORE) (32), which enhanced the association (RR=2.05; p=0.0003, binomial test; Fig. 4B). While a trend towards enrichment in cases is observed in the core promoter (Fig. 4A and 4B), we do not see enrichment for motifs associated with RNA polymerase II (e.g. TATA; Table S6). Looking at the enrichment in cases across the promoter in 200bp sliding windows (Fig. 4C and 4D), the strongest enrichment is observed between 750bp and 2,000bp.
Figure 4. Relationship of conserved promoter mutations to the Transcription Start Site.
A) Frequency of Conserved Loci promoter mutations in cases and controls across the promoter region. B) Frequency of Conserved Loci promoter mutations in cases and controls at JASPAR transcription factor binding sites (TFBS) across the promoter region. C) Enrichment of Conserved Loci promoter mutations in cases, shown as relative risk, in sliding windows of 200bp across the promoter region. The purple line is the generalized additive model fit for relative risk and the 95% confidence interval is in grey. Ticks under the plot show individual mutations in cases (red) and controls (blue). D) The plot in ‘C’ is repeated for Conserved Loci promoter mutations at JASPAR TFBS. Statistical tests: A, B) Binomial, two-sided. Error bars show the 95% confidence interval (95%CI). TFBS: transcription factor binding sites; TSS: transcription start site.
Discussion
These analyses leverage WGS from 7,608 individuals with an unbiased genome-wide association framework to demonstrate that de novo noncoding mutations alter risk for a complex neurodevelopmental disorder (Fig. 2). In a recent study (15), we highlighted the importance of genome-wide analyses with appropriate correction for multiple testing to identify noncoding regions robustly associated with ASD. Following this principle, no single noncoding annotation category was significant after conservative correction for multiple testing (Fig. 1E). Similarly, we could not replicate candidate noncoding hypotheses described in previous analyses of ASD and developmental delay cohorts (Table S4) (12–14, 22, 33). However, a “de novo risk score”, developed from a genome-wide Lasso analysis of multiple noncoding annotation categories, was a significant predictor of ASD risk (Fig. 2A). Such scores are routinely used in genomic analyses, including polygenic risk scores of common variants and, recently, a rare variant risk score for coding mutations in schizophrenia (34). Consistent with expectations, the magnitude of the contribution from noncoding mutations is smaller than that of the coding region, even having excluded de novo PTVs (Fig. 2A). Yet, this early iteration of a de novo risk score could underestimate the true risk conferred by all noncoding mutations, as has been seen for polygenic risk score from common variants in successively larger cohorts (35).
Enrichment of annotation terms in the de novo risk score reveals that it is mutations in promoter regions, defined as 2,000bp upstream of the TSS, that underlie this noncoding association with ASD (Fig. 2A) and the risk score continues to demonstrate ASD association when considering only promoter categories (45/163 categories; Fig. 2A). A consistent association signal can be observed across all 1,855 promoter categories (Fig. 2B) and for 931 mutations at conserved loci (Fig. 3E). Notably, ASD cases with conserved promoter mutations have lower nonverbal IQ scores compared to those without (Fig. 3F), an effect also observed in children with ASD-associated PTV mutations and missense mutations (1). Within promoters, the most robust association is observed for promoter mutations at Conserved Loci (Table 1), particularly at known transcription factor binding sites (Fig. 4B) (32). At Conserved Loci, the relative risk is similar to that observed for de novo damaging missense mutations (Fig. 3E), though we expect that the true relative risk is likely to be lower due to the winner’s curse. Surprisingly, the strongest signal was not at the TSS and core promoter, but rather in the distal promoter, 750–2,000bp away from the TSS (Fig. 4). As expected for the distal promoter, the mutations in cases are frequently at experimentally-defined transcription factor binding sites (Fig. 4D).
A key question is whether these promoter regions target a common or distinct set of genes as those identified by variants in protein-coding regions. We favor the former possibility, though we cannot definitively exclude the later. Our evidence for this is: 1) The enrichment for GO-terms relating to transcriptional regulation and cell differentiation in the genes targeted by Conserved Loci mutations, terms that are also enriched in ASD-associated genes (1); 2) The trend towards enrichment for ASD-associated genes and several other gene sets previously implicated in ASD (Fig. S9); and 3) The detection of clusters defined by developmental delay genes and CHD8 binding targets (Fig. 3A; Table 1), both of which are enriched for ASD risk genes.
Our analysis establishes a specific hypothesis that can be tested for replication in future ASD cohorts and assessed in developmental and neuropsychiatric disorder cohorts: de novo mutations at conserved loci (46 vertebrate species PhastCons ≥ 0.2 and/or 46 vertebrate species PhyloP ≥ 2) in promoter regions (2,000bp upstream of the TSS based on GENCODEv27 annotation with VEP) are associated with risk. To facilitate such analyses by others, we have generated a file of loci that meet these criteria (Table S9). Despite these promising insights, we cannot yet identify which of the 522 conserved promoter mutations in cases truly confer risk, nor can we be confident which of the remaining 126,031 noncoding case mutations do not. Instead, our results demonstrate that elucidation of the contribution of de novo noncoding mutations to human disorders is feasible, and that the yields are likely to improve substantially with increases in cohort sizes (10, 15).
That conserved loci are one of the major factors underlying the promoter association could be interpreted to mean that nonhuman models can be used to assay noncoding function in humans, although parallel work in humans will be required to show that the specific regulatory effects are also conserved. Enrichment at transcription factor binding sites is also promising. If ASD association can be detected for specific transcription factors or loci, it raises the prospect of high-resolution neurobiological insights into spatiotemporal development, especially when, where, and in which cell type typical development is disrupted in ASD. Such insights will require detailed functional data on transcription factors, a goal of PsychENCODE and other groups, and how they relate to mutations found in ASD.
The association that we observe from these data represents the integration of work from multiple fields, including human cohort collections (2, 18), gene definitions (26), comparative genomics (23, 24), and functional genomics (32, 36). The methods and infrastructure necessary to replicate and refine this association, identify specific loci, or extend beyond promoters, are being developed: larger cohorts with consistently analyzed WGS data (e.g. the WGSPD consortium (10)), refined annotation of noncoding regions in the human brain (e.g. the PsychENCODE consortium (36)), WGS-tailored analytical methods (15, 25), and large-scale functional assays (e.g. massively parallel reporter assays (37)). The evolving results from these fields provide a path to improving diagnosis and novel therapeutic strategies that could benefit a wide range of human disorders.
Materials and Methods
Detection and annotation of de novo mutations
WGS data were generated by the New York Genome Center with a mean coverage of 35.5 in 1,902 ASD quartet families. Previously described variant filtering criteria were applied (15) to identify 255,106 high quality de novo mutations. These mutations were annotated using the Ensembl Variant Effect Predictor (VEP; version 90.4a44397) with GENCODE v27 gene definitions. Nucleotide sequence conservation across 46 vertebrate species (PhyloP, PhastCons), and regulatory regions (e.g. transcription factor binding sites, chromatin states) were annotated using VEP. In addition to 424 previously validated loci, 45 de novo mutations in promoter regions with two or more mutations in different samples were validated as de novo by analyzing all four family members with PCR and Sanger sequencing.
Category Wide Association Study (CWAS)
To assess multiple hypotheses, we implemented the CWAS method, described previously (15). Considering 70 annotation terms from five groups in combination defined 55,143 non-redundant categories for downstream analysis. ASD association was tested for each category by comparing the burden of case and control mutations with a two-sided binomial test, having corrected the rate of de novo mutations for paternal age. To estimate the penalty of multiple comparisons, the number of effective tests was estimated using Eigen decomposition of p-values in 10,000 simulated datasets. Each simulated dataset contained 255,106 random variants and maintained the GC bias and proportion of SNVs to indels observed in the original data.
De novo risk score analysis
To build a de novo risk score, we excluded all categories that could contain de novo PTVs, then selected 8,418 rare annotation categories with ≤3 mutations in controls. From the training dataset of 519 families described previously (15), we used a Lasso regression with 5-fold cross-validation to estimate the regularization parameter, and then applied this fitted prediction model to the remaining 1,383 new families to estimate the predictive power of the risk score. The significance of the prediction was calculated from 1,000 permutations with case-control status swapped in 50% of families selected at random. The frequency of the 62 noncoding annotation terms was compared between the 36,828 non-redundant noncoding categories and the 163 noncoding categories in the de novo risk score. A binomial test was used to assess the enrichment of these terms, corrected for 62 comparisons.
DAWN clustering analysis of promoter categories
The DAWN hidden Markov random field model (25) was used to assess the risk factors underlying ASD association of promoters. Clusters of individual promoter categories were defined by K-means (K=70) based on the p-value correlation network generated from 10,000 simulated datasets. Of these 70 clusters, 47 had at least 20 mutations and 2 categories and were considered further. Observed p-values were transformed to z-scores and sparse PCA analysis was used to estimate the p-value and relative risk per cluster. Using a Hidden Markov Random Field model, these estimates were modified to yield a posterior probability based on enrichment in neighboring clusters in the simulated p-value correlation network.
Detailed information for materials and methods can be found in the supplementary document (19).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all the families participating in this research, including the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC) and Korean cohort. We would like to thank the SSC principal investigators (AL Beaudet, R Bernier, J Constantino, EH Cook, Jr, E Fombonne, D Geschwind, DE Grice, A Klin, DH Ledbetter, C Lord, CL Martin, DM Martin, R Maxim, J Miles, O Ousley, B Peterson, J Piggot, C Saulnier, MW State, W Stone, JS Sutcliffe, CA Walsh, and E Wijsman) and the coordinators and staff at the SSC clinical sites; the SFARI staff, in particular N Volfovsky; DB Goldstein and KC Eggan for contributing to the experimental design; the Rutgers University Cell and DNA repository for accessing biomaterials; the New York Genome Center for generating the WGS data; the reviewers for their constructive feedback; and the editorial staff for their assistance throughout the editorial process. Annotation data was generated as part of the PsychENCODE Consortium, supported by: U01MH103392, U01MH103365, U01MH103346, U01MH103340, U01MH103339, R21MH109956, R21MH105881, R21MH105853, R21MH103877, R21MH102791, R01MH111721, R01MH110928, R01MH110927, R01MH110926, R01MH110921, R01MH110920, R01MH110905, R01MH109715, R01MH109677, R01MH105898, R01MH105898, R01MH094714, R01MH109901, P50MH106934, 5R24HD000836 and SFARI #307705 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke University), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (University of Southern California), Mark Gerstein (Yale University), Daniel Geschwind (University of California, Los Angeles), Ian Glass (Washington University), Fernando Goes (Johns Hopkins University), Thomas M. Hyde (Lieber Institute for Brain Development), Andrew Jaffe (Lieber Institute for Brain Development), James A. Knowles (University of Southern California), Chunyu Liu (SUNY Upstate Medical University), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Panos Roussos (Icahn School of Medicine at Mount Sinai), Stephan Sanders (University of California, San Francisco), Nenad Sestan (Yale University), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (University of California, San Francisco), Patrick Sullivan (University of North Carolina), Flora Vaccarino (Yale University), Daniel Weinberger (Lieber Institute for Brain Development), Sherman Weissman (Yale University), Kevin White (University of Chicago), Jeremy Willsey (University of California, San Francisco), and Peter Zandi (Johns Hopkins University).
Funding: This work was supported by grants from the Simons Foundation for Autism Research Initiative (SFARI #402281 to SJS, MWS, BD, KR; SFARI #385110 to SJS, MWS, AJW, NS; SFARI #574598 to SJS; SFARI #385027 to MET, BD, KR, JB, MJD; SFARI #346042 to MET; SFARI #575097 to BD, KR; SFARI #573206 to MET; SFARI #513631 to GTM; #388196 to HC, GTM), the National Institute for Health (U01 MH105575 to MWS; U01 MH100239–03S1 to MWS, SJS, AJW; R01 MH110928 to SJS, MWS, AJW; R01 MH109901 to SJS, MWS, AJW; R37 MH057881 to BD, KR; R01 HD081256 to MET; R01 MH115957 to MET; R01 MH049428 to JLR; R01 MH107649–03 to BMN; R01 MH094400 to HC), and the Stanley Center for Psychiatric Genetics.
Footnotes
Competing interests: Gabor T. Marth (GTM) is co-founder and Chief Scientific Officer of Frameshift Labs, Inc. Benjamin M. Neale (BMN) is a member of the Deep Genomics Scientific Advisory Board and a consultant for Camp4 Therapeutics Corporation, Merck & Co., and Avanir Pharmaceuticals. Matthew W. State (MWS) is on the Scientific Advisory Boards for ArRett Pharmaceuticals and BlackThorn Therapeutics and holds stock options in ArRett Pharmaceuticals.
Data and code availability: All sequencing and phenotype data are hosted by the Simons Foundation for Autism Research Initiative (SFARI) and are available for approved researchers at SFARIbase (https://base.sfari.org/, Accession ID: SFARI_SSC_WGS_p, SFARI_SSC_WGS_1, and SFARI_SSC_WGS_2). Methods for de novo SNV and indel annotation and statistical analyses in WGS data can be replicated using code hosted on Amazon Web Services on a publicly available Amazon Machine Image (for the most current AMI ID, see https://github.com/sanderslab/cwas). For methods for estimating the de novo risk score, clustering of annotation categories, and estimating the significance of clusters of annotation categories see https://github.com/lingxuez/WGS-Analysis.
REFERENCES
- 1.Sanders SJ et al. , Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McRae JF et al. , Prevalence and architecture of de novo mutations in developmental disorders. Nature (2017), doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heyne HO et al. , De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet 50, 1048–1053 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Jin SC et al. , Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet 49, 1593–1601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redin C et al. , The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet 49, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willsey AJ et al. , Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Shalom R et al. , Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol. Psychiatry (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugathan A et al. , CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A. 111, E4468–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotney J et al. , The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 6, 6404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders SJ et al. , Whole Genome Sequencing in Psychiatric Disorders: the WGSPD Consortium Whole Genome Sequencing for Psychiatric Disorders (WGSPD). Nat. Neurosci 20, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Lupski JR, Non-coding genetic variants in human disease. Hum. Mol. Genet 24, R102–R110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Short PJ et al. , De novo mutations in regulatory elements in neurodevelopmental disorders. Nature, 112896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner TNN et al. , Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am. J. Hum. Genet 98, 58–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner TN et al. , Genomic Patterns of De Novo Mutation in Simplex Autism. Cell. 171, 710–722.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werling D et al. , An analytical framework for whole genome sequence data and its implications for autism spectrum disorder. Nat. Genet 50, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandler WM et al. , Paternally inherited cis-regulatory structural variants are associated with autism. Science. 360, 327–331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudbridge F, Gusnanto A, Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol 32, 227–234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischbach GD, Lord C, The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 68, 192–195 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supplementary materials.
- 20.Jónsson H et al. , Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 549, 519–522 (2017). [DOI] [PubMed] [Google Scholar]
- 21.De Rubeis S et al. , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munoz A et al. , De novo indels within introns contribute to ASD incidence. bioRxiv (2017) (available at http://biorxiv.org/content/early/2017/05/24/137471.abstract).
- 23.Siepel A et al. , Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A, Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L et al. , DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics. Mol Autism. 5, 1–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrow J et al. , GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M et al. , Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosmicki JA et al. , Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet 49, 504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HJ et al. , Spatio-temporal transcriptome of the human brain. Nature. 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darnell JC et al. , FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak H, Fuda NJ, Core LJ, Lis JT, Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science. 339, 950–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan A et al. , JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuen RKC et al. , Genome-wide characteristics of de novo mutations in autism. npj Genomic Med. 1, 16027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell SM et al. , A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 506, 185–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterjee N et al. , Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet 45, 400–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbarian S et al. , The PsychENCODE project. Nat Neurosci. 18, 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue F et al. , A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 27, 38–52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.