Abstract
Studies of the genetics of psychiatric disorders has become one of the most exciting and fast-moving areas in human genetics. A decade ago, there were few reproducible findings and now there are hundreds. In this review, we focus on the findings that have illuminated the genetic architecture of psychiatric disorders and the challenges of using these findings to inform our understanding of pathophysiology. The evidence is now overwhelming that psychiatric disorders are “polygenic”, that many genetic loci contribute to risk. With the exception of a subset of those with ASD, few individuals with a psychiatric disorder have a single, deterministic genetic cause; rather, developing a psychiatric disorder is influenced by hundreds of different genetic variants, consistent with a polygenic model. As progressively larger studies have uncovered more about their genetic architecture, the need to elucidate additional architectures has become clear. Even if we were to have complete knowledge of the genetic architecture of a psychiatric disorder, full understanding requires deep knowledge of the functional genomic architecture – the implicated loci impact regulatory processes that influence gene expression and the functional coordination of genes that control biological processes. Following from this is cellular architecture: of all brain regions, cell types, and developmental stages, where and when are the functional architectures operative? Given that the genetic architectures of different psychiatric disorders often strongly overlap, we are challenged to re-evaluate and refine the diagnostic architectures of psychiatric disorders using fundamental genetic and neurobiological data.
Introduction
Psychiatric disorders are the most enigmatic maladies in medicine. Although their existence has been known for millennia (Porter, 2002) and their impact on the public health well-documented, remarkably little is known about their causal risk factors and fundamental neurobiology despite a considerable corpus of research. In the past century, many have applied the best tools then available but, until recently, without reproducible successes. The lack of success using approaches that were fruitful elsewhere is attributable an inadequate toolkit and the intrinsic complexity of the brain. Psychiatric disorders impact higher cortical functions (mood, behavior, perception, and cognition), which are far more difficult to localize, quantify, and model than more basic neurological functions. In addition, psychiatric disorders are defined based on self-report and observation of cognition and behavior rather than on direct measurement of an etiological factor, making them syndromes rather than single diseases. These features strongly suggest diverse and complex etiologies.
Despite these challenges, there has been remarkable progress in the past decade in elucidating the genetic underpinnings of psychiatric disorders with numerous findings that meet modern criteria for significance and reproducibility (Geschwind and Flint, 2015; Sullivan et al., 2018). In this review, we focus on the findings that have illuminated the genetic architecture of psychiatric disorders and the challenges of using these findings to inform our understanding of pathophysiology. Genetic architecture refers to the overall composition of the implicated risk variants in the population – the total number of variants and, for each, the frequencies in those afflicted and in the general population, and the degree of risk conferred (Timpson et al., 2018). The concept of genetic architecture is applicable to any trait (e.g., Huntington’s disease is caused by a rare, deterministic variant). Knowledge of genetic architecture can help optimize gene discovery (e.g., study design, ascertainment, and choice of genotyping technology) (Timpson et al., 2018; Visscher et al., 2012). Genetic architecture can inform prospects for clinical utility: although many deterministic monogenetic conditions are predicted or diagnosed using genetic testing, application to most psychiatric disorders traverses far more murky, probabilistic terrain (Timpson et al., 2018).
The evidence is now overwhelming that psychiatric disorders have a “polygenic” basis – that many genetic loci mostly with small effect sizes contribute to risk (Visscher et al., 2017). In this respect, psychiatric disorders are broadly similar to other common biomedical diseases. The polygenic concept allows for the fact that some individuals can harbor genetic variants of far larger effects. This is particularly salient for ASD where a large effect variant is present in ~15% of cases along with smaller proportions of individuals with TS, ADHD, and SCZ (Iossifov et al., 2012; Sanders et al., 2012; Satterstrom et al., 2018c; Singh et al., 2016; Willsey et al., 2017). A polygenic model can include weak and strong genetic effects as well as non-genetic influences (e.g., the impact of environmental exposures, life events (e.g., chronic fear), and the impact of individual choices). A key empirical finding is that genetic risk can be non-specific and shared to varying extents across many adult and childhood onset psychiatric disorders (Brainstorm Consortium, 2018; Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013b; Schork et al., In press).
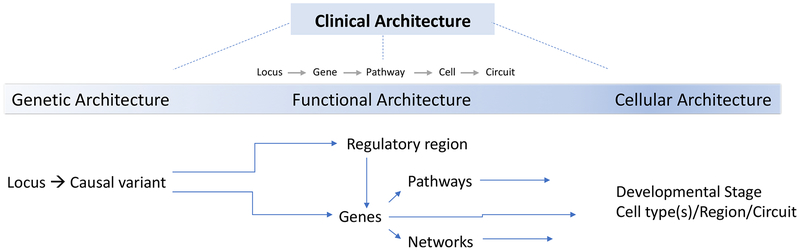
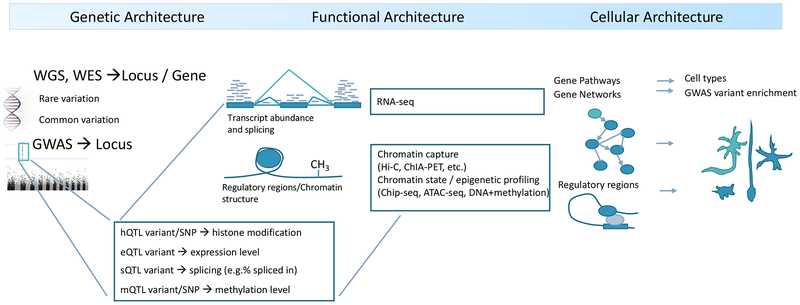
As progressively larger studies of psychiatric disorders have uncovered increasingly more about their genetic architecture, the need to elucidate additional “architectures” has become clear (Figure 1). Even if we were to have complete knowledge of the genetic architecture of a psychiatric disorder, full understanding requires deep knowledge of the functional genomic architecture – how these loci interact in the nucleus (often across large distances), how gene and isoform expression are coordinated for many genes, and how these affect networks. Second, following from this, is cellular architecture: of all brain regions, cell types, and developmental stages, where and when are the functional architectures operative and what circuits do they influence? Finally, the data used to diagnose psychiatric disorders consist of signs and symptoms determined during patient-clinician interactions that infrequently have recourse to objective biomarkers to support or refute a diagnosis. Furthermore, the internationally accepted definitions of psychiatric disorders were crafted by experts and influenced by traditions dating back a century or more. Given that the genetic architectures of different psychiatric disorders can strongly overlap, we are challenged to re-evaluate and refine the diagnostic architectures of psychiatric disorders with respect to fundamental genetic and neurobiological data.
Figure 1: Relationship of the levels of disease architecture to different stages of analysis.
Genetic studies identify the loci and causal variants that impact disease and thereby its genetic architecture. The subset of causal variants in coding regions are typically directly assignable to genes. As many loci are non-coding, regulatory regions and the genes they regulate need to be empirically defined and identified – such studies render the functional architecture of disease. As psychiatric disorders all appear to be polygenic, it is also necessary to consider the implicated genes in the context of biological networks and pathways. Sets of genes and networks can be places in specific developmental stages and cell types to generate more precise understanding their effects on brain regions and circuits. Clinical architecture – the “structure” of the interrelationships between psychiatric syndromes – is subsequently refined by increased knowledge at each of these levels.
Psychiatric disorders and genetics
Definitions.
Many psychiatric disorders are internationally recognized (World Health Organization, 1993). In this review, we focus on the ten psychiatric disorders that have been the subject of the greatest scrutiny by geneticists, and all are the focus of working groups in the Psychiatric Genomics Consortium (PGC) (Sullivan et al., 2018). We do not cover dementia and intellectual disability which are often considered neurological conditions with prominent psychiatric manifestations, but recognize the inherent arbitrariness of following this conventional delineation. Table 1 contains brief definitions of each condition along with lifetime prevalence rates and twin-heritabilities. The essence of each disorder is a persistent, pervasive, and pathological pattern of abnormal mood (as in mania or major depression), perception (e.g., auditory hallucinations in SCZ or bizarrely distorted body image in AN), behavior (e.g., repetitive hand-washing in OCD or injurious ethanol consumption in ALC), or higher-level cognition (e.g., delusions in SCZ). People with serious psychiatric disorders are often acutely aware that their symptoms and behaviors “don’t make sense,” and have made exhaustive attempts to ameliorate their illness.
Table 1.
Descriptive features of 10 psychiatric disorders
| Abbrev. | Name | Lifetime prevalence | Twin-heritability | SNP-heritability | GWAS cases | GWAS loci | Essential characteristics | Notable impacts |
|---|---|---|---|---|---|---|---|---|
| ADHD | Attention-deficit hyperactivity disorder | 0.053 | 0.76 | 0.216 | 20,183 | 12 | Persistent inattention, hyperactivity, impulsivity | Costs estimated at ~$US 100 billion/year |
| ALC | Alcohol dependence | 0.125 | 0.51 | 0.090 | 14,904 | 2 | Persistent ethanol use despite tolerance, withdrawal, dysfunction | Most expensive psychiatric disorder (total costs >$US 225 billion/year) |
| AN | Anorexia nervosa | 0.009 | 0.58 | 0.110 | 16,992 | 8 | Dangerously low weight from self-starvation | Notably high standardized mortality ratio |
| ASD | Autism spectrum disorder | 0.017 | 0.74 | 0.118 | 18,381 | 5 | Abnormal social interaction and communication beginning before age 3 | Wide range of function, from complete care to exceptional achievement |
| BIP | Bipolar disorder | 0.010 | 0.85 | 0.213 | 29,764 | 30 | Manic-depressive illness, episodes of mania usually with depressive episodes | Nearly as disabling as SCZ |
| MDD | Major depressive disorder | 0.162 | 0.37 | 0.087 | 135,458 | 44 | Unipolar depression, persistent dysphoria with physical/cognitive symptoms | Top five in burden of disease globally |
| OCD | Obsessive-compulsive disorder | 0.011 | 0.47 | 0.280 | 2,688 | 0 | Uncontrollable, persistent, thoughts (obsessions) and repetitive behaviors (compulsions) | Top 10 globally for lost income and decreased quality of life |
| PTSD | Post-traumatic stress disorder | 0.068 | 0.46 | 0.038 | 23,212 | 2 | Trauma-related re-experiencing, avoidance, negative thoughts, and hyperarousal † | High medical and psychiatric comorbidities (suicide, substance depdence). |
| SCZ | Schizophrenia | 0.004 | 0.81 | 0.244 | 40,675 | 145 | Long-standing delusions and hallucinations | Life expectancy decreased by 12–15 years |
| TS | Tourette’s syndrome | 0.005 | 0.37 ‡ | 0.350 | 4,819 | 1 | Vocal or motor tics (stereotyped, involuntary movement and utterances) | Comorbid psychiatric disorders cause more disability than tics. |
All definitions are made more restrictive by requiring persistence over time (e.g., the criteria for SCZ require ≥6 months of symptoms), presence in different contexts (e.g., for ADHD, inattention at home, school, and in peer interactions), and significant impairment. See Table S1 for data and citations. Updated from (Sullivan et al., 2012).
PTSD is distinctive in requiring traumatic exposure to death, injury, or sexual violence.
Heritability from national pedigrees is higher (0.77).
Each of these disorders has an explicit operational definition based on symptoms (reported by a person or an informant) and signs (observed by a clinician). Many diagnostic features from laboratory testing, brain imaging, or pathology have been evaluated but few have acceptable positive and negative predictive values to support routine clinical use. One exception is the measurement of intelligence which defines intellectual disability and which is an important clinical stratifier for many psychiatric disorders (particularly ADHD and ASD). Thus, these conditions are “disorders” or syndromes not “diseases” due to their descriptive/syndromic definitions without objective defining features based on etiology. All are idiopathic with rare exceptions (single-gene disorders with prominent ASD features like MECP2 and Rett Syndrome).
Impact.
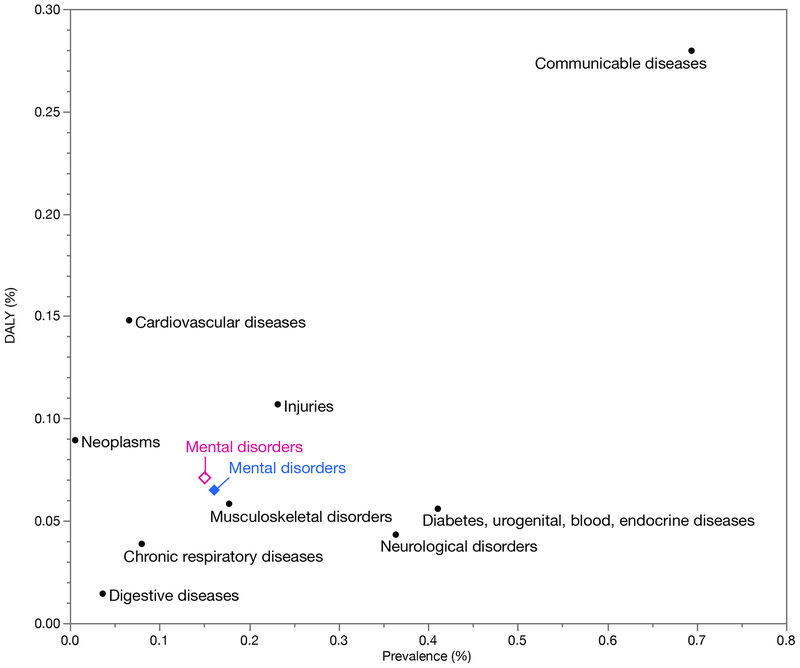
Psychiatric disorders are among the conditions with the greatest impacts (Global Burden of Disease Collaborative Network, 2017), ranking fifth globally in causes of disability (Figure 2). These disorders are associated with considerable morbidity and increased rates of mortality due to suicide and ill health (e.g., 10–15 year reduction in life expectancy for SCZ), and cost (due to health care, disability, and lost income). The human impact of a severe mental illness on the lives of the people afflicted and their families and communities is not readily condensed into a statistic but is nonetheless often profound. In addition, empirical studies have demonstrated the effectiveness of social, psychological, and/or pharmacological therapies for all of these disorders. These are treatable conditions and treatment often leads to marked improvements in symptoms and quality of life. However, particularly for severe psychiatric disorders, current therapies may only mitigate symptoms. Therapeutic failure is common.
Figure 2:
Prevalence and impact of psychiatric disorders compared to other major diseases. Looking at both measures allows evaluation of both how common and how impactful a psychiatric disorder is. These data are from global surveys, and we have included other major classes of disease. Prevalence (X-axis) and disability-adjusted life years (DALYs, Y-axis) for ten major classes of disorders. DALYs are a measure of overall disease burden due to the number of years lost due to poor health, disability, or premature mortality, here expressed as the proportion of total global DALYs. Psychiatric disorders rank fifth and accounted for 6.7% (females are the open diamond and males the closed diamond) (Global Burden of Disease Collaborative Network, 2017).
Commonalities.
Four clinical features of psychiatric disorders are notable. First, there is considerable clinical variability. For example, individuals with ADHD or OCD can have mild, transient symptoms in childhood or lifelong, incapacitating symptoms. People with ASD can have profound impairment requiring lifelong care, as well as high academic/occupational achievement (despite impairments in social relations and behavioral flexibility). Features of many psychiatric disorders are on a continuum: depressed mood is a normal human experience, but becomes MDD if present continuously for weeks or months. Second, many psychiatric disorders are chronic illnesses: MDD often begins in adolescence and recurs throughout adulthood. SCZ frequently begins in early adulthood and is often life-altering. Most people with ASD in childhood continue to have ASD in adulthood (Billstedt et al., 2007; Howlin and Magiati, 2017). Third, given the syndromic nature of the definitions, it is unsurprising that these conditions are commonly comorbid (e.g., many people with AN or ALC also meet criteria for MDD, AN overlaps considerably with MDD and OCD, and about half of people with ASD have ADHD symptoms (de Bruin et al., 2007).
Finally, the neurological impact of psychiatric disorders can be subtle. Some individuals have important neurological impairments (e.g., epilepsy and motor or sensory abnormalities) or neurological “soft-signs” (deficits in sensory integration, coordination, and complex motor sequencing). However, most people with a severe psychiatric disorder have little if any neurological impairment (e.g., consciousness, sensation, motor function, language, and many aspects of memory). Individuals who are at the worst point in their illnesses – floridly hallucinating, severely manic, profoundly melancholic, or starved down to a body mass index of 10 – usually have normal neurological exams and unremarkable or only non-specific structural and functional brain imaging findings. This again suggests relatively subtle and heterogeneous etiological processes.
A brief history of genetic studies.
For over 150 years, researchers have applied the best available methods to try to find the causes of serious psychiatric disorders. Many of these methods had been informative for other medical disorders but unsuccessful for psychiatric disorders. The most reproducible single clue that emerged was the tendency for psychiatric disorders to “run” in families – as established by 50+ years of twin, family, and adoption studies (summarized in Table 1) (Polderman et al., 2015). This observation logically led to attempts to identify the specific locations in the genome conferring risk. The progression of genetic studies mirrors technology development since the 1960s: single protein biomarkers, small numbers of restriction fragment length polymorphisms, genome-wide panels of microsatellite markers for linkage analysis, small numbers of selected single nucleotide polymorphisms (SNPs), arrays containing 105-106 genome-wide SNPs, and resequencing of genes and then exomes and whole genomes. Whenever a new technology emerged, a prominent early success was strongly influential. Examples include: identification of a genomic region for Huntington’s disease using linkage analysis of 12 markers in 1983, the association of common variation in APOE with Alzheimer’s disease in a candidate gene study of 30 cases in 1993, identification of CFH as a risk factor for age-related macular degeneration using SNP arrays in 96 cases in 2005, and exome sequencing identifying the cause of Miller syndrome in four cases in 2009.
These early successes were a form of “winner’s curse” (Ioannidis, 2005) that led to gross underestimation of the efforts that would ultimately be required (we note that geneticists working on most other complex human diseases were similarly misled). Linkage analysis is poorly powered for complex traits (Risch and Merikangas, 1996). Compared to current knowledge, the reproducible yield of candidate gene association studies is negligible (Farrell et al., 2015). Linkage and candidate gene studies led to many claims of gene discovery (e.g., COMT, DISC1, DTNBP1, and NRG1 for SCZ) that were not subsequently supported (Border et al., In press; Farrell et al., 2015). Psychiatric genetics was bedeviled by reproducibility problems.
Global consortia.
The failure of simple models led to widespread acknowledgement of a need for sample sizes that were beyond the reach of any single group to achieve power to detect generalizable findings. The need for unprecedented levels of cooperation became widely recognized (Fischbach and Lord, 2010; Geschwind et al., 2001; Lajonchere and Consortium, 2010; Moldin, 2003; Psychiatric GWAS Consortium Coordinating Committee, 2009; Psychiatric GWAS Consortium Steering Committee, 2009). Many consortia emerged to combine efforts across research groups to elucidate reproducible genetic risk factors for psychiatric disorders. For adult onset disorders, this began with transient efforts (e.g., GAIN, ISC, and SGENE). For childhood onset disorders, these efforts began in ASD with smaller consortia such as the IMGSAC and PARIS during the linkage era (International Molecular Genetic Study of Autism Consortium, 2001; Philippe et al., 1999). Subsequently, the formation of the Autism Genetic Resource Exchange (AGRE) enabled expansion to include multiplex families (Geschwind et al., 2001; Lajonchere and Consortium, 2010). The largest consortium in psychiatric genetics is the PGC which began in 2007 (URLs) (Sullivan et al., 2018), and has spearheaded many of the major genetic advances in the field. The PGC has 800+ members from 40+ countries and working groups for 11 psychiatric disorders. The PGC is a mega-analysis consortium that allows highly harmonized analyses, rigorous quality standards, and significance thresholds that maximize reproducibility. A feature of most consortia is making summary results freely available along with paths for other researchers to get access to individual data or biological samples for independent research.
As whole exome and whole genome sequencing (WES, WGS) have become mainstream, the Whole Genome Sequencing for Psychiatric Disorders consortium is adopting a similar approach as the PGC, but for modern resequencing (Sanders et al., 2018). Investigators conducting WES for ASD have formed multiple consortia. The Simons Simplex collection focused on discovery of de novo variation via WES and played a major role in accelerating gene discovery in ASD (Fischbach and Lord, 2010). The SPARK initiative is conducting WES on 50,000 ASD cases a rapid data-release policy (Spark Consortium, 2018). The Autism Sequencing Consortia rigorously harmonizes ASD resequencing data from multiple studies (Buxbaum et al., 2012), and combining data from multiple cohorts has enabled major advances (De Rubeis et al., 2014; Sanders et al., 2015).
Genetic architecture
We review here what we have learned about the genetic architectures of the 10 psychiatric disorders in Table 1. Because some of the basic techniques may be unfamiliar, we provide in Table 2 brief definitions and accessible introductions to these topics that are beyond the scope of this review. The results to date indicate that psychiatric disorder risk is imparted by many common variants of individually small effects, and several disorders also have contributions from rarer variants with larger impact on risk (Geschwind and Flint, 2015; Sullivan et al., 2018).
Table 2:
Brief list of “omic” technologies used to understand psychiatric disorders.
| Initialism or acronym | Reversed | Description |
|---|---|---|
| GWAS | Genome-wide association study | Genomics: usually a case-control comparison of common genetic variation revealed by SNP arrays (pre-specified set of reliably measured biallelic genetic markers selected for good performance and coverage of the genome). Can achieve coverage of >90% of common variants in the genome. Can also identify rare copy number variants. Many studies of psychiatric disorders. Reviews PMID 19895722 28969442 28686856. |
| WGS | Whole genome sequencing | Genomics: ab initio resequencing of the genome. In concept, can identify all types of genetic variation. Increasingly used clinically for rare genetic syndromes. Few studies of psychiatric disorders to date. Review PMID 29700468. |
| WES | Whole exome sequencing | Genomics: a version of WGS focused on the protein-coding parts of the genome (~3% using one of several methods to “pull-down” all known exons. This provides a focused and more inexpensive way to identify gene-disrupting or missense variants in exons. WES has identified ~100 genes for ASD, and an increased “burden” of rare, protein-altering genetic variation differing between cases and controls in SCZ and a few other disorders. Reviews PMID 24941179 26139844. |
| - | Epigenomics | Unlike the (usually) static, body-wide nature of genomics (GWAS, WGS, WES), multiple readouts that capture changes that do not affect DNA sequence, but act to alter the functional state of cells and tissues. These include DNA methylation, histone tail modifications, etc. Initial approaches required large numbers of cells but improved versions can increasingly be applied to single cells. Epigenomic changes can be highly specific to a cell or tissue or common across the body; generally reflect cell differentiation and function. Review PMID 22955614. |
| OC | Open chromatin | Epigenomic: regions of the genome that are not histone-bound in cell nuclei and thus “open” to gene regulatory processes. Main methods are ATAC-seq and DNase-seq. Review PMID 22955614. |
| ChIP-seq | Chromatin immunoprecipitation sequencing | Epigenomic: a class of methods to identify functional modifications to specific genomic regions. Many focus on changes to the N-terminus tails of histone proteins. Such changes are part of the “histone code” that can dramatically alter gene expression. Examples of histone marks strongly associated with functional chromatin states include acetylation at the 27th lysine of the histone H3 protein (H3K27ac) and trimethylation of the 4th lysine of the histone H3 protein (H3K4me3). Review PMID 22955614. |
| Hi-C | None | Epigenomic: One of several chromosome conformation capture methods that can capture genomic regions that are near each other in cell nuclei. Hi-C does this in an “all-to-all” manner, whereas other methods target more specific interactions. A subset of these DNA-DNA contacts these can mediate regulatory interactions between regions that are located far apart. Reviews PMID 28905911 30367165. |
| RNA-seq | RNA sequencing | Epigenomic: identify the amount of all RNA molecules in a cell or tissue, a transcriptomic technology. RNA-seq can also capture splicing and isoform level information. Review PMID 28626224. |
| eQTL | Expression quantitative trait loci | Genomic & epigenomic: identify genetic predictors of gene expression. Essentially, GWAS for every variable transcript in a tissue (~50,000) to identify genetic variants associated with RNA abundance. Many are highly tissue- or stage-specific. Reviews PMID 23650636 26813401. |
Background.
Knowledge of genetic architecture is fundamental to rational study design and genotyping technology choice. For many decades, this was debated with various authors speculating architectures inferred from indirect clinical or epidemiological data. The extreme positions were the common disease/common variant model (psychiatric disorders result from the cumulative effect of many common variants of small effect) and the multiple rare variant model (strong genetic impacts on single genes cause psychiatric disorders with each case having a different causal mutation). Neither model can explain all of the genetic risk, and many possible genetic architectures lie between these extremes.
Genetic variation lies on a continuum from common to extremely rare: a risk variant might be present on half the chromosomes in a population or be observed only once in 1,000,000 people. We can consider a frequency continuum from ultra-rare (present once in a large sample, frequency <0.001%) to rare (present in a pedigree or in descendants of a recent ancestor, <0.1%) to uncommon (0.1% to 1%) to common (>1%). In general, rare variants arose recently and common variants are far older. Given what we know now, common variants generally have small effects on disease risk (OR <1.15), and rare variants typically have larger effect sizes (>2.0), are more likely to be deleterious, and tend to be removed by natural selection (Fu et al., 2013; Nelson et al., 2012; Zeng et al., 2018). This is not an invariant rule, as rare variants may have a continuum of risk (Marouli et al., 2017), and a fraction of common variants have large effects (e.g., APOE and Alzheimer’s disease).
Technology.
Two main technologies have emerged for capturing germline genetic variation in individual subjects, resequencing and SNP arrays. Resequencing determines anew many types of genetic variation in the immediately accessible genome. It captures many types of genetic variation – SNPs, insertion-deletions, copy number variation – across the frequency spectrum, from ultra-rare to common. In concept, resequencing is the method of choice for psychiatric genomics. Costs for WGS have declined considerably (US $800/sample) but analyzing WGS data remains challenging. Most resequencing studies to data used WES, reducing expense and analytic burden via a focus on protein-coding regions where the functional impact of variants is easier to interpret than in the non-coding genome. Study designs are usually either standard case-control comparisons or family-based methods. For the latter, trios of unaffected parents and an affected offspring are popular as they enable identification of de novo variation (i.e., present in an affected child but absent in parents), which can improve power to detect high impact variants. Other resequencing technologies can focus on the less accessible parts of the genome (repetitive regions or regions with variable structure and gene content). Although very expensive and technically complex, single-cell resequencing of nuclei from a tissue can identify somatic mutations that arose during development (these changes are not heritable, but may contribute to illness in some individuals) (Evrony et al., 2016; McConnell et al., 2017). The largest resequencing studies of psychiatric disorders have fewer than 25,000 cases but this will change in the next few years.
SNP arrays commonly include 700,000 or more readily genotyped biallelic genetic variants. These SNPs are preselected for reliability and capacity to capture 90% or more of common genetic variation in a population either directly or indirectly by capitalizing on linkage disequilibrium (LD, the strong tendency for nearby genetic variants to be co-inherited). In effect, direct assessment of <1 million SNPs can be leveraged to accurately estimate genotypes for 10 million or more common, uncommon, and even rare genetic variants. SNP arrays are inexpensive ($35/sample) and have been applied to very large numbers of people. Arrays technology can also identify large, rare copy number variants (CNV Working Group of the Psychiatric Genomics, 2017; Luo et al., 2012; Sanders et al., 2011; Sebat et al., 2007). WGS provides substantial more genome coverage and resolution, especially with regards to certain forms of chromosomal structural variation (Redin et al., 2017), but at an order of magnitude cost more than SNP arrays. SNP array studies of readily measured human traits (e.g., height, educational attainment, lipid levels) now routinely exceed sample sizes of 1 million, and studies of psychiatric disorders have 10,000–130,000 cases. These studies do not directly capture genetic variation in well-defined functional areas of the genome, necessitating substantial follow up to identify the causal variants and genes affected (Gusev et al., 2018; Sekar et al., 2016; Wang et al., 2018a).
Key issues in all of these studies are rigorous quality control, careful assessment and control for multiple types of bias, and correction for multiple comparisons. A large number of statistical tests are conducted requiring correction for multiple comparisons. For example, for SNP array studies an accepted threshold is P<5×10−8, akin to correcting a=0.05 for 1 million comparisons.
Common variant association studies of psychiatric disorders.
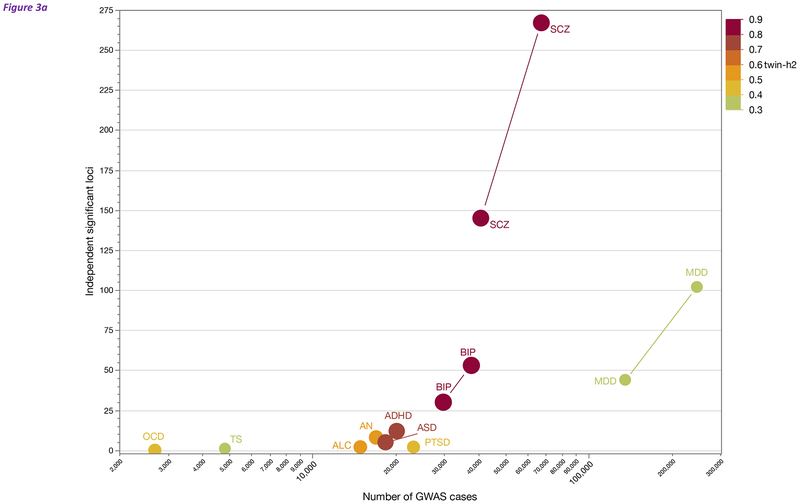
Most genetic studies of psychiatric disorders in the past decade have used SNP arrays to assess the role of accessible common variation (also known as GWAS, genome-wide association studies). The common variant findings for psychiatric disorders are summarized in Figure 3a. Studies in SCZ and MDD have yielded >100 loci, BIP has 53 loci, and ADHD, AN, and ASD have from 5–12 loci. The crucial determinant of the number of loci discovered is the number of cases; as sample sizes increase, more loci will be identified (Geschwind and Flint, 2015; Sullivan et al., 2018). As a common disorder with relatively low twin-heritability (Levinson et al., 2014), MDD has had notable difficulties with genetic discovery, but focusing on severe cases (CONVERGE Consortium, 2015) and increasing sample sizes has been particularly fruitful (Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, 2018).
Figure 3:
(a) Overview of common variant gene discovery for the psychiatric disorders in Table 1. Sources and label definitions are in Table 1. The X-axis is the log10 of the number of cases in the largest current GWAS. The Y-axis is the number of genome-wide significant and LD-independent loci. The color of each point reflects twin-heritability per the scale on the right. For BIP, MDD, and SCZ, the graph includes published and in preparation/in press results (connected by a line). Sample size is the major determinant of discovery. We thank PGC colleagues for allowing us to present pre-publication results.
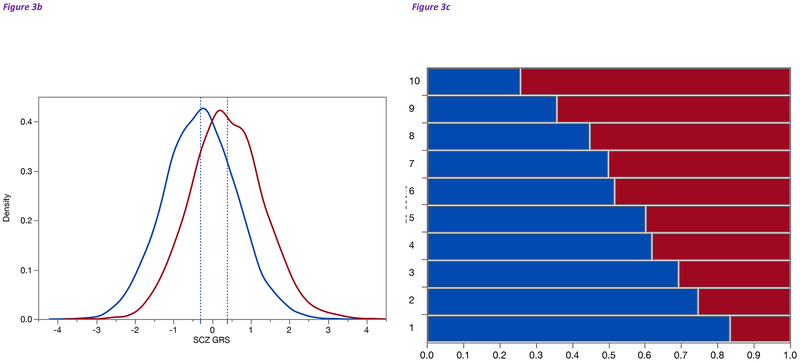
(b) Density plot of genetic risk scores (GRS) in 4,932 SCZ cases (red) and 6,210 controls (blue) from Sweden (training set is from the PGC 2014 SCZ paper excluding Swedish samples) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). The X-axis shows the standardized GRS and the Y-axis shows the smoothed density, a prediction of the proportion of cases or controls with a given GRS value. The dashed vertical lines show the means of each group. The group means differ by over ⅔ of a standard deviation (0.686), and are highly significantly different (P=1.1e-254, controlling for genotyping array and 5 ancestry PCs). The two curves overlap substantially but there are 48 controls with GRS > 2 and 24 cases with GRS < −2.
(c) Depiction of GRS described in Figure 3c but showing the proportions of cases (red) and controls (blue) in each SCZ GRS decile (Y-axis, 1=lowest 10%, 10=highest 10% GRS). X-axis is the proportion within each decile. The proportions of cases increase steadily from lowest to highest. However, there are substantial numbers of cases in the lowest decile and controls in the highest decile.
Across all disorders, 241 loci have a significant association with the 10 psychiatric disorders in Table 1 with 22 loci associated with ≥2 psychiatric disorders. Although most loci are disease specific, many loci increase risk for multiple disorders. These loci together implicate ~76 Mb of the genome as containing common genetic variants involved in the etiology of these disorders. We speculate that many loci contain multiple functional elements that contribute to risk. Around 400 protein-coding genes lie in these loci. Traditionally, genomic location is used to assign “SNPs-to-genes”; however, as discussed more fully below, this practice yields an incomplete portrait. If we overlay functional genomic data from human brain (e.g., eQTL or regulatory chromatin interactions), about 50% of the time genes located in loci are also implicated by functional data. Crucially, recent studies have shown that genes located far outside of a locus are often implicated (see functional architecture section below) (Wang et al., 2018a; Won et al., 2016).
Although the effects of any individual variant may be small, they can nonetheless point to biological processes that may be highly relevant to therapeutics. For example, GWAS results for SCZ (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and MDD (Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, 2018) are enriched for genes that encode proteins known to interact with pharmacological targets of antipsychotics and antidepressants.
Genetic risk scores (GRS), SNP-heritability, genetic correlations.
In the past decade, GWAS provided the impetus for several methodological developments. These methods were partly motivated by the failure of early genetic studies to identify common variant associations with SCZ (sample sizes 250–1000 cases).
First, based on ideas from livestock genetics, GRS initially appeared as part of a SCZ GWAS (International Schizophrenia Consortium, 2009). A GRS captures the number of inherited common risk variants as a normally distributed number and can be compared to a population mean (e.g., a person might have a standardized SCZ GRS of 2 indicating inheritance of SCZ risk alleles in the top 2–3 percentiles). Computing a GRS requires a sizable external training set and can be applied to new individuals of similar ancestry. GRS can use significant, nearly significant, and non-significant SNP associations, and have clearly indicated that more common variants will be discovered as sample sizes increase (International Schizophrenia Consortium, 2009). Indeed, GRS differences between cases and controls are now so widely replicated that GRS are used for quality control (the absence of a difference often indicates a basic problem with a dataset). Inheriting a notably large number of SCZ risk alleles (e.g., being in the top vs bottom decile for GRS) carries more than a 10-fold increased risk of SCZ (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014).
For example, Figure 3b shows the distributions of SCZ GRS in a set of SCZ cases and controls. There is a highly significant mean difference between groups but the distributions overlap substantially. Figure 3c depicts the same data but shows the proportions of cases and controls in each GRS decile. Cases in the top decile have 15 fold increased risk for schizophrenia compared to the lowest decile (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Intriguingly, there are many controls in the top decile and many cases in the lowest decile. Detailed investigations of these observations are underway (e.g., do controls in the upper decile have a sub-clinical form of SCZ or have strong protective factors? Are cases in the lower decile phenocopies or more likely to have a strong-effect genetic variants?).
GRS have emerged as a potentially important output from psychiatric genetics and may help guide future precision medicine approaches. In other areas of medicine, GRS provide new ways to evaluate risk and to stratify patients – e.g., for prostate cancer, breast cancer, cardiovascular disease, and type 2 diabetes mellitus (Gronberg et al., 2015; Khera et al., 2018; McCarthy and Mahajan, 2018; Shieh et al., 2016). For psychiatric disorders, considerable research is in progress; the potential is that, for the cost of an inexpensive SNP array, GRS could assist in differential diagnosis, therapeutic selection, outcome prediction, and patient stratification. Multiple clinical questions could be addressed: for an individual with multiple comorbidities (ADHD, ASD, OCD), do the three GRS indicate that one is the logical focus of treatment? Should this person with MDD and a high BIP GRS receive a mood stabilizer as well as an antidepressant? Can we identify people with PTSD at first presentation who are at high risk of a pernicious course of illness? Can information from genes in biological pathways be used to develop “mechanistic GRS” that could then be used to identify an antipsychotic with the greatest chance of clinical response? We would like to add an important caveat: although GRS are conceptually straightforward, their creation and use requires considerable care and sophistication to derive secure and reproducible findings (Lewis and Vassos, 2017; Torkamani et al., 2018). As just one example, incorrect inference can readily occur if the GRS training and target datasets are from different ancestries (Martin et al., 2017).
Second, several methods can calculate the heritability of a trait using SNP array data (Bulik-Sullivan et al., 2015; Yang et al., 2011). These provide assessments of heritability based on genome-wide genotypes, and improves upon traditional heritability measurements given their basis in direct genetic measurements. SNP-heritability can be estimate for traits that are difficult or impossible to assess using twins (e.g., antipsychotic adverse drug reactions). Indeed, SNP-heritability estimates are available for thousands of traits (Zheng et al., 2017). Table 1 shows SNP-heritability estimates, and these tend to follow traditional heritability. These provide exceptionally strong indications that common variation genetic variation is important for all complex psychiatric disorders, and more will be discovered with increasing sample sizes.
In almost all instances, SNP-heritability is less than twin/pedigree heritability. If reviewed critically, indirect twin/pedigree heritability estimates are often upwardly biased, and the degree to which SNP-heritability is different from indirect measures is unclear. For any real difference between SNP- and twin/pedigree-heritability, the major reasons are: (a) imperfect assessment of common variation (i.e., missing common variation in hard to genotype or impute regions); (b) complex, non-SNP common genetic variation whose identification requires resequencing or specialized methods; and/or (c) poor measurement of rare genetic variation with current sample sizes and technologies. It is important to note that the goal of genetic studies of psychiatric disorders is to generate clinical and biological insights and not to align different conceptualizations of heritability.
Third, we can now readily estimate the genetic correlations between traits using SNP array data (Bulik-Sullivan et al., 2015; Yang et al., 2011). These methods have provided insights into the fundamental basis of these disorders. A similar construct could be assessed using twin or pedigree data but with lesser power and precision. Notably, the major psychiatric disorders have significant and often sizable genetic correlations (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013a). A more comprehensive effort of 25 psychiatric and neurological disorders showed that most psychiatric disorders had significant genetic inter-correlations, but there were far fewer for neurological conditions (Brainstorm Consortium, 2018). Importantly, comparison of SCZ results between European and East Asian samples indicated that the genetic correlation was indistinguishable from one, strongly indicating that the common variant genetic basis of SCZ is highly similar across these global populations (Lam et al., Submitted). Under a set of specific assumptions, we can also apply Mendelian Randomization (MR) to suggest causality; for two traits with sufficient numbers of significant associations, MR can assess the plausibility of whether one trait has a causal relation with another (e.g., lower educational attainment and higher body mass were putatively causal for MDD) (Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, 2018).
Rare variant association studies of psychiatric disorders.
Resequencing studies that implicate ultra-rare and de novo variation have the major advantage of pinpointing risk variants in specific genes. Compelling results can leverage the extensive neuroscience toolkit for experimental modeling of specific genes. Until relatively recently, identifying rare variants for psychiatric disorders was mainly limited to large structural variants (CNV Working Group of the Psychiatric Genomics, 2017; De Rubeis et al., 2014; Iossifov et al., 2012; Sebat et al., 2007). As noted above, resequencing technologies enable rare variant discovery in ever larger samples, and we know now that ultra-rare and de novo single nucleotide variants contribute to risk (Genovese et al., 2016; Sanders et al., 2015; Satterstrom et al., 2018b; Singh et al., 2016; Wang et al., 2018b; Willsey et al., 2017). Rare variant association tests require the aggregation of rare, deleterious mutations at a particular locus (usually in protein-coding exons or annotated regulatory regions) in cases compared to controls (Zuk et al., 2014).
At present, the largest resequencing efforts are for ASD and SCZ. Rare variant discovery has been most successful in ASD where WES for rare, de novo, protein truncating variants (PTVs) in mutation intolerant genes has identified around 100 high-confidence connections to specific genes (Satterstrom et al., 2018b). Although each gene accounts for only a small fraction of cases, rare de novo variation is predicted to account for ~15% of ASD cases (Iossifov et al., 2014). Most of these mutations also decrease IQ and ID is an important comorbidity of ASD (Buja et al., 2018; Iossifov et al., 2014), which is consistent with previous work identifying dozens of known severe, rare medical genetic syndromes associated with ASD (reviewed in (Abrahams and Geschwind, 2008; Geschwind, 2009)).
The yield of resequencing in ASD is markedly higher than for other psychiatric disorders. WES has implicated only two genes for SCZ (Singh et al., 2016; Steinberg et al., 2017) at sample sizes that yielded dozens of associations for ASD. In TS, a role for de novo gene disrupting and missense variants has been established (Willsey et al., 2017) and two high-confidence genes for TS were recently identified (Wang et al., 2018b). Sizable WES of ADHD and BIP are underway. For the other psychiatric disorders in Table 1, major resequencing efforts are at more nascent stages. There is debate in the field as to whether resequencing efforts are worth the 10–15× greater cost, particularly for later-onset disorders that are not associated with ID or neurological impairment. The sobering experiences in SCZ and type 2 diabetes suggest a limited role of large-scale resequencing in adult disorders until the costs decline substantially.
Identification of rare, genic mutations can be extremely informative. They directly implicate specific genes and are amenable to experimental modeling. At the same time, interpretation of these models is a formidable task. While some of these genes are relatively specific to a disorder like ASD, many confer broader phenotype risks (Abrahams and Geschwind, 2008; Ronemus et al., 2014; Satterstrom et al., 2018b). Pleiotropy is more the rule: most mutations increase risk for a range of neurodevelopmental outcomes (e.g., ID, ASD, epilepsy, or psychosis). These pleiotropic large effect mutations may work by disrupting key neurodevelopmental processes rather than specifically causing one defined clinical disorder (Geschwind and Levitt 2007). Even for a highly significant gene identified from resequencing, precisely which human phenotype is being modeled and with what specificity may be uncertain (i.e., ID and/or ASD). Another question from these findings is whether genes that harbor large effect mutations causing ASD and ID affect biological processes different from those that cause ASD that is not comorbid with ASD. Indeed, some gene network analyses suggest the existence of molecular processes that distinguish ASD from ID (Parikshak et al., 2013; Satterstrom et al., 2018b).
The relative contributions of rare de novo missense or inherited mutations to psychiatric disorders are not quite as well established as de novo protein truncating variants. However, both rare missense and inherited mutations have been shown to contribute to ASD, simply with smaller effect sizes than de novo variants (Ruzzo et al., 2018; Sanders et al., 2015). Furthermore, the effects of PTVs can be assessed in a functional and evolutionary context (loss of one copy of the gene and the degree of constraint) (Samocha et al., 2014), while the functional impact of individual missense mutations is harder to determine. One approach to this problem integrates prior information such as gene or PPI networks to boost the signal of missense variation (Parikshak et al. 2013). The detection of inherited variation may be further hindered by ascertainment bias from study designs that favor detection of de novo variants. It is illustrative that studying families having multiple children with ASD significantly reduced the signal from de novo variation compared to singleton families, while enhancing that from inherited variation to identify risk genes (Ruzzo et al., 2018). Consistent with a role of rare, inherited variation in risk also comes from a recent WES of ASD and ADHD that excluded cases with ID or comorbidity (Satterstrom et al., 2018a). These investigators found that rare PTV in mutation intolerant genes occurred with equal frequency in both ASD and ADHD and that the genes impacted significantly overlapped. Larger samples are needed to determine if genes with statistically significant association with each disorder are shared, and whether the mutations have similar molecular impact. For example, even if mutations increasing risk for ASD and ADHD were in the same gene, they might impact different isoforms that could have different functional consequences. Emerging data from RNA sequencing from brain shows remarkable isoform diversity in parallel with distinct protein interactions and cell type specificity, further highlighting the importance of understanding mutational consequences in an isoform context.
Copy number variation (CNVs)
refers to structural chromosomal variants greater than 1 kb in size that lead to an increase or decrease in the DNA sequences encompassed by the CNV (e.g., fewer or more than two copies of an autosomal region). Approximately 4% of the genome comprises such structural variation, much of which is common, inherited, and relatively benign with regards to imparting disease risk (Brand et al., 2014; Conrad et al., 2010; Mills et al., 2011; Sebat et al., 2004). Larger de novo CNVs, especially ones that disrupt genes or change gene dosages, can carry major risks particularly for neurodevelopmental disorders (Malhotra and Sebat, 2012; Sebat et al., 2007).
Several dozen rare CNVs are known to confer relatively strong risks for psychiatric disorders, most commonly in ASD and SCZ and less frequently in BIP, TS, and ADHD. Most known pathogenic CNVs increase risk for multiple disorders (de la Torre-Ubieta et al., 2016; Kirov, 2015; Lowther et al., 2017; Malhotra and Sebat, 2012). These recurrent CNVs usually arise de novo, mainly via non-allelic homologous recombination in regions flanked by low copy number repeats.
CNVs associated with psychiatric disorders share several commonalities: (a) they usually contain multiple genes (with a few exceptions (Bucan et al., 2009; Talkowski et al., 2011)); (b) are usually >500 kb in size (although many expect that smaller CNVs will be found using WGS) and the major pathogenic mechanism is presumed to be dosage-sensitivity of genes in the CNV although distal regulatory effects on genes outside of the CNV are also plausible (de la Torre-Ubieta et al., 2018); (c) many CNVs are associated with partial disruption of a range of developmental programs and impact multiple organs (cardiac, gut, immune, and endocrine as well as brain); (d) most CNVs confer increased risk for multiple psychiatric disorders including ID, ASD, ADHD, and psychotic disorders (Kirov, 2015; Lowther et al., 2017); (e) penetrance can be highly variable, ranging from subtle effects detectable by neuropsychological tests to mild degrees of anxiety/ADHD to co-occurrence of severe psychiatric disorders (Kendall et al., 2017; Stefansson et al., 2014; Ulfarsson et al., 2017). Emerging evidence suggests that among other factors, some of this pleiotropy may be due to modification GRS because even in those with ASD or SCZ carrying large effect de novo mutations, there appears to be an additive effect of common variation on phenotypic expression (Tansey et al., 2016; Weiner et al., 2017).
Synthesis.
In the past decade, major papers from the PGC and other consortia have conclusively shown that all of the psychiatric disorders in Table 1 have an important contribution from hundreds or thousands of common genetic variants of relatively subtle effect. Exactly how these variants influence gene expression in the context of biological networks is generally unknown but has highlighted critical gaps in our knowledge of gene regulation. Work in progress on the functional architecture and cellular/tissue architecture will, we believe, yield the needed insights. The impact of rare variation is less well studied. Empirical data show that rare genetic variation plays a role in some of these psychiatric disorders (ASD and SCZ in particular but also for TS and ADHD). However, direct comparisons of the contributions of common and rare genetic variation show that common variation dominates heritable risk for SCZ and ASD (Gaugler et al., 2014; Purcell et al., 2014). Still, rare variants that disrupt genes provide a clear starting point for mechanistic studies, and identification of large effect mutations in patients is of substantial clinical utility. Finally, many disorders are early in the discovery process. Consistent with the documented clinical and epidemiological comorbidity, there is also important genetic overlap, including substantial components of genetic variation that increase risk for multiple disorders – both of which necessitate consideration of diagnostic architecture.
Functional architecture
Moving from common variant findings to genes, molecular pathways, and cells requires in genomic analysis. Table 2 contains additional definitions and references to important background that is beyond the scope of this review. Figure 4 presents a schematic of how we can systematically evaluate the implications and impacts of genetic architecture findings.
Figure 4: Establishing the functional and cellular architectures based on genetic findings.
To begin, genetic analyses identify highly confident associations with one or more psychiatric disorders. Common variation is usually detected using GWAS and SNP array technologies. Rare variation capitalizes on CNVs or resequencing via WES or WGS. Some causal variants alter protein structure or function and thereby directly point at specific genes. However, most genetic variation discovered to date is in non-coding regions which can have highly diverse regulatory functions (e.g., enhancer or repressor activity or regulation of splicing or alternative promotor usage). Assigning non-coding regulatory variants to genes is imprecise as gene regulation often occurs at a distance and does not necessarily involve the nearest gene. Instead, one can identify candidate target genes impacted by non-coding disease associated genetic variation using a range of functional genomic data. For example, quantitative mapping approaches can identify how a particular variant effects open chromatin, histone tail modifications, gene expression, splicing, and DNA methylation. These methods integrate DNA-based genetic variation with multi-level “omic” data – RNA sequencing (eQTL or sQTL), methylation analysis (mQTL), or ChIP-seq (hQTL) – to identify the quantitative impact of genetic variation on these molecular phenotypes. Other biochemical methods identify active/open chromatin (ATAC-seq. DNase-seq) or 3D chromatin structures such as enhancer-promotor loops (Hi-C, ChIA-PET), which provide additional information on the relationship between regulatory regions and specific genes with which they interact. Many functional genomic readouts are tissue-specific highlighting the need for comprehensive studies of the human brain across development. When combined, these methods can identify the likely functional impact of disease associated variation on specific genes, which can then be experimentally validated. Molecular pathways can be identified using pathway or gene network analysis. Sets of disease-associated candidate genes can be tested for cell type enrichment to define the cellular architecture. A similar approach applied to identified regulatory regions to define functional regulatory networks or the cell types impacted by disease associated regulatory variation.
From variant to gene.
Because most genetic variation that contributes to common psychiatric disorders is not in protein-coding regions, a crucial step in understanding disease mechanisms is pinpointing the genes impacted by risk variants (Thurman et al., 2012; Visel et al., 2009). This requires functional annotation of non-coding regions, the goal of consortia like ENCODE (ENCODE Project Consortium, 2011), Roadmap (Roadmap Epigenomics Consortium, 2015), and GTEx (GTEx Consortium, 2017), which produced comprehensive initial regulatory maps and transcriptional profiles across spectrum of cells and tissues. However, around half of non-coding regions have regulatory functions that are shared across tissues meaning that half of the regulatory elements in a given tissue may be relatively specific to a tissue, cell type, or developmental stage (Liu et al., 2017a; Roadmap Epigenomics Consortium, 2015; Won et al., 2016). This is particularly important for brain which has higher cellular heterogeneity and longer developmental trajectories compared to other tissues. The need for brain-specific functional genomic data led to PsychENCODE (PsychENCODE Consortium, 2018) (URLs) which has produced and integrated multiple types of functional genomic data from human brain (Gandal et al., 2018b; Li et al., 2018; Wang et al., 2018a). Its goals are to complement the work of these other consortia by producing accurate regional, cell type, and stage-specific annotation of gene regulation and transcription at tissue and cellular levels in brain from healthy individuals and cases with major psychiatric disorders. This effort is complemented by the BRAIN single cell atlas of cell types and gene expression in human and mouse (Ecker et al., 2017).
These resources essentially provide maps for interpretation of genetic variation implicated in psychiatric disorders in the context of genes, their regulation, and the effects on biological pathways. A complicating factor is that assigning even well-annotated genomic regions to specific genes is not as simple as choosing the closest gene or genes containing variation that is highly correlated with the associated SNPs which is usually the default approach (Whalen et al., 2016; Won et al., 2016). Rather as suggested by studies of brain eQTL (GTEx Consortium, 2017; Hauberg et al., 2017) and chromatin structure (de la Torre-Ubieta et al., 2018; Won et al., 2016), nearly half of the target genes of human regulatory variation are not in genomic loci defined by LD (Whalen and Pollard, 2018) (Table 3). Thus, “4D mapping” of chromatin interactions (i.e., brain regions across developmental time) is critical for understanding the functional relationships of regulatory regions to genes (Dekker et al., 2017).
Table 3:
Many regulatory interactions are distal
| Distance from Regulatory Element to TSS | ||
| eQTL | ATAC-seq | HI-C |
| 127 kb | 394 kb* | |
| Distribution of eQTL Distance from TSS | ||
| < 10kb | > 10kb | >100kb |
| 24% | 76% | 29% |
Average distance from regulatory elements defined by eQTL, ATAC-seq and Hi-C in fetal brain is shown as well as percentage of eQTL in >10kb (distal) and <10kb (proximal) bins from the TSS of genes in fetal brain. Data from (de la Torre-Ubieta et al., 2018; Polioudakis et al., 2018; Won et al., 2016) and Walker and Geschwind (unpublished). eQTL are generally closer to the TSS than the biochemically defined putative regulatory regions which is expected especially given the limited (10 kb) resolution of Hi-C.
Functional genomic data include gene expression surveys, open chromatin, eQTLs, chromatin QTLs, methylation QTLs, histone marks, and regulatory chromatin interactions, initially for bulk tissues or sorted types of cells but increasingly at the single-cell level. As illustrated in Figure 4, these data can be combined to define candidate enhancer-promoter interactions (from locus to gene) whose accuracy can then be assessed in a biological system. Published brain eQTL data have N<1000 and contain only a fraction of presumed regulatory relationships. Chromatin capture methods such as Hi-C can define chromatin structure in brain nuclei (Dekker et al., 2013) and can predict functional interactions defined by eQTL and enhancer-mRNA relationships (Won et al., 2016). Although integration of functional genomic data from brain yields empirically-based hypotheses about regulatory relationships, experimental validation is required. Techniques like STARR-seq permit large scale validation (which suggests enhancer functionality) (Arnold et al., 2013; Liu et al., 2017b), while analysis in an appropriate cell type with epigenome editing technologies can confirm target identity (de la Torre-Ubieta et al., 2018; Won et al., 2016). Currently, it is wise to be conservative and rely on regulatory interactions identified by multiple methods (e.g., eQTL/Hi-C (Gusev et al., 2018) or ATAC-seq/Hi-C (de la Torre-Ubieta et al., 2018). These distinct data types – often derived in different laboratories in different samples- show significant overlap in regulatory predictions (Gusev et al., 2018). This is in contrast to comparisons relying on LD blocks or the assignment by the closest gene, where the overlaps with methods that directly assess chromatin are less substantial (Short et al., 2018; Whalen and Pollard, 2018).
Application of functional genomic approaches to define regulatory regions and target genes has yielded important albeit tentative clues as to the developmental and cell type architecture of psychiatric disorders. One example comes from studies that partition disease heritability defined by genome-wide SNP genotyping, or by mapping putative causal variants across the genome, to identify regions of enrichment, and ask in what tissues and what stages are these regions active (de la Torre-Ubieta et al., 2018; Finucane et al., 2018; Skene et al., 2018; Won et al., 2016). As discussed more fully below, these studies have implicated specific development epochs and brain regions in risk for several psychiatric disorders and cognitive phenotypes. These initial studies demonstrate that creation of these gene regulatory maps with multiple methods that address different molecular processes, developmental stages and brain regions is a critical step in understanding how disease risk biologically unfolds.
From genes to networks.
To understand how genes contribute to psychiatric disorders, we are faced with the task of measuring and understanding phenotypes across a hierarchically organized complex system, connecting genes to behavior. Few genes act in isolation but rather affect the function of other genes to influence a particular phenotype via in cellular networks or pathways (Barabasi et al., 2011; Geschwind and Konopka, 2009). This challenge is exacerbated by the polygenic nature of psychiatric disorders. To understand how genes contribute to CNS phenotypes, many groups have applied an analytical framework at a gene-network level involving coordinated regulation of gene expression (Parikshak et al., 2015; Parikshak et al., 2013). Network analysis can interrogate multiple levels of molecular organization and enable integration with other information including known pathway annotations. Furthermore, when hundreds of genes are involved, network analysis provides an organizing framework that can divide large gene sets into biologically coherent modules for prioritization (Parikshak et al., 2015; Parikshak et al., 2013), or add power to GWAS (Horn et al., 2018). Combining network approaches with systems neuroscience permits the methodical connection of heterogeneous genetic risk factors to brain mechanisms (Gandal et al., 2016; Geschwind and Konopka, 2009).
Two general network approaches have been used in psychiatric genomics based on literature-curated pathway databases (e.g., Gene Ontology or, KEGG) or data-driven tissue specific approaches based on transcriptomic, proteomic, or other “omic” data (Parikshak et al., 2015). The former approach has many biases, including weighting highly studied genes, non-CNS functional annotations, or very non-specific annotations (e.g. “synaptic function”), and lack of tissue specificity (missing tissue specific interactions or emphasizing those observed in other tissues). Curated pathway-based studies using combinations of multiple methods and data sources are far more convincing than those using single sources, and have yielded evidence for common pathways across psychiatric disorders (Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015), but still do not fully overcome biases inherent in literature curation. This illustrates a weakness in current functional annotations that are broad and biased with regards to how the neuronal annotation method. Gene network approaches can identify presumed functional modules in an unbiased manner but understanding what these modules mean beyond broad annotations remains a major stumbling block for the field, and will require efforts connecting gene expression to neural cell biology and physiology.
Despite these limitations, several studies in ASD and SCZ highlight the power of using transcriptional networks based on normal human brain tissue across development or brain regions, or more generalized protein-protein interactions (Hormozdiari et al., 2015; Li et al., 2014; Lin et al., 2015) to identify molecular pathways, developmental epochs, or brain circuits enriched for genetic variation. Despite clear genetic heterogeneity, both ASD and SCZ risk converge on shared molecular pathways (Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015; Parikshak et al., 2015). In ASD, these pathways involve regulation of transcription and chromatin structure during neurogenesis, and subsequent processes of synaptic development and function during early fetal cortical development (Parikshak et al., 2015). A small study implicated similar stages during the developmental of the prefrontal cortex in SCZ risk (Gulsuner et al., 2013) consistent with several decades of neuroanatomical studies (Glausier and Lewis, 2018; Piper et al., 2012). Importantly, these findings are emerge from different methods including protein-protein interactions PPI (Lin et al., 2015; O’Roak et al., 2012), integration of protein, gene expression, and phenotype data (Gilman et al., 2011; Hormozdiari et al., 2015), and chromatin marks (Sun et al., 2016). These efforts point at similar pathways and/or convergence of risk loci on similar biological processes (Corominas et al., 2014; Gilman et al., 2012; Li et al., 2014).
One caveat in the interpretation of these studies is that they are based on current knowledge of genetic contributions. In ASD, this is heavily biased towards rare, de novo PTV identified in simplex families which could impact different pathways than those affected by inherited variation. Although there is likely a role for rare inherited variation in ASD (Krumm et al., 2015), few studies have identified significant signals based on inherited risk variants for specific genes. A recent study of multiplex ASD families found that inherited risk impacts pathways similar to those for de novo variation (Ruzzo et al., 2018). Similarly, the developmental trajectories of risk genes implicated by rare and common variation appear to overlap, particularly for the fetal period for ASD risk.
Transcriptomic networks define disorder-associated molecular pathology.
Psychiatric disorders are not generally associated with brain pathology on gross or microscopic examination. The development of methods to capture the brain transcriptome led to studies of differential expression in cases versus controls and evaluation of convergent molecular pathology (Parikshak et al., 2015). To organize these data, network/pathway approaches have been applied to brain tissue from subjects with most major psychiatric disorders including SCZ, MDD, and ASD (Parikshak et al., 2015). However, any changes detected in postmortem brain could be causal or reflect reverse causation. Integration of these data with genetic risk variants provides an opportunity to identify a causal foothold. In ASD, these analyses, replicated using different methods and samples, implicate synaptic and neuronal signaling pathways overlapping with other causal gene-based network methods (Parikshak et al., 2016; Voineagu et al., 2011). Similar network analysis based on gene co-expression identifies transcriptional networks dysregulated in SCZ, including co-expressed neuronal genes enriched for both common and rare SCZ-associated variants (Fromer et al., 2016). Transcriptomic findings for ASD, SCZ, BIP, and MDD suggested shared and disorder-specific gene expression changes (Gandal et al., 2018a). Notably, cross-disorder transcriptome correlations parallel genetic correlations, consistent with common biology processes (Gandal et al., 2018a).
Several genes that cause rare forms of ASD (e.g., FMR1, CACNA1C, and TCF4) regulate expression or splicing of many genes associated with psychiatric disorders (Tian et al., 2014; Weyn-Vanhentenryck et al., 2014). FMR1 in particular interacts with the mRNA of many ASD and SCZ risk genes (Iossifov et al., 2014; Parikshak et al., 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Analysis of transcriptional (Cotney et al., 2015; Sugathan et al., 2014), splicing (Berto et al., 2016; Fogel et al., 2012; Weyn-Vanhentenryck et al., 2014) or signaling (Tian et al., 2014) networks indicates that at least some of the rare major gene forms of psychiatric disorders impact pathways that are more generally related to risk in the population. This highlights the relevance of rare forms of psychiatric disorders to understanding common genetic variation.
Tissue and cellular architecture
As with gene network analyses to identify biological pathways, it is possible to apply similar methods to identify empirically the brain regions and developmental stages in which the genetic findings are enriched. These analyses are important in a general sense – are these disorders rooted in early fetal development, childhood, adolescence, or adulthood? – but also because of neuroscience tools that can manipulate increasingly specific brain cell types in space and time.
Two general approaches are used to determine cell type or stage specificity. The first assigns genes implicated by risk variants directly to cell types based on transcriptomics (Polioudakis et al., 2018; Skene et al., 2018). The second partitions genetic risk across non-coding regions and compares the predicted activity of these regions across cell types and developmental stages, which to date has been primarily based on tissue level open chromatin rather than single cells (de la Torre-Ubieta et al., 2018). Development of robust single cell methods for chromatin analysis promises to be important (Cusanovich et al., 2018a; Cusanovich et al., 2018b). At present, many psychiatric GWAS are under-powered to accomplish these intentions (Skene et al., 2018).
This lack of power for common variant analyses is certainly the case for ASD, where studies have relied primarily on measuring expression enrichment or de novo PTVs. These studies have demonstrated that ASD risk variants are enriched in cortical glutamatergic neurons expressed during neurogenesis and neuronal migration during fetal cortical development in human and mouse (Parikshak et al., 2013; Willsey et al., 2013). Examination of the laminar patterns of expression in primate indicated that ASD risk genes are enriched in upper relative to lower layer neurons. This may be important for understanding circuit level architecture because upper layer neurons form the primary direct connections between cerebral hemispheres and cortical regions (Parikshak et al., 2013) and lower layer neurons primarily, but not exclusively, project to subcortical regions.
A recent study of single-nuclei RNA sequencing from human fetal and adult brain has validated the enrichment of genes harboring large effect de novo mutations associated with ASD in fetal glutamatergic neurons (Polioudakis et al., 2018). These detailed transcriptomic profiles provide nuance, especially for individual genes, identifying genes expressed broadly across neurons or with relative specificity for inhibitory neurons, neural progenitors, or non-neural cells (Polioudakis et al., 2018). The importance of the fetal period for ASD is supported by GWAS results integrated with regulatory chromatin interactions and gene expression, which show enrichment of enhancer marks in the fetal brain and higher expression of ASD target genes during fetal corticogenesis (Grove et al., In press). Comparisons across brain regions, both prenatally and in adult, confirms prenatal cerebral cortical enrichment over other brain regions both prenatally, and relative to adult expression levels.
For SCZ, although earlier developmental stages are important for risk, considerable cell-type specificity emerges in the adult brain. The most comprehensive analysis to date used single-cell and single-nuclei RNA-seq from multiple brain regions in mouse and human (Skene et al., 2018). Distinct patterns of enrichment were identified for different disorders, often mirroring known biology (e.g., multiple sclerosis and Alzheimer’s disease risk were enriched in microglia). Common variant genetic findings for SCZ showed enrichment in a limited set of major cell types: pyramidal neurons in cortex and hippocampal CA1, striatal medium spiny neurons, and cortical interneurons. MDD risk was clustered in cortical interneurons and embryonic midbrain neurons (these findings replicate in multiple new datasets, in preparation). Orthogonal functional genomic data are consistent with these finding as open chromatin in neuronal nuclei (NeuN+) from 14 regions from human adult brain showed significant enrichment of SCZ GWAS findings in cortex and striatum (Fullard et al., 2018), and open chromatin in mouse cortical layers showed SCZ enrichment in excitatory neurons in layer V (Hook and McCallion, 2018).
Although these studies are not yet definitive, we highlight emerging points of consistency. Genetic risk for SCZ appears to be more widespread in “4D” (Li et al., 2018) and somewhat more specific to adult brain (particularly pyramidal neurons, striatal medium spiny neurons, and cortical interneurons), but also with effects during fetal cortical development (de la Torre-Ubieta et al., 2018; Won et al., 2016). MDD risk is enriched in adult cortical interneurons (Skene et al., 2018), but also with fetal enrichment in midbrain neurons (Skene et al., 2018) (consistent with theories of catecholaminergic cortically-projecting brainstem systems in MDD). Genetic risk for ASD appears act primarily in fetal periods, involving cortical glutamatergic neurogenesis and early development. While ASD risk converges on glutamatergic neuron development, by no means is every risk gene expressed exclusively in these neurons (Polioudakis et al., 2018). These findings broadening and refining the neuronal classes where ASD risk genes act are supported by other analyses (Satterstrom et al., 2018b). The implication of fetal neurogenesis in childhood and adult-onset disorders may highlight a critical period in early brain development for multiple psychiatric disorders (Geschwind and Rakic, 2013). As knowledge of gene regulation at a single cell level increases, the precision of assigning of genetic risk to specific cell types will establish a solid framework for the circuit architecture of these disorders.
Diagnostic architecture
Psychiatry is one of the few areas in medicine that lack of objective biomarkers of illness. Other areas of medicine have frequently updated diagnostic classifications as new biological data and increased understanding of etiopathology emerge. In the absence of objective diagnostic features from laboratory testing, brain imaging, or pathology, the definitions of psychiatric disorders are necessarily based on descriptive data collected via human interactions and organized by expert panels. A long-standing tension is whether psychiatric disorders are better considered as fewer broad categories or more numerous refined categories. In the past 30 years, psychiatric nosology has tended toward the latter position.
For almost all psychiatric disorders, genetic data are the most fundamental biomarker yet discovered (recalling that humans are “exposed” to their genomes from conception and given the plausible absence of reverse causation). Given the well-documented and extensive patterns of comorbidity, it is perhaps unsurprising that genetic results show fundamental overlaps between many adult and childhood disorders. For common variation, SCZ has significant positive genetic correlations with BIP, MDD, ADHD, ASD, and AN (Brainstorm Consortium, 2018), and MDD has significant positive genetic correlations with anxiety disorders, ASD, ADHD, BIP, and AN (Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, 2018). Neurological conditions in contrast have far fewer significant genetic correlations (and largely for clinical subtypes like migraine with/without aura). Moreover, the lifetime presence or absence of many psychiatric disorders have positive genetic correlations with quantitative measures of symptoms – e.g., lifetime MDD has a genetic correlation of 0.98 with depressive symptoms (Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, 2018). Similar results have been reported for ASD, ADHD, and OCD (Martin et al., 2018a). Similarly, for rare variation, as described above, there are pleiotrophic effects for most rare CNV and exon variants of strong effect as many such variants increase risk for multiple neurodevelopmental conditions.
Given the emerging genetic findings, one might naturally wonder about clinical genetic testing – what are the standards for technological readiness and precisely which findings are ready for clinical use in psychiatry? A full treatment of this complex topic is beyond the scope of this review, and the answers also depend on national laws, local ethical standards, and access to genetic testing technologies. On the scientific side, we think that the available data support three uses in clinical psychiatry. (a) For severe, childhood onset neurodevelopmental disorders (particularly severe ID and ASD), one can argue for genetic evaluation of large CNVs and rare mutations that disrupt the protein sequence of genes important to neurodevelopment. We note that this is now done in many academic centers. The utility is mostly diagnostic for the child and relevant to family planning for the parent; some variants will also be medically important and lead to a change in clinical management. (b) Large CNVs in severe psychotic disorders (SCZ and schizoaffective disorder) will be present in 3–5% of cases. The utility is diagnostic and in ameliorating medical morbidity given that most CNVs are multi-system disorders carrying additional medical risks. (c) Unusual cases: individuals with a wide range of single-gene disorders can initially present with prominent psychiatric features. Instead of a primary psychiatric disorder, the behavioral features are secondary to a biological process that has been disrupted by a strong-effect mutation. Classic examples include Wilson’s disease and Huntington’s disease which can present with psychotic or mood symptoms. The utility here is diagnostic and possibly therapeutic (e.g., copper chelation therapy for Wilson’s disease can markedly improve outcomes if not delayed due to a missed diagnosis).
In many countries, genetic tests can be used by consumers without having rigorous evaluation of analytical validity, clinical validity, and clinical utility (again there are complex and country-dependent issues). However, there are abundant examples of genetic tests that are now being used clinically that have minimal or no scientific basis. This is obviously highly troubling and problematic but such testing has been allowed to occur due to failures of regulatory processes.
The fundamental database is not sufficiently complete to draw unambiguous conclusions – research in progress by many groups is combining epidemiological, clinical, and genetic risk factors in historically large samples. However, we posit that the genetic results are consistent with a tentative position: based on pervasive genetic overlap between most childhood and adult-onset psychiatric disorders and their inter-correlations with cognitive ability and personality, a central part of the inherited liability is shared by many psychiatric disorders. There may well be additional genetic factors that increase risk for specific disorders. When the scientific database is more mature, revision of psychiatric nosology based on combining clinical with rare and common variant genetic results may well be warranted.
Conclusions & future directions
Complete genetic discovery.
In the past decade, genetic approaches to psychiatric disorders have yielded more reproducible insights into etiology than any other prior approach. We now know vastly more about the fundamental causes of these impactful disorders than ever before. What we know now is incomplete and inadequate. We need to complete genetic discovery, and we believe that this should be an international priority in this area. Inexpensive SNP arrays can measure the contributions of the vast majority of common variation and be efficiently assessed in large populations now. To measure the full spectrum of rare variation and less accessible common variation, we will need resequencing efforts of similar magnitude but this will likely have to wait for more efficient platforms and improved functional annotations.
Genetic architecture in individuals.
We have described multiple architectures for psychiatric disorders. The ultimate goal is understanding as fully as possible the etiological process in individuals with a severe psychiatric disorder. How does knowledge derived from large populations contribute to illness in an individual? For example, in those with ASD who harbor a rare de novo PTV, is that variant sufficient to cause the disorder or are additional genetic or environmental risk factors required? The answers will likely vary depending on the gene, but already there are multiple hints that risk profiles in individuals are likely to be complex, even in those harboring large effect mutations. For instance, few large effect mutations are specific to a disorder suggesting a possible role for additional genetic, environmental, and stochastic factors. Although some large effect CNVs associated with ASD or SCZ have effects on fecundity, when discovered in population surveys in individuals without regard to disease status, many have relatively modest effects on the ability to have offspring, compared with those having the disease diagnosis (Stefansson et al., 2014). In the instances where this has been studied directly, polygenic risk acts additively with major mutational burdens (Gaugler et al., 2014; Niemi et al., 2018; Purcell et al., 2014; Weiner et al., 2017).
The model that we prefer is that many (but not all) large effect mutations sensitize an individual to manifest a developmental neuropsychiatric disorder. We recognize that there are rare large effect mutations that show clear preferential effects towards a disorder as is the case for some CNVs and rare protein altering mutations. However, the effects on brain development and function of many de novo or Mendelian mutations are so large as to be non-specific with respect to any single disorder (e.g. epilepsy, ASD, SCZ, ID). The resultant phenotype in an individual is dependent on the impact of environmental factors, and/or the additive effects of other rare variation and polygenic risk. This model may also help explain the high unaffected carrier rate for some inherited mutations, if one presumes that the parent carrying the mutation lacks the polygenic risk that has accumulated in the child. However, we are still a long way from being able to confidently predict disorder phenotypes from measurement of genetic risk. As noted above, the basic data remain incomplete and our – further genetic discovery efforts are needed to derive secure and enduring answers to these fundamental questions. We do note that the concept of individual architecture also spans multiple the other architectures described in this review, which will be essential to understanding mechanisms and focused therapeutics in the individual.
Sex differences.
The genetic and pathophysiological explanations for sex differences in psychiatric disorders remain poorly understood, but the advances in gene discovery described here provide a new foundation to fuel studies in this important area. Many psychiatric disorders show a different prevalence or onset in males and females (Seedat et al., 2009). For example, marked sex differences in lifetime risk are apparent for ADHD, AN, ASD, MDD, and SCZ (Hudson et al., 2007; Martin et al., 2018b; Philippe et al., 1999). Whether sex differences are due to differential vulnerability, diverging behavioral/cognitive manifestations, and/or observer bias is not known with clarity but is likely to differ across diagnoses. For example, in ASD, genetic and functional genomic evidence suggests the presence of female protective factors based in brain function and structure (Robinson et al., 2013; Werling and Geschwind, 2013; Werling et al., 2016), whereas in MDD or ALC, social factors likely may have a larger role (Riecher-Rossler, 2017). Understanding the basis of sex differences may provide critical clues for pathophysiology and could inform diagnosis and treatment.
What’s the end game?
It is essential to consider what is required to improve the diagnosis and treatment of individuals with severe psychiatric disorders. An extreme possibility is that achieving this intention could require a full understanding of the development of the human brain, the most complicated machine known to us. Achieving this intention is unlikely to occur in the foreseeable future. However, there are indications from other areas of medicine that full understanding of a pathological process is not required to improve therapeutics. For instance, the causes of melanoma are not fully worked out but the advent of checkpoint inhibitors – based on several key pieces of the melanoma puzzle — has markedly improved outcomes for disseminated disease.
A reasonable, and not overly optimistic, answer is that a solid beachhead is needed, a definite, reproducible, and clear identification of a neurobiological process conferring risk or protection for a psychiatric disorder. With such knowledge, the field changes markedly: beachheads become lodgements, lodegments become full theaters of engagement, and manifest progress becomes achievable. Instead of discovering medicines by accident and happenstance (as with virtually all prototypic medications used in clinical psychiatry), the power of modern rational drug design can be implemented.
To achieve this end, we suggest the need for a concerted global effort. We are far from being able to confidently predict disorder phenotypes from measurement of genetic risk. Given the marked progress to date, we believe it sensible to continue large and comprehensive gene discovery efforts. Such efforts are now clearly incomplete, but definable stopping points can be articulated (e.g., where genetic discovery asymptotes or if new discoveries only replicate known functional and cellular architectures). This will require working with groups traditionally underrepresented in psychiatric research to attain an inclusive and complete understanding of the contribution to disease in individuals with non-European ancestries. Discovery efforts across multiple architectures are warranted to understand the individual architecture that underlies disease risk and pathophysiology.
Supplementary Material
Acknowledgements
We thank two anonymous reviewers for helpful critiques. PFS was supported by the Swedish Research Council (Vetenskapsrådet, award D0886501), the Horizon 2020 Program of the European Union (COSYN, RIA grant agreement n° 610307), and US NIMH (U01 MH109528 and R01 MH077139). The PGC Substance Use Disorders Working Group receives support from the National Institute on Drug Abuse and the National Institute of Mental Health (U01 MH109532). DHG was supported by US NIMH (R01 MH100027, R01 MH081754, U01 MH115746U01, MH105991, R01 MH100028, R01 MH109912, R01 MH110927, P50 MH106438, R01 MH100900, R01 MH094714, R33 MH087898), NICHD (P50 HD055784 and R01 HD065280), NINDS (R01 NS073871), the Hartwell Foundation, the Simons Foundation (grants 206744 and 239766), CIRM (GC1R-0667-A), and the Paul G Allen Family Foundation. For assistance with Table 1, Table S1, and Figure 3a, we thank the PGC BIP group (Ole Andreassen, Eli Stahl, Andreas Forstner), PGC eating disorders (Cynthia Bulik), PGC MDD group (Cathryn Lewis, Andrew McIntosh), PGC OCD/Tourette’s group (Carol Mathews, Jeremiah Scharf, Dongmei Yu, Manuel Mattheisen, James Crowley), PGC PTSD group (Caroline Nievergelt), PGC SCZ group (Mick O’Donovan), and PGC SUD (Arpana Agrawal, Howard Edenberg, Joel Gelernter). We thank Damon Pouliadakis for assistance with Figure 4.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts
PF Sullivan reports the following potentially competing financial interests. Current: Lundbeck (advisory committee, grant recipient). Past three years: Pfizer (scientific advisory board), Element Genomics (consultation fee), and Roche (speaker reimbursement). DH Geschwind has the following disclosures: Research funding from Takeda pharmaceuticals, and serving as a scientific advisor for Falcon Computing, Ovid Therapeutics, Axial Biosciences, Acurastem, and Third Rock Ventures.
URLs
Psychiatric Genomics Consortium, http://pgc.unc.edu
PsychENCODE Consortium, https://www.synapse.org//#!Synapse:syn4921369/wiki/235539
Simons Simplex Collection, https://www.sfari.org/resource/sfari-base
Autism Whole Genome Sequence (AGRE, iHART), http://www.ihart.org/data; https://research.mss.ng
References
- Abrahams BS, and Geschwind DH (2008). Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 9, 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, and Stark A (2013). Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Gulbahce N, and Loscalzo J (2011). Network medicine: a network-based approach to human disease. Nat Rev Genet 12, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berto S, Usui N, Konopka G, and Fogel BL (2016). ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum Mol Genet 25, 2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billstedt E, Gillberg IC, and Gillberg C (2007). Autism in adults: symptom patterns and early childhood predictors. Use of the DISCO in a community sample followed from childhood. J Child Psychol Psychiatry 48, 1102–1110. [DOI] [PubMed] [Google Scholar]
- Border R, Johnson EC, Evans LM, Smolen A, Berley N, Sullivan PF, and Keller MC (In press). No support for candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. American Journal of Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainstorm Consortium (2018). Analysis of shared heritability in common disorders of the brain. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand H, Pillalamarri V, Collins RL, Eggert S, O’Dushlaine C, Braaten EB, Stone MR, Chambert K, Doty ND, Hanscom C, Rosenfeld JA, Ditmars H, Blais J, Mills R, Lee C, Gusella JF, McCarroll S, Smoller JW, Talkowski ME, and Doyle AE (2014). Cryptic and complex chromosomal aberrations in early-onset neuropsychiatric disorders. Am J Hum Genet 95, 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, Kim C, Gidaya NB, Lindquist I, Hutman T, Sigman M, Kustanovich V, Lajonchere CM, Singleton A, Kim J, Wassink TH, McMahon WM, Owley T, Sweeney JA, Coon H, Nurnberger JI, Li M, Cantor RM, Minshew NJ, Sutcliffe JS, Cook EH, Dawson G, Buxbaum JD, Grant SF, Schellenberg GD, Geschwind DH, and Hakonarson H (2009). Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet 5, e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja A, Volfovsky N, Krieger AM, Lord C, Lash AE, Wigler M, and Iossifov I (2018). Damaging de novo mutations diminish motor skills in children on the autism spectrum. Proc Natl Acad Sci U S A 115, E1859–E1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics, C., Patterson N, Daly MJ, Price AL, and Neale BM (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, State MW, and Autism Sequencing, C. (2012). The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron 76, 1052–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNV Working Group of the Psychiatric Genomics, C. (2017). Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Wellcome Trust Case Control, C., Tyler-Smith C, Carter NP, Lee C, Scherer SW, and Hurles ME (2010). Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE Consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, Broly M, Rodriguez M, Tam S, Trigg SA, Fan C, Yi S, Tasan M, Lemmens I, Kuang X, Zhao N, Malhotra D, Michaelson JJ, Vacic V, Calderwood MA, Roth FP, Tavernier J, Horvath S, Salehi-Ashtiani K, Korkin D, Sebat J, Hill DE, Hao T, Vidal M, and Iakoucheva LM (2014). Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun 5, 3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, Reilly SK, Tebbenkamp AT, Bichsel C, Pletikos M, Sestan N, Roeder K, State MW, Devlin B, and Noonan JP (2015). The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun 6, 6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium (2013a). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium (2013b). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, Lee C, Regalado SG, Read DF, Steemers FJ, Disteche CM, Trapnell C, and Shendure J (2018a). A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 174, 1309–1324 e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Reddington JP, Garfield DA, Daza RM, Aghamirzaie D, Marco-Ferreres R, Pliner HA, Christiansen L, Qiu X, Steemers FJ, Trapnell C, Shendure J, and Furlong EEM (2018b). The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EI, Ferdinand RF, Meester S, de Nijs PF, and Verheij F (2007). High rates of psychiatric comorbidity in PDD-NOS. J Autism Dev Disord 37, 877–886. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, and Geschwind DH (2018). The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell 172, 289–304 e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won H, Stein JL, and Geschwind DH (2016). Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimaki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnstrom K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Ruther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, Study, D.D.D., Homozygosity Mapping Collaborative for, A., Consortium, U.K., Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, and Buxbaum JD (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, Politz JCR, Shendure J, Zhong S, and Network, D.N. (2017). The 4D nucleome project. Nature 549, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, and Mirny LA (2013). Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 14, 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, and Zeng H (2017). The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron 96, 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2011). A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9, e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Lee E, Park PJ, and Walsh CA (2016). Resolving rates of mutation in the brain using single-neuron genomics. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’Donovan MC, Corvin A, Cichon S, and Sullivan PF (2015). Evaluating historical candidate genes for schizophrenia. Mol Psychiatry 20, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh PR, Lareau C, Shoresh N, Genovese G, Saunders A, Macosko E, Pollack S, Brainstorm C, Perry JRB, Buenrostro JD, Bernstein BE, Raychaudhuri S, McCarroll S, Neale BM, and Price AL (2018). Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach GD, and Lord C (2010). The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195. [DOI] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, and Geschwind DH (2012). RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet 21, 4171–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gumus ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, Readhead B, Stahl EA, Xiao J, Parvizi M, Hamamsy T, Fullard JF, Wang YC, Mahajan MC, Derry JM, Dudley JT, Hemby SE, Logsdon BA, Talbot K, Raj T, Bennett DA, De Jager PL, Zhu J, Zhang B, Sullivan PF, Chess A, Purcell SM, Shinobu LA, Mangravite LM, Toyoshiba H, Gur RE, Hahn CG, Lewis DA, Haroutunian V, Peters MA, Lipska BK, Buxbaum JD, Schadt EE, Hirai K, Roeder K, Brennand KJ, Katsanis N, Domenici E, Devlin B, and Sklar P (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Project, N.E.S., and Akey JM (2013). Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullard JF, Hauberg ME, Bendl J, Egervari G, Cirnaru MD, Reach SM, Motl J, Ehrlich ME, Hurd YL, and Roussos P (2018). An atlas of chromatin accessibility in the adult human brain. Genome Res 28, 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP, CommonMind, C., Psych, E.C., i, P.-B.W.G., Horvath S, and Geschwind DH (2018a). Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Leppa V, Won H, Parikshak NN, and Geschwind DH (2016). The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci 19, 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H, Varghese M, Wang Y, Shieh AW, Haney J, Parhami S, Belmont J, Kim M, Moran Losada P, Khan Z, Mleczko J, Xia Y, Dai R, Wang D, Yang YT, Xu M, Fish K, Hof PR, Warrell J, Fitzgerald D, White K, Jaffe AE, Psych EC, Peters MA, Gerstein M, Liu C, Iakoucheva LM, Pinto D, and Geschwind DH (2018b). Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, Ripke S, Sandin S, Sklar P, Svantesson O, Reichenberg A, Hultman CM, Devlin B, Roeder K, and Buxbaum JD (2014). Most genetic risk for autism resides with common variation. Nat Genet 46, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M, Moran JL, Purcell SM, Sklar P, Sullivan PF, Hultman CM, and McCarroll SA (2016). Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci 19, 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH (2009). Advances in autism. Annu Rev Med 60, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, and Flint J (2015). Genetics and genomics of psychiatric disease. Science 349, 1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, and Konopka G (2009). Neuroscience in the era of functional genomics and systems biology. Nature 461, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, and Rakic P (2013). Cortical evolution: judge the brain by its cover. Neuron 80, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, and Committee AS (2001). The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet 69, 463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, and Vitkup D (2012). Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci 15, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, and Vitkup D (2011). Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, and Lewis DA (2018). Mapping pathologic circuitry in schizophrenia. Handb Clin Neurol 150, 389–417. [DOI] [PubMed] [Google Scholar]
- Global Burden of Disease Collaborative Network (2017). Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronberg H, Adolfsson J, Aly M, Nordstrom T, Wiklund P, Brandberg Y, Thompson J, Wiklund F, Lindberg J, Clements M, Egevad L, and Eklund M (2015). Prostate cancer screening in men aged 50–69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol 16, 1667–1676. [DOI] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters R, Won H, Pallesen J, Agerbo E, Andreassen OA, Anney R, Belliveau R, Bettella F, Buxbaum JD, Bybjerg-Grauholm J, Bækved-Hansen M, Cerrato F, Chambert K, Christensen JH, Churchhouse C, Dellenvall K, Demontis D, De Rubeis S, Devlin B, Djurovic S, Dumont A, Goldstein J, Hansen CS, Hauberg ME, Hollegaard MV, Hope S, Howrigan DP, Huang H, Hultman C, Klei L, Maller J, Martin J, Martin AR, Moran J, Nyegaard M, Nærland T, Palmer DS, Palotie A, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Pourcain BS, Qvist P, Rehnström K, Reichenberg A, Reichert J, Robinson E, Roeder K, Roussos P, Saemundsen E, Sandin S, Satterstrom FK, Smith GD, Stefansson H, Stefansson K, Steinberg S, Stevens C, Sullivan PF, Turley P, Walters GB, Xu X, Geschwind D, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Neale BM, Daly MJ, and Børglum AD (In press). Common risk variants identified in autism spectrum disorder. Nature Genetics, 224774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2017). Genetic effects on gene expression across human tissues. Nature 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Consortium on the Genetics of, S., Group, P.S., Nimgaonkar VL, Go RC, Savage RM, Swerdlow NR, Gur RE, Braff DL, King MC, and McClellan JM (2013). Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A, Schizophrenia Working Group of the Psychiatric Genomics, C., McCarroll S, Neale BM, Ophoff RA, O’Donovan MC, Crawford GE, Geschwind DH, Katsanis N, Sullivan PF, Pasaniuc B, and Price AL (2018). Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 50, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauberg ME, Zhang W, Giambartolomei C, Franzen O, Morris DL, Vyse TJ, Ruusalepp A, CommonMind, C., Sklar P, Schadt EE, Bjorkegren JLM, and Roussos P (2017). Large-Scale Identification of Common Trait and Disease Variants Affecting Gene Expression. Am J Hum Genet 100, 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook P, and McCallion A (2018). Heritability enrichment in open chromatin reveals cortical layer contributions to schizophrenia. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Penn O, Borenstein E, and Eichler EE (2015). The discovery of integrated gene networks for autism and related disorders. Genome Res 25, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H, Lawrence MS, Chouinard CR, Shrestha Y, Hu JX, Worstell E, Shea E, Ilic N, Kim E, Kamburov A, Kashani A, Hahn WC, Campbell JD, Boehm JS, Getz G, and Lage K (2018). NetSig: network-based discovery from cancer genomes. Nat Methods 15, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlin P, and Magiati I (2017). Autism spectrum disorder: outcomes in adulthood. Curr Opin Psychiatry 30, 69–76. [DOI] [PubMed] [Google Scholar]
- Hudson JI, Hiripi E, Pope HG Jr., and Kessler RC (2007). The prevalence and correlates of eating disorders in the National Comorbidity Survey Replication. Biol Psychiatry 61, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (2001). A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69, 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP (2005). Why most published research findings are false. PLoS Med 2, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, and Wigler M (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB, Mardis ER, Wilson RK, Schatz MC, McCombie WR, and Wigler M (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall KM, Rees E, Escott-Price V, Einon M, Thomas R, Hewitt J, O’Donovan MC, Owen MJ, Walters JTR, and Kirov G (2017). Cognitive Performance Among Carriers of Pathogenic Copy Number Variants: Analysis of 152,000 UK Biobank Subjects. Biol Psychiatry 82, 103–110. [DOI] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, and Kathiresan S (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G (2015). CNVs in neuropsychiatric disorders. Hum Mol Genet 24, R45–49. [DOI] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, and Eichler EE (2015). Excess of rare, inherited truncating mutations in autism. Nat Genet 47, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajonchere CM, and Consortium, A. (2010). Changing the landscape of autism research: the autism genetic resource exchange. Neuron 68, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Chen C-Y, Li Z, Martin AR, Bryois J, Ma X, Gaspar H, Ikeda M, Benyamin B, Brown BC, Liu R, Zhou W, Guan L, Kamatani Y, Kim S-W, Kubo M, Kusumawardhani A, Liu C-M, Ma H, Periyasamy S, Takahashi A, Wang Q, Xu Z, Yu H, Zhu F, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Indonesia Schizophrenia Consortium, Genetic REsearch on schizophreniA neTwork-China and Netherland (GREAT-CN), Chen WJ, Faraone S, Glatt SJ, He L, Hyman SE, Hwu H-G, Li T, McCarroll S, Neale BM, Sklar P, Wildenauer D, Yu X, Zhang D, Mowry B, Lee J, Xu S, Sullivan PF, Ripke S, O’Donovan M, Daly MJ, Qin S, Sham P, Iwata N, Hong KS, Schwab SG, Yue W, Tsuang M, Liu J, Ma X, Kahn RS, Shi Y, and Huang H (Submitted). Comparative genetic architectures of schizophrenia in East Asian and European populations. [Google Scholar]
- Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR, and Sullivan PF (2014). Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry 76, 510–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, and Vassos E (2017). Prospects for using risk scores in polygenic medicine. Genome Med 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shi M, Ma Z, Zhao S, Euskirchen G, Ziskin J, Urban A, Hallmayer J, and Snyder M (2014). Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol 10, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, Sousa AMM, Werling DM, Kitchen RR, Kang HJ, Pletikos M, Choi J, Muchnik S, Xu X, Wang D, Lorente-Galdos B, Liu S, Giusti-Rodriguez P, Won H, de Leeuw CA, Pardinas AF, BrainSpan C, Psych EC, Psych EDS, Hu M, Jin F, Li Y, Owen MJ, O’Donovan MC, Walters JTR, Posthuma D, Levitt P, Weinberger DR, Hyde TM, Kleinman JE, Geschwind DH, Hawrylycz MJ, State MW, Sanders SJ, Sullivan PF, Gerstein MB, Lein ES, Knowles JA, and Sestan N (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, Vidal M, Sebat J, and Iakoucheva LM (2015). Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron 85, 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Finucane HK, Gusev A, Bhatia G, Gazal S, O’Connor L, Bulik-Sullivan B, Wright FA, Sullivan PF, Neale BM, and Price AL (2017a). Functional Architectures of Local and Distal Regulation of Gene Expression in Multiple Human Tissues. Am J Hum Genet 100, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu S, Dhiman VK, Brunetti T, Eckart H, and White KP (2017b). Functional assessment of human enhancer activities using whole-genome STARR-sequencing. Genome Biol 18, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther C, Costain G, Baribeau DA, and Bassett AS (2017). Genomic Disorders in Psychiatry-What Does the Clinician Need to Know? Curr Psychiatry Rep 19, 82. [DOI] [PubMed] [Google Scholar]
- Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, Chu SH, Klei L, Cai C, Ou J, Lowe JK, Hurles ME, Devlin B, State MW, and Geschwind DH (2012). Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am J Hum Genet 91, 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, and Sebat J (2012). CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148, 1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, Rueger S, Thorleifsson G, Justice AE, Lamparter D, Stirrups KE, Turcot V, Young KL, Winkler TW, Esko T, Karaderi T, Locke AE, Masca NG, Ng MC, Mudgal P, Rivas MA, Vedantam S, Mahajan A, Guo X, Abecasis G, Aben KK, Adair LS, Alam DS, Albrecht E, Allin KH, Allison M, Amouyel P, Appel EV, Arveiler D, Asselbergs FW, Auer PL, Balkau B, Banas B, Bang LE, Benn M, Bergmann S, Bielak LF, Bluher M, Boeing H, Boerwinkle E, Boger CA, Bonnycastle LL, Bork-Jensen J, Bots ML, Bottinger EP, Bowden DW, Brandslund I, Breen G, Brilliant MH, Broer L, Burt AA, Butterworth AS, Carey DJ, Caulfield MJ, Chambers JC, Chasman DI, Chen YI, Chowdhury R, Christensen C, Chu AY, Cocca M, Collins FS, Cook JP, Corley J, Galbany JC, Cox AJ, Cuellar-Partida G, Danesh J, Davies G, de Bakker PI, de Borst GJ, de Denus S, de Groot MC, de Mutsert R, Deary IJ, Dedoussis G, Demerath EW, den Hollander AI, Dennis JG, Di Angelantonio E, Drenos F, Du M, Dunning AM, Easton DF, Ebeling T, Edwards TL, Ellinor PT, Elliott P, Evangelou E, Farmaki AE, Faul JD, Feitosa MF, Feng S, Ferrannini E, Ferrario MM, Ferrieres J, Florez JC, Ford I, Fornage M, Franks PW, Frikke-Schmidt R, Galesloot TE, Gan W, Gandin I, Gasparini P, Giedraitis V, Giri A, Girotto G, Gordon SD, Gordon-Larsen P, Gorski M, Grarup N, Grove ML, Gudnason V, Gustafsson S, Hansen T, Harris KM, Harris TB, Hattersley AT, Hayward C, He L, Heid IM, Heikkila K, Helgeland O, Hernesniemi J, Hewitt AW, Hocking LJ, Hollensted M, Holmen OL, Hovingh GK, Howson JM, Hoyng CB, Huang PL, Hveem K, Ikram MA, Ingelsson E, Jackson AU, Jansson JH, Jarvik GP, Jensen GB, Jhun MA, Jia Y, Jiang X, Johansson S, Jorgensen ME, Jorgensen T, Jousilahti P, Jukema JW, Kahali B, Kahn RS, Kahonen M, Kamstrup PR, Kanoni S, Kaprio J, Karaleftheri M, Kardia SL, Karpe F, Kee F, Keeman R, Kiemeney LA, Kitajima H, Kluivers KB, Kocher T, Komulainen P, Kontto J, Kooner JS, Kooperberg C, Kovacs P, Kriebel J, Kuivaniemi H, Kury S, Kuusisto J, La Bianca M, Laakso M, Lakka TA, Lange EM, Lange LA, Langefeld CD, Langenberg C, Larson EB, Lee IT, Lehtimaki T, Lewis CE, Li H, Li J, Li-Gao R, Lin H, Lin LA, Lin X, Lind L, Lindstrom J, Linneberg A, Liu Y, Liu Y, Lophatananon A, Luan J, Lubitz SA, Lyytikainen LP, Mackey DA, Madden PA, Manning AK, Mannisto S, Marenne G, Marten J, Martin NG, Mazul AL, Meidtner K, Metspalu A, Mitchell P, Mohlke KL, Mook-Kanamori DO, Morgan A, Morris AD, Morris AP, Muller-Nurasyid M, Munroe PB, Nalls MA, Nauck M, Nelson CP, Neville M, Nielsen SF, Nikus K, Njolstad PR, Nordestgaard BG, Ntalla I, O’Connel JR, Oksa H, Loohuis LM, Ophoff RA, Owen KR, Packard CJ, Padmanabhan S, Palmer CN, Pasterkamp G, Patel AP, Pattie A, Pedersen O, Peissig PL, Peloso GM, Pennell CE, Perola M, Perry JA, Perry JR, Person TN, Pirie A, Polasek O, Posthuma D, Raitakari OT, Rasheed A, Rauramaa R, Reilly DF, Reiner AP, Renstrom F, Ridker PM, Rioux JD, Robertson N, Robino A, Rolandsson O, Rudan I, Ruth KS, Saleheen D, Salomaa V, Samani NJ, Sandow K, Sapkota Y, Sattar N, Schmidt MK, Schreiner PJ, Schulze MB, Scott RA, Segura-Lepe MP, Shah S, Sim X, Sivapalaratnam S, Small KS, Smith AV, Smith JA, Southam L, Spector TD, Speliotes EK, Starr JM, Steinthorsdottir V, Stringham HM, Stumvoll M, Surendran P, t Hart LM, Tansey KE, Tardif JC, Taylor KD, Teumer A, Thompson DJ, Thorsteinsdottir U, Thuesen BH, Tonjes A, Tromp G, Trompet S, Tsafantakis E, Tuomilehto J, Tybjaerg-Hansen A, Tyrer JP, Uher R, Uitterlinden AG, Ulivi S, van der Laan SW, Van Der Leij AR, van Duijn CM, van Schoor NM, van Setten J, Varbo A, Varga TV, Varma R, Edwards DR, Vermeulen SH, Vestergaard H, Vitart V, Vogt TF, Vozzi D, Walker M, Wang F, Wang CA, Wang S, Wang Y, Wareham NJ, Warren HR, Wessel J, Willems SM, Wilson JG, Witte DR, Woods MO, Wu Y, Yaghootkar H, Yao J, Yao P, Yerges-Armstrong LM, Young R, Zeggini E, Zhan X, Zhang W, Zhao JH, Zhao W, Zhao W, Zheng H, Zhou W, Consortium, E.P.-I., Consortium, C.H.D.E., Exome, B.P.C., Consortium, T.D.-G., Go, T.D.G.C., Global Lipids Genetics, C., ReproGen, C., Investigators, M., Rotter JI, Boehnke M, Kathiresan S, McCarthy MI, Willer CJ, Stefansson K, Borecki IB, Liu DJ, North KE, Heard-Costa NL, Pers TH, Lindgren CM, Oxvig C, Kutalik Z, Rivadeneira F, Loos RJ, Frayling TM, Hirschhorn JN, Deloukas P, and Lettre G (2017). Rare and low-frequency coding variants alter human adult height. Nature 542, 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, and Kenny EE (2017). Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet 100, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Taylor MJ, and Lichtenstein P (2018a). Assessing the evidence for shared genetic risks across psychiatric disorders and traits. Psychol Med 48, 1759–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Walters RK, Demontis D, Mattheisen M, Lee SH, Robinson E, Brikell I, Ghirardi L, Larsson H, Lichtenstein P, Eriksson N, andMe Research, T., Psychiatric Genomics Consortium, A.S., i, P.-B.A.W., Werge T, Mortensen PB, Pedersen MG, Mors O, Nordentoft M, Hougaard DM, Bybjerg-Grauholm J, Wray NR, Franke B, Faraone SV, O’Donovan MC, Thapar A, Borglum AD, and Neale BM (2018b). A Genetic Investigation of Sex Bias in the Prevalence of Attention-Deficit/Hyperactivity Disorder. Biol Psychiatry 83, 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, and Mahajan A (2018). The value of genetic risk scores in precision medicine for diabetes\. Expert Review of Precision Medicine and Drug Development 3, 279–281. [Google Scholar]
- McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D, Ganz J, Jaffe AE, Kwan KY, Kwon M, Lodato MA, Mills RE, Paquola ACM, Rodin RE, Rosenbluh C, Sestan N, Sherman MA, Shin JH, Song S, Straub RE, Thorpe J, Weinberger DR, Urban AE, Zhou B, Gage FH, Lehner T, Senthil G, Walsh CA, Chess A, Courchesne E, Gleeson JG, Kidd JM, Park PJ, Pevsner J, Vaccarino FM, and Brain Somatic Mosaicism, N. (2017). Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HY, Leng J, Li R, Li Y, Lin CY, Luo R, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi X, Stromberg MP, Stutz AM, Urban AE, Walker JA, Wu J, Zhang Y, Zhang ZD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Ye K, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO, and Genomes P (2011). Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldin SO (2003). NIMH Human Genetics Initiative: 2003 update. Am J Psychiatry 160, 621–622. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zollner S, Whittaker JC, Chissoe SL, Novembre J, and Mooser V (2012). An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium (2015). Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 18, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, McAloney K, McRae J, Radford EJ, Yu S, Gecz J, Martin NG, Wright CF, Fitzpatrick DR, Firth HV, Hurles ME, and Barrett JC (2018). Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature 562, 268–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, and Eichler EE (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Gandal MJ, and Geschwind DH (2015). Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat Rev Genet 16, 441–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, and Geschwind DH (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LT, Huang J, Lowe JK, Blencowe BJ, Horvath S, and Geschwind DH (2016). Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, and Leboyer M (1999). Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet 8, 805–812. [DOI] [PubMed] [Google Scholar]
- Piper M, Beneyto M, Burne TH, Eyles DW, Lewis DA, and McGrath JJ (2012). The neurodevelopmental hypothesis of schizophrenia: convergent clues from epidemiology and neuropathology. Psychiatr Clin North Am 35, 571–584. [DOI] [PubMed] [Google Scholar]
- Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, and Posthuma D (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709. [DOI] [PubMed] [Google Scholar]
- Polioudakis D, de la Torre-Ubieta L,LJ, Elkins AG, Stein JL, Vuong CK, Opland CK, Lu D, Connell W, Ruzzo EK, Lowe JK, Hadzic T, Hinz FI, Sabri S, Lowry WE, Plath K, and Geschwind D (2018). A single cell transcriptomic analysis of human neocortical development. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R (2002). Madness: A Brief History (Oxford: Oxford University Press; ). [Google Scholar]
- PsychENCODE Consortium (2018). Revealing the brain’s molecular architecture. Science 362, 1262–1263. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Coordinating Committee (2009). Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry 166, 540–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Steering Committee (2009). A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry 14, 10–17. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, and Sklar P (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, Hanscom C, Pillalamarri V, Seabra CM, Abbott MA, Abdul-Rahman OA, Aberg E, Adley R, Alcaraz-Estrada SL, Alkuraya FS, An Y, Anderson MA, Antolik C, Anyane-Yeboa K, Atkin JF, Bartell T, Bernstein JA, Beyer E, Blumenthal I, Bongers EM, Brilstra EH, Brown CW, Bruggenwirth HT, Callewaert B, Chiang C, Corning K, Cox H, Cuppen E, Currall BB, Cushing T, David D, Deardorff MA, Dheedene A, D’Hooghe M, de Vries BB, Earl DL, Ferguson HL, Fisher H, FitzPatrick DR, Gerrol P, Giachino D, Glessner JT, Gliem T, Grady M, Graham BH, Griffis C, Gripp KW, Gropman AL, Hanson-Kahn A, Harris DJ, Hayden MA, Hill R, Hochstenbach R, Hoffman JD, Hopkin RJ, Hubshman MW, Innes AM, Irons M, Irving M, Jacobsen JC, Janssens S, Jewett T, Johnson JP, Jongmans MC, Kahler SG, Koolen DA, Korzelius J, Kroisel PM, Lacassie Y, Lawless W, Lemyre E, Leppig K, Levin AV, Li H, Li H, Liao EC, Lim C, Lose EJ, Lucente D, Macera MJ, Manavalan P, Mandrile G, Marcelis CL, Margolin L, Mason T, Masser-Frye D, McClellan MW, Mendoza CJ, Menten B, Middelkamp S, Mikami LR, Moe E, Mohammed S, Mononen T, Mortenson ME, Moya G, Nieuwint AW, Ordulu Z, Parkash S, Pauker SP, Pereira S, Perrin D, Phelan K, Aguilar RE, Poddighe PJ, Pregno G, Raskin S, Reis L, Rhead W, Rita D, Renkens I, Roelens F, Ruliera J, Rump P, Schilit SL, Shaheen R, Sparkes R, Spiegel E, Stevens B, Stone MR, Tagoe J, Thakuria JV, van Bon BW, van de Kamp J, van Der Burgt I, van Essen T, van Ravenswaaij-Arts CM, van Roosmalen MJ, Vergult S, Volker-Touw CM, Warburton DP, Waterman MJ, Wiley S, Wilson A, Yerena-de Vega MC, Zori RT, Levy B, Brunner HG, de Leeuw N, Kloosterman WP, Thorland EC, Morton CC, Gusella JF, and Talkowski ME (2017). The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet 49, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riecher-Rossler A (2017). Sex and gender differences in mental disorders. Lancet Psychiatry 4, 8–9. [DOI] [PubMed] [Google Scholar]
- Risch N, and Merikangas K (1996). The future of genetic studies of complex human diseases. Science 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, Lichtenstein P, Anckarsater H, Happe F, and Ronald A (2013). Examining and interpreting the female protective effect against autistic behavior. Proc Natl Acad Sci U S A 110, 5258–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M, Iossifov I, Levy D, and Wigler M (2014). The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet 15, 133–141. [DOI] [PubMed] [Google Scholar]
- Ruzzo E, Perez-Cano L, Jung J, Wang L-K, Kashef-Haghighi D, Hartl C, Hoekstra J, Leventhal O, Gandal MJ, Paskov K, Stockham N, Polioudakis D, Lowe JK, Geschwind DH, and Wall DP (2018). Whole genome sequencing in multiplex families reveals novel inherited and de novo genetic risk in autism. BioRxiv. [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnstrom K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH Jr., Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, and Daly MJ (2014). A framework for the interpretation of de novo mutation in human disease. Nat Genet 46, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O’Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Gunel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH Jr., Geschwind D, Roeder K, Devlin B, and State MW (2011). Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF 3rd, Klei L, Mandell JD, Moreno-De-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW, Autism Sequencing C, Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, and State MW (2015). Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Gunel M, Roeder K, Geschwind DH, Devlin B, and State MW (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Neale BM, Huang H, Werling DM, An JY, Dong S, Abecasis G, Arguello PA, Blangero J, Boehnke M, Daly MJ, Eggan K, Geschwind DH, Glahn DC, Goldstein DB, Gur RE, Handsaker RE, McCarroll SA, Ophoff RA, Palotie A, Pato CN, Sabatti C, State MW, Willsey AJ, Hyman SE, Addington AM, Lehner T, Freimer NB, and Whole Genome Sequencing for Psychiatric, D. (2018). Whole genome sequencing in psychiatric disorders: the WGSPD consortium. Nat Neurosci 21, 1017. [DOI] [PubMed] [Google Scholar]
- Satterstrom F, Walters R, Singh T, Wigdor E, Lescai F, Demontis D, Kosmicki J, Grove J, Stevens C, Bybjerg-Grauholm J, Børglum A, and Daly M (2018a). ASD and ADHD have a similar burden of rare protein-truncating variants. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, Peng M, Collins RL, Grove J, Klei L, Stevens C, Reichert J, Mulhern M, Artomov M, Gerges S, Sheppard B, Xu X, Bhaduri A, Norman U, Brand H, Schwartz G, Nguyen R, Guerrero E, Dias C, Aleksic B, Anney RJL, Barbosa M, Bishop S, Brusco A, Bybjerg-Grauholm J, Carracedo A, Chan MCY, Chiocchetti A, Chung B, Coon H, Cuccaro M, Curró A, Dalla Bernardina B, Doan R, Domenici E, Dong S, Fallerini C, Fernández-Prieto M, Ferrero GB, Freitag CM, Fromer M, Gargus JJ, Geschwind D, Giorgio E, González-Peñas J, Guter S, Halpern D, Hassen-Kiss E, He X, Herman G, Hertz-Picciotto I, Hougaard DM, Hultman CM, Ionita-Laza I, Jacob S, Jamison J, Jugessur A, Kaartinen M, Knudsen GP, Kolevzon A, Kushima I, Lee SL, Lehtimäki T, Lim ET, Lintas C, Lipkin WI, Lopergolo D, Lopes F, Ludena Y, Maciel P, Magnus P, Mahjani B, Maltman N, Manoach DS, Meiri G, Menashe I, Miller J, Minshew N, Montenegro M de Souza E, Moreira D, Morrow E, Mors O, Mortensen PB, Mosconi M, Muglia P, Neale B, Nordentoft M, Ozaki N, Palotie A, Parellada M, Passos-Bueno MR, Pericak-Vance M, Persico A, Pessah I, Puura K, Reichenberg A, Renieri A, Riberi E, Robinson E, Samocha KE, Sandin S, Santangelo SL, Schellenberg G, Scherer S, Schlitt S, Schmidt R, Schmitt L, Silva IMW, Singh T, Siper P, Smith M, Soares G, Stoltenberg C, Suren P, Susser E, Sweeney J, Szatmari P, Tang L, Tassone F, Teufel K, Trabetti E, Trelles M.d.P., Walsh C, Weiss L, Werge T, Werling D, Wigdor EM, Wilkinson E, Willsey JA, Yu T, Yu MHC, Yuen R, Zachi E, Betancur C, Cook EH, Gallagher L, Gill M, Lehner T, Senthil G, Sutcliffe JS, Thurm A, Zwick ME, Børglum AD, State MW, Cicek AE, Talkowski ME, Cutler DJ, Devlin B, Sanders SJ, Roeder K, Buxbaum JD, and Daly MJ (2018b). Novel genes for autism implicate both excitatory and inhibitory cell lineages in risk. BioRxiv, 484113. [Google Scholar]
- Satterstrom FK, Walters RK, Singh T, Wigdor EM, Lescai F, Demontis D, Kosmicki JA, Grove J, Stevens C, Bybjerg-Grauholm J, Bækvad-Hansen M, Palmer DS, Maller JB, Nordentoft M, Mors O, Robinson EB, Hougaard DM, Werge TM, Bo Mortensen P, Neale BM, Børglum AD, and Daly MJ (2018c). ASD and ADHD have a similar burden of rare protein-truncating variants. BioRxiv, 277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, Hougaard D, Baekved-Hansen M, Bybjerg-Grauholm J, Pedersen MG, Pedersen CB, Neale BM, Daly MJ, Nordentoft M, Mors O, Boerglum AD, Mortensen PB, Buil A, Thompson WK, Geschwind D, and Werge T (In press). A genome-wide association study for shared risk across major psychiatric disorders in a nation-wide birth cohort implicates fetal neurodevelopment as a key mediator. Nature Neuroscience, 240911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, and Wigler M (2007). Strong association of de novo copy number mutations with autism. Science 316, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, and Wigler M (2004). Large-scale copy number polymorphism in the human genome. Science 305, 525–528. [DOI] [PubMed] [Google Scholar]
- Seedat S, Scott KM, Angermeyer MC, Berglund P, Bromet EJ, Brugha TS, Demyttenaere K, de Girolamo G, Haro JM, Jin R, Karam EG, Kovess-Masfety V, Levinson D, Medina Mora ME, Ono Y, Ormel J, Pennell BE, Posada-Villa J, Sampson NA, Williams D, and Kessler RC (2009). Cross-national associations between gender and mental disorders in the World Health Organization World Mental Health Surveys. Arch Gen Psychiatry 66, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Schizophrenia Working Group of the Psychiatric Genomics, C., Daly MJ, Carroll MC, Stevens B, and McCarroll SA (2016). Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh Y, Hu D, Ma L, Huntsman S, Gard CC, Leung JW, Tice JA, Vachon CM, Cummings SR, Kerlikowske K, and Ziv E (2016). Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res Treat 159, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, Wright CF, Firth HV, FitzPatrick DR, Barrett JC, and Hurles ME (2018). De novo mutations in regulatory elements in neurodevelopmental disorders. Nature 555, 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Suvisaari J, Chheda H, Blackwood D, Breen G, Pietilainen O, Gerety SS, Ayub M, Blyth M, Cole T, Collier D, Coomber EL, Craddock N, Daly MJ, Danesh J, DiForti M, Foster A, Freimer NB, Geschwind D, Johnstone M, Joss S, Kirov G, Korkko J, Kuismin O, Holmans P, Hultman CM, Iyegbe C, Lonnqvist J, Mannikko M, McCarroll SA, McGuffin P, McIntosh AM, McQuillin A, Moilanen JS, Moore C, Murray RM, Newbury-Ecob R, Ouwehand W, Paunio T, Prigmore E, Rees E, Roberts D, Sambrook J, Sklar P, St Clair D, Veijola J, Walters JT, Williams H, Swedish Schizophrenia Study, Interval Study, D. D. D. Study, UK10K Consortium, Sullivan PF, Hurles ME, O’Donovan MC, Palotie A, Owen MJ, and Barrett JC (2016). Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 19, 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Munoz-Manchado AB, O’Donovan MC, Owen MJ, Pardinas AF, Ryge J, Walters JTR, Linnarsson S, Lein ES, Major Depressive Disorder Working Group of the Psychiatric Genomics, C., Sullivan PF, and Hjerling-Leffler J (2018). Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spark Consortium (2018). SPARK: A US Cohort of 50,000 Families to Accelerate Autism Research. Neuron 97, 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, Bjornsdottir G, Walters GB, Jonsdottir GA, Doyle OM, Tost H, Grimm O, Kristjansdottir S, Snorrason H, Davidsdottir SR, Gudmundsson LJ, Jonsson GF, Stefansdottir B, Helgadottir I, Haraldsson M, Jonsdottir B, Thygesen JH, Schwarz AJ, Didriksen M, Stensbol TB, Brammer M, Kapur S, Halldorsson JG, Hreidarsson S, Saemundsen E, Sigurdsson E, and Stefansson K (2014). CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505, 361–366. [DOI] [PubMed] [Google Scholar]
- Steinberg S, Gudmundsdottir S, Sveinbjornsson G, Suvisaari J, Paunio T, Torniainen-Holm M, Frigge ML, Jonsdottir GA, Huttenlocher J, Arnarsdottir S, Ingimarsson O, Haraldsson M, Tyrfingsson T, Thorgeirsson TE, Kong A, Norddahl GL, Gudbjartsson DF, Sigurdsson E, Stefansson H, and Stefansson K (2017). Truncating mutations in RBM12 are associated with psychosis. Nat Genet 49, 1251–1254. [DOI] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, Miles J, Sheridan SD, Stortchevoi A, Kellis M, Haggarty SJ, Katsanis N, Gusella JF, and Talkowski ME (2014). CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A 111, E4468–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, Cichon S, Edenberg HJ, Faraone SV, Gelernter J, Mathews CA, Nievergelt CM, Smoller JW, O’Donovan MC, and Psychiatric Genomics C. (2018). Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry 175, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, and O’Donovan M (2012). Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 13, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Poschmann J, Cruz-Herrera Del Rosario R, Parikshak NN, Hajan HS, Kumar V, Ramasamy R, Belgard TG, Elanggovan B, Wong CCY, Mill J, Geschwind DH, and Prabhakar S (2016). Histone Acetylome-wide Association Study of Autism Spectrum Disorder. Cell 167, 1385–1397 e1311. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Mullegama SV, Rosenfeld JA, van Bon BW, Shen Y, Repnikova EA, Gastier-Foster J, Thrush DL, Kathiresan S, Ruderfer DM, Chiang C, Hanscom C, Ernst C, Lindgren AM, Morton CC, An Y, Astbury C, Brueton LA, Lichtenbelt KD, Ades LC, Fichera M, Romano C, Innis JW, Williams CA, Bartholomew D, Van Allen MI, Parikh A, Zhang L, Wu BL, Pyatt RE, Schwartz S, Shaffer LG, de Vries BB, Gusella JF, and Elsea SH (2011). Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet 89, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey KE, Rees E, Linden DE, Ripke S, Chambert KD, Moran JL, McCarroll SA, Holmans P, Kirov G, Walters J, Owen MJ, and O’Donovan MC (2016). Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry 21, 1085–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, and Stamatoyannopoulos JA (2012). The accessible chromatin landscape of the human genome. Nature 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Voineagu I, Pasca SP, Won H, Chandran V, Horvath S, Dolmetsch RE, and Geschwind DH (2014). Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, and Richards JB (2018). Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet 19, 110–124. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Wineinger NE, and Topol EJ (2018). The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19, 581–590. [DOI] [PubMed] [Google Scholar]
- Ulfarsson MO, Walters GB, Gustafsson O, Steinberg S, Silva A, Doyle OM, Brammer M, Gudbjartsson DF, Arnarsdottir S, Jonsdottir GA, Gisladottir RS, Bjornsdottir G, Helgason H, Ellingsen LM, Halldorsson JG, Saemundsen E, Stefansdottir B, Jonsson L, Eiriksdottir VK, Eiriksdottir GR, Johannesdottir GH, Unnsteinsdottir U, Jonsdottir B, Magnusdottir BB, Sulem P, Thorsteinsdottir U, Sigurdsson E, Brandeis D, Meyer-Lindenberg A, Stefansson H, and Stefansson K (2017). 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Transl Psychiatry 7, e1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, and Pennacchio LA (2009). Genomic views of distant-acting enhancers. Nature 461, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Goddard ME, Derks EM, and Wray NR (2012). Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Mol Psychiatry 17, 474–485. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, and Geschwind DH (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, Xu M, Gandal MJ, Lou S, Zhang J, Park JJ, Yan C, Rhie SK, Manakongtreecheep K, Zhou H, Nathan A, Peters M, Mattei E, Fitzgerald D, Brunetti T, Moore J, Jiang Y, Girdhar K, Hoffman GE, Kalayci S, Gumus ZH, Crawford GE, Psych EC, Roussos P, Akbarian S, Jaffe AE, White KP, Weng Z, Sestan N, Geschwind DH, Knowles JA, and Gerstein MB (2018a). Comprehensive functional genomic resource and integrative model for the human brain. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Mandell JD, Kumar Y, Sun N, Morris MT, Arbelaez J, Nasello C, Dong S, Duhn C, Zhao X, Yang Z, Padmanabhuni SS, Yu D, King RA, Dietrich A, Khalifa N, Dahl N, Huang AY, Neale BM, Coppola G, Mathews CA, Scharf JM, Tourette International Collaborative Genetics Study, Tourette Syndrome Genetics, S., Eastern Europe, I., Tourette Association of America International Consortium for Genetics, Fernandez TV, Buxbaum JD, De Rubeis S, Grice DE, Xing J, Heiman GA, Tischfield JA, Paschou P, Willsey AJ, and State MW (2018b). De Novo Sequence and Copy Number Variants Are Strongly Associated with Tourette Disorder and Implicate Cell Polarity in Pathogenesis. Cell Rep 24, 3441–3454 e3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein JI, Okbay A, Bybjerg-Grauholm J, Werge T, Hougaard DM, Taylor J, iPsych-Broad Autism Group, Psychiatric Genomics Consortium Autism Group, Skuse D, Devlin B, Anney R, Sanders SJ, Bishop S, Mortensen PB, Borglum AD, Smith GD, Daly MJ, and Robinson EB (2017). Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 49, 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, and Geschwind DH (2013). Understanding sex bias in autism spectrum disorder. Proc Natl Acad Sci U S A 110, 4868–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Parikshak NN, and Geschwind DH (2016). Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat Commun 7, 10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, and Zhang C (2014). HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S, and Pollard K (2018). Most regulatory interactions are not in linkage disequilibrium. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S, Truty RM, and Pollard KS (2016). Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet 48, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Fernandez TV, Yu D, King RA, Dietrich A, Xing J, Sanders SJ, Mandell JD, Huang AY, Richer P, Smith L, Dong S, Samocha KE, Tourette International Collaborative, G., Tourette Syndrome Association International Consortium for, G., Neale BM, Coppola G, Mathews CA, Tischfield JA, Scharf JM, State MW, and Heiman GA (2017). De Novo Coding Variants Are Strongly Associated with Tourette Disorder. Neuron 94, 486–499 e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, Murtha MT, Bichsel C, Niu W, Cotney J, Ercan-Sencicek AG, Gockley J, Gupta AR, Han W, He X, Hoffman EJ, Klei L, Lei J, Liu W, Liu L, Lu C, Xu X, Zhu Y, Mane SM, Lein ES, Wei L, Noonan JP, Roeder K, Devlin B, Sestan N, and State MW (2013). Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, Lee C, Eskin E, Voineagu I, Ernst J, and Geschwind DH (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (1993). The ICD-10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research (Geneva: ). [Google Scholar]
- Yang J, Lee SH, Goddard ME, and Visscher PM (2011). GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, de Vlaming R, Wu Y, Robinson MR, Lloyd-Jones LR, Yengo L, Yap CX, Xue A, Sidorenko J, McRae AF, Powell JE, Montgomery GW, Metspalu A, Esko T, Gibson G, Wray NR, Visscher PM, and Yang J (2018). Signatures of negative selection in the genetic architecture of human complex traits. Nat Genet 50, 746–753. [DOI] [PubMed] [Google Scholar]
- Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Early G, Lifecourse Epidemiology Eczema C., Pourcain BS, Warrington NM, Finucane HK, Price AL, Bulik-Sullivan BK, Anttila V, Paternoster L, Gaunt TR, Evans DM, and Neale BM (2017). LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, and Lander ES (2014). Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A 111, E455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.