Abstract
Recent developments in genetics and genomics are providing a detailed and systematic characterisation of the genetic underpinnings of common metabolic diseases and traits, highlighting the inherent complexity within systems for homeostatic control, and the many ways in which that control can fail. The genetic architecture underlying these common metabolic phenotypes is complex, with each trait influenced by hundreds of loci spanning a range of allele frequencies and effect sizes. Here, we review the growing appreciation of this complexity and how this has fostered the implementation of genome-scale approaches that deliver robust mechanistic inference, and unveil new strategies for translational exploitation.
Introduction
One of the key objectives of human and medical genetics is to provide insights into disease aetiology. Early studies focused on establishing the genetic basis of Mendelian disorders, for which, typically, a single causal mutation can explain the recurrence of disease within each affected family. However, transfer of the family-based linkage and candidate gene approaches which had been successful in Mendelian diseases – including many rare inherited metabolic disorders - to the identification of genetic variants influencing common, multifactorial diseases proved an unrewarding task generating very few robust findings (Hani et al. 1998, Altshuler et al. 2000, Hugot et al. 2001, Ogura et al. 2001, Grant et al. 2006). This failure of methods best-suited to the detection of rare, high-impact variants, indicated a need for approaches tuned to the detection of common variants with more modest effects. It was only with the advent of genome-wide association studies (GWAS) in 2005 that the fortunes of prospectors of complex genetics improved. The use of genome-wide genetic surveys conducted in large samples has now become the workhorse of complex disease/trait genetics, and the number of discoveries has exploded.
The major challenge nowadays is to extract the biological “gold” from the thousands of discoveries that have been made. In contrast to Mendelian diseases, most complex trait associations map to non-coding regions of the genome, and it has proven difficult to define the genes and pathways through which they operate. This has led some to question the value of these approaches for aetiological insight (Dickson et al. 2010, McClellan et al. 2010), and their potential to deliver biological and translational value (Visscher et al. 2012, Visscher et al. 2017). In recent years, a shift towards the generation of genome-scale functional data has ensured that major inroads into the biological interpretation of the signals are being made. These are exposing a more nuanced view of the relationships between genetic variation and clinical phenotype, transcending simple reductionist notions that each genetic locus would resolve to a single genetic variant, impacting a single gene in a specific tissue, to influence a single disease or trait.
In this review we will summarise progress made, and the insights gained from broad application of GWAS across a range of highly prevalent metabolic diseases (type 2 diabetes, obesity and cardiovascular disease) and related traits (for example, fat distribution, glycemic and lipid measurements). We will end by reviewing some of the early translational applications based on findings from these studies and discussing future developments in the field.
The genetic architecture of metabolic traits
A multiplicity of loci
A key observation arising from GWAS is that common metabolic phenotypes, whether binary (e.g. diabetes status) or continuous (e.g. circulating triglycerides), are influenced by a broad diversity of DNA sequence variants.
At one end of the spectrum are rare (allele frequencies <0.5%), high-impact variants shown to be causal for monogenic and syndromic forms of metabolic disease, such as maturity onset diabetes of the young (MODY) or familial hypercholesterolaemia (Defesche et al. 2017, Hattersley et al. 2017). These are typically expressed in early life, with clear familial segregation. Historically, the causal variants responsible were detected through linkage analysis within affected pedigrees, but diagnosis and classification now largely proceeds through whole exome or genome sequencing.
At the other end of the spectrum are the common variants (allele frequencies >5%) which account for much of the predisposition to more prevalent, later-onset metabolic diseases. GWAS have provided an increasingly-complete inventory of these variants, revealing that the inherited contribution to disease risk is influenced by many hundreds (if not thousands) of such common variants, each with modest impact on disease-risk (Scott et al. 2012, Locke et al. 2015, Klarin et al. 2018, Mahajan et al. 2018a). Early GWAS were underpowered, and detected only a few of the larger signals (WTCCC 2007). Furthermore, because the assays used preferentially captured common variation, early discoveries were similarly restricted. These factors contributed to an apparent disparity between estimates of overall heritability derived from twin and family studies, and the fraction of that heritability attributed to the discovered risk-variants (Manolio et al. 2009). Various explanations for this “missing heritability” were offered, with strong advocacy for the contribution of rare variants (Lupski et al. 2011). However, as GWAS sample sizes increased, and with sequence data now enabling systematic interrogation of rare variants, it has become clear that genetic predisposition to prevalent, late-onset metabolic traits is mostly attributable to a long tail of common, progressively-smaller allele effects (Yang et al. 2015, Fuchsberger et al. 2016). The latest GWAS for T2D, for example, demonstrated that common variants explain approximately 20% of overall T2D-risk, which equates to at least half the estimated overall heritability (Mahajan et al. 2018a). Sequence-based analyses have attributed a far smaller contribution of lower frequency variants to T2D-risk.
This pattern is consistent with the expectation that the variants underlying late-onset metabolic diseases will have had limited impact on reproductive fitness during human prehistory, and some may even have been beneficial (Neel 1962). In such a situation, most disease-associated variation will occur at sites of common, shared variation. (This does not preclude evidence for selection at a subset of loci – see below). Though there will also be many thousands of rare alleles contributing to variation in disease risk, most people are “wild-type” homozygotes at almost all of them, and their overall contribution to population risk is limited.
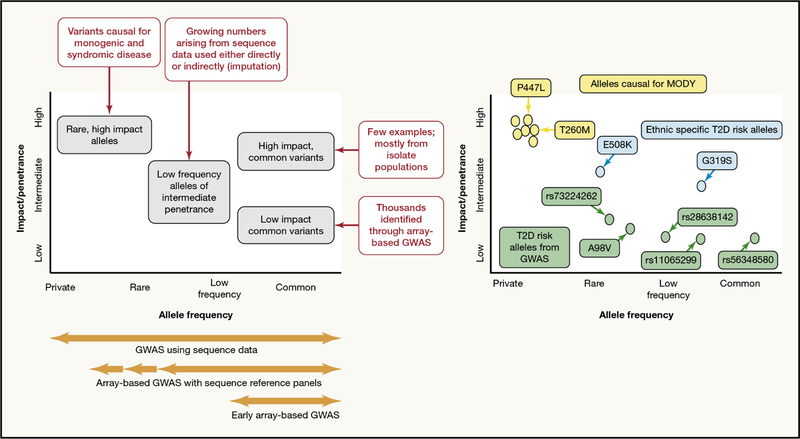
Historically, human genetics has been dominated by two models – monogenic and polygenic – which mapped onto the approaches used for risk-variant detection (family-based vs population based). There is however, no biological reason why the variants that influence the function or expression of a given susceptibility-gene should fall neatly into two extreme groups (rare, impactful, vs. common, weak). Detailed analyses that expose the full spectrum of variation increasingly highlight the diversity of coding and non-coding risk-variants in genes like HNF1A (Hegele et al. 1999, Flannick et al. 2013, Estrada et al. 2014, Mahajan et al. 2018a). These represent a subset of the natural variation seen in and around the coding or regulatory circuitry of each gene, much of it (irrespective of their allele frequency) with zero impact on disease risk. One common fallacy is to believe that, because most high-impact alleles are rare, due to their adverse effect on survival, that most rare alleles are likely to have a deleterious phenotypic effect. In fact, the low frequency of most rare alleles merely reflects their recent origin, rather than indicating serious clinical impact (Figure 1).
Figure 1 -. The allelic spectrum of effects contributing to disease risk.
The left panel highlights that contributions to variance in a trait of interest may come from variants across the allele spectrum. Most of the common variants identified by GWAS for complex multifactorial have modest effects, though there are some exceptions. Rare, high impact alleles are most relevant to monogenic and syndromic forms of disease but contribute to some extent to phenotypic variance at the population-level. The right panel illustrates the range of variants in one gene (HNF1A) with a proven or potential impact on diabetes risk: this includes rare variants causal for monogenic forms of diabetes (yellow), common and low-frequency variants of modest effect influencing T2D-risk (green), and additional ethnic-specific variants with more substantial effects on T2D-risk in selected populations (blue).
Genetic heterogeneity across ethnic groups
Most metabolic traits of biomedical significance affect diverse ethnic groups, though disease presentation and population prevalence often vary. Given the multifactorial nature of these conditions, it is important to understand genetic and non-genetic risk factors underlying this heterogeneity.
Rare variants, mostly of recent origin, tend to be restricted in their geographic and ethnic representation. In contrast, common variants are older, often predating the expansion out of Africa, with cosmopolitan, often global, reach, and many common variant associations with metabolic phenotypes have broadly similar effects on disease across a wide range of ethnic groups (Mahajan et al. 2014).
This does not necessarily mean that all common variants with robust association in one population are detectable in others, though such disparities usually reflect ethnic differences in allele frequency. The principal T2D-risk allele at the TCF7L2 locus, for example (rs7903146), is much less frequent in East Asians (2%) than Europeans (30%) (Grant et al. 2006). Other risk-variants are restricted to African-descent populations, either because they were absent from early waves of “out-of-Africa” migration, or because of region-specific selective pressures. An example of the latter involves the G6PD p.G202A variant (associated with HbA1c) which maintains a high frequency in African-descent individuals due to relative protection against malaria (Wheeler et al. 2017).
Human population expansion has driven other ethnic-specific association signals, increasing risk-allele frequency through a combination of chance (“genetic drift”) and positive selection. T2D provides multiple examples, including the Arg193His variant in PAX4 (East Asian specific) (Fuchsberger et al. 2016), the E508K in HNF1A (Latinos) (Estrada et al. 2014) and an interesting variant near SLC16A11, frequent in those of East Asian and Native American descent and thought to have arisen in the Neanderthal genome (Williams et al. 2014).
Whilst these ethnic-specific risk variants can be highly informative from a biological perspective and allow the exploration of gene-environment effects (see below), marked ethnic heterogeneity is relatively uncommon and does not explain observed differences in disease prevalence or presentation. Some of the rare ethnic-specific variants emerging from ongoing sequencing efforts may be more relevant in this respect.
Genetic signals for other relevant biomedical phenotypes
Progress in characterising the genetic basis of many clinically-relevant biomedical traits – such as the complications of diabetes, or response to bariatric surgery - has been far slower (Dahlstrom et al. 2017). Whilst this may reflect differences in trait heritability, genetic architecture, and phenotypic precision (all influencing the power to detect robust association signals), the biggest obstacle has been limited sample size. These disease traits are often rare, or difficult to diagnose (e.g. diabetic neuropathy), and uncertainties over phenotypic criteria may have compromised efforts to assemble adequate samples for genetic analysis (e.g. polycystic ovarian syndrome, diabetic kidney disease).
Different traits may have different “tipping points” in terms of the sample sizes required before GWAS discoveries accumulate: schizophrenia and hypertension provide examples of common diseases where early GWAS results were disappointing, but increasing sample size has now delivered scores of established loci (Schizophrenia Working Group of the Psychiatric Genomics 2014, Kraja et al. 2017). Equivalent efforts are likely to prove fruitful for those metabolic traits that have thus far produced limited numbers of loci. Concomitant improvements in phenotypic precision are also valuable, as aetiologically homogenous samples will also bolster discovery power. However, clinical concepts of disease classification may not map precisely onto molecular aetiology, and in many settings, expansion of sample size has proven the more rewarding strategy. GWAS analyses for declining renal function have proven surprisingly effective despite the wide range of underlying disease pathologies: most of the strongest effects influence renal function irrespective of the original cause of the renal insult, and power to detect genetic signals is maximised when the initiating disease state is ignored (Chasman et al. 2012).
Biological Insights
Most association signals map to regulatory sequence
To gain mechanistic insights from association signals, a typical first step involves identification of the causal variant (or variants) responsible for the signal. Humans are a young species, and the consequent local correlation of variants (“linkage disequilibrium”), though advantageous for association signal discovery, often frustrates efforts to home in on the specific causal variant(s). Since these correlation patterns often differ between ethnic groups, trans-ethnic association studies can allow causal variants to be distinguished from their non-functional neighbours (Mahajan et al. 2014, Wheeler et al. 2017, Mahajan et al. 2018b).
Even within a single ethnic group, the combination of massive sample sizes, and high quality imputation (which uses sequence data to infer genotypes at untyped variants), can tease apart local correlation and highlight the most likely causal variant(s) at a non-trivial proportion of association signals [Figure 2] (Mahajan et al. 2018a, Mahajan et al. 2018b). Analyses such as these have shown that only 5–10% of the common risk variants influencing common metabolic phenotypes can be attributed to variation in coding sequence (Mahajan et al. 2018b). Given that coding sequence accounts for only ~1.5% of genome sequence, this still represents an enrichment over expectation: nevertheless, it contrasts with the predominant role played by coding variants in Mendelian disease.
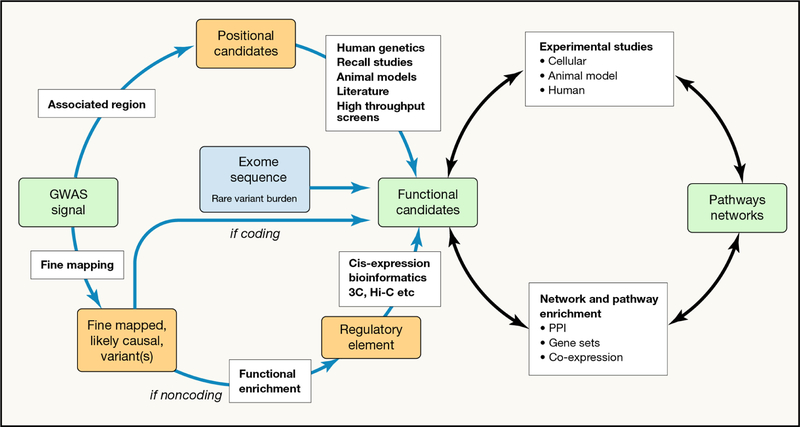
Figure 2 -. A schema for generating biological and clinical insights from human genetic findings.
Given a set of (largely non-coding) GWAS signals for a phenotype of interest, the aim is generally to identify the genes through which the effect is likely to be mediated (“positional candidates”) and the networks and pathways which are implicated. The first step (from signals to positional candidates) proceeds through a combination of evaluating nearby genes for their biological relevance to the trait of interest (upper left) and of linking fine-mapped causal variants to their transcriptional targets (lower left). Once candidates have been prioritised, experimental validation (through perturbation experiments in cellular and animal models) is essential, and the information gathered examined for evidence of networks and pathways causally implicated in disease pathogenesis (or trait variance). Abbreviations: PPI, protein-protein interaction; 3C, Chromosome conformation capture; Hi-C, a derivative of 3C methods.
The remaining association signal for common metabolic phenotypes maps to non-coding sequence. The variants responsible are likely to act by influencing transcription, through effects on promoter or enhancer function of nearby genes or through disturbing non-coding RNAs or microRNAs. Incomplete understanding of the “grammar” of the non-coding genome has complicated efforts to define the mechanisms whereby such variants influence disease risk [Figure 2].
The closest gene is not always the right answer
Arguably, the most important step in mechanistic inference at GWAS loci involves the identification of the “effector transcript”, that is, the gene (or genes) through which associated variant(s) exert their effects on disease predisposition. Links between the causal variants and the regulatory elements, and the genes that they regulate can be established by testing the effects of those variants on the expression of local genes, or by looking for evidence of loops in the DNA sequence that bring the implicated regulatory elements into contact with nearby gene promoters. The impact of those candidate effector transcripts on the development of disease can then be further validated by their manipulation in cellular and animal models.
At many loci, these studies suggest the nearest gene as the most likely culprit but this is not always the case [Figure 3]. A prime example involves the FTO locus, where the impact of intronic FTO variant(s) on obesity is now thought to reflect regulatory effects on distally-mapped genes [Figure 3]. Early bioinformatic and functional studies at this locus focused on the FTO gene itself (Gerken et al. 2007). Knockout and overexpression studies in mice supported the idea that alterations in Fto expression were causally related to variation in body weight (Fischer et al. 2009, Church et al. 2010, Tung et al. 2014). However, the human relevance of these findings was questioned when individuals carrying a homozygous missense mutation (R316Q), which rendered the FTO enzymatically null, were shown to have a complex malformation syndrome characterised by childhood mortality but no particular weight-related phenotype in homozygous or heterozygous carriers (Boissel et al. 2009).
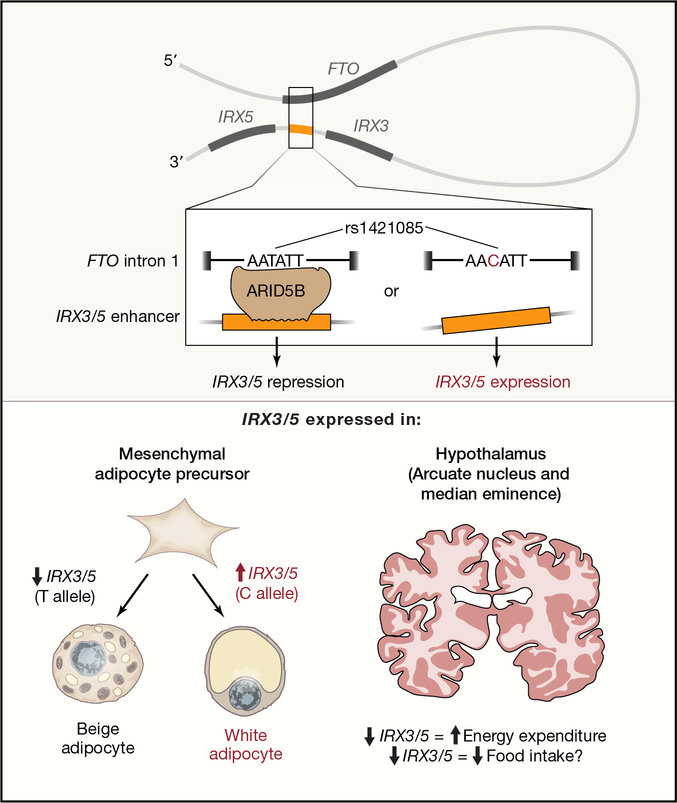
Figure 3 -. Causal variants and proposed mechanism at the FTO locus.
Variants within the first intron of FTO (top) make contact with regulatory regions close to the IRX3 and IRX5 genes. The BMI-raising allele (C), is proposed to disrupt the ARID5B repressor biding site leading to overexpression of IRX3/5. Consequently, differentiating adipocytes are directed towards white lipid storing, instead of beige, adipocytes (bottom panel). In the hypothalamus (right bottom), overexpression of IRX3/5 could lead to increased food intake and decreased energy expenditure. Adapted from Herman, M. A. and E. D. Rosen (2015). “Making Biological Sense of GWAS Data: Lessons from the FTO Locus.” Cell Metab 22(4): 538–539.
More recently, the region harbouring the BMI-associated variants was shown to act as a long range enhancer for the IRX3 gene (Smemo et al. 2014). Irx3-null mice were shown to have a 25–30% reduction in body weight, associated with increased basal metabolic rate and browning of white adipose tissue, through a proposed hypothalamic effect (Smemo et al. 2014). Using a combination of epigenomics, comparative genomics, human genetics and gene editing approaches, Claussnitzer and colleagues (Claussnitzer et al. 2015) concluded instead that the FTO BMI-increasing allele (rs1421085) disrupts the ARID5B repressor, leading to overexpression of IRX3 and IRX5 during early adipocyte differentiation. This consequently appears to shift energy-dissipating beige adipocytes toward energy-storing white adipocytes, with a reduction in mitochondrial thermogenesis, and an increase in lipid storage. Neither of these studies have evaluated all regional variants and in all tissues, so it is possible, even likely, given the unusually large impact of FTO variants on BMI, that there are other contributory mechanisms, including hypothalamic impact on central control of energy expenditure [Figure 3].
One locus, multiple signals/causal variants
Early GWAS follow-up studies focused on the idea of finding the single “causal” variant that was driving predisposition at each associated locus. With increased sample size, and higher variant density (including targeted arrays with dense imputation or sequence data), has come a higher resolution view of these associations that highlights their complexity. Multiple distinct signals (each driven by a different causal variant) may occur at a given locus (Mahajan et al. 2015, Wessel et al. 2015), including a mix of coding and non-coding variants, across a range of allele frequencies and effect sizes.
At some loci, several variants on the same haplotype are likely to be contributing to functional impact. At others, the alignment of coding and regulatory risk-alleles results in a more subtle arrangement of effects. For example, at the G6PC2/ABCB11 locus associated with fasting glucose, there are multiple distinct G6PC2 coding variant signals (p.Val291Leu, p.His177Tyr, p.Arg283X, P.Ser324Pro and p.Tyr207Ser) (Mahajan et al. 2015, Wessel et al. 2015) in addition to the non-coding GWAS variant (Bouatia-Naji et al. 2008, Chen et al. 2008). Detailed haplotype and functional analysis showed that it was only possible to reconcile genetic and functional data if one considered the haplotypic phase of the non-coding and coding variants [Figure 4]. In Europeans, the allele at the common p.Val219Leu variant associated in vitro with reduced G6PC2 protein expression (which would be anticipated to reduce glucose levels), appears to have a paradoxical population-level association with higher glucose. The explanation is that the same glucose-lowering coding allele is restricted to a haplotype that carries the glucose-raising allele of the regulatory GWAS variant, thought to upregulate G6PC2 expression through an impact on pre-mRNA splicing, which “masks” the Val219Leu true functional direction of effect.
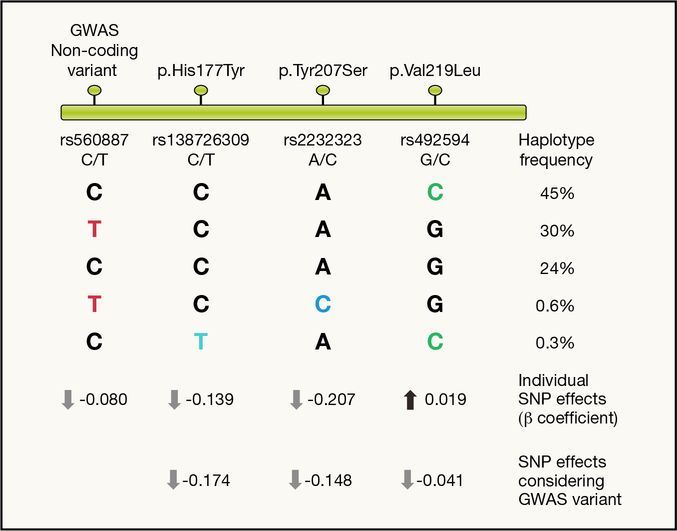
Figure 4 -. Coding variants at G6PC2/ABCB11 locus.
Schematic representation of GWAS and coding variants mapping along the chromosome around the locus (top). Common haplotypes formed by the four variants are represented with their frequency. Coloured letters represent the glucose lowering allele based on in vitro assay results (black letters refer to the glucose raising allele). Estimating the effect of each SNP individually (black/coloured letter in each column) ignoring the background haplotype may lead to incorrect inferences regarding the effect of that variant on glucose levels. This effect is particularly evident for the p.Val219Leu (MAF=48%) where single SNP analysis estimates the effect of this variant to increase glucose levels, whereas functional data show it decreases glucose levels. When the effect of this variant is estimated conditioning on the effect of the GWAS index variant (i.e. taking account of its effect on glucose levels) it becomes apparent that Leu219 decreases glucose levels. These results are in agreement with in vitro results where p.Val219Leu was shown to decrease protein expression levels by 49%, in comparison to a 99% reduction (p.His177Tyr) and 100% reduction (Tyr207Ser) for the other two coding variants. This striking difference on protein expression levels is in agreement with the much more modest effects on fasting glucose for p.Val219Leu compared to the two other variants.
From associated loci to convergence of biological pathways
With the number of genetic loci influencing many metabolic phenotypes (such as T2D, HDL-cholesterol levels, or urate) extending into the hundreds, if not thousands, such complexity raises questions about their biological interpretation. Is it even worth finding more and more loci and trying to understand the biological processes they implicate, if it transpires there are almost as many signals as there are genes? However, such a negative perspective ignores the subtlety of genetic regulation: even if dysregulation of the same gene is implicated in multiple diseases, the directions of effect, tissue-specificity, and developmental window may well differ. The recently-proposed omnigenic model (Boyle et al. 2017) argues that many of the lesser genetic effects that contribute to the long polygenic tail of association feed into regulatory modulation of disease-specific core pathways which are the main pathophysiological drivers (Boyle et al. 2017, Wray et al. 2018).
Growing evidence for a convergence of disease-specific biology within the association signals supports this notion. For many complex diseases, it is increasingly clear that the distribution of association signals across the non-coding space is non-uniform, with substantial signal enrichment within enhancers that show tissue-specific activity, and which regulate genes central to tissue-specific functions and identity. In T2D, those signals point to the pancreatic islet, and to a lesser degree to fat and liver (Mahajan et al. 2018a); in hyperlipidaemia, they point to the liver (Willer et al. 2013).
GWAS signals are also non-random with respect to the genes which map nearby. Even when specific downstream genes have not been precisely-determined, it is possible to demonstrate enrichment for disease processes that fit within the scope of known disease biology. Association signals for hyperlipidaemia, for example, show substantial co-localisation with genes coding for known components of lipid metabolic pathways, including known targets for cholesterol-lowering medication (statins and ezetimibe) (Willer et al. 2013, Lange et al. 2014, Stitziel et al. 2014). There are marked differences between the distributions of association signals influencing overall obesity (as measured by BMI) and fat distribution (as measured by waist:hip ratio). The former map to regions and genes implicated in central hypothalamic, hippocampal and limbic processes involved in appetite control and energy balance (Locke et al. 2015), and the latter show enrichment for peripheral pathways linked to adipogenesis and insulin signalling (Shungin et al. 2015).
Similar analyses for glycemic traits have revealed enrichment of association signals for genes involved in multiple expected pathways, including glucose metabolism, insulin secretion and processing (Morris et al. 2012, Scott et al. 2012), as well as enrichment for binding sites for transcription factors with a role in pancreatic development (e.g. highlighted by loci PDX1, FOXA2) (Gaulton et al. 2015). There have been unexpected findings too, such as a link between genetic variants influencing circadian rhythm genes (e.g. MTNR1B, CRY2) and fasting glucose (Dupuis et al. 2010) and the role of cell cycle regulation and T2D risk (Morris et al. 2012).
Genes and Environment
Joint (additive) effects and interaction (multiplicative) effects are not the same
Metabolic diseases, including obesity, dyslipidaemias and diabetes, as well as variation in glucose and lipid levels in non-diseased individuals, represent prime examples of the consequences of the joint effects of genes and environment [Figure 5]. For example, the phenotypic consequences of a BMI-influencing allele on obesity will be dependent on food availability. However, joint effects themselves do not provide evidence for a statistical interaction between genetic and environmental effects [Figure 5]. The presence of a gene-by-environment (or more accurately, genotype-by-environment; GxE) interaction requires that the consequences of the risk allele vary between individuals exposed to different environments [Figure 5]. One could imagine, for example, that a sedentary lifestyle or diet rich in saturated fats carries a particularly high risk of promoting obesity in individuals carrying a particular variant allele (or set of such alleles). Knowledge of GxE could prove helpful in identifying individuals who might be especially likely to benefit from lifestyle modification, as well as providing new insights into biological pathways underpinning disease.
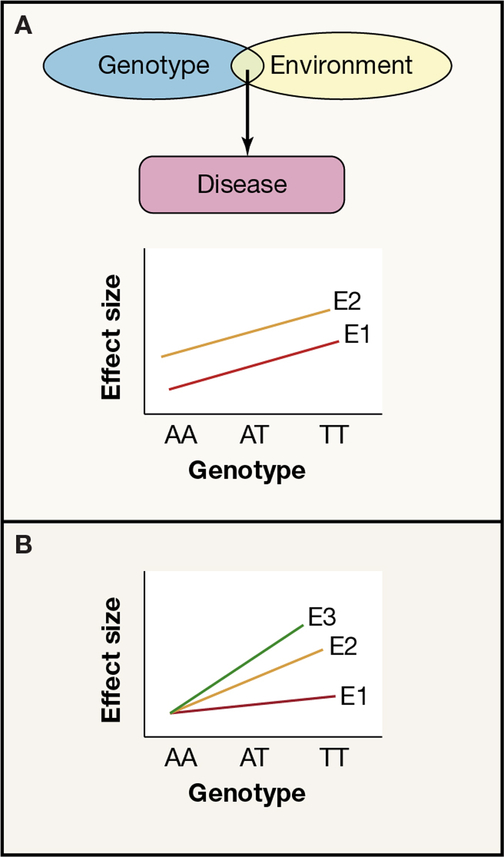
Figure 5 -. Effects of genes and environment on disease risk.
Panel a) shows joint effects of genes and environment, but no GxE interaction. Both genotype and environment are needed to cause disease (above) but the effect of the genotype is the same across all environments (below- gradient of the lines E1 and E2 is the same), environment E2 increases the risk across all genotypes by the same amount. Panel b) illustrates a genotype x environment interaction (GxE) where the effect of the genotype on disease risk varies between different environments E1, E2, E3 (gradient of the lines is different).
Metabolic traits have been the subject of many GxE studies, but most of these have been limited in scale and scope, featuring small sample sizes, and a limited range of genetic and environmental exposures. Robust evidence of GxE effects has been scant (Franks et al. 2016): limited power, incomplete adjustment for multiple testing, and weak measures of exposure have all contributed. Some of the widely-used “environmental” effects, such as food choice, turn out to be highly genetic (Pallister et al. 2015).
The strongest evidence for GxE effects have emanated from obesity. BMI-influencing variants near FTO have shown interaction effects with a variety of factors including physical activity, alcohol consumption, socioeconomic status, diet, smoking and sleep duration (Sonestedt et al. 2009, Ahmad et al. 2011, Corella et al. 2011, Corella et al. 2012, Phillips et al. 2012, Richardson et al. 2014, Reddon et al. 2016, Young et al. 2016, Graff et al. 2017, Rask-Andersen et al. 2017, Moore et al. 2018). Other studies have attempted to bolster the power to detect interactions by combining genetic evidence from multiple variants into a genetic risk score (GRS), and GRS composed of obesity- and BMI-increasing alleles have revealed nominal interactions with a range of environments (Li et al. 2010, Elks et al. 2012, Ahmad et al. 2013, Johnson et al. 2014). However, the magnitude of these interaction effects tends to relatively modest, and clinical utility is unclear. Ongoing efforts to provide more accurate measurement of environmental exposures in larger studies, allied to the application of more powerful statistical methods should render a much fuller understanding of the contribution of GxE to trait variance (Moore; et al. 2018, Young et al. 2018).
Selection shapes our genome
Environment can also shape the impact of genetic variation over many generations, through an impact on selection, be that positive (where the derived allele gradually increases in frequency due to beneficial effects on fitness), negative (where reduced fitness tends to keep allele frequencies low) or balancing (where the derived allele is maintained due to opposing positive and negative pressures). The “thrifty” genotype hypothesis (Neel 1962), for example, proposed that the high prevalence of BMI-raising alleles in contemporary populations could be the consequence of a historical selective advantage associated with enhanced fat deposition during periods of erratic food availability.
Although the evidence of strong genome-wide selection for cardiometabolic traits is limited (Ayub et al. 2014, Xue et al. 2018), there are isolated variants where selective pressures do seem to have been operating. Cold exposure and a diet characterised by high intake of animal fats and limited access to plant foods may, in populations living in the Arctic, have led to a selective sweep (where a newly-derived allele replaces the ancestral allele almost completely) at the CPT1 p.Pro479Leu variant (rs80356779, G>A, [Table 1]). The derived (A) allele at this variant appears to have arisen 6,000–23,000 years ago (Raghavan et al. 2014, Rasmussen et al. 2014) and then spread through positive selection (Cardona et al. 2014) through Eskimo-Aleutian and Siberian populations reaching a frequency exceeding 40% in current populations (Greenberg et al. 2009, Rajakumar et al. 2009, Collins et al. 2010, Clemente et al. 2014). CPT1A encodes a protein responsible for importing long chain fatty acids into mitochondria for fatty acid oxidation, helping to maintain energy homeostasis and normoglycaemia in diets low in carbohydrates. By decreasing CPT1A activity, this allele may have protected against excess ketone body production in populations adapted to extreme cold where the diet is rich in marine-based n-3 polyenoic fatty acids (Greenberg et al. 2009). The same allele is now associated with more adverse effects -- sudden infant death syndrome and hypoketotic hypoglycaemia – possibly reflecting a switch towards a more carbohydrate-rich diet (Greenberg et al. 2009).
Table 1 -.
Allele frequencies of variants expanded in specific populations
| VARIANT | Frequency in local populations | Worldwide frequencies (gnomAD, 1000 genomes for FADS1/2 reference data) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | AFR | AMR | ASJ | CEU | CHB | EAS | EUR | FIN | NFE | OTH | SAS | ||
|
CPT1 rs80356779 (G>A; p. Pro479Leu) |
North eastern Siberians = 68% Chukchi = 90% Eskimo = 87.5% Koryak = 56.25% Greenlandic Inuit = 73% Canadian Inuit = 73% Nunavut = 44% |
0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |||
|
FADS1, FADS2 -rs74771917 DAF (T) |
Greenlandic Inuit = 98% | 14% | 7% | 35% | 2.5% | 16% | 24% | 3% | 9% | ||||
|
CREBPF rs373863828 (G>A; p.Arg457Gln) |
Samoans = 25.9% Tongans = 15% Melanesians = 2% Micronesians = 6% |
0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |||
|
TBC1D4, rs61736969 (C.2050C>T; P.Arg684Ter) |
Greenlandic Inuit = 17% | 0.0012% (3) |
0% | 0% | 0% | 0.0058% (1) |
0% | 0.0012% (2) |
0% | 0% | |||
A similar story likely explains the positive selection of alleles around the fatty acid desaturase gene cluster in Inuit (Fumagalli et al. 2015) [Table 1]. FADS1 encodes delta-5, and FADS2 encodes delta-6 desaturase, both rate-limiting enzymes in the conversion of short-chain linoleic (omega-6) and α-linolenic (omega-3) acids to longer, less-saturated forms. Selection here favoured increased levels of short-chain forms which may have provided compensation for the high dietary intake of long-chain eicosapentaenoic acid from Inuit diet: this region (but different alleles) has been shown to be under positive selection in other populations undergoing adaptation to diet (Ameur et al. 2012, Mathieson et al. 2015, Amorim et al. 2017, Buckley et al. 2017). The alleles concerned are associated with increased glucose and cholesterol levels (but reduced triglycerides), and an increase in cardiovascular risk, a plausible example of a “thrifty” genotype (Kathiresan et al. 2009, Dupuis et al. 2010).
Diet may also have been an important selective force for the “A” allele (rs373863828; G>A) of CREBRF p.Arg457Gln variant, present in about one-half of Samoans (Minster et al. 2016) [Table 1], and associated with increased risk of obesity (but intriguingly a reduced risk of diabetes). Overexpression of the CREBRF gene in an adipocyte model has been shown to reduce energy use and increase fat storage (Loos 2016). One explanation is that, by promoting fat storage, the 457Gln allele enhanced survival during long oceanic journeys and the settlement of Pacific Islands. The variant has its highest frequency in Samoa, but is also seen in other Pacific populations (Naka et al. 2017) (Berry et al. 2018, Ohashi et al. 2018). It is however, absent from other parts of the world.
A final example of the potential impact of selection involves the TBC1D4 p.Arg684Ter variant (c.2050C>T, rs61736969) which has a frequency of 17% in Greenlandic individuals (Moltke et al. 2014) but is virtually absent in all other populations [Figure 6]. Despite evidence of positive selection, it is unclear what the selective advantage this variant confers. Homozygous variant carriers have raised glucose and insulin levels after oral glucose testing, and a ~10-fold increased risk of T2D. The T2D-risk allele is associated with reduced expression of the long isoform of TBC1D4 in skeletal muscle, which appears to compromise insulin–stimulated glucose uptake into muscles leading to hyperglycaemia.
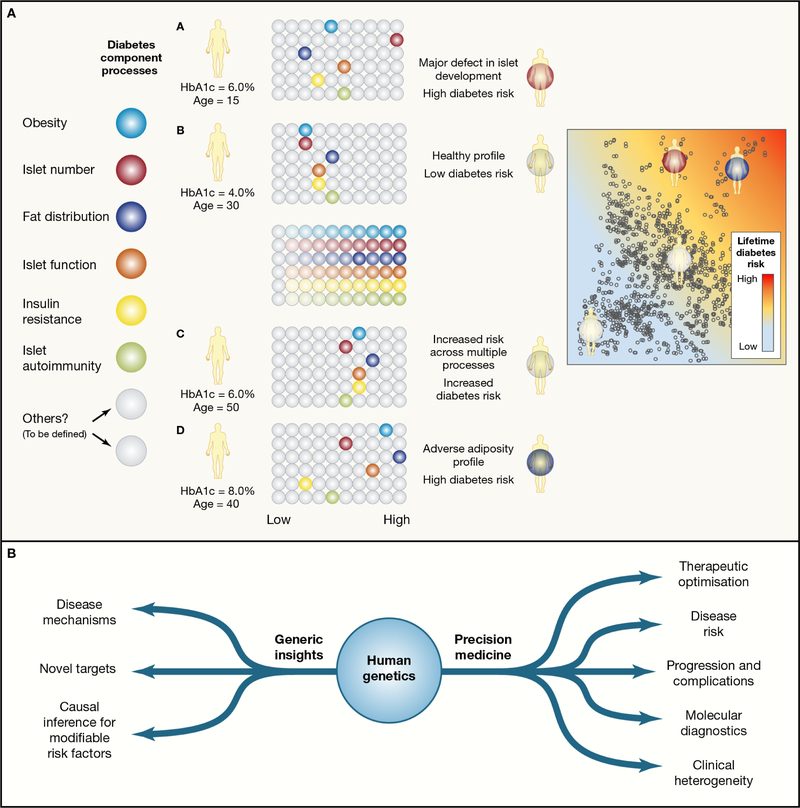
Figure 6 -. Translational Opportunities.
Panel a) Partitioned risk scores and the delivery of precision medicine (using type 2 diabetes as an example).
For many complex diseases, individual predisposition reflects the aggregation of risk for multiple intermediary processes that contribute to the phenotype. A subset of those relevant to T2D risk are shown on the left, with each represented in terms of a base color. Each of these processes is subject to multifactorial (genetic and non genetic) influences. For any given individual, loadings for each of these processes may range from low- to high-risk. Overall risk of T2D will depend on how many are registering as “high risk”, but phenotypic presentation and clinical course may be more dependent on the patterns of risk across the processes. Note that individuals at high risk may have their disease profile dominated by a single process (such as individual A), or may simply have above average risk loadings across multiple processes (individuals C and D).
Panel b) The translational value of human genetic data for complex metabolic phenotypes.
Translational opportunities can be broadly divided into those that are related to providing generic insights into the processes underlying disease predisposition (or trait variance), and those that can be exploited to deliver information at the level of the individual, and support precision medicine approaches. Some examples of each are provided.
Internal environments
Epidemiological associations between events in early life and later disease risk (Harder et al. 2007, Risnes et al. 2011, Lawn et al. 2014) are in part mediated through the joint effects of maternal genotype and nutritional state on intrauterine environment, acting in concert with offspring genotype (Horikoshi et al. 2016). Two competing hypotheses prevail: the fetal programming hypothesis (Hales 1994) argues that exposure in utero to maternal malnutrition could lead to smaller babies through fetal programming. In contrast, the fetal insulin hypothesis holds that an inherited predisposition to reduced insulin secretion or action within the fetal genotype can lead to lower birthweight given insulin’s important influence on fetal growth (Hattersley et al. 1999). Maternal genotype also contributes directly, since exposure to high glucose concentrations in utero (for example, in the pregnancies of diabetic mothers) drives higher birth weights through the consequent increase in fetal insulin secretion. The contributions of both maternal and fetal genotype were first illustrated in the context of rare alleles implicated in monogenic forms of diabetes (notably associated with mutations in the glucokinase gene) (Hattersley et al. 1998), but have since extended to common variants that have joint effects on early growth and later disease risk. There is, for example, substantial overlap between variants implicated in T2D-risk and those influencing birthweight, though those relationships are complex. A subset of common T2D-risk variants (such as at MTNR1B) act predominantly via their impact on maternal glycaemia and are associated with increased offspring birthweight; others (such as CDKAL1) exert their main effect on fetal insulin secretion and are associated with reduced birthweight (Beaumont et al. 2018).
The gut microbiome can be considered as another example of an “internal” environmental exposure, and there is growing evidence of a relationship between gut microflora and metabolic and anthropometric phenotypes (Ridaura et al. 2013, Bonder et al. 2016, Pedersen et al. 2016) (SANNA). However, the precise mechanisms involved, and the nature of the causal relationships remain in doubt. Recent studies have suggested that most diversity in individual microbiome content reflects environmental influences such as diet and medication (Rothschild et al. 2018, Weissbrod et al. 2018). An important corollary is that microbiome alterations aimed at improving clinical outcomes will likely work across different ancestral groups (Rothschild et al. 2018). Nonetheless, there is growing evidence that variation in host genotype also modulates microbiome content, although some of these effects may reflect genotype-diet interaction rather than direct host-microbiome effects, as in the case of variants in the gene encoding lactase (LCT, rs4988235) (Bonder et al. 2016). Genetic signals directly influencing the microbiome can serve as useful instrumental variables, and early indications from MR analyses support a causal role of microbiome diversity with respect to T2D and obesity risk (Pedersen et al. 2016) (SANNA NG in press). It has been suggested that the rapid success of bariatric surgery in restoring metabolic health may be linked to changes in gut microbiota, though this interpretation has been questioned (Aron-Wisnewsky et al. 2018, Pucci et al. 2018). Changes in gut hormones, bile acid availability, amongst other possible mechanisms, may be more important factors (reviewed in (Pucci et al. 2018)).
Translation
Translating genetics: better management of rare monogenic diseases
For those presenting with monogenic forms of metabolic disease (such as neonatal diabetes, familial hyperlipidaemias, extreme early-onset obesity), it is now standard-of-care to establish a molecular diagnosis, and to modify management accordingly. Children with neonatal diabetes due to mutations in the KNCJ11 gene (encoding part of the pancreatic beta-cell KATP channel) can usually be managed with oral sulfonylureas rather than insulin injections (Bowman et al. 2018). A molecular diagnosis of familial hypercholesterolemia should initiate intensive management of cardiovascular risk and cascade testing of relatives (Sturm et al. 2018). The lives of children with severe childhood hyperphagia and obesity attributable to mutations in the leptin gene have been transformed by leptin replacement treatment (Farooqi et al. 2014). These examples of genetically-driven personalised medicine demonstrate the benefits of targeted management in those at greatest genetic risk of disease. However, key to early adoption in these examples has been the combination of extreme, easily recognisable, clinical phenotypes matched to the presence of rare, high-impact alleles.
Conventional wisdom assumes that rare alleles have almost complete penetrance. However, growing use of exome and genome sequencing has demonstrated that, for many monogenic traits, historical estimates of penetrance were inflated by the selective ascertainment of multiply-affected pedigrees, and that alleles considered “causal” for monogenic disease can also be compatible with lifelong health (Minikel et al. 2016). For example, HNF1A alleles which, in some families, drive the segregation of MODY are also present in individuals resolutely non-diabetic into later life (Flannick et al. 2013).
These observations are consistent with the variable phenotypic expression of monogenic disease and highlight the role of other genetic and non-genetic modifiers. From a clinical perspective, incomplete penetrance complicates the task of predicting individual clinical outcome given a specific genetic result, especially when the individual concerned has not presented with the classical extreme phenotype.
Several errors have compounded this diagnostic imprecision. There has been a tendency to overattribute causal roles to any biologically-plausible allele detected through sequencing of individuals with an extreme phenotype. Some of these genes considered causal for a monogenic phenotype have subsequently been shown to play no such role (Walsh et al. 2017). Even when there is strong evidence linking some variants in a gene to a causal role in disease pathogenesis, it is naïve to assume that all variants in that gene (particularly novel alleles detected on sequencing) have equivalent functional and phenotypic impacts. Unfortunately, errors such as these, once made, tend to be propagated through the literature and clinical databases (Steele et al. 2011). The rollout of high-throughput approaches for capturing the functional impact of all possible coding alleles within a gene should provide a more accurate basis for attributing variant-level causation (Majithia et al. 2016).
Translating genetics: new targets for common multifactorial diseases
For common, multifactorial metabolic disease, individual predisposition reflects the combined effects of hundreds of genetic risk loci, and diverse, often pervasive, environmental exposures and lifestyle decisions. The genetic discoveries of the past decade are, as described above, gradually being converted into a deeper understanding of the processes involved in disease pathogenesis.
Once the genes mediating GWAS signals are identified (see above), their protein products provide novel therapeutic opportunities, their potential as targets endorsed by evidence that perturbation in humans (rather than preclinical models) is causally related to variation in the disease phenotype. Many of the known drug targets for type 2 diabetes (KCNJ11 [sulfonylureas]; PPARG [thiazolidinediones], GLP1R [GLP1 receptor agonists]) (Mahajan et al. 2018a) and hyperlipidemia (HMGCR [statins]; NPC1L1 [ezetimibe]) are detected by these analyses (Willer et al. 2013, Lange et al. 2014, Stitziel et al. 2014), reinforcing the view that this approach should provide clues to entirely novel future therapies, or support drug repositioning. The identification of individuals who carry rare loss-of-function alleles that result in abrogation or abolition of gene activity can be particularly informative in this regard, and there is major investment (from pharma and academia) in their detection through sequencing in samples from diverse ancestries (Saleheen et al. 2017). Genetics can also provide insights that discourage target development: for example, evidence either of non-efficacy or adverse effects. The most celebrated instance of the former relates to genetic data indicating that levels of HDL-cholesterol are not causally related to coronary artery disease risk (Voight et al. 2012). This finding prefigured disappointing results from randomised cardiovascular clinical trials for CETP inhibitor drugs developed specifically to elevate HDL levels (Schwartz et al. 2012).
Translating genetics: identifying modifiable non-genetic risk factors
Paradoxically, one of the most powerful translational outcomes from human genetics may be the characterisation of potentially modifiable, non-genetic factors that influence predisposition to common complex diseases. The key environmental and lifestyle factors that drive the exploding prevalence of these conditions lie somewhere in the complex nexus of global changes in nutrition, activity, and other environmental exposures and behaviours. Strong interdependencies between these diverse influences have frustrated efforts to determine whether some exposures are especially pernicious, impeding the promotion of effective public health measures that could have an impacton disease prevention and treatment (Franks et al. 2016, Moore et al. 2018).
Human genetic approaches to address these questions use a Mendelian randomisation (MR) strategy, essentially the genetic analogue of a “randomised control trial” with randomisation to different risk genotypes occurring at conception (Zheng et al. 2017). There has, for example, been a longstanding controversy (as yet not entirely resolved) concerning the causal contribution of low vitamin D levels to T2D development. The epidemiological evidence for the relationship is strong, but there are multiple confounders. The genetic approach “randomizes” individuals according to inferred differences in lifelong exposure to vitamin D using variants that influence vitamin D metabolism and action, and asks whether those groups differ in T2D-risk. Most available evidence suggests they do not (Ye et al. 2015), a result consistent with clinical trials of vitamin D supplementation (Moreira-Lucas et al. 2017). Analogous approaches are being deployed to characterise the causal consequences of a wider range of exposures including links between early growth and metabolic disease and between microbiome diversity and obesity (Bonder et al. 2016, Horikoshi et al. 2016).
Translating genetics: towards precision medicine
The preceding examples illustrate how human genetics can provide “generic” mechanistic insights that offer translational opportunities to all with the disease concerned. The focus is now turning to the value of genetics (in combination with other genomic and clinical information) to deliver precision (or personalised) medicine. This could operate across a range of clinical outcomes: from targeted prevention in those at greatest risk of future disease, to better ways of detecting those likely to have the poorest prognosis, and more effective approaches for optimising therapeutic choices. However, in comparison with the gathering momentum behind precision approaches to manage rare diseases (using germline sequencing) or cancers (tumour sequencing), precision medicine applications in common disease are currently limited.
More promising strategies are emerging. Individual variants contributing to common disease risk may have modest impact, but combined they have profound effects on personal risk. This information can be converted into polygenic risk scores capable of identifying individuals with particularly high (or low) genetic risk of disease. For example, in recent large-scale T2D association analyses, individuals with the highest 2.5% of such scores had almost ten times the prevalence of T2D as those with the lowest 2.5% (Mahajan et al. 2018a). At the population-level, the number of individuals with extreme polygenic risk of T2D far exceeds the number with similar lifetime risk from monogenic causes. Similar gradations in risk have been observed for coronary artery disease using the appropriate risk score (Khera et al. 2018). The potential for early risk stratification is clear, but, before deploying risk scores in clinical practice, we need to better understand their applicability across populations, and the extent to which the risk they capture overlaps with that enumerated using classical demographic and clinical information, such as family history, anthropometry and glycaemic measures (McCarthy et al. 2018, Torkamani et al. 2018).
Clinical application of a polygenic risk score for type 1 diabetes (T1D) has gone one step further. Recent studies have demonstrated that a T1D-risk score can identify individuals with late-onset diabetes who have type 1, rather than type 2, diabetes: these individuals are likely to suffer rapid, progressive loss of beta-cell function and require early recourse to insulin therapy (Thomas et al. 2018).
There are additional opportunities in using genetic risk scores (together with lifestyle and biomarker information) to capture the clinical and phenotypic heterogeneity evident within disease, in ways that support stratification for important clinical outcomes including disease progression and therapeutic optimisation. The approach involves generating a series of “partitioned” polygenic risk scores, each representing the subset of disease-risk variants that act through a given pathophysiological mechanism (Mahajan et al. 2018b, Udler et al. 2018). In T2D, for example, those risk scores capture the specific contributions of T2D-associated genetic variation to intermediary processes such as islet development, islet function, insulin sensitivity, and obesity. Early applications have demonstrated that partitioning of aetiological heterogeneity in this way exposes differential effects on complication risk (Udler et al. 2018). Similar approaches may also support personalised therapeutic choices based on predictions of individual efficacy and side effect profile.
Conclusion
Over the past 15 years there has been a massive growth in the discovery of genetic variants influencing a range of metabolic diseases. These have collectively highlighted the complexity of human biology: multiple variants, genes, and tissues may be involved in mediating the phenotypic effects of a single association signal. Pleiotropy is pervasive and associated variants often influence multiple traits, often in unexpected ways (Dewey et al. 2016). The upscaling of the functional assessment of variants, sequences and genes to the genome-scale (through projects such as GTEx, ENCODE and the Roadmap Epigenome) provides increasingly powerful tools for the interrogation of these signals and delivering mechanistic insights (Mahajan et al. 2018a). High-throughput tools for more precise interrogation of function are following close behind, with massively parallel reporter assays, CRISPR screens and deep mutational scans being applied at scale (Majithia et al. 2014, Findlay et al. 2018, Roman et al. 2018). Novel statistical and inferential frameworks are required for robust and accurate annotation of variant and gene function and to allow for seamless navigation from association signals to disease mechanism.
Over the next decade, we can anticipate huge advances in capitalising on the biological promise of human genetics for complex diseases, delivering causal pathways and mechanisms for diseases that are still, in many respects, mysterious. These advances will provide the platform for improvements in disease diagnosis, prevention and management. However, so that the benefits of precision medicine are available to all, there needs to be a redoubling of efforts to ensure better representation of diversity in genetic studies and more equitable access to healthcare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad S, Rukh G, Varga TV, Ali A, Kurbasic A, Shungin D, Ericson U, Koivula RW, Chu AY, Rose LM, et al. (2013). “Gene x physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry.” PLoS Genet 9(7): e1003607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad T, Lee IM, Pare G, Chasman DI, Rose L, Ridker PM and Mora S (2011). “Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women.” Diabetes Care 34(3): 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al. (2000). “The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes.” Nat Genet 26(1): 76–80. [DOI] [PubMed] [Google Scholar]
- Ameur A, Enroth S, Johansson A, Zaboli G, Igl W, Johansson AC, Rivas MA, Daly MJ, Schmitz G, Hicks AA, et al. (2012). “Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids.” Am J Hum Genet 90(5): 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim CE, Nunes K, Meyer D, Comas D, Bortolini MC, Salzano FM and Hunemeier T (2017). “Genetic signature of natural selection in first Americans.” Proc Natl Acad Sci U S A 114(9): 2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, Verger EO, Hedjazi L, Bouillot JL, Chevallier JM, et al. (2018). “Major microbiota dysbiosis in severe obesity: fate after bariatric surgery.” Gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayub Q, Moutsianas L, Chen Y, Panoutsopoulou K, Colonna V, Pagani L, Prokopenko I, Ritchie GR, Tyler-Smith C, McCarthy MI, et al. (2014). “Revisiting the thrifty gene hypothesis via 65 loci associated with susceptibility to type 2 diabetes.” Am J Hum Genet 94(2): 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, Geller F, Myhre R, Richmond RC, Paternoster L, et al. (2018). “Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics.” Hum Mol Genet 27(4): 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SD, Walker CG, Ly K, Snell RG, Atatoa Carr PE, Bandara D, Mohal J, Castro TG, Marks EJ, Morton SMB, et al. (2018). “Widespread prevalence of a CREBRF variant amongst Maori and Pacific children is associated with weight and height in early childhood.” Int J Obes (Lond) 42(4): 603–607. [DOI] [PubMed] [Google Scholar]
- Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari F, Kadhom N, et al. (2009). “Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations.” Am J Hum Genet 85(1): 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, et al. (2016). “The effect of host genetics on the gut microbiome.” Nat Genet 48(11): 1407–1412. [DOI] [PubMed] [Google Scholar]
- Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proenca C, Marchand M, Hartikainen AL, Sovio U, De Graeve F, et al. (2008). “A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels.” Science 320(5879): 1085–1088. [DOI] [PubMed] [Google Scholar]
- Bowman P, Sulen A, Barbetti F, Beltrand J, Svalastoga P, Codner E, Tessmann EH, Juliusson PB, Skrivarhaug T, Pearson ER, et al. (2018). “Effectiveness and safety of long-term treatment with sulfonylureas in patients with neonatal diabetes due to KCNJ11 mutations: an international cohort study.” Lancet Diabetes Endocrinol 6(8): 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI and Pritchard JK (2017). “An Expanded View of Complex Traits: From Polygenic to Omnigenic.” Cell 169(7): 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley MT, Racimo F, Allentoft ME, Jensen MK, Jonsson A, Huang H, Hormozdiari F, Sikora M, Marnetto D, Eskin E, et al. (2017). “Selection in Europeans on Fatty Acid Desaturases Associated with Dietary Changes.” Mol Biol Evol 34(6): 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Pagani L, Antao T, Lawson DJ, Eichstaedt CA, Yngvadottir B, Shwe MT, Wee J, Romero IG, Raj S, et al. (2014). “Genome-wide analysis of cold adaptation in indigenous Siberian populations.” PLoS One 9(5): e98076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Fuchsberger C, Pattaro C, Teumer A, Boger CA, Endlich K, Olden M, Chen MH, Tin A, Taliun D, et al. (2012). “Integration of genome-wide association studies with biological knowledge identifies six novel genes related to kidney function.” Hum Mol Genet 21(24): 5329–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, Silver KD, Timpson NJ, Hansen T, Orru M, Grazia Piras M, et al. (2008). “Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels.” J Clin Invest 118(7): 2620–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Bruning JC, Nolan PM, Ashcroft FM, et al. (2010). “Overexpression of Fto leads to increased food intake and results in obesity.” Nat Genet 42(12): 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. (2015). “FTO Obesity Variant Circuitry and Adipocyte Browning in Humans.” N Engl J Med 373(10): 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente FJ, Cardona A, Inchley CE, Peter BM, Jacobs G, Pagani L, Lawson DJ, Antao T, Vicente M, Mitt M, et al. (2014). “A Selective Sweep on a Deleterious Mutation in CPT1A in Arctic Populations.” Am J Hum Genet 95(5): 584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SA, Sinclair G, McIntosh S, Bamforth F, Thompson R, Sobol I, Osborne G, Corriveau A, Santos M, Hanley B, et al. (2010). “Carnitine palmitoyltransferase 1A (CPT1A) P479L prevalence in live newborns in Yukon, Northwest Territories, and Nunavut.” Mol Genet Metab 101(2–3): 200–204. [DOI] [PubMed] [Google Scholar]
- Corella D, Arnett DK, Tucker KL, Kabagambe EK, Tsai M, Parnell LD, Lai CQ, Lee YC, Warodomwichit D, Hopkins PN, et al. (2011). “A high intake of saturated fatty acids strengthens the association between the fat mass and obesity-associated gene and BMI.” J Nutr 141(12): 2219–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corella D, Ortega-Azorin C, Sorli JV, Covas MI, Carrasco P, Salas-Salvado J, Martinez-Gonzalez MA, Aros F, Lapetra J, Serra-Majem L, et al. (2012). “Statistical and biological gene-lifestyle interactions of MC4R and FTO with diet and physical activity on obesity: new effects on alcohol consumption.” PLoS One 7(12): e52344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom E and Sandholm N (2017). “Progress in Defining the Genetic Basis of Diabetic Complications.” Curr Diab Rep 17(9): 80. [DOI] [PubMed] [Google Scholar]
- Defesche JC, Gidding SS, Harada-Shiba M, Hegele RA, Santos RD and Wierzbicki AS (2017). “Familial hypercholesterolaemia.” Nat Rev Dis Primers 3: 17093. [DOI] [PubMed] [Google Scholar]
- Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, O’Dushlaine C, Van Hout CV, Staples J, Gonzaga-Jauregui C, et al. (2016). “Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study.” Science 354(6319). [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H and Goldstein DB (2010). “Rare variants create synthetic genome-wide associations.” PLoS Biol 8(1): e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. (2010). “New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk.” Nat Genet 42(2): 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks CE, Loos RJ, Hardy R, Wills AK, Wong A, Wareham NJ, Kuh D and Ong KK (2012). “Adult obesity susceptibility variants are associated with greater childhood weight gain and a faster tempo of growth: the 1946 British Birth Cohort Study.” Am J Clin Nutr 95(5): 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Aukrust I, Bjorkhaug L, Burtt NP, Mercader JM, Garcia-Ortiz H, Huerta-Chagoya A, Moreno-Macias H, Walford G, Flannick J, et al. (2014). “Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population.” JAMA 311(22): 2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi IS and O’Rahilly S (2014). “20 years of leptin: human disorders of leptin action.” J Endocrinol 223(1): T63–70. [DOI] [PubMed] [Google Scholar]
- Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD, Huang X, Starita LM and Shendure J (2018). “Accurate classification of BRCA1 variants with saturation genome editing.” Nature 562(7726): 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC and Ruther U (2009). “Inactivation of the Fto gene protects from obesity.” Nature 458(7240): 894–898. [DOI] [PubMed] [Google Scholar]
- Flannick J, Beer NL, Bick AG, Agarwala V, Molnes J, Gupta N, Burtt NP, Florez JC, Meigs JB, Taylor H, et al. (2013). “Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes.” Nat Genet 45(11): 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PW and McCarthy MI (2016). “Exposing the exposures responsible for type 2 diabetes and obesity.” Science 354(6308): 69–73. [DOI] [PubMed] [Google Scholar]
- Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ, et al. (2016). “The genetic architecture of type 2 diabetes.” Nature 536(7614): 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. (2015). “Greenlandic Inuit show genetic signatures of diet and climate adaptation.” Science 349(6254): 1343–1347. [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Magi R, Reschen ME, Mahajan A, Locke A, Rayner NW, Robertson N, et al. (2015). “Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci.” Nat Genet 47(12): 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. (2007). “The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase.” Science 318(5855): 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, Winkler TW, Chu AY, Mahajan A, Hadley D, et al. (2017). “Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults.” PLoS Genet 13(4): e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. (2006). “Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes.” Nat Genet 38(3): 320–323. [DOI] [PubMed] [Google Scholar]
- Greenberg CR, Dilling LA, Thompson GR, Seargeant LE, Haworth JC, Phillips S, Chan A, Vallance HD, Waters PJ, Sinclair G, et al. (2009). “The paradox of the carnitine palmitoyltransferase type Ia P479L variant in Canadian Aboriginal populations.” Mol Genet Metab 96(4): 201–207. [DOI] [PubMed] [Google Scholar]
- Hales CN (1994). “The pathogenesis of NIDDM.” Diabetologia 37 Suppl 2: S162–168. [DOI] [PubMed] [Google Scholar]
- Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G and Froguel P (1998). “Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians.” Diabetologia 41(12): 1511–1515. [DOI] [PubMed] [Google Scholar]
- Harder T, Rodekamp E, Schellong K, Dudenhausen JW and Plagemann A (2007). “Birth weight and subsequent risk of type 2 diabetes: a meta-analysis.” Am J Epidemiol 165(8): 849–857. [DOI] [PubMed] [Google Scholar]
- Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R and Ellard S (1998). “Mutations in the glucokinase gene of the fetus result in reduced birth weight.” Nat Genet 19(3): 268–270. [DOI] [PubMed] [Google Scholar]
- Hattersley AT and Patel KA (2017). “Precision diabetes: learning from monogenic diabetes.” Diabetologia 60(5): 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley AT and Tooke JE (1999). “The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease.” Lancet 353(9166): 1789–1792. [DOI] [PubMed] [Google Scholar]
- Hegele RA, Cao H, Harris SB, Hanley AJ and Zinman B (1999). “Hepatocyte nuclear factor-1 alpha G319S. A private mutation in Oji-Cree associated with type 2 diabetes.” Diabetes Care 22(3): 524. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, Grarup N, et al. (2016). “Genome-wide associations for birth weight and correlations with adult disease.” Nature 538(7624): 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. (2001). “Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease.” Nature 411(6837): 599–603. [DOI] [PubMed] [Google Scholar]
- Johnson W, Ong KK, Elks CE, Wareham NJ, Wong A, Muniz-Terrera G, Hardy R, M. N. scientific and t. data collection (2014). “Modification of genetic influences on adiposity between 36 and 63 years of age by physical activity and smoking in the 1946 British Birth Cohort Study.” Nutr Diabetes 4: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. (2009). “Common variants at 30 loci contribute to polygenic dyslipidemia.” Nat Genet 41(1): 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. (2018). “Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations.” Nat Genet 50(9): 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, et al. (2018). “Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program.” Nat Genet 50(11): 1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraja AT, Cook JP, Warren HR, Surendran P, Liu C, Evangelou E, Manning AK, Grarup N, Drenos F, Sim X, et al. (2017). “New Blood Pressure-Associated Loci Identified in Meta-Analyses of 475 000 Individuals.” Circ Cardiovasc Genet 10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange LA, Hu Y, Zhang H, Xue C, Schmidt EM, Tang ZZ, Bizon C, Lange EM, Smith JD, Turner EH, et al. (2014). “Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol.” Am J Hum Genet 94(2): 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn JE, Blencowe H, Oza S, You D, Lee AC, Waiswa P, Lalli M, Bhutta Z, Barros AJ, Christian P, et al. (2014). “Every Newborn: progress, priorities, and potential beyond survival.” Lancet 384(9938): 189–205. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, Wareham NJ and Loos RJ (2010). “Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study.” PLoS Med 7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. (2015). “Genetic studies of body mass index yield new insights for obesity biology.” Nature 518(7538): 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ (2016). “CREBRF variant increases obesity risk and protects against diabetes in Samoans.” Nat Genet 48(9): 976–978. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Belmont JW, Boerwinkle E and Gibbs RA (2011). “Clan genomics and the complex architecture of human disease.” Cell 147(1): 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, et al. (2014). “Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility.” Nat Genet 46(3): 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, Highland HM, Locke AE, Grarup N, Im HK, Cingolani P, et al. (2015). “Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus.” PLoS Genet 11(1): e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. (2018a). “Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps.” Nat Genet 50(11): 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Wessel J, Willems SM, Zhao W, Robertson NR, Chu AY, Gan W, Kitajima H, Taliun D, Rayner NW, et al. (2018b). “Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes.” Nat Genet 50(4): 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majithia AR, Flannick J, Shahinian P, Guo M, Bray MA, Fontanillas P, Gabriel SB, T. D. C. Go, N. J. F. A. S. Project, S. T. D. Consortium, et al. (2014). “Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes.” Proc Natl Acad Sci U S A 111(36): 13127–13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majithia AR, Tsuda B, Agostini M, Gnanapradeepan K, Rice R, Peloso G, Patel KA, Zhang X, Broekema MF, Patterson N, et al. (2016). “Prospective functional classification of all possible missense variants in PPARG.” Nat Genet 48(12): 1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. (2009). “Finding the missing heritability of complex diseases.” Nature 461(7265): 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. (2015). “Genome-wide patterns of selection in 230 ancient Eurasians.” Nature 528(7583): 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI and Mahajan A (2018). “The value of genetic risk scores in precision medicine for diabetes.” Expert Review of Precision Medicine and Drug Development 3(5): 279–281. [Google Scholar]
- McClellan J and King MC (2010). “Genetic heterogeneity in human disease.” Cell 141(2): 210–217. [DOI] [PubMed] [Google Scholar]
- Minikel EV, Vallabh SM, Lek M, Estrada K, Samocha KE, Sathirapongsasuti JF, McLean CY, Tung JY, Yu LP, Gambetti P, et al. (2016). “Quantifying prion disease penetrance using large population control cohorts.” Sci Transl Med 8(322): 322ra329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minster RL, Hawley NL, Su CT, Sun G, Kershaw EE, Cheng H, Buhule OD, Lin J, Reupena MS, Viali S, et al. (2016). “A thrifty variant in CREBRF strongly influences body mass index in Samoans.” Nat Genet 48(9): 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltke I, Grarup N, Jorgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, Korneliussen TS, Andersen MA, Nielsen TS, Krarup NT, et al. (2014). “A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes.” Nature 512(7513): 190–193. [DOI] [PubMed] [Google Scholar]
- Moore R, Casale FP, Jan Bonder M, Horta D, Heijmans BT, C.’t Hoen PA, van Meurs J, Isaacs A, Jansen R, Franke L, et al. (2018). “A linear mixed-model approach to study multivariate gene–environment interactions.” Nature Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R, Barroso I; and Stegle O (2018). PgmNr 2170: Phenome-wide patterns of genotype-environment (GxE) interactions. American Society Human Genetics. [Google Scholar]
- Moreira-Lucas TS, Duncan AM, Rabasa-Lhoret R, Vieth R, Gibbs AL, Badawi A and Wolever TM (2017). “Effect of vitamin D supplementation on oral glucose tolerance in individuals with low vitamin D status and increased risk for developing type 2 diabetes (EVIDENCE): A double-blind, randomized, placebo-controlled clinical trial.” Diabetes Obes Metab 19(1): 133–141. [DOI] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. (2012). “Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes.” Nat Genet 44(9): 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka I, Furusawa T, Kimura R, Natsuhara K, Yamauchi T, Nakazawa M, Ataka Y, Ishida T, Inaoka T, Matsumura Y, et al. (2017). “A missense variant, rs373863828-A (p.Arg457Gln), of CREBRF and body mass index in Oceanic populations.” J Hum Genet 62(9): 847–849. [DOI] [PubMed] [Google Scholar]
- Neel JV (1962). “Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”?” Am J Hum Genet 14: 353–362. [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. (2001). “A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease.” Nature 411(6837): 603–606. [DOI] [PubMed] [Google Scholar]
- Ohashi J, Naka I, Furusawa T, Kimura R, Natsuhara K, Yamauchi T, Nakazawa M, Ishida T, Inaoka T, Matsumura Y, et al. (2018). “Association study of CREBRF missense variant (rs373863828:G > A; p.Arg457Gln) with levels of serum lipid profile in the Pacific populations.” Ann Hum Biol 45(3): 215–219. [DOI] [PubMed] [Google Scholar]
- Pallister T, Sharafi M, Lachance G, Pirastu N, Mohney RP, MacGregor A, Feskens EJ, Duffy V, Spector TD and Menni C (2015). “Food Preference Patterns in a UK Twin Cohort.” Twin Res Hum Genet 18(6): 793–805. [DOI] [PubMed] [Google Scholar]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G, et al. (2016). “Human gut microbes impact host serum metabolome and insulin sensitivity.” Nature 535(7612): 376–381. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Kesse-Guyot E, McManus R, Hercberg S, Lairon D, Planells R and Roche HM (2012). “High dietary saturated fat intake accentuates obesity risk associated with the fat mass and obesity-associated gene in adults.” J Nutr 142(5): 824–831. [DOI] [PubMed] [Google Scholar]
- Pucci A and Batterham RL (2018). “Mechanisms underlying the weight loss effects of RYGB and SG: similar, yet different.” J Endocrinol Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, DeGiorgio M, Albrechtsen A, Moltke I, Skoglund P, Korneliussen TS, Gronnow B, Appelt M, Gullov HC, Friesen TM, et al. (2014). “The genetic prehistory of the New World Arctic.” Science 345(6200): 1255832. [DOI] [PubMed] [Google Scholar]
- Rajakumar C, Ban MR, Cao H, Young TK, Bjerregaard P and Hegele RA (2009). “Carnitine palmitoyltransferase IA polymorphism P479L is common in Greenland Inuit and is associated with elevated plasma apolipoprotein A-I.” J Lipid Res 50(6): 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M, Karlsson T, Ek WE and Johansson A (2017). “Gene-environment interaction study for BMI reveals interactions between genetic factors and physical activity, alcohol consumption and socioeconomic status.” PLoS Genet 13(9): e1006977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW Jr., Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, et al. (2014). “The genome of a Late Pleistocene human from a Clovis burial site in western Montana.” Nature 506(7487): 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddon H, Gerstein HC, Engert JC, Mohan V, Bosch J, Desai D, Bailey SD, Diaz R, Yusuf S, Anand SS, et al. (2016). “Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study.” Sci Rep 6: 18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AS, North KE, Graff M, Young KM, Mohlke KL, Lange LA, Lange EM, Harris KM and Gordon-Larsen P (2014). “Moderate to vigorous physical activity interactions with genetic variants and body mass index in a large US ethnically diverse cohort.” Pediatr Obes 9(2): e35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. (2013). “Gut microbiota from twins discordant for obesity modulate metabolism in mice.” Science 341(6150): 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risnes KR, Vatten LJ, Baker JL, Jameson K, Sovio U, Kajantie E, Osler M, Morley R, Jokela M, Painter RC, et al. (2011). “Birthweight and mortality in adulthood: a systematic review and meta-analysis.” Int J Epidemiol 40(3): 647–661. [DOI] [PubMed] [Google Scholar]
- Roman TS and Mohlke KL (2018). “Functional genomics and assays of regulatory activity detect mechanisms at loci for lipid traits and coronary artery disease.” Curr Opin Genet Dev 50: 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. (2018). “Environment dominates over host genetics in shaping human gut microbiota.” Nature 555(7695): 210–215. [DOI] [PubMed] [Google Scholar]
- Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA, Won HH, Karczewski KJ, O’Donnell-Luria AH, Samocha KE, et al. (2017). “Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity.” Nature 544(7649): 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. (2014). “Biological insights from 108 schizophrenia-associated genetic loci.” Nature 511(7510): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, et al. (2012). “Effects of dalcetrapib in patients with a recent acute coronary syndrome.” N Engl J Med 367(22): 2089–2099. [DOI] [PubMed] [Google Scholar]
- Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Magi R, Strawbridge RJ, Rehnberg E, Gustafsson S, et al. (2012). “Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways.” Nat Genet 44(9): 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al. (2015). “New genetic loci link adipose and insulin biology to body fat distribution.” Nature 518(7538): 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. (2014). “Obesity-associated variants within FTO form long-range functional connections with IRX3.” Nature 507(7492): 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E and Orho-Melander M (2009). “Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity.” Am J Clin Nutr 90(5): 1418–1425. [DOI] [PubMed] [Google Scholar]
- Steele AM, Tribble ND, Caswell R, Wensley KJ, Hattersley AT, Gloyn AL and Ellard S (2011). “The previously reported T342P GCK missense variant is not a pathogenic mutation causing MODY.” Diabetologia 54(8): 2202–2205. [DOI] [PubMed] [Google Scholar]
- Stitziel NO, Won HH, Morrison AC, Peloso GM, Do R, Lange LA, Fontanillas P, Gupta N, Duga S, Goel A, et al. (2014). “Inactivating mutations in NPC1L1 and protection from coronary heart disease.” N Engl J Med 371(22): 2072–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm AC, Knowles JW, Gidding SS, Ahmad ZS, Ahmed CD, Ballantyne CM, Baum SJ, Bourbon M, Carrie A, Cuchel M, et al. (2018). “Clinical Genetic Testing for Familial Hypercholesterolemia: JACC Scientific Expert Panel.” J Am Coll Cardiol 72(6): 662–680. [DOI] [PubMed] [Google Scholar]
- Thomas NJ, Jones SE, Weedon MN, Shields BM, Oram RA and Hattersley AT (2018). “Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank.” Lancet Diabetes Endocrinol 6(2): 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Wineinger NE and Topol EJ (2018). “The personal and clinical utility of polygenic risk scores.” Nat Rev Genet 19(9): 581–590. [DOI] [PubMed] [Google Scholar]
- Tung YCL, Yeo GSH, O’Rahilly S and Coll AP (2014). “Obesity and FTO: Changing Focus at a Complex Locus.” Cell Metab 20(5): 710–718. [DOI] [PubMed] [Google Scholar]
- Udler MS, Kim J, von Grotthuss M, Bonas-Guarch S, Cole JB, Chiou J, D. A. o. b. o. M. Christopher, I. the, Boehnke M, Laakso M, et al. (2018). “Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: A soft clustering analysis.” PLoS Med 15(9): e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI and Yang J (2012). “Five years of GWAS discovery.” Am J Hum Genet 90(1): 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA and Yang J (2017). “10 Years of GWAS Discovery: Biology, Function, and Translation.” Am J Hum Genet 101(1): 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Holm H, Ding EL, Johnson T, et al. (2012). “Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study.” Lancet 380(9841): 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, et al. (2017). “Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples.” Genet Med 19(2): 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrod O, Rothschild D, Barkan E and Segal E (2018). “Host genetics and microbiome associations through the lens of genome wide association studies.” Curr Opin Microbiol 44: 9–19. [DOI] [PubMed] [Google Scholar]
- Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, Dauriz M, Hivert MF, Raghavan S, Lipovich L, et al. (2015). “Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility.” Nat Commun 6: 5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler E, Leong A, Liu CT, Hivert MF, Strawbridge RJ, Podmore C, Li M, Yao J, Sim X, Hong J, et al. (2017). “Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis.” PLoS Med 14(9): e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. (2013). “Discovery and refinement of loci associated with lipid levels.” Nat Genet 45(11): 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, Garcia-Ortiz H, Gomez-Vazquez MJ, Burtt NP, Aguilar-Salinas CA, et al. (2014). “Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico.” Nature 506(7486): 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Wijmenga C, Sullivan PF, Yang J and Visscher PM (2018). “Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model.” Cell 173(7): 1573–1580. [DOI] [PubMed] [Google Scholar]
- WTCCC, W. T. C. C. C. (2007). “Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls.” Nature 447(7145): 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE, Zheng Z, Yengo L, Lloyd-Jones LR, Sidorenko J, Wu Y, et al. (2018). “Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes.” Nat Commun 9(1): 2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, Robinson MR, Perry JR, Nolte IM, van Vliet-Ostaptchouk JV, et al. (2015). “Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index.” Nat Genet 47(10): 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Sharp SJ, Burgess S, Scott RA, Imamura F, InterAct C, Langenberg C, Wareham NJ and Forouhi NG (2015). “Association between circulating 25-hydroxyvitamin D and incident type 2 diabetes: a mendelian randomisation study.” Lancet Diabetes Endocrinol 3(1): 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AI, Wauthier F and Donnelly P (2016). “Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index.” Nat Commun 7: 12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AI, Wauthier FL and Donnelly P (2018). “Identifying loci affecting trait variability and detecting interactions in genome-wide association studies.” Nat Genet 50(11): 1608–1614. [DOI] [PubMed] [Google Scholar]
- Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, Evans DM and Smith GD (2017). “Recent Developments in Mendelian Randomization Studies.” Curr Epidemiol Rep 4(4): 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]