Abstract
MicroRNAs of the miR-16 and miR-34 families have been reported to inhibit cell cycle progression, and their loss has been linked to oncogenic transformation. Utilizing a high-throughput, genome-wide screen for miRNAs and mRNAs that are differentially regulated in osteosarcoma (OS) cell lines, we report that miR-449a and miR-424, belonging to the miR-34 and miR-16 families, respectively, target the major S/G2 phase cyclin, cyclin A2 (CCNA2), in a bipartite manner. We found that the 3′-UTR of CCNA2 is recognized by miR-449a, whereas the CCNA2 coding region is targeted by miR-424. Of note, we observed loss of both miR-449a and miR-424 in OS, resulting in derepression of CCNA2 and appearance of aggressive cancer phenotypes. Ectopic expression of miR-449a and miR-424 significantly decreased cyclin A2 levels and inhibited proliferation rate, migratory potential, and colony-forming ability of OS cells. To further probe the roles of miR-449a and miR-424 in OS, we developed an OS mouse model by intraosseous injection of U2OS cells into the tibia bone of NOD-scid mice, which indicated that miR-449a and miR-424 co-expression suppresses tumor growth. On the basis of this discovery, we analyzed the gene expression of human OS biopsy samples, revealing that miR-449a and miR-424 are both down-regulated, whereas cyclin A2 is significantly up-regulated in these OS samples. In summary, the findings in our study highlight that cyclin A2 repression by miRNAs of the miR-16 and miR-34 families is lost in aggressive OS.
Keywords: microRNA (miRNA), cell cycle, cyclin, tumor suppressor gene, osteosarcoma (OS), bone cancer, CCNA2, cyclin A2, non-coding RNA, oncogenesis, posttranscriptional regulation
Introduction
Osteosarcoma (OS)2 is the most common malignant bone tumor in children and adolescents (1). Despite advancements in aggressive OS treatment, the prognosis has not significantly improved, and thus there is a need for alternate molecular therapies (2). Extensive molecular studies have deciphered that genomic aberrations, such as amplification of genes coding for MYC or CDK4 or mutations in genes coding for retinoblastoma or p53 proteins, are commonly associated with the oncogenesis of osteosarcoma (3). Apart from the alterations in protein-coding genes, various studies have also demonstrated the dysregulation of miRNAs as a characteristic feature of OS, highlighting the potential of miRNAs as possible diagnostic, prognostic, and therapeutic candidates for OS treatment (4, 5). However, only a few of these dysregulated miRNAs, such as miR-199a and miR-874, have been thoroughly investigated for their specific role in osteosarcomas (6–8).
The miRNAs of the miR-16 and miR-34a families are among the well-established tumor-suppressive miRNAs, which induce cell cycle arrest and apoptosis by targeting growth-regulating genes such as MYC, CCNE1, CCND1, CDC25A, CDK6, and BCL2 (9, 10). The members of the miR-16 family include miR-15a, miR-15b, miR-16, miR-497, miR-195, miR-424, and miR-503, whereas miR-34a, miR-34b, miR-34c, miR-449a, miR-449b, and miR-449c comprise the miR-34 family. Although all of the miRNAs of both the miR-16 and miR-34 families are characterized for their role in tumor suppression, not much has been reported regarding combinatorial effects of these miRNA families (7, 11, 12, 14–19). A comprehensive analysis of previous studies alludes to a functional synergy between these miRNAs. For instance, it has been reported that miR-15b and miR-16, which target the anti-apoptotic gene BCL2, are lost in gastric cancers, but the increase in BCL2 was more than what could be explained by the loss of miR-15/16, suggesting an additional mechanism of BCL2 regulation in cancers (20). Subsequent studies demonstrating the down-regulation of BCL2 by miRNAs of the miR-34 family, including miR-34a and miR-449a, highlighted that the individual studies may not have completely discerned the contribution of multiple growth-inhibitory miRNAs in cell cycle regulation (7, 21). A similar inference can be drawn from the individual studies reporting the regulation of another cell cycle regulator, cyclin D1 by either miR-16 or miR-34 family miRNAs (10, 22). Overall, these results emphasize a lacuna in our understanding of simultaneous regulation of key cell cycle genes by miR-16 and miR-34 family miRNAs, and hence there is a need to address the combinatorial regulation of critical cell cycle genes by these tumor-suppressor miRNA families.
In the present study, we set out to explore the key regulatory miRNAs and their target gene networks associated with the progression of osteosarcoma. We have focused on identifying the miRNAs that regulate the growth and proliferation of OS cells by targeting cell cycle–associated genes. In this study, we performed a high-throughput screening of genome-wide miRNAs as well as mRNAs, which are differentially regulated in two OS cell lines, U2OS and HAL, classified as aggressive and nonaggressive, respectively, based on their proliferative, invasive, and colony-forming capacities (23, 24). In-depth analysis revealed a weak expression of miR-449a and miR-424 family miRNAs in aggressive osteosarcoma, in contrast to the expression of their putative target genes. Our results demonstrate that there is a simultaneous inhibition of miR-16 and miR-34 family miRNAs leading to derepression of their common target, CCNA2, in aggressive osteosarcoma, and restoration of these miRNAs results in suppression of CCNA2 and cancer-associated phenotypes.
Results
Integrated analysis of miRNA and mRNA expression profiles and their interactions in osteosarcoma
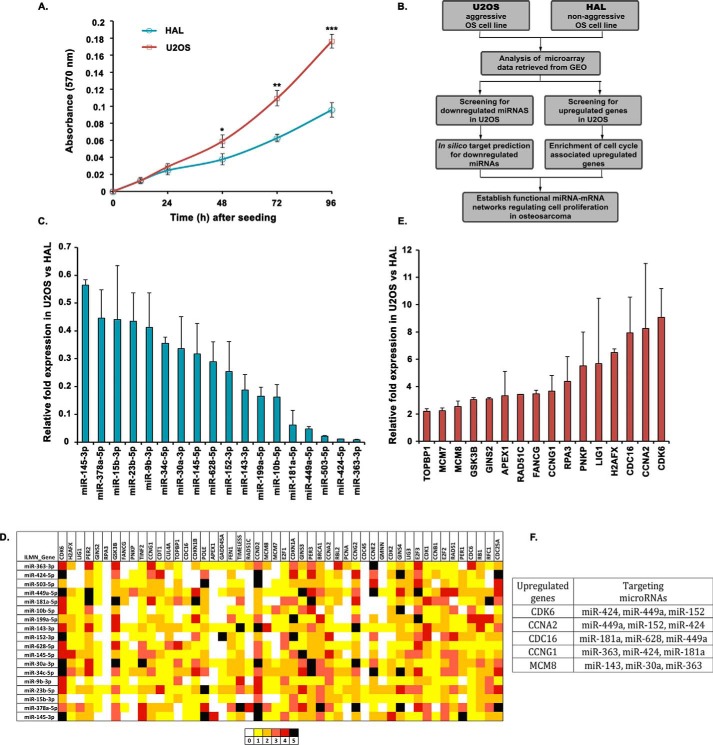
To identify the miRNAs regulating cell cycle and proliferation during oncogenesis, we compared their expression in two osteosarcoma cell lines, U2OS and HAL, which differ in their aggressiveness. We observed that the proliferation rate of U2OS cells was significantly higher than that of HAL cells (Fig. 1A). By comparing the miRNA expressions of U2OS and HAL retrieved from GEO (GSE28425), we attempted to identify the novel miRNAs regulating cell proliferation (Fig. 1B). From the microarray data analysis of 820 miRNAs, we found that 152 miRNAs were down-regulated, whereas 142 miRNAs were up-regulated, by more than 2-fold, in U2OS cells. From the list of 152 miRNAs, we selected 40 most down-regulated miRNAs for validation by individual real-time quantitative RT-PCR and observed that 18 miRNAs were significantly down-regulated in U2OS cells (Fig. 1C). Our strategy as described in Fig. 1B assumes that miRNAs which are down-regulated in aggressive osteosarcoma are more likely to regulate the expression of growth-supporting genes. Few of these miRNAs, including miR-181, miR-199, and miR-10b, have been previously documented for their tumor-suppressive role. Next, we obtained the mRNA microarray data of U2OS and HAL cell lines from GEO and listed top 50 cell cycle–associated genes that were up-regulated. To predict whether the up-regulated cell cycle–associated genes were the targets of the down-regulated miRNAs in U2OS cells, we used five different miRNA target prediction algorithms, namely TargetScan, miRanda, miRWalk, RNAhybrid, and RNA22. Thus, a pairwise matching of down-regulated miRNAs with up-regulated genes on the basis of predicted target binding sites identified several proto-oncogenes as putative targets of miRNAs down-regulated in U2OS (Fig. 1D). Individual qRT-PCR validation confirmed significant up-regulation of 16 cell cycle–associated genes, including CDK6 and CCNA2, in U2OS that were observed as common targets for several down-regulated miRNAs (Fig. 1, E and F).
Figure 1.
A screen to identify novel miRNA–mRNA interactions in osteosarcomas. A, MTT proliferation assay to compare the growth rate of U2OS and HAL cell lines. The absorbance at 570 nm reflects the proportion of viable cells for each cell line at the indicated time interval. The data represent the mean of three technical replicates ± S.D. (error bars). p values calculated using a two-tailed t test show that U2OS samples are significantly different from HAL samples (*, p < 0.05; **, p < 0.01; ***, p < 0.001). B, schematic outline describing the strategy for identifying novel miRNAs regulating cell proliferation by comparing the expression of miRNAs as well as cell cycle–associated genes in two osteosarcoma cell lines, U2OS and HAL, differing in their growth aggressiveness. C, relative expression of 18 selected miRNAs in U2OS cells as compared with HAL cells, evaluated by individual quantitative real-time PCR. Small nucleolar RNA, RNU66, was used as the endogenous control for normalization of miRNA expression in the two cell lines. D, in silico analysis for identification of novel miRNA–mRNA pairs regulating cell proliferation in aggressive osteosarcoma. The heat map represents a matching of the up-regulated cell cycle genes with down-regulated miRNAs in U2OS using five different bioinformatics tools for miRNA target predictions, including TargetScan, miRanda, miRWalk, RNA22, and RNAhybrid. The colors represent the number of algorithms that predict a specific gene (arranged horizontally) as a target of the respective miRNA (arranged vertically). E, relative expression of cell cycle–associated genes in U2OS cells as compared with HAL cells, evaluated by individual quantitative real-time PCR. Housekeeping gene, β2-microglobulin, was used as the endogenous control for normalization of gene expression in the two cell lines. F, list of key cell cycle regulatory genes that are overexpressed in an aggressive osteosarcoma cell line, U2OS, and the miRNAs that are predicted to target them. For expression levels of miRNA/mRNA, the data are represented as the mean of two experiments, each with two technical replicates, ± S.D.
miR-449a, a miR-34 family miRNA, is down-regulated in aggressive osteosarcoma
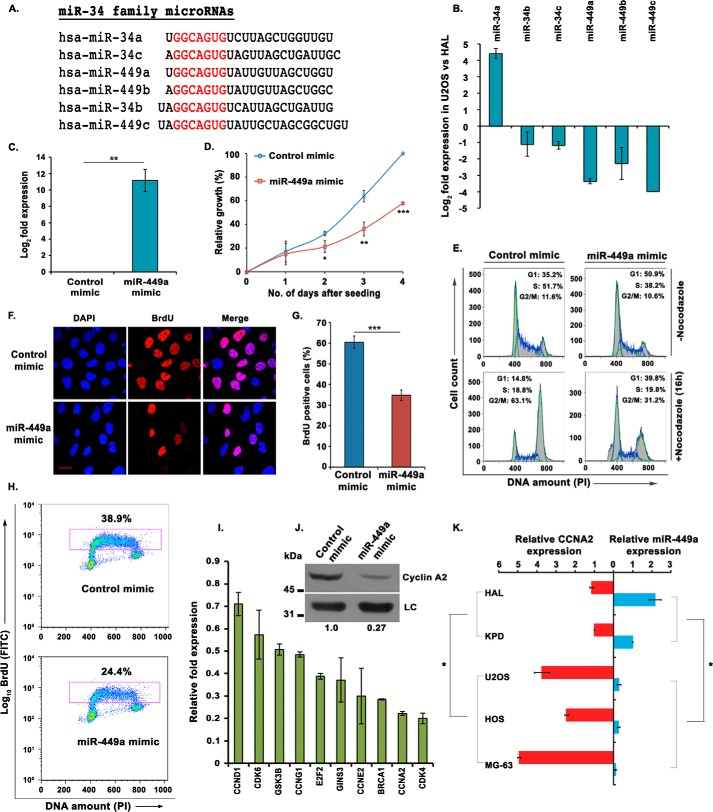
Microarray analysis of miRNA expression in differentially proliferating human OS cell lines, U2OS and HAL, revealed that miR-34c and miR-449a, and not other members of miR-34 family, figured in the list of significantly down-regulated miRNAs in U2OS, as reported previously (7, 8, 14). The miR-34 family has six members, including miR-34a, miR-34b, miR-34c, miR-449a, miR-449b, and miR-449c, each of them sharing the same seed sequence GGCAGUG, and because microarray data sets may suffer from errors, we ascertained the levels of other members of miR-34 family (Fig. 2A). We observed that miR-449a, miR-449b, and miR-449c, transcribed in a cluster from chromosomal position 5q11.2, were significantly down-regulated in aggressive osteosarcoma, whereas polycistronic miRNAs miR-34b and miR-34c, transcribed in another cluster from chromosome 11q23.1, were moderately down-regulated (Fig. 2B). miR-449a is expressed across different tissue samples, and thus we selected it to understand its role in the regulation of cell proliferation in osteosarcoma.
Figure 2.
miR-449a, a miR-34 family miRNA, is down-regulated in aggressive osteosarcoma. A, alignment of the mature sequences of miR-34 family miRNAs; the seed sequences of all miRNAs are highlighted in red. miR-34b and miR-449c are shown along with the miR-34 family, as their seed sequences are very similar. B, relative expression of all mature miRNAs of miR-34 family in U2OS, compared with HAL. The expression was determined by qRT-PCR and normalized with the expression of small nucleolar RNA, RNU48. C, relative expression of miR-449a in U2OS cells transfected with control or miR-449a mimic for 3 consecutive days and harvested after 24 h of the third transfection. D, MTT assay displaying the growth rates of U2OS cells transfected three times with either control or miR-449a mimic as described in C and seeded at equal numbers after transfection. The graph represents the growth rate with respect to control mimic–transfected cells at the indicated days after seeding the cells. The data are represented as mean of three experiments ± S.D. (error bars). E, U2OS cells transfected for 3 consecutive days with either control or miR-449a mimic followed by either treatment with nocodazole for 16 h or no treatment were stained with propidium iodide (PI) for analysis of cell cycle distribution by flow cytometry. The inset shows the distribution of cells in different phases of the cell cycle. F, U2OS cells transfected with control or miR-449a mimic were pulsed with BrdU for 30 min followed by staining with anti-BrdU antibody (red), whereas DNA was stained with DAPI (blue). Scale bar, 10 μm. G, quantification of F, which shows that miR-449a overexpression significantly decreases S phase population. The data are represented as the mean of three experiments ± S.D. H, flow cytometry of control or miR-449a mimic–transfected U2OS cells, pulsed with BrdU followed by staining with FITC-conjugated anti-BrdU antibody along with propidium iodide. The dot plot displays BrdU incorporation (y axis) and DNA content (x axis), and the inset shows the cells incorporating BrdU. I, relative expression of putative target genes after three consecutive transfections of U2OS cells with miR-449a mimic. The -fold expression is represented compared with control mimic–transfected cells and normalized with the expression of the β2-microglobulin gene. J, immunoblotting of cyclin A2 in U2OS cells transfected with either control mimic or miR-449a mimic as described in C. LC, loading control, showing equal protein load in different lanes. K, relative expression of miR-449a and CCNA2 mRNA in different OS cell lines with respect to the KPD cell line. The expression of miR-449a in both of the nonaggressive OS cell lines, HAL and KPD, is significantly different from the aggressive cell lines including U2OS, HOS, and MG63. Similarly, the levels of CCNA2 were significantly different between these two groups of OS cell lines. For expression levels of miRNA/mRNA, the data are represented as the mean of two experiments, each with two technical replicates, ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To study the effect of miR-449a in osteosarcoma, we transfected U2OS cells with synthetic mimic of mature miR-449a and performed a proliferation assay, where we observed that miR-449a significantly reduced the rate of cell proliferation (Fig. 2, C and D). To delineate the mechanism behind regulation of cell proliferation by miR-449a, we determined the cell cycle distribution of miR-449a–overexpressing U2OS cells and found that miR-449a leads to an accumulation of cells in the G1 phase (Fig. 2E). To clearly demonstrate a G1 accumulation, miR-449a mimic–transfected cells were treated with nocodazole to block the cells in G2/M phase, before evaluating the cell cycle distribution by flow cytometry. Nocodazole treatment markedly reduced the G1 phase population of control mimic–transfected cells by blocking the majority of cell population in the G2/M phase; however, the percentage of the G1 phase population was significantly higher in miR-449a mimic–transfected cells, thus demonstrating a G1 arrest caused by the overexpression of miR-449a. We next evaluated the rate of DNA synthesis by measuring the incorporation of nucleotide analog, 5-bromo-2-deoxyuridine (BrdU), in an immunofluorescence assay, where we observed that it was reduced to 34% in cells transfected with miR-449a mimic as compared with 60% in control cells (Fig. 2, F and G). Similarly, in the flow cytometry assay, BrdU-incorporating cells decreased from 38 to 24%, further establishing that miR-449a impedes S phase progression (Fig. 2H).
Having identified that miR-449a is significantly down-regulated in aggressive osteosarcoma and that the overexpression of miR-449a in U2OS cells leads to a G1 arrest, S phase reduction, and slower proliferation rate, we focused on determining its target gene. Although CLIP-Seq or RNA-Seq would identify new targets of miR-449a that would elucidate its novel physiological functions, in this study, we focused on key cell cycle regulators. Utilizing a combination of five prediction algorithms, we identified several key cell cycle genes as putative targets of miR-449a (Fig. 1D). Of the total 10 growth-regulating genes that were predicted as targets of miR-449a by two or more algorithms, CCNE2, CCNA2, and CDK4 were most significantly down-regulated upon miR-449a overexpression in U2OS cells, whereas other putative growth-promoting target genes, including CCND1, GSK3B, and CDK6, did not show significant alteration (Fig. 2I). Additionally, we observed a significant decrease in cyclin A2 protein levels after miR-449a overexpression in U2OS cells (Fig. 2J).
We now ascertained whether there is a pattern of expression of miR-449a in other aggressive OS cell lines. We tested the levels of miR-449a in OS cell lines displaying varying aggressiveness of cancer phenotypes; cell lines MG63, HOS, and U2OS are reported to be highly proliferating, whereas HAL and KPD are poorly proliferating OS cell lines (23). We observed that miR-449a was significantly down-regulated in the group of aggressively proliferating OS cells lines (i.e. U2OS, MG63, and HOS) compared with less proliferating OS cell lines, such as HAL and KPD (Fig. 2K). In contrast to miR-449a, CCNA2 displays an inverse pattern; it is up-regulated in aggressively proliferating OS cell lines (U2OS, MG63, and HOS), compared with HAL and KPD. Thus, we have observed down-regulation of miR-449a and up-regulation of CCNA2 as a common phenomenon in aggressively proliferating osteosarcomas. We focused on CCNA2 regulation by miR-449a, as the role of cyclin A2 has been well-established in mediating progression of cells through both G1/S and S/G2 phases of the cell cycle.
miR-449a directly targets the 3′-UTR of CCNA2, impeding DNA synthesis
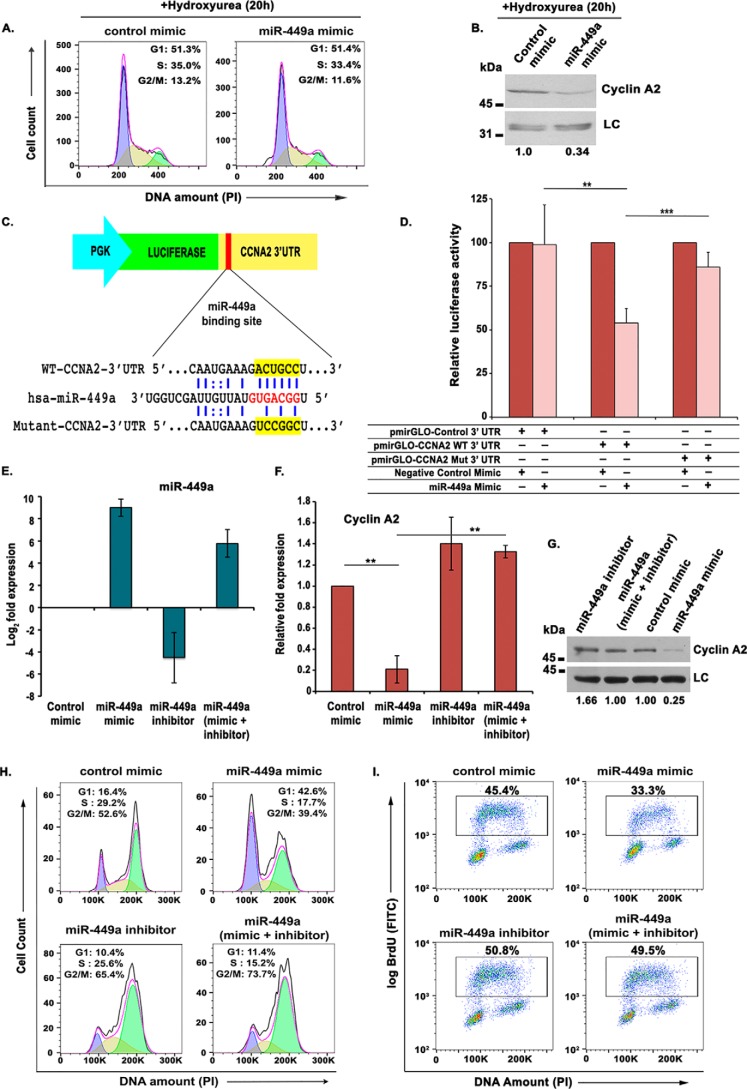
In view of the fact that miR-449a is predicted to target several other cell cycle genes, including CCND1 and CDK6, we wanted to ascertain that CCNA2 down-regulation is a not an indirect effect caused by accumulation of cells in G1 phase due to targeting of other G1/S-regulating genes by miR-449a. To test whether the changes in cell cycle distribution were the reason for cyclin A2 decrease, we transfected asynchronous U2OS cells twice with either miR-449a or control mimic, and after 4 h, hydroxyurea was added to synchronize the transfected cells at the G1/S transition. We observed that after hydroxyurea treatment, miR-449a mimic– and control mimic–transfected cells displayed identical cell cycle distribution, but the levels of cyclin A2 were significantly reduced in miR-449a–transfected cells, ruling out that cell cycle changes are the reason for cyclin A2 decrease (Fig. 3, A and B).
Figure 3.
miR-449a targets the 3′-UTR of CCNA2, impeding DNA synthesis. A, U2OS cells transfected twice with either control or miR-449a mimic followed by incubation with 2 mm hydroxyurea for 20 h and PI staining for analysis of cell cycle distribution by flow cytometry. The different colors mark the G1, S, and G2/M phases, whereas the inset shows the distribution of cells in different phases of the cell cycle. B, immunoblotting of cyclin A2 in U2OS cells transfected with either control mimic or miR-449a mimic as described in A. C, an illustration of miR-449a binding with WT and mutant CCNA2 3′-UTR, cloned downstream of the firefly luciferase gene under the control of the PGK promoter in pmirGLO Dual-Luciferase vector. D, CCNA2 3′-UTR possessing the WT or mutant (Mut) binding site for miR-449a cloned in pmirGLO Dual-Luciferase vector was cotransfected in U2OS cells with control or miR-449a mimic, as indicated, and the relative luciferase activity was determined. The data are represented as the mean of six experiments ± S.D. (error bars). E–G, U2OS cells were transfected on 3 consecutive days with control mimic, miR-449a mimic, or miR-449a inhibitor, as indicated, and miR-449a, CCNA2 mRNA, and protein levels were analyzed. LC, loading control, a nonspecific band that displays equal protein load in different lanes. The numbers indicate levels of cyclin A2 relative to control mimic–transfected cells. H, flow cytometry of propidium iodide-stained transfected cells demonstrates that restoration of CCNA2 by miR-449a inhibitor releases the G1 arrest caused by miR-449a mimic. U2OS cells transfected on three consecutive days, as described in E, were treated with nocodazole for 16 h before staining with propidium iodide followed by a flow cytometry assay for determining the cell cycle phase distribution of transfected cells. I, U2OS cells transfected on 3 consecutive days were pulsed with BrdU for 30 min after 24 h of the third transfection and stained with FITC-conjugated anti-BrdU antibody along with propidium iodide, followed by a flow cytometry assay for BrdU incorporation. The dot plot displays BrdU incorporation (y axis) and DNA content (x axis), and the inset shows the percentage of cells incorporating BrdU. For expression levels of miRNA/mRNA, the data are represented as the mean of two experiments, each with two technical replicates, ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To further evaluate whether the CCNA2 gene is a direct target of miR-449a, we cloned 3′-UTR of CCNA2 carrying either the WT or mutated binding site of miR-449a in pmirGLO Dual-Luciferase reporter vector and assayed the effect of miR-449a overexpression on luciferase activity (Fig. 3C). When the WT CCNA2 3′-UTR was introduced into U2OS cells along with miR-449a mimic, it exhibited a significant reduction in the luciferase activity, but the mutation in the binding site at the 3′-UTR suppressed the inhibition of luciferase activity caused by miR-449a overexpression (Fig. 3D). Thus, miR-449a inhibits CCNA2 expression through direct interaction with its 3′-UTR.
After determining the effect of miR-449a overexpression on cyclin A2 levels, we next assessed the effect of loss-of-function of miR-449a. We transfected U2OS cells with miR-449a mimic or single-stranded antisense inhibitor of miR-449a, alone or in combination, and evaluated the effect on CCNA2 expression. Transfection of miR-449a inhibitor alone led to a significant decrease of miR-449a levels and also suppressed the miR-449a increase observed after miR-449a mimic transfection (Fig. 3E). As expected, we observed a marked decrease in CCNA2 mRNA as well as protein levels after miR-449a overexpression, whereas inhibition of miR-449a only marginally increased CCNA2 levels (Fig. 3, F and G). However, transfection of miR-449a inhibitor reversed the CCNA2 down-regulation caused by miR-449a mimic expression, confirming that miR-449a regulates endogenous levels of CCNA2. We have previously shown that miR-449a overexpression leads to an accumulation in the G1 phase. However, when miR-449a inhibitor was transfected along with miR-449a mimic, the G1 block was suppressed, confirming that miR-449a overexpression was the cause of the observed G1 arrest (Fig. 3H). Similarly, the decrease in rate of DNA synthesis due to overexpression of miR-449a was reversed by miR-449a inhibitor (Fig. 3I).
miR-449a restrains tumorigenic phenotypes in osteosarcoma
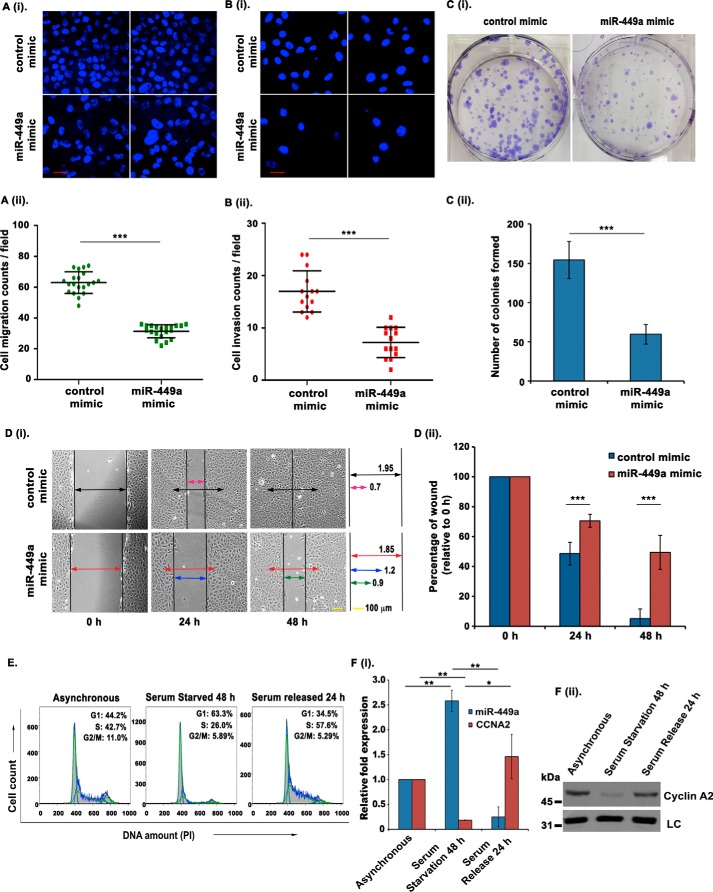
To verify the functional significance of miR-449a expression, we investigated the effect of miR-449a overexpression on various tumor-associated phenotypes of U2OS cells. We utilized in vitro transwell migration and invasion assays to assess the effects of miR-449a overexpression on cell migration and invasion ability. We observed that miR-449a overexpression inhibited the cell migration ability of U2OS cells (Fig. 4A, i and ii). Furthermore, the invasion of U2OS cells through a Matrigel-coated synthetic membrane was significantly suppressed upon miR-449a overexpression (Fig. 4B, i and ii). Next, we determined the effect of miR-449a on the clonogenic ability of U2OS cells after transfecting with miR-449a or control mimic followed by crystal violet staining and colony counting after 12 days. The ectopic expression of miR-449a led to a marked reduction in the colony-forming ability of U2OS cells (Fig. 4C, i and ii). Finally, we also performed a scratch-wound assay to demonstrate the role of miR-449a in modulating the healing ability of U2OS cells. The assay was carried out by scratching a wound in the monolayer of confluent miR-449a mimic– or control mimic–transfected U2OS cells and then monitoring the size of the wound for 48 h. It was found that control cells displayed an absolute healing of the wound within 48 h, whereas in miR-449a mimic–transfected cells, around two-thirds of the damaged region could be healed in the same duration (Fig. 4D, i and ii). Overall, we have established that the ectopic expression of CCNA2–regulating miRNA, miR-449a, intrudes on the oncogenic features of U2OS cells.
Figure 4.
miR-449a overexpression inhibits tumorigenic phenotypes. A (i and ii), U2OS cells were transfected on 3 consecutive days with either control mimic or miR-449a mimic, and the cells migrating through a microporous membrane were counted 24 h after the third transfection. i, representative fields showing DAPI-stained nuclei of migrated cells. ii, quantification of the migratory cells observed in i. Each point refers to the number of cells in an individual field, whereas long and short horizontal bars represent the mean and S.D. (error bars) of all fields, respectively. Scale bar, 20 μm. B (i and ii), U2OS cells were transfected as described in A, and the cells invading through a Matrigel-coated membrane were counted at 24 h after the third transfection. i, representative fields of each sample showing the DAPI-stained nuclei of invading cells. ii, quantification of the invading cells as described in A. C (i and ii), clonogenic assay to evaluate the effect of miR-449a expression. i, U2OS cells were transfected on 3 consecutive days with either control or miR-449a mimic, allowed to grow for 12 days, and stained with crystal violet, after which the colonies were counted. ii, quantification of i, which demonstrates that the colony-forming ability is significantly reduced upon miR-449a overexpression. The data are represented as the mean of six experiments ± S.D. D (i and ii), wound-healing assay to evaluate the effect of miR-449a overexpression on cell migration: U2OS cells were transfected on two consecutive days with either control or miR-449a mimic and grown to confluence, after which a wound was created using a micropipette tip. The extent of wound healing was monitored every 24 h. Arrows of different colors represent the relative width of the wound at different time points in different samples. ii, quantification of i (units), which demonstrates that the wound-healing capability is profoundly reduced upon miR-449a overexpression. The data are represented as the mean of two experiments ± S.D. E and F, U2OS cells were serum-starved for 48 h, followed by resupplementation with serum for 24 h. E, flow cytometry of PI-stained DNA representing the cell cycle phase distribution after serum starvation and resupplementation in U2OS cells. F (i), relative expression of miR-449a and CCNA2 mRNA in asynchronous, serum-starved, and serum-resupplemented U2OS cells determined using qRT-PCR and normalized with the expression of RNU48 and BMG, respectively. F (ii), immunoblotting of the same samples as described in E shows differential expression of CCNA2 after serum starvation and serum resupplementation. For expression levels of miRNA/mRNA, the data are represented as the mean of two experiments, each with two technical replicates, ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Next, we wanted to evaluate the physiological role of miR-449a in the tumor microenvironment; growth factor deficiency, a characteristic of the tumor microenvironment, has been used to understand the adaptation of cancer cells and subsequent tumor progression (25, 26). Thus, to determine the role of miR-449a in mediating quiescence in the tumor microenvironment, we analyzed the levels of miR-449a and CCNA2 in serum-starved and serum-resupplemented U2OS cells. Serum starvation was induced in U2OS cells by growing the cells in serum-free medium for 48 h and then resupplementing with fresh medium containing 10% fetal bovine serum for 24 h following starvation. Flow cytometry for propidium iodide–stained DNA established that serum starvation for 48 h led to an increase in the G0/G1 population, indicating cell cycle exit, whereas serum restimulation for 24 h resulted in cell cycle reentry, seen as a clear accumulation in the S phase (Fig. 4E). Interestingly, we observed that serum starvation in U2OS induces miR-449a expression, and serum restimulation restores the normal levels of the miRNA, whereas the expression of CCNA2 was inversely regulated during the same conditions (Fig. 4F, i and ii). Thus, the growth factor deficiency observed in the tumor microenvironment would result in miR-449a mediated repression of CCNA2, leading to cell cycle arrest.
Restoration of CCNA2 rescues miR-449a–mediated inhibition of DNA replication but not the G1 block
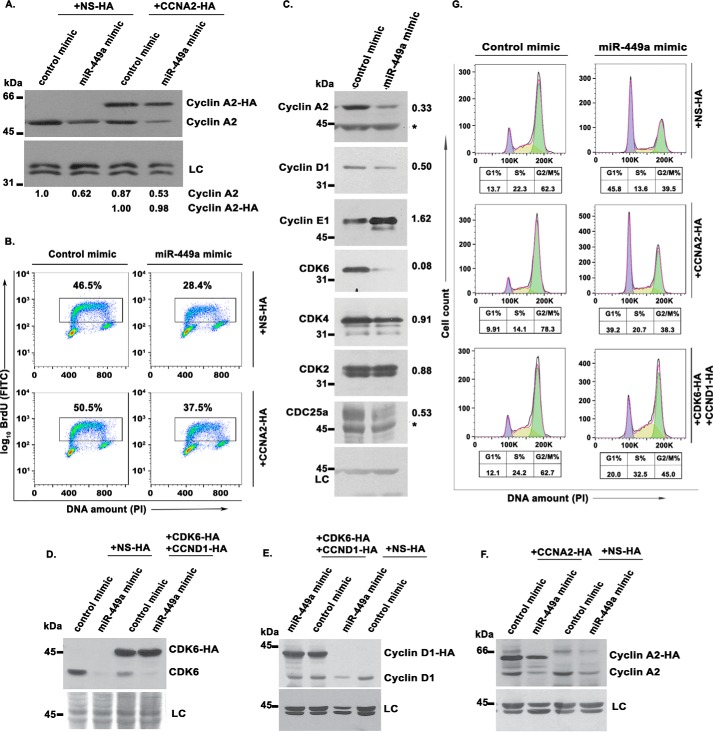
We have shown that miR-449a inhibits S phase progression and cell proliferation by targeting CCNA2. To further establish that the delay in S phase progression was primarily due to miR-449a–mediated down-regulation of CCNA2, we cloned the coding region of the CCNA2 gene, which lacks the site for miR-449a binding, into a retroviral vector. U2OS cells stably expressing CCNA2 were then transfected with miR-449a, followed by incubation with BrdU, to evaluate the rate of DNA synthesis. We found that the endogenous cyclin A2 protein levels were down-regulated in both samples transfected with miR-449a mimic, whereas the exogenously expressed CCNA2 remained unaffected upon miR-449a overexpression (Fig. 5A). Ectopic expression of miR-449a significantly reduced the population of BrdU-positive cells, but co-expression of miR-449a–resistant CCNA2 partially restored the DNA synthesis, demonstrating that the effect of miR-449a on the S phase progression was primarily due to down-regulation of CCNA2 (Fig. 5B).
Figure 5.
Restoration of CCNA2 rescues miR-449a–mediated inhibition of DNA replication. A, U2OS cells stably expressing exogenous HA-tagged CCNA2 without the 3′-UTR or control protein (NS) were transfected on 3 consecutive days with either control or miR-449a mimic. Immunoblotting with anti-cyclin A2 demonstrates a decrease in the endogenous cyclin A2 levels by miR-449a overexpression, whereas the levels of HA-tagged cyclin A2 are unaltered. LC, loading control, a nonspecific band that displays equal protein load in different lanes. The numbers indicate levels of endogenous and HA-tagged cyclin A2 relative to control mimic–transfected cells. B, flow cytometry of BrdU-labeled transfected cells, as described in A. Cells were pulsed with BrdU, followed by staining with anti-BrdU antibody conjugated to FITC along with PI. The dot plot displays BrdU incorporation (y axis) and DNA content (x axis), and the inset shows the percentage of cells incorporating BrdU. The data demonstrate that the effect of miR-449a on S phase progression was primarily due to inhibition of cyclin A2. C, immunoblotting of different cell cycle regulatory proteins in U2OS cells transfected with either control mimic or miR-449a mimic. The numbers indicate levels of specific proteins after miR-449a mimic transfection relative to control mimic–transfected cells. Nonspecific bands have been marked by asterisks. D–F, U2OS cells stably expressing exogenous HA-tagged cyclin A2, both HA-tagged cyclin D1 and HA-tagged CDK6 or control protein (NS), as indicated in different panels, were transfected on 3 consecutive days with either control or miR-449a mimic. Immunoblotting with anti-CDK6 (D), anti-cyclin D1 (E), and anti-cyclin A2 (F) demonstrates a decrease in the endogenous protein levels by miR-449a overexpression, whereas the HA-tagged exogenous proteins remained stable. G, after the third transfection, as described in D–F, the cells were treated with nocodazole for 16 h and stained with PI for analysis of cell cycle distribution by flow cytometry. The different colors mark the G1, S, and G2/M phases, whereas the numbers show the distribution of cells in different phases of the cell cycle.
Apart from CCNA2, miR-449a is predicted to target other key cell cycle regulators, and we wanted to identify the protein whose inhibition is key for the observed cell cycle retardation by miR-449a. First, we assayed the levels of major cyclins and Cdks mediating the G1-S progression (Fig. 5C). Overexpression of miR-449a led to the down-regulation of cyclin D1 and CDK6 in U2OS cells, which can be the most likely cause of G1 phase arrest. On the other hand, CDK4 and CDK2, which associate with cyclin D and cyclin E, respectively, for mediating Rb phosphorylation were not decreased. A marginal increase in the levels of cyclin E1 was observed, probably because of accumulation of cyclin E–positive cells in late G1–early S phases. We observed that Cdc25A phosphatase was down-regulated, and because its activity is critical for CDK2 activation, we believe that miR-449a targets cyclin A directly and CDK2 indirectly to impede S phase progression. Further, to address the relative contribution of the inhibited genes in causing a G1 block, we constructed retroviral vectors that express the coding region of CCND1 or CDK6, which are resistant to miR-449a–mediated degradation. Control or miR-449a mimic were transfected for 3 consecutive days in U2OS cells stably expressing either CCNA2 or both CCND1 and CDK6. Endogenous cyclin A2 and CDK6 were significantly, whereas cyclin D1 was mildly, down-regulated after miR-449a overexpression, but the exogenously expressed HA-tagged proteins remained stable (Fig. 5, D–F). The transfected cells were treated with nocodazole for 16 h that causes a G2/M phase block and thus makes any G1 accumulation due to miR-449a overexpression easily discernible (Fig. 5G). As previously observed, miR-449a mimic transfection led to an increase in G1 phase, but complementation with CCNA2 did not relieve the G1 block, indicating that the miR-449a–induced G1 block was not primarily due to cyclin A2 inhibition. On the other hand, the accumulation of cells in G1 phase from 13 to 45% caused by miR-449a overexpression was significantly reversed to 20% by complementation with CCND1 and CDK6. Thus, CCND1 and CDK6 are the key targets of miR-449a for causing G1 arrest, whereas miR-449a–mediated down-regulation of CCNA2 is pivotal for causing S phase arrest. In conclusion, inhibition of CCNA2 as well as CCND1 and CDK6, leading to arrest in different phases of the cell cycle, contributes to the retardation of cell cycle caused by miR-449a overexpression.
miR-424, a miR-16 family miRNA, targets CCNA2 and is down-regulated in aggressive osteosarcoma
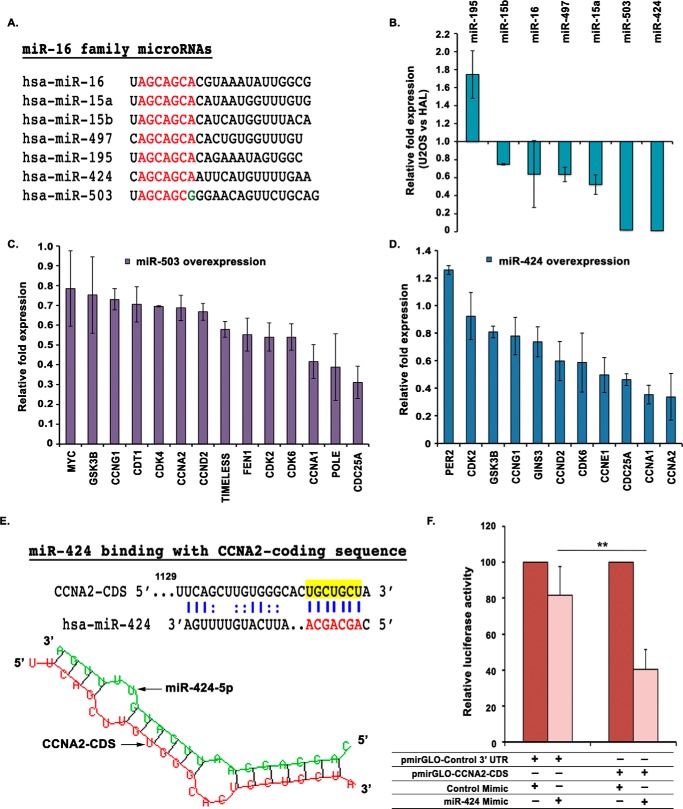
Our initial screen of differentially regulated miRNAs in OS also identified miR-16 family miRNAs, which were significantly down-regulated (Fig. 1C). The members of the miR-16 family include miR-15a, miR-15b, miR-16, miR-497, miR-195, and miR-424, which share the same seed sequence, AGCAGCA, with miR-503, an extended member of miR-16 family (Fig. 6A). Real-time qRT-PCR analysis demonstrated that in aggressive osteosarcoma, miR-503 and miR-424, transcribed in a cluster from chromosomal position Xq26.3, were significantly down-regulated, whereas polycistronic miRNAs miR-15b and miR-16, transcribed in another cluster from chromosome 3q25.33, were moderately down-regulated (Fig. 6B). To understand the role of these miRNAs, we transfected synthetic mimics of either miR-503 or miR-424 in U2OS cells and analyzed their effect on putative cell cycle target genes (Fig. 6, C and D). It was found that overexpression of miR-503 led to a significant down-regulation of several key cell cycle genes, including CDC25A. Similarly, ectopic expression of miR-424 resulted in a marked decrease in the mRNA levels of CDC25A, CCNA2, and CCNE1 among other critical cell cycle regulators. Having observed a decrease in cyclin A2 mRNA levels after miR-424 overexpression, we next wanted to investigate whether CCNA2 was a direct target of miR-424. Although we could not find a binding site for miR-424 in CCNA2 3′-UTR, we identified a putative binding site in the coding region of CCNA2 by using the RNA hybrid miRNA target prediction tool (Fig. 6E). Next, we cloned the complete coding region of CCNA2 into the pmirGLO Dual-Luciferase reporter vector and cotransfected it with miR-424 mimic in U2OS cells followed by a luciferase assay. We observed that overexpression of miR-424 significantly reduced the luciferase activity, confirming that miR-424 targets CCNA2 directly by binding to its coding region (Fig. 6F). Our previous results have demonstrated that miR-449a targets CCNA2 in its 3′-UTR, whereas miR-424 inhibits CCNA2 by binding to its coding region. It is thus a classic example of how the two tumor suppressor miRNA families target the coding region and the 3′-UTR of a target gene to achieve effective inhibition.
Figure 6.
miR-424, a miR-16 family miRNA, targets CCNA2 and is down-regulated in aggressive osteosarcoma. A, alignment of the mature sequences of miR-16 family miRNAs; the seed sequences of all miRNAs are highlighted in red. miR-503 is shown along with miR-16 family, as their seed sequences are very similar. B, relative expression of all mature miRNAs of miR-16 family in rapid-proliferating osteosarcoma cell line, U2OS, compared with slow-proliferating cell line, HAL. The expression was determined by qRT-PCR and normalized with the expression of small nucleolar RNA, RNU48. C and D, relative expression of putative target genes after three consecutive transfections of U2OS cells with either miR-503 or miR-424 mimic. The -fold expression of target genes is represented compared with control mimic–transfected cells and normalized with the expression of the β2-microglobulin gene. For expression levels of miRNA/mRNA, the data are represented as the mean of two experiments, each with two technical replicates, ± S.D. (error bars). E, an illustration of miR-424 binding within the CCNA2-coding sequence, which was cloned downstream of the firefly luciferase gene in pmirGLO Dual-Luciferase vector. miR-424 binding to CCNA2 as per RNAhybrid has been depicted. F, U2OS cells were transfected with pmirGLO Dual-Luciferase vector in which either the coding sequence of CCNA2 or a nonspecific 3′-UTR has been cloned downstream of the firefly luciferase gene. The cells were co-transfected with either miR-424 or control mimic, and the relative firefly/Renilla luciferase activity was determined for each sample. Data are represented as the mean of four experiments ± S.D. **, p < 0.01.
Combinatorial effect of miR-449a and miR-424 on CCNA2
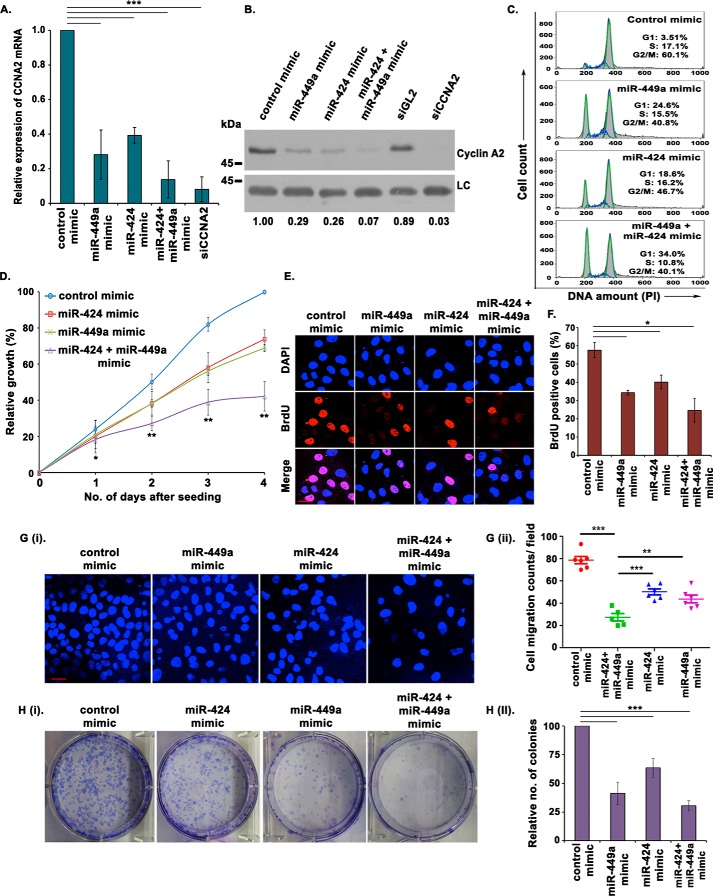
In this study, we have identified that CCNA2 is inhibited by a miR-34 family microRNA, miR-449a, as well as a miR-16 family microRNA, miR-424, which led us to postulate a synergistic regulation of CCNA2 by these two miRNAs in osteosarcomas. To determine the combined tumor-suppressive effect of miR-449a and miR-424 in osteosarcomas, we overexpressed the two miRNAs individually as well as in combination, in U2OS cells. The co-expression of miR-449a and miR-424 mimics resulted in a decrease in CCNA2 mRNA as well as protein levels that was more than what was observed after overexpression of either of the two miRNAs individually (Fig. 7, A and B). The individual expression of miR-424 resulted in G1 arrest and a decreased rate of cell proliferation, as has been described previously for miR-449a (Fig. 7, C and D). Co-expression of miR-449a and miR-424 further enhanced the accumulation of cells in G1 phase and reduced the cell proliferation. Similar effects were observed on the rate of BrdU incorporation, demonstrated by a decreased rate of DNA synthesis, after co-expression of miR-449a and miR-424 (Fig. 7, E and F). An in vitro transwell migration assay revealed that the decrease in the migration ability after simultaneous overexpression of miR-449a and miR-424 was more than what was observed after overexpression of either of the two miRNAs individually (Fig. 7G, i and ii). The colony-forming ability was also found to be significantly reduced in U2OS cells after co-expression of miR-449a and miR-424 (Fig. 7H, i and ii).
Figure 7.
Combinatorial effect of miR-449a and miR-424 on CCNA2. A and B, U2OS cells were transfected on 3 consecutive days with either miR-449a or miR-424 mimic individually or in combination, as indicated, and CCNA2 transcript (A) and protein (B) levels were analyzed. The numbers indicate levels of cyclin A2 relative to control mimic–transfected cells. Data are represented as the mean of three experiments ± S.D. (error bars). C, U2OS cells transfected for 3 consecutive days, as described in A, were treated with nocodazole for 16 h and stained with PI for analysis of cell cycle distribution by flow cytometry. The inset shows the percentage of cells in each phase, indicating an enhanced G1 arrest in cells co-transfected with both miR-424 and miR-449a. D, MTT assay displaying the growth rate of U2OS cells, as described in A. The graph represents the average percentage of cell growth at the indicated days. Data are represented as the mean of three experiments ± S.D. E, U2OS cells transfected for 3 consecutive days, as described in A, were pulsed with BrdU for 30 min followed by staining with α-BrdU (red) antibody. Immunofluorescence images display BrdU incorporation, whereas DNA has been stained with DAPI (blue). Scale bar, 10 μm. F, quantification of E shows that miR-449a and miR-424 co-expression significantly decreases S phase population. Data are represented as the mean of two experiments, with 25 fields in each sample, ± S.D. G (i and ii), U2OS cells were transfected for 3 consecutive days, as described in A, and the cells migrating through a microporous membrane were counted at 24 h after the third transfection. i, representative fields showing DAPI-stained nuclei of migrated cells. Scale bar, 20 μm. ii, quantification of the migratory cells observed in i. Each point refers to the number of cells in an individual field, whereas long and short horizontal bars represent the mean and S.D. of all fields, respectively. H (i and ii), U2OS cells were transfected for 3 consecutive days, as described in A, allowed to grow for 12 days, and stained with crystal violet, after which the colonies were counted. ii, quantification of i, which demonstrates that the colony-forming ability is significantly decreased upon simultaneous expression of miR-449a and miR-424. The data are represented as the mean of five experiments ± S.D. *, p < 0.05; **p < 0.01; ***p < 0.001.
Mouse model of osteosarcoma demonstrates tumor-suppressive effect of miR-449a and miR-424
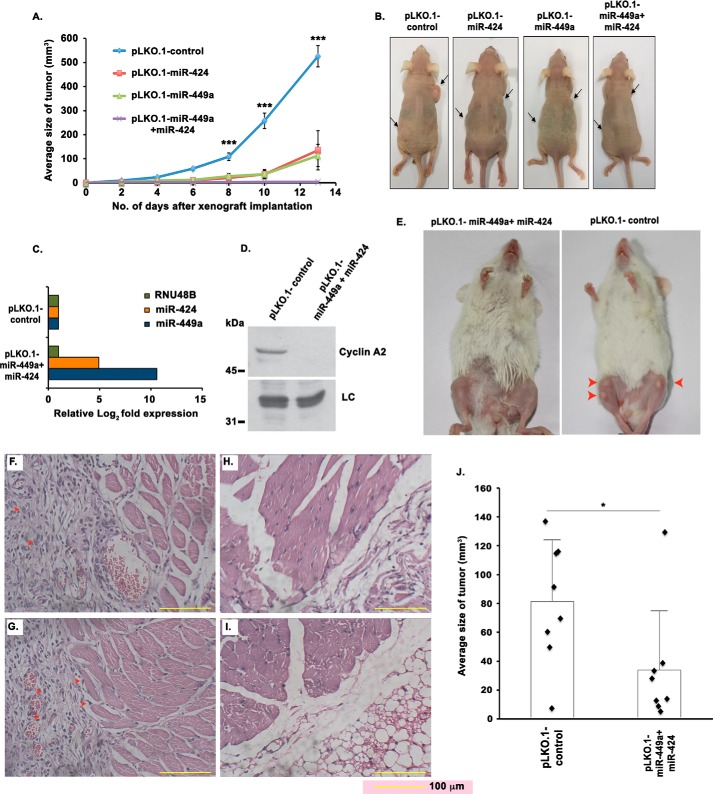
We investigated the effect of either individual or simultaneous expression of these miRNAs on tumor formation in nude mice. We could not obtain measurable tumors by subcutaneous injection of OS cells like U2OS, HOS, and MG-63, and thus we carried out the in vivo functional study using HCT116-derived tumors in nude mice. HCT116 cells were infected with lentiviral particles harboring pLKO.1-miR-424 or pLKO.1-miR-449a individually or in combination. We observed a significant increase in the levels of respective miRNAs, whereas the levels of endogenous cyclin A2 protein were considerably reduced after transduction. HCT116 cells stably expressing miR-449a and miR-424, either individually or simultaneously, were injected subcutaneously in nude mice aged between 4 and 6 weeks. Tumor volume was measured every second day, and mice were sacrificed after 13 days, followed by excision of tumors. As compared with the control group, mice injected with cells expressing miR-449a or miR-424 displayed reduced tumor growth from the sixth day onward (Fig. 8, A and B). Tumors excised from the implanted site of xenografts expressing both miR-449a and miR-424 were highly diminished in size and weight when compared with the control tumors.
Figure 8.
Mouse model of osteosarcoma demonstrates tumor-suppressive effect of miR-449a and miR-424. A and B, tumor-suppressive activity of miR-449a and miR-424 demonstrated in nude mice by subcutaneous injection of colorectal cancer cells (HCT116). A, tumor volume in nude mice subcutaneously injected with HCT116 cells expressing pLKO.1-control, pLKO.1-miR-424, pLKO.1-miR-449a, or both pLKO.1-miR-424 and pLKO.1-miR-449a. Tumor volume was measured on alternate days in 5 mice/group, and the mean tumor volume of each group ± S.D. (error bars) is represented. B, representative photographs of nude mice injected with HCT116 cells as described in A. Arrowheads point to the site of tumor injection. C–J, tumor-suppressive activity of miR-449a and miR-424 demonstrated in NOD-scid mice by intraosseous injection of U2OS cells. C, levels of miR-449a and miR-424 in U2OS cells stably transduced with lentiviral vectors either expressing nonspecific miRNA (pLKO.1-control) or co-expressing both miR-449a and miR-424 (pLKO.1-miR-449a and pLKO.1-miR-424). D, cyclin A2 protein levels in samples described in C. E, tumor development in NOD-scid mice on intraosseous injection of 5 million U2OS cells into the tibial tuberosity of U2OS stable cells as described in C. After 16 days of injection, mice were photographed. Arrows indicate the tumors. F and G, H&E-stained sections (×20 magnification) of tissue excised from the site of injection as described in C (two representative images shown in F and G). Arrows in red point to pleomorphic, poorly differentiated sarcomatous neoplastic cells that are aggressively proliferating and filling in the periosteum space between the bone and muscular structure. H and I, H&E-stained tissue sections from the limbs of normal mice (two representative images shown in H and I). J, tumor volume in NOD-scid mice on intraosseous injection of 5 million U2OS cells into the tibial tuberosity of U2OS stable cells as described in C. Tumor volume was measured after 16 days of injection in four mice per group that were each injected in both the left and right tibia. Each point refers to the tumor volume in a single tibia bone, whereas long and short horizontal bars represent the mean and S.D., respectively. ***, p < 0.001; *, p < 0.05.
Next, we attempted to develop a xenograft mouse model of human osteosarcoma by intraosseous injection of U2OS cells into the tibia bone of NOD-scid. In brief, a sub-centimeter infero-lateral incision was made to expose the tibia of NOD-scid mice, and the cortical bone was punctured with a needle by a rotary movement into which the cells were injected. The mice were administered an intraosseous injection into the tibial tuberosity with U2OS cells either stably expressing nonspecific miRNA (pLKO.1_control) or co-expressing both miR-449a and miR-424 (pLKO.1-miR-449a and pLKO.1-miR-424). qRT-PCR confirmed a significant increase in the mature miRNA levels after transduction with the respective miRNA lentivirus, whereas the levels of endogenous cyclin A2 protein were considerably reduced after expression of miR-449a and miR-424 (Fig. 8, C and D). After 16 days of intraosseous injection, small bumps were visible at the site of injection, which were larger in mice injected with control miRNA compared with miR-424– and miR-449a–expressing U2OS cells (Fig. 8E). We carried out histological analysis of tissues excised from the xenograft sites, which indicated pleomorphic, poorly differentiated neoplastic cells, filling the periosteum space between the bone and muscular structure, confirming cancerous tissue growth (Fig. 8, F and G). This is in contrast to normal tissue, where the periosteum and surrounding muscular layer appeared normal, and no invasion of neoplastic cells was observed (Fig. 8, H and I). As compared with the control group, mice injected with cells expressing both miR-449a and miR-424 displayed a reduced average tumor size (Fig. 8J).
Inverse pattern of miR-449a and CCNA2 expression in human osteosarcomas
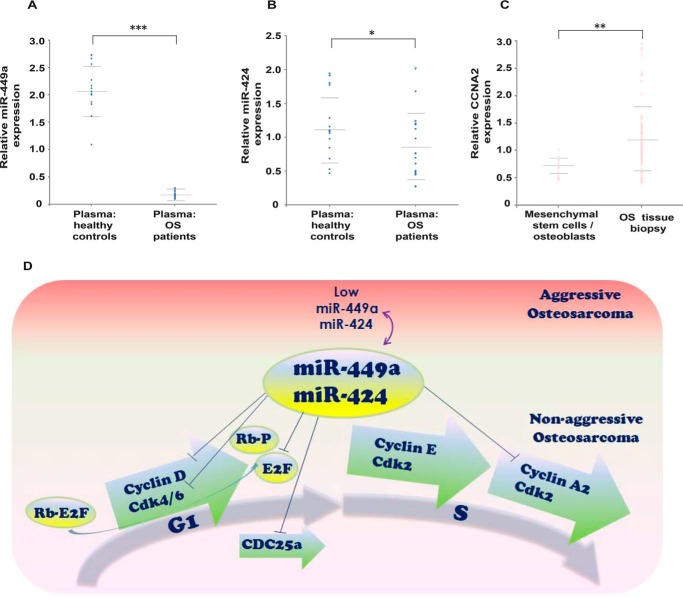
In this study, we have demonstrated that miR-449a inhibition of CCNA2 is lost in osteosarcoma, and finally, we wanted to check whether human biopsy samples display the same pattern. For evaluating the predicted levels of the miRNAs and target genes in patient biopsy samples, we obtained the data from NCBI GEO data set GSE65071 and compared the levels of miR-449a and miR-424 in plasma of OS patients and healthy controls. We observed that miR-449a was significantly down-regulated, whereas miR-424 was moderately down-regulated in the sample set of 20 OS patients compared with 15 healthy controls (Fig. 9, A and B). Similar results were obtained with another patient data set, GSE64915 (data not shown). We also analyzed the levels of CCNA2 in biopsies of 84 patients of high-grade OS and compared them with its putative progenitor cells (i.e. osteoblasts derived from osteogenic differentiated bone marrow–derived mesenchymal stem cells, NCBI GEO data set GSE33383) and concluded that CCNA2 was significantly up-regulated in human osteosarcomas (Fig. 9C). Thus, analysis of human biopsy samples confirms that miR-449a and CCNA2 exhibit an inverse pattern of expression in human osteosarcomas.
Figure 9.
Clinical data demonstrate an inverse expression pattern of miR-449a and CCNA2 in human osteosarcomas. A and B, relative expression of miR-449a and miR-424 in plasma of 20 osteosarcoma patients and 15 healthy controls. Each point refers to the miR-449a (A) or miR-424 (B) levels in one sample, whereas long and short horizontal bars represent the mean and S.D., respectively. C, expression of CCNA2 in biopsies from 84 patients of high-grade osteosarcoma, which is compared with its putative progenitor cells (i.e. mesenchymal stem cells (n = 12) and osteoblasts (n = 3)). Each point refers to CCNA2 levels in one sample, whereas long and short horizontal bars represent the mean and S.D., respectively. *, p = 0.05; **, p < 0.01; ***, p < 0.001. D, model depicting miR-449a and miR-424 control of the cell cycle progression. The G1 phase of the cell cycle is driven by activities of cyclin D/CDK4/6 and cyclin E/CDK2, which phosphorylate retinoblastoma (Rb), whereas S phase progression is driven by the activities of cyclin E/CDK2 and cyclin A2/CDK2. In nonaggressive osteosarcomas, miR-449a and miR-424 regulate G1 phase progression by targeting cyclin D as well as CDK6 (50, 51). It has also been shown that miR-449a targets transcription factor E2F3 as well as Cdc25A phosphatase whose activity is critical for CDK2 activation (13). Our study demonstrates that miR-449a and miR-424 regulate the S phase progression by targeting CCNA2. We show that loss of miR-449a and miR-424 expression in aggressive osteosarcomas results in high CCNA2 levels and development of oncogenic phenotypes, such as increased proliferation, migration, and invasion.
Discussion
The literature is replete with reports that demonstrate the role of miR-16 and miR-34 family miRNAs on similar growth pathways, but the coalescence between them has been addressed far less. For instance, overexpression of miR-34 family miRNAs, including miR-34a and miR-449a, has been demonstrated to trigger G1 phase accumulation by inhibiting the cell cycle regulators, such as CCND1, CDK4, and CDK6 (10, 27). Similarly, miR-16 family miRNAs have also been independently reported to suppress the expression of CCND1, CDC25A, CCNE1, and CDK6 (9, 28). However, the concordance is not apparent, most likely because the most extensively studied miRNAs from each of these two families, miR-34a and miR-16, do not display an enhanced tumor-suppressive effect. There are, however, many reports that allude to such an effect; in bladder cancer, it was claimed that loss of miR-16 led to an increase of CCND1, but a closer examination revealed that moderate changes in miR-16 activity cannot completely account for the increased CCND1, and thus, most likely, other factors are simultaneously regulating CCND1 (22). Thus, it is logical to assume that one or more members of miR-34 and miR-16 families probably act together or in combination with other miRNAs to target a common repertoire of growth-associated genes, an issue that has not been addressed extensively in the literature. Our study demonstrating the inhibition of CCNA2 by miR-449a and miR-424 strengthens the evidence of combinatorial activity of miR-34 and miR-16 family miRNAs to control cell cycle and proliferation.
The cooperative effects of miRNAs have been described by multiple studies in diverse physiological conditions (29). The literature indicates that the effect of a single miRNA on its target genes can range from mild suppression to complete knockdown, whereas binding of more than one miRNA to a common target often enhances the effect on target gene regulation (30). Thus, the miRNA-mediated regulation of genes may not always be a binary on-off mechanism, whereas most of the miRNA–mRNA interactions result in fine-tuning of gene expression, depending on the intracellular or extracellular signals. Moreover, the ectopic expression of miRNAs that target multiple genes of a common regulatory pathway has been found to generate a more pronounced effect on the overall phenotype when compared with the effect of single target gene knockdown (31). Thus, our study demonstrating the suppression of a key cell cycle regulator establishes combinatorial regulation of cell cycle and proliferation by multiple tumor-suppressive miRNAs. We propose that inhibition of several cell cycle genes, including CCNA2, CCND1, and CDK6, leading to arrest in different phases of the cell cycle, contributes to the cell cycle retardation by miR-449a and miR-424 (Fig. 9D).
In our study, we have identified cyclin A2 as a common target of miR-449a and miR-424. The kinase activity of CCNA2–Cdk2 is imperative for initiation of DNA replication and S phase progression, whereas later in the G2 phase, cyclin A–Cdk1 activity controls the entry of cells into mitosis (32, 33). Extensive studies have demonstrated that increased activity of cyclin A–dependent kinases cause aberrant phosphorylation of various oncoproteins and tumor suppressors, thus leading to uncontrolled progression of cell cycle and stimulating tumorigenesis (34). Elevated expression of CCNA2 has been detected in a variety of cancers, and its dysregulation has also been associated with poor prognosis (35, 36). Whereas the increased expression of CCNA2 has been primarily shown to result from gene amplification and loss of transcriptional regulation, a few studies have also reported the role of noncoding RNAs in the derepression of CCNA2 in human cancers (37). Therefore, our identification of CCNA2 as a target of tumor-suppressive miRNAs, miR-449a and miR-424, raises the prospects of utilization of clinically significant miRNAs working in coordination with other miRNAs of similar activity to control cancer progression.
Although our study emphasizes the tumor-suppressive role of miR-449a/424–CCNA2 interaction, the miRNAs of miR-34 and miR-16 families are likely to have diverse physiological consequences (38, 39). It has been reported that miR-424 is up-regulated during differentiation of myofibroblasts, and it is thus likely that the miR-424–CCNA2 regulatory arc contributes to the differentiation of myoblast cells (40). Similarly, overexpression of miR-449a has been reported to induce neuroblastoma cell differentiation by causing cell cycle arrest, which, in the light of our discovery, could be due to CCNA2 inhibition (41). Apart from cell cycle regulation, miR-16 and miR-34 family miRNAs influence other cellular processes, such as differentiation, epithelial-to-mesenchymal transition, and drug sensitivity of cancer cells; it has been reported that miR-16 and miR-34 modulate the chemosensitivity of tumor cells toward anticancer drugs such as paclitaxel by controlling the expression of genes regulating drug response pathways (14, 42, 43). Our findings allude to the role of other members of miR-34 and miR-16 families in altering the chemoresistance and differentiation of different cancer cells, and it will also be interesting to decipher a combinatorial role of miR-449a/b/c and miR-424/503 in modulating these pathways.
Our study assumes significance in the light of efforts that evaluate miR-16 and miR-34a individually for treatment of human cancers (44, 45). It may be noted that a study that utilized a combinatorial approach for co-delivery of miR-34a and let-7 in preclinical mouse models has achieved a significantly higher reduction in tumor growth compared with individual miRNAs (46). Therefore, we suggest that the functional similarity of miR-449a and miR-424 could be exploited for therapies of human cancers in which CCNA2 is overexpressed and predicted to have an oncogenic activity. The ability of miRNAs to target multiple genes suggests their functional regulation of several compensatory pathways, making it crucial to draw a complete picture of miRNA targetome before testing their clinical implications. In summation, we demonstrate that miR-449a and miR-424, belonging to miR-34 and miR-16 families, respectively, both target the major S/G2 phase cyclin, CCNA2, and prevent cancer progression.
Experimental procedures
Acquisition and analysis of microarray data and miRNA target prediction
Publicly available microarray data sets (GSE28425) of mRNA and miRNA expression in osteosarcoma cell lines were downloaded from the Gene Expression Omnibus (GEO) database. Data were analyzed by calculating the -fold change in expression of genome-wide mRNAs and miRNAs in U2OS with respect to HAL using the background-corrected and log2-normalized intensity values for both cell lines. The functions of the identified up-regulated genes were further annotated using the online Gene Ontology tools of the DAVID (Database for Annotation, Visualization, and Integrated Discovery) and KEGG pathway databases. The mRNA–miRNA interaction network was constructed by using a combination of five different target prediction algorithms, including TargetScan, miRanda, miRWalk, RNAhybrid, and RNA22.
Cell culture, irradiation, cell synchronization, and cloning
U2OS, HAL, KPD, HOS, MG-63 (human osteosarcoma cell lines), HEK293T (human embryonic kidney cells with SV40 large T antigen cell line), and HCT116 (human colorectal cell line) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% antibiotic and antimycotic (100×) solution at 37 °C in a humidified atmosphere with 5% CO2. For cloning of the human CCNA2 coding sequence, CCNA2 cDNA was amplified by PCR and cloned into pMX-puro-3NLS-GST-HA plasmid. HEK293T cells were then transfected with pMX-puro-CCNA2 along with helper plasmids expressing the Gag, Pol, and viral VSV-G envelope proteins to generate complete viral particles. To obtain the U2OS cell line stably expressing CCNA2, U2OS cells were infected with the viral particles and selected with 1 μg/ml of puromycin after 48 h.
Transfection
For RNAi-mediated gene silencing, siRNAs against GL2 and CCNA2 were custom-synthesized by Dharmacon. For gain- and loss-of-function studies of miRNAs, miRIDIAN miRNA mimics and inhibitors were used. Cells were transfected with either 100 nm miRNA inhibitor or 80 nm siRNA or miRNA mimic using Lipofectamine 2000 reagent (Invitrogen) for 3 consecutive days. The cells were harvested 24 h after the last transfection for immunoblotting, flow cytometric analysis, or RT-PCR. The siRNA sequences used are as follows: GL2, 5′-CGUACGCGGAAUACUUCGA-3′; CCNA2, 5′-AAGGCAGCGCCCGUCCAACAA-3′.
Cell cycle analysis and flow cytometry
For cell cycle analysis, U2OS cells transfected with miRNA or control mimic on 3 consecutive days were harvested after 24 h of the third transfection and fixed with 70% ethanol at 4 °C for 1 h. For arresting the cells at the G2/M transition, the transfected cells were incubated with nocodazole (100 ng/ml) for 16 h before harvesting and fixation with 70% ethanol. Following fixation, the cells were washed with 1× PBS, and the cell pellet was resuspended in 1× PBS with 0.1% Triton X-100, 20 mg/ml RNase A, and 70 mg/ml propidium iodide, and then the stained cells were analyzed by flow cytometry. The flow cytometry data were acquired on a BD Biosciences FACSCalibur machine by Cell Quest Pro software. Cell cycle distribution was evaluated by the Dean/Jett/Fox method using FlowJo software. To study the BrdU incorporation, transfected cells were cultured in medium containing 100 μm BrdU (BD Biosciences) for 30 min, prior to harvesting. After fixation, cells were treated with 2 n HCl for 15–20 min to denature the DNA, followed by neutralization with 0.1 m sodium tetraborate (pH 8.5) for 5 min at room temperature. Cells were then washed with a blocking solution comprising 3% BSA in PBS containing 0.1% Triton X-100 followed by incubation with mouse anti-BrdU antibody (dilution 1:10 in blocking solution) conjugated to FITC for 1 h. After antibody staining, cells were washed with 1× PBS, and DNA was stained with propidium iodide and run on a FACS machine as described above.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) and reverse-transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen). For miRNA analysis, total RNA was poly(A)-tailed using Escherichia coli poly(A) polymerase (New England Biolabs) before reverse transcription as described previously (6). The qRT-PCR reactions were carried out in duplicates in a 10-μl volume for the expression analysis. The reaction mixture contained SYBR Select master mix (1×, Applied Biosystems), cDNA template, and forward and reverse gene/miRNA-specific primers (0.1 μm each). The target sequence amplification temperature profile followed was as follows: initial denaturation for 10 min at 95 °C, followed by 40 cycles of 10 s at 95 °C and amplification for 30 s at an annealing temperature of 60 °C. Finally, a melt curve analysis was carried out at a temperature range of 60–95 °C for 20 min. The small nucleolar RNAs, RNU48 and RNU66, and housekeeping gene, BMG (β2-microglobulin), were used as internal controls for miRNA and mRNA quantification, respectively. Results were calculated using the ΔΔCt method to determine the -fold change in expression between the experimental and control groups.
Indirect immunofluorescence and microscopy
To study the BrdU incorporation, U2OS cells grown on coverslips were cultured in medium containing 100 μm BrdU for 30 min prior to harvesting. Cells were fixed with 4% formaldehyde for 10 min, followed by denaturation with 2 n HCl for 15–20 min and neutralization with 0.1 m sodium tetraborate (pH 8.5) for 2 min. Cells were then blocked with 3% BSA for 30 min and stained with mouse anti-BrdU antibody (BD Biosciences) for 1 h, followed by incubation with secondary antibody for 1 h. Finally, the cells were visualized under the microscope after mounting with Vectashield mounting reagent containing 4′,6-diamidino-2-phenylindole (DAPI) that stains the nucleus. The secondary antibody used was anti-mouse conjugated to Alexa Fluor 488 or Alexa Fluor 555 (Invitrogen). For co-immunofluorescence of HA and BrdU, cells were incubated with mouse anti-BrdU antibody and rabbit anti-HA antibody (final dilution of each antibody, 1:250 in blocking solution) after blocking with 3% BSA. The secondary antibodies used were anti-mouse conjugated to Alexa Fluor 488 and anti-rabbit conjugated to Alexa Fluor 555 (Invitrogen).
Immunoblotting and antibodies
For Western blotting, the whole-cell lysates from cells of equal confluence were prepared in a proportionate volume of Laemmli buffer and denatured at 95 °C, followed by SDS-PAGE. The gel was transferred onto a nitrocellulose membrane, blocked with 3% BSA prepared in 1× TBST. The membrane was then incubated with the appropriate antibody, washed, and probed with horseradish peroxidase-conjugated secondary antibody. Enhanced chemiluminescence was used to visualize the protein bands. Quantity One Software was utilized to evaluate the levels of specific proteins, which were expressed after normalization with the protein-loading control. The following antibodies were used for Western blotting. Antibodies against cyclin A2, cyclin E1, CDK2, CDK4, CDK6, and β-actin were obtained from Santa Cruz Biotechnology, Inc. Antibodies against cyclin D1 and Cdc25A were obtained from Abcam. Antibody used to detect the HA epitope was purchased from Sigma.
Luciferase reporter assay
To construct luciferase reporter plasmids, CCNA2 full-length coding sequence, CCNA2 full-length 3′-UTR, and CCNA2 3′-UTR mutant for the miR-449a–binding site were cloned individually downstream to the firefly luciferase into pmirGLO Dual-Luciferase miRNA target expression vector (Promega). For the luciferase reporter assay, U2OS cells were co-transfected with 750 ng of luciferase reporter plasmid harboring the intact or mutant binding site of the gene along with 80 nm miRNA mimic using Lipofectamine 2000 reagent. After 48 h of transfection, cells were harvested, and luciferase activity was measured in a Lumicount Luminometer (Packard) using the Dual-Luciferase reporter assay kit (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell proliferation assays
For proliferation assay, cells were seeded in triplicates: 10,000 cells in 200 μl of medium/well in a 96-well plate or 25,000 cells in 500 μl of medium/well in a 24-well plate. The tetrazolium dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to cells in culture at a final concentration of 0.5 mg/ml and incubated at 37 °C. After 3–4 h, the cells were resuspended in isopropyl alcohol/DMSO (9:1) solution (100 μl in a 96-well plate and 250 μl in a 24-well plate). The quantity of MTT reduction to formazan by active NADP(H) molecules was measured by recording changes in absorbance at 570 and 630 nm (reference wavelength) using a microplate reader (BiotekPowerwave XS).
Wound-healing assay
U2OS cells seeded in 6-well plates with an approximate confluence of 30–40% were transfected with 80 nm miRNA mimic using Lipofectamine 2000 reagent for 2 consecutive days and were cultured until confluence. A wound was then created by manually scraping the cell monolayer with a 200-μl pipette tip. The cultures were washed twice with 1× PBS and supplemented with fresh medium. Cell movement into the wound was observed at three preselected time points (0, 24, and 48 h) in three randomly selected microscopic fields for each condition and time point. Images were acquired with a Nikon TE2000-S inverted fluorescence microscope. The distance traveled by the cells was determined by measuring the wound width at different time points.
Clonogenic assay
U2OS cells transfected with miRNA mimic were counted, and 1000 cells were seeded in a 6-well culture dish in triplicates. After 11–12 days of incubation, plates were gently washed with 1× PBS and stained with 0.1% crystal violet. Colonies with over 50 cells were manually counted.
Invasion and migration assays
U2OS cells (6 × 104) resuspended in 250 μl of serum-free medium were seeded into the upper well of the Matrigel-coated membrane of a transwell chamber (8-μm pore size; Corning), for assaying cell invasion. For migration assays, the cells were seeded into the upper well of a Millicell hanging cell culture insert of a transwell chamber (8-μm pore size, Millipore). Serum-containing medium was added to the lower chamber, and cells were incubated at 37 °C for 24 h. Subsequently, cells in the upper chamber were removed, and the cells migrating to or invading the bottom of the membrane were fixed with cold methanol and visualized under the microscope after mounting with Vectashield mounting reagent containing DAPI that stains the nucleus. Eight random fields of each membrane were photographed and counted for statistical analysis.
Generation of lentivirus and infection into mammalian cell lines
For lentivirus preparation, lentiviral vector pLKO.1 containing mature miRNA sequences was co-transfected with packaging vector pMD2.G and envelope vector psPAX2 at a 4:3:1 ratio using Lipofectamine 2000 in HEK293T cells. Supernatant was collected after 48 h of transfection, pooled, and filtered using 0.45-μm filters. The filtered supernatant was used along with 1 μg/ml Polybrene for infecting U2OS or HCT116 cells. After 24 h of infection, cells were grown in selection medium containing 1 μg/ml puromycin.
Animals
Nude and NOD-scid (NOD.CB17-Prkdcscid) mice were maintained in a pathogen-free environment and were housed in individually ventilated cages containing sterilized feed, water, and autoclaved bedding. All experimental procedures were performed as per approval of the Animal Ethics Committee of the National Institute of Immunology and in accordance with the Guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Ministry of Environment, Government of India.
In vivo tumorigenicity model developed by subcutaneous injection of HCT116 cells in nude mice
A total of 20 female nude mice, aged between 6 and 8 weeks were randomly divided into four groups. Five million HCT116 cells stably expressing either control_pLKO.1, miR-449a_pLKO.1, or miR-424_pLKO.1 or co-expressing both miR-449a_pLKO.1 and miR-424_pLKO.1 were suspended in 100 μl of PBS along with 10% Matrigel and injected subcutaneously in the right fore flank and left hind flank of each nude mice. Tumor growth was measured every alternate day using a vernier caliper, and the mice were euthanized after 13 days. The relative tumor volume was calculated using the formula, tumor volume = width × length × height. Next, the tumors were extracted and weighed.
In vivo tumorigenicity model of osteosarcoma developed by intraosseous injection of U2OS cells in NOD-scid mice
Five million U2OS cells either stably expressing nonspecific miRNA (pLKO.1_control) or co-expressing both miR-424 and miR-449a (pLKO.1-miR-424 and pLKO.1-miR-449a) were suspended in 20 μl of PBS along with 20% Matrigel. Six- to seven-week-old NOD-scid (NOD.CB17-Prkdcscid) mice were shaved, a sub-centimeter infero-lateral incision was made to expose the tibia, and the cortical bone was punctured with a needle by a rotary movement. The mice were then administered intraosseous injection under general anesthesia (isoflurane, 1.5–4%) into the tibial tuberosity. Each group had four mice that were injected in both the left and right tibia. The tumor growth was assessed after 16 days of injection using a vernier caliper. Pictures of tumor growth were taken at the end of the experiment. Fresh tissue samples were fixed in 10% formalin and embedded in paraffin before sectioning and staining. Tissue sections (3 μm) were deparaffinized in xylene and rehydrated in an ethanol series. Hematoxylin and eosin (H&E) staining was performed according to standard protocol.
Clinical data
The expression levels of miR-449a and miR-424 were obtained from Gene Expression Omnibus data repository accession number GSE65071, where a quantitative PCR–based platform was used to evaluate the levels of 750 miRNAs from the plasma of 20 osteosarcoma patients and 15 healthy controls (47). The Ct values were converted to relative gene expression, and the difference in the levels of miR-449a between 20 osteosarcoma patients and 15 healthy controls was evaluated by the 2−ΔΔCt method. The expression levels of CCNA2 were obtained from GEO GSE33383, in which a microarray (Human-6 version 2.0) was used to evaluate the gene expression in biopsies from 84 patients of high-grade osteosarcoma and compared with its putative progenitor cells (i.e. mesenchymal stem cells (n = 12) and osteoblasts (n = 3)) (48). To calculate relative CCNA2 expression, the log2 transformation intensity values in each sample were expressed as a ratio of the average of the log2 transformation intensity values of all samples.
Statistical analysis
The miRNA/mRNA -fold increase data were calculated according to the ΔΔCT method (49). The results were presented as mean ± S.D. and analyzed with Student's t test. A p value of 0.05 or less was considered significant unless noted otherwise.
Author contributions
R. S. conceived the project, conducted the experiments, analyzed the data, and wrote the manuscript. P. P. conducted miRNA and phenotype assays. P. K., T. G., M. M. K., P. N., and S. S. contributed to the manuscript.
Acknowledgments
We thank Shailendra K. Arindkar for subcutaneous injections in the nude mice and administration of injections for general anesthesia during radiography. We thank Akhil Varshney and Sunder Bisht for helping in various parts of this paper.
This study was supported in part by Government of India Projects BT/PR22824/BRB/10/1578/2016 and EMR/2016/001702. The authors declare that they have no conflicts of interest with the contents of this article.
- OS
- osteosarcoma
- qRT-PCR
- quantitative RT-PCR
- BrdU
- 5-bromo-2-deoxyuridine
- miRNA
- microRNA
- DAPI
- 4′,6-diamidino-2-phenylindole
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI
- propidium iodide.
References
- 1. Picci P. (2007) Osteosarcoma (osteogenic sarcoma). Orphanet J. Rare Dis. 2, 6 10.1186/1750-1172-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lewis I. J., Nooij M. A., Whelan J., Sydes M. R., Grimer R., Hogendoorn P. C., Memon M. A., Weeden S., Uscinska B. M., van Glabbeke M., Kirkpatrick A., Hauben E. I., Craft A. W., Taminiau A. H., MRC BO06 and EORTC 80931 collaborators, and European Osteosarcoma Intergroup (2007) Improvement in histologic response but not survival in osteosarcoma patients treated with intensified chemotherapy: a randomized phase III trial of the European Osteosarcoma Intergroup. J. Natl. Cancer Inst. 99, 112–128 10.1093/jnci/djk015 [DOI] [PubMed] [Google Scholar]
- 3. Miller C. W., Aslo A., Won A., Tan M., Lampkin B., and Koeffler H. P. (1996) Alterations of the p53, Rb and MDM2 genes in osteosarcoma. J. Cancer Res. Clin. Oncol. 122, 559–565 10.1007/BF01213553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lulla R. R., Costa F. F., Bischof J. M., Chou P. M., de F. Bonaldo M., Vanin E. F., and Soares M. B. (2011) Identification of differentially expressed microRNAs in osteosarcoma. Sarcoma 2011, 732690 10.1155/2011/732690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kobayashi E., Hornicek F. J., and Duan Z. (2012) MicroRNA involvement in osteosarcoma. Sarcoma 2012, 359739 10.1155/2012/359739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghosh T., Varshney A., Kumar P., Kaur M., Kumar V., Shekhar R., Devi R., Priyanka P., Khan M. M., and Saxena S. (2017) MicroRNA-874-mediated inhibition of the major G1/S phase cyclin, CCNE1, is lost in osteosarcomas. J. Biol. Chem. 292, 21264–21281 10.1074/jbc.M117.808287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J., Zhou J., Chen X., Yang B., Wang D., Yang P., He X., and Li H. (2015) miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumor Biol. 36, 8221–8229 10.1007/s13277-015-3568-y [DOI] [PubMed] [Google Scholar]
- 8. van der Deen M., Taipaleenmäki H., Zhang Y., Teplyuk N. M., Gupta A., Cinghu S., Shogren K., Maran A., Yaszemski M. J., Ling L., Cool S. M., Leong D. T., Dierkes C., Zustin J., Salto-Tellez M., et al. (2013) MicroRNA-34c inversely couples the biological functions of the runt-related transcription factor RUNX2 and the tumor suppressor p53 in osteosarcoma. J. Biol. Chem. 288, 21307–21319 10.1074/jbc.M112.445890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Q., Fu H., Sun F., Zhang H., Tie Y., Zhu J., Xing R., Sun Z., and Zheng X. (2008) miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 36, 5391–5404 10.1093/nar/gkn522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun F., Fu H., Liu Q., Tie Y., Zhu J., Xing R., Sun Z., and Zheng X. (2008) Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 582, 1564–1568 10.1016/j.febslet.2008.03.057 [DOI] [PubMed] [Google Scholar]
- 11. Li Q., Li H., Zhao X., Wang B., Zhang L., Zhang C., and Zhang F. (2017) DNA methylation mediated downregulation of miR-449c controls osteosarcoma cell cycle progression by directly targeting oncogene c-Myc. Int. J. Biol. Sci. 13, 1038–1050 10.7150/ijbs.19476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng N., Li L., Gao J., Zhou J., Wang Y., Wang C., and Liu Y. (2018) Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem. Biophys. Res. Commun. 495, 189–196 10.1016/j.bbrc.2017.11.028 [DOI] [PubMed] [Google Scholar]
- 13. Ren X. S., Yin M. H., Zhang X., Wang Z., Feng S. P., Wang G. X., Luo Y. J., Liang P. Z., Yang X. Q., He J. X., and Zhang B. L. (2014) Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett. 344, 195–203 10.1016/j.canlet.2013.10.031 [DOI] [PubMed] [Google Scholar]
- 14. Xu M., Jin H., Xu C. X., Bi W. Z., and Wang Y. (2014) MiR-34c inhibits osteosarcoma metastasis and chemoresistance. Med. Oncol. 31, 972 10.1007/s12032-014-0972-x [DOI] [PubMed] [Google Scholar]
- 15. Long X. H., Mao J. H., Peng A. F., Zhou Y., Huang S. H., and Liu Z. L. (2013) Tumor suppressive microRNA-424 inhibits osteosarcoma cell migration and invasion via targeting fatty acid synthase. Exp. Ther. Med. 5, 1048–1052 10.3892/etm.2013.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai C. K., Zhao G. Y., Tian L. Y., Liu L., Yan K., Ma Y. L., Ji Z. W., Li X. X., Han K., Gao J., Qiu X. C., Fan Q. Y., Yang T. T., and Ma B. A. (2012) miR-15a and miR-16–1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol. Rep. 28, 1764–1770 10.3892/or.2012.1995 [DOI] [PubMed] [Google Scholar]
- 17. Mao J. H., Zhou R. P., Peng A. F., Liu Z. L., Huang S. H., Long X. H., and Shu Y. (2012) microRNA-195 suppresses osteosarcoma cell invasion and migration in vitro by targeting FASN. Oncol. Lett. 4, 1125–1129 10.3892/ol.2012.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lv T., Liu Y., Li Z., Huang R., Zhang Z., and Li J. (2018) miR-503 is down-regulated in osteosarcoma and suppressed MG63 proliferation and invasion by targeting VEGFA/Rictor. Cancer Biomark. 23, 315–322 10.3233/CBM-170906 [DOI] [PubMed] [Google Scholar]
- 19. Ge L., Zheng B., Li M., Niu L., and Li Z. (2016) MicroRNA-497 suppresses osteosarcoma tumor growth in vitro and in vivo. Oncol. Lett. 11, 2207–2212 10.3892/ol.2016.4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xia L., Zhang D., Du R., Pan Y., Zhao L., Sun S., Hong L., Liu J., and Fan D. (2008) miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 123, 372–379 10.1002/ijc.23501 [DOI] [PubMed] [Google Scholar]
- 21. Yin D., Ogawa S., Kawamata N., Leiter A., Ham M., Li D., Doan N. B., Said J. W., Black K. L., and Phillip Koeffler H. (2013) miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene 32, 1155–1163 10.1038/onc.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Q. Q., Liu B., and Yuan T. (2013) MicroRNA-16 inhibits bladder cancer proliferation by targeting cyclin D1. Asian Pac. J. Cancer Prev. 14, 4127–4130 10.7314/APJCP.2013.14.7.4127 [DOI] [PubMed] [Google Scholar]
- 23. Lauvrak S. U., Munthe E., Kresse S. H., Stratford E. W., Namløs H. M., Meza-Zepeda L. A., and Myklebost O. (2013) Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer 109, 2228–2236 10.1038/bjc.2013.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mohseny A. B., Machado I., Cai Y., Schaefer K. L., Serra M., Hogendoorn P. C., Llombart-Bosch A., and Cleton-Jansen A. M. (2011) Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab. Invest. 91, 1195–1205 10.1038/labinvest.2011.72 [DOI] [PubMed] [Google Scholar]
- 25. Hanahan D., and Coussens L. M. (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 26. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Achari C., Winslow S., Ceder Y., and Larsson C. (2014) Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer 14, 538 10.1186/1471-2407-14-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Linsley P. S., Schelter J., Burchard J., Kibukawa M., Martin M. M., Bartz S. R., Johnson J. M., Cummins J. M., Raymond C. K., Dai H., Chau N., Cleary M., Jackson A. L., Carleton M., and Lim L. (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 27, 2240–2252 10.1128/MCB.02005-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bueno M. J., Gómez de Cedrón M., Gómez-López G., Pérez de Castro I., Di Lisio L., Montes-Moreno S., Martínez N., Guerrero M., Sánchez-Martínez R., Santos J., Pisano D. G., Piris M. A., Fernández-Piqueras J., and Malumbres M. (2011) Combinatorial effects of microRNAs to suppress the Myc oncogenic pathway. Blood 117, 6255–6266 10.1182/blood-2010-10-315432 [DOI] [PubMed] [Google Scholar]
- 30. Wu S., Huang S., Ding J., Zhao Y., Liang L., Liu T., Zhan R., and He X. (2010) Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 29, 2302–2308 10.1038/onc.2010.34 [DOI] [PubMed] [Google Scholar]
- 31. Perdigão-Henriques R., Petrocca F., Altschuler G., Thomas M. P., Le M. T., Tan S. M., Hide W., and Lieberman J. (2016) miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 35, 158–172 10.1038/onc.2015.69 [DOI] [PubMed] [Google Scholar]
- 32. Resnitzky D., Hengst L., and Reed S. I. (1995) Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol. Cell Biol. 15, 4347–4352 10.1128/MCB.15.8.4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fotedar A., Cannella D., Fitzgerald P., Rousselle T., Gupta S., Dorée M., and Fotedar R. (1996) Role for cyclin A-dependent kinase in DNA replication in human S phase cell extracts. J. Biol. Chem. 271, 31627–31637 10.1074/jbc.271.49.31627 [DOI] [PubMed] [Google Scholar]
- 34. Gopinathan L., Tan S. L., Padmakumar V. C., Coppola V., Tessarollo L., and Kaldis P. (2014) Loss of Cdk2 and cyclin A2 impairs cell proliferation and tumorigenesis. Cancer Res. 74, 3870–3879 10.1158/0008-5472.CAN-13-3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J. Q., Miki H., Wu F., Saoo K., Nishioka M., Ohmori M., and Imaida K. (2002) Cyclin A correlates with carcinogenesis and metastasis, and p27(kip1) correlates with lymphatic invasion, in colorectal neoplasms. Hum. Pathol. 33, 1006–1015 10.1053/hupa.2002.125774 [DOI] [PubMed] [Google Scholar]
- 36. Yasmeen A., Berdel W. E., Serve H., and Müller-Tidow C. (2003) E- and A-type cyclins as markers for cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 3, 617–633 10.1586/14737159.3.5.617 [DOI] [PubMed] [Google Scholar]
- 37. Yang F., Hu Y., Liu H. X., and Wan Y. J. (2015) MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J. Biol. Chem. 290, 6507–6515 10.1074/jbc.M114.620369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Otto T., Candido S. V., Pilarz M. S., Sicinska E., Bronson R. T., Bowden M., Lachowicz I. A., Mulry K., Fassl A., Han R. C., Jecrois E. S., and Sicinski P. (2017) Cell cycle-targeting microRNAs promote differentiation by enforcing cell-cycle exit. Proc. Natl. Acad. Sci. U.S.A. 114, 10660–10665 10.1073/pnas.1702914114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bae Y., Yang T., Zeng H. C., Campeau P. M., Chen Y., Bertin T., Dawson B. C., Munivez E., Tao J., and Lee B. H. (2012) miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 21, 2991–3000 10.1093/hmg/dds129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao X., Huang C., Zhao C., Gou X., Senavirathna L. K., Hinsdale M., Lloyd P., and Liu L. (2015) Regulation of myofibroblast differentiation by miR-424 during epithelial-to-mesenchymal transition. Arch. Biochem. Biophys. 566, 49–57 10.1016/j.abb.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao Z., Ma X., Sung D., Li M., Kosti A., Lin G., Chen Y., Pertsemlidis A., Hsiao T. H., and Du L. (2015) microRNA-449a functions as a tumor suppressor in neuroblastoma through inducing cell differentiation and cell cycle arrest. RNA Biol. 12, 538–554 10.1080/15476286.2015.1023495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patel N., Garikapati K. R., Pandita R. K., Singh D. K., Pandita T. K., Bhadra U., and Bhadra M. P. (2017) miR-15a/miR-16 down-regulates BMI1, impacting Ub-H2A mediated DNA repair and breast cancer cell sensitivity to doxorubicin. Sci. Rep. 7, 4263 10.1038/s41598-017-02800-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu Y., Yang Q., Wang L., Wang S., Sun F., Xu D., and Jiang J. (2018) Knockdown of the oncogene lncRNA NEAT1 restores the availability of miR-34c and improves the sensitivity to cisplatin in osteosarcoma. Biosci. Rep. 38, BSR20180375 10.1042/BSR20180375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nana-Sinkam S. P., and Croce C. M. (2013) Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 93, 98–104 10.1038/clpt.2012.192 [DOI] [PubMed] [Google Scholar]
- 45. Beg M. S., Brenner A. J., Sachdev J., Borad M., Kang Y. K., Stoudemire J., Smith S., Bader A. G., Kim S., and Hong D. S. (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188 10.1007/s10637-016-0407-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stahlhut C., and Slack F. J. (2015) Combinatorial action of microRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle 14, 2171–2180 10.1080/15384101.2014.1003008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Allen-Rhoades W., Kurenbekova L., Satterfield L., Parikh N., Fuja D., Shuck R. L., Rainusso N., Trucco M., Barkauskas D. A., Jo E., Ahern C., Hilsenbeck S., Donehower L. A., and Yustein J. T. (2015) Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 4, 977–988 10.1002/cam4.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuijjer M. L., Rydbeck H., Kresse S. H., Buddingh E. P., Lid A. B., Roelofs H., Bürger H., Myklebost O., Hogendoorn P. C., Meza-Zepeda L. A., and Cleton-Jansen A. M. (2012) Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer 51, 696–706 10.1002/gcc.21956 [DOI] [PubMed] [Google Scholar]
- 49. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 50. Noonan E. J., Place R. F., Basak S., Pookot D., and Li L. C. (2010) miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget 1, 349–358 10.18632/oncotarget.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang X., Feng M., Jiang X., Wu Z., Li Z., Aau M., and Yu Q. (2009) miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 23, 2388–2393 10.1101/gad.1819009 [DOI] [PMC free article] [PubMed] [Google Scholar]