Abstract
Alzheimer's disease (AD) is pathologically characterized by the deposition of the β-amyloid (Aβ) peptide in senile plaques in the brain, leading to neuronal dysfunction and eventual decline in cognitive function. Genome-wide association studies have identified the bridging integrator 1 (BIN1) gene within the second most significant susceptibility locus for late-onset AD. BIN1 is a member of the amphiphysin family of proteins and has reported roles in the generation of membrane curvature and endocytosis. Endocytic dysfunction is a pathological feature of AD, and endocytosis of the amyloid precursor protein is an important step in its subsequent cleavage by β-secretase (BACE1). In vitro evidence implicates BIN1 in endosomal sorting of BACE1 and Aβ generation in neurons, but a role for BIN1 in this process in vivo is yet to be described. Here, using biochemical and immunohistochemistry analyses we report that a 50% global reduction of BIN1 protein levels resulting from a single Bin1 allele deletion in mice does not change BACE1 levels or localization in vivo, nor does this reduction alter the production of endogenous murine Aβ in nontransgenic mice. Furthermore, we found that reduction of BIN1 levels in the 5XFAD mouse model of amyloidosis does not alter Aβ deposition nor behavioral deficits associated with cerebral amyloid burden. Finally, a conditional BIN1 knockout in excitatory neurons did not alter BACE1, APP, C-terminal fragments derived from BACE1 cleavage of APP, or endogenous Aβ levels. These results indicate that BIN1 function does not regulate Aβ generation in vivo.
Keywords: Alzheimer disease, amyloid precursor protein (APP), beta-amyloid (AB), beta-secretase 1 (BACE1), pathogenesis, 5XFAD, APP, BACE1, BIN1, GWAS, amphiphysin, neurodegeneration, endocytosis, secretase, amyloid
Introduction
Overwhelming evidence places the generation and eventual deposition of the β-amyloid peptide in the brain parenchyma as a key pathological driver of Alzheimer's disease (AD)2 (1). Targeting its production, which occurs through the sequential proteolysis of the amyloid precursor protein (APP) by the β-site APP cleaving enzyme 1 (BACE1) and γ-secretase complex, remains an important therapeutic target in the Alzheimer's disease field. Although early onset, familial forms of AD are driven by excess Aβ accumulation as a result of mutations in the vicinity of the Aβ coding region of the APP gene or mutations in genes encoding the catalytic subunits of the γ-secretase complex, the molecular mechanisms that contribute to aberrant Aβ production and accumulation in late-onset AD (LOAD) remain largely unknown.
Recent genome-wide association studies (GWAS) have identified a range of genetic loci that are linked to LOAD risk, implicating several molecular mechanisms through which genetic LOAD risk may be conferred. The bridging integrator 1 (BIN1) gene was identified as the second most prevalent susceptibility locus for LOAD alongside several other genes with known functions in the regulation of endocytic trafficking including SORL1, PICALM, and CD2AP (2–4). BIN1 belongs to the Bin-Amphiphysin-Rvs (BAR) family of adaptor proteins which are implicated in the production of membrane curvature and endocytosis in various cellular functions (5, 6). BIN1 mRNA undergoes alternative splicing to produce multiple protein isoforms in a tissue- and cell-specific manner (7). Several BIN1 isoforms, including those expressed in the brain, contain a clathrin and AP-2 adaptor complex binding (CLAP) domain, which is involved in clathrin-mediated endocytosis (CME) (5, 8). The exact consequence of the LOAD-associated SNPs identified at the BIN1 locus remains unknown. Previous studies have reported conflicting findings on BIN1 protein expression levels in AD (9–12) and how it correlates with amyloid and tau pathology (9, 10, 13–15), whereas BIN1 mRNA levels have been shown to be altered in disease and correlate with age of disease onset and disease duration (16).
Endocytic dysfunction has been described in AD (17), and numerous in vitro studies highlight the importance of correct endosomal regulation in the spatial separation of APP and BACE1, which significantly impacts Aβ production. Following maturation in the constitutive secretory pathway, APP reaches the cell surface. After internalization from the surface, APP is sorted to early endosomes and can be recycled or targeted for lysosomal degradation (18, 19). The acidic pH of the early endosomes provides optimal conditions for BACE1 proteolytic activity, and the majority of amyloidogenic proteolysis of APP and Aβ generation is thought to occur in this subcellular environment (20, 21). Although many of these trafficking pathways have been extensively studied in immortalized cells lines, they fail to consider the additional complexities of APP and BACE1 trafficking in neurons and in vivo (22–25). In vitro evidence exists for a role for BIN1 in the regulation of the membrane trafficking of BACE1 (26, 27), but a role for BIN1 in this process in vivo is yet to be demonstrated. Here, we report that a 50% reduction in BIN1 expression does not alter BACE1 or APP localization or endogenous Aβ production in WT mice. Similarly, 50% reduction of BIN1 did not modify amyloid pathology and behavioral deficits in a mouse model of AD. Furthermore, neuron-specific conditional knockout (cKO) of Bin1 did not alter BACE1 or APP localization to membrane fractions generated in the mouse brain or affect Aβ production.
Results
A 50% global loss of Bin1 does not alter APP or BACE1 expression or localization or Aβ production
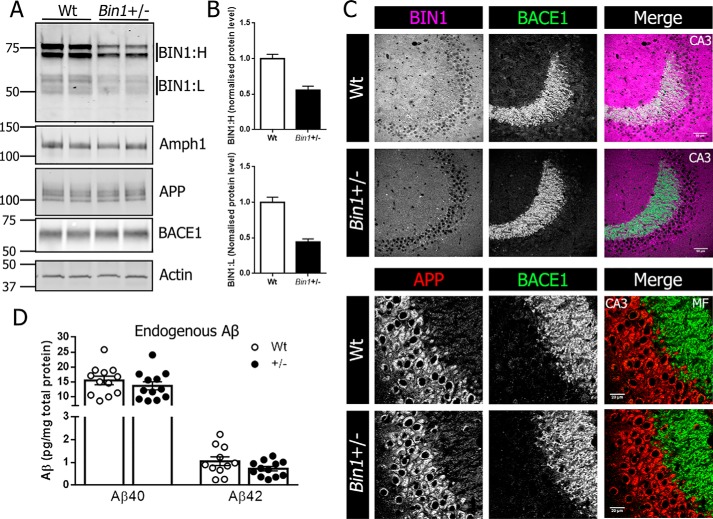
The role of BIN1 in the brain remains largely unknown, but several lines of evidence indicate its expression in a range of brain cell subtypes (11, 12, 28). Furthermore, evidence exists for noncell-autonomous deposition of Aβ in the absence of PSEN1 expression in excitatory neurons (29). To explore the possible contribution of Bin1 expression in Aβ production, we sought to generate a model of global Bin1 knockdown. Because homozygous KO of Bin1 in mice results in perinatal lethality due to severe cardiomyopathy (30), we generated mice with germline deletion of a single Bin1 allele by crossing male Bin1Fl/Fl:Syn-Cre mice to female C57BL/6 mice. Synapsin expression in the testes causes recombination of the floxed gene in the sperm (31) resulting in progeny with an inherited recombined floxed allele. The resulting progeny had a global single-allele deletion of Bin1 (Bin1+/−), and pups were viable and had no obvious phenotypic abnormalities. Loss of a single Bin1 allele caused a 50% reduction in forebrain BIN1 levels in Bin1+/− mice compared with WT littermate controls, but no significant differences were observed in the steady-state levels of APP, BACE1, or the BIN1 paralog amphiphysin 1 (Amph1) (Fig. 1A). Quantification showed a 45% reduction in high molecular weight BIN1 isoforms (BIN1:H) and 55% reduction in low molecular weight BIN1 isoforms (BIN1:L) (Fig. 1B). Prominent BACE1 localization in hippocampal mossy fibers has been widely reported, with its absence in BACE1 KO mice resulting in disorganized infrapyramidal mossy fiber bundles (32). The 50% reduction of BIN1 in Bin1+/− mice did not cause any overt mislocalization of BACE1 or APP with similar BACE1 localization of BACE1 to hippocampal mossy fibers and APP to the CA3 neuronal cell layer in WT and Bin1+/− mice (Fig. 1C). To assess whether the diminution in BIN1 expression affected amyloidogenic processing of APP, we quantified endogenous Aβ levels. Analysis of homogenates from 4-month-old WT and Bin1+/− mice showed no significant differences in the steady-state levels of Aβ40 or Aβ42 (Fig. 1D).
Figure 1.
Single allele deletion of Bin1 does not alter APP or BACE localization or Aβ levels. A, forebrain homogenates from 4-month-old Bin1+/− mice and WT littermate controls were analyzed by immunoblotting for BIN1, APP, BACE1, and Amph1 levels. B, quantitative analysis of BIN1:H (top) and BIN1:L (bottom) in forebrain lysates from WT and Bin1+/− mice. C, top panel, immunofluorescent staining for BIN1 (magenta) and BACE1 (green) in the hippocampal CA3 region from WT and Bin1+/− mice. Scale bar, 50 μm. Bottom panel, higher magnification of immunofluorescent staining for APP (red) and BACE1 (green) in CA3 neurons and mossy fibers (MF) of the mouse hippocampus. Scale bar, 20 μm. D, forebrain lysates from WT and Bin1+/− mice were analyzed for steady-state levels of endogenous Aβ40 and Aβ42 using a V-PLEX 4G8 immunoassay (Aβ40, n = 12 per genotype; Aβ42, n = 11 WT, and 12 Bin1+/−).
Reduction of BIN1 expression does not alter cerebral amyloid burden in 5XFAD mice
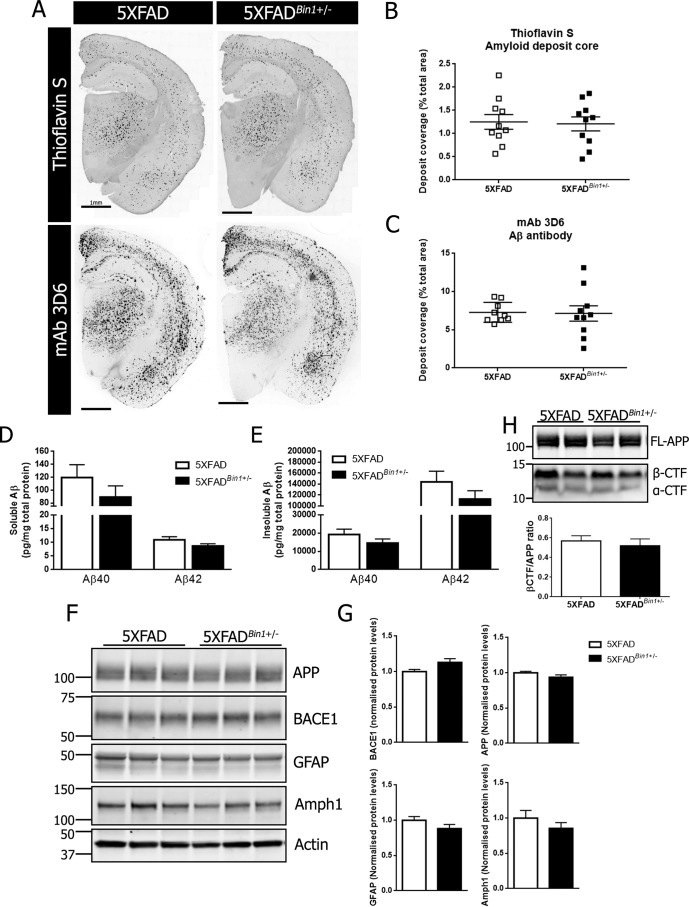
To determine whether a 50% reduction in BIN1 expression influenced cerebral amyloid burden, we crossed Bin1+/− mice to the 5XFAD line to generate 5XFAD mice with a single Bin1 allele deletion (5XFADBin1+/−) and littermates with normal BIN1 levels (5XFAD). The cerebral amyloid burden was assessed in females (Fig. 2A) and males (Fig. S2A) at 4 months of age with mAb 3D6 and thioflavin S staining in the hemibrain sections and in smaller hippocampal and cortex regions. It has previously been reported that in the 5XFAD line, females develop pathology at an earlier time point than males because of the presence of an estrogen response element in the transgene Thy-1 promoter (33, 34) and thus only mice of the same sex were directly compared. Quantitative analysis of hemibrains from female 5XFAD and 5XFADBin1+/− mice revealed no significant difference in the area covered with thioflavin S–positive fibrillary Aβ deposits (1.25 ± 0.161% in 5XFAD versus 1.208 ± 0.151% in 5XFADBin1+/−) (Fig. 2B). Similarly, no significant difference was observed in the area covered by mAb 3D6–positive deposits (7.29 ± 0.437% in 5XFAD versus 7.14 ± 0.998% in 5XFADBin1+/−) (Fig. 2C). Separate regional analysis of the hippocampus and cortex showed no significant difference in deposit coverage with thioflavin S (Fig. S1A) or mAb 3D6 immunofluorescence (Fig. S1B). A small but statistically significant difference was observed in average mAb 3D6 deposit size between female 5XFAD and 5XFADBin1+/− mice (931.7 ± 62.21 μm2 versus 746 ± 32.42 μm2) (Fig. S1C). Consistent with previous reports, male 5XFAD mice showed reduced amyloid burden as compared with female littermates. However, no significant differences were observed in the amyloid burden of male 5XFAD and 5XFADBin1+/− mice when thioflavin S– (0.68 ± 0.089% versus 0.53 ± 0.111%) (Fig. S2B) or mAb 3D6–positive deposits were quantified (5.79 ± 0.73% versus 4.22 ± 0.84%) (Fig. S2C) in the whole hemisphere or in hippocampal or cortex subregions. ELISA analysis of homogenates from female mice revealed no significant differences in soluble Aβ40 (119.6 ± 19.9 pg/mg in 5XFAD versus 89.24 ± 17.37 pg/mg in 5XFADBin1+/−) or Aβ42 levels (10.95 ± 1.143 pg/mg in 5XFAD versus 8.66 ± 0.78 pg/mg in 5XFADBin1+/−) (Fig. 2D). Similarly, no significant difference was observed in soluble Aβ40 (69.05 ± 15.75 pg/mg in 5XFAD versus 60.67 ± 7.894 pg/mg in 5XFADBin1+/−) or Aβ42 (9.28 ± 1.418 pg/mg in 5XFAD versus 8.592 ± 0.7668 in 5XFADBin1+/−) in male cohorts (Fig. S2D). Levels of soluble Aβ38 were below the detection threshold for the assay employed. Furthermore, no significant differences were observed in insoluble Aβ40 (19,194 ± 2954 pg/mg in 5XFAD versus 14,518 ± 2339 pg/mg in 5XFADBin1+/−) or in Aβ42 (14,4273 ± 19,325 pg/mg in 5XFAD versus 112,617 ± 15,263 in 5XFADBin1+/−) in female mice (Fig. 2E). Similarly, no significant differences were observed in insoluble Aβ40 (7115 ± 1040 pg/mg in 5XFAD versus 7046 ± 1353 in 5XFADBin1+/−) or Aβ42 (62,495 ± 8385 pg/mg in 5XFAD versus 66,481 ± 10,572 in 5XFADBin1+/−) in male mice (Fig. S2D). Forebrain lysates from female (Fig. 2F) or male (Fig. S2E) mice were subjected to immunoblot analysis for APP, BACE1, GFAP, and Amph1. No significant differences were observed in protein levels when comparing female 5XFAD and 5XFADBin1+/− mice (Fig. 2G). A statistically significant 16% increase in BACE1 protein levels was observed in male 5XFADBin1+/− compared with 5XFAD controls, but no significant change in APP, Amph1, or GFAP was observed (Fig. S2F). Similarly, no significant differences were observed in the levels of C-terminal fragments derived from BACE1 cleavage of APP (β-CTF) in female (Fig. 2H) or male (Fig. S2G) mice.
Figure 2.
Reduction of BIN1 expression does not alter amyloid deposition in female 5XFAD mice. A, representative images of Aβ deposit core staining with thioflavin S and mAb 3D6 immunostaining in 4-month-old female 5XFAD and 5XFADBin1+/− mice at 4 months of age. B, quantification of amyloid burden identified by thioflavin S staining (n = 10 per genotype). C, quantification of amyloid burden identified by mAb 3D6 immunostaining (n = 9 5XFAD and 10 5XFADBin1+/−). Forebrain tissue from 4-month-old female 5XFAD and 5XFADBin1+/− mice was sequentially extracted in TBS and formic acid to generate soluble and insoluble Aβ fractions. D and E, the levels of soluble and insoluble Aβ40 and Aβ42 were measured by V-PLEX 6E10 immunoassay (n = 10 per genotype). F, immunoblot analysis of APP, BACE1, GFAP, and Amph1 levels in 5XFAD and 5XFADBin1+/− mice. G, quantitative analysis of the levels of BACE1, APP, GFAP, and Amph1, normalized to actin (n = 8 per genotype). H, immunoblot analysis of full-length APP (FL-APP) and APP C-terminal fragments (CTF) and quantification of β-CTF normalized to FL-APP (n = 4 per genotype).
50% reduction in BIN1 does not modify the behavior in 5XFAD mice
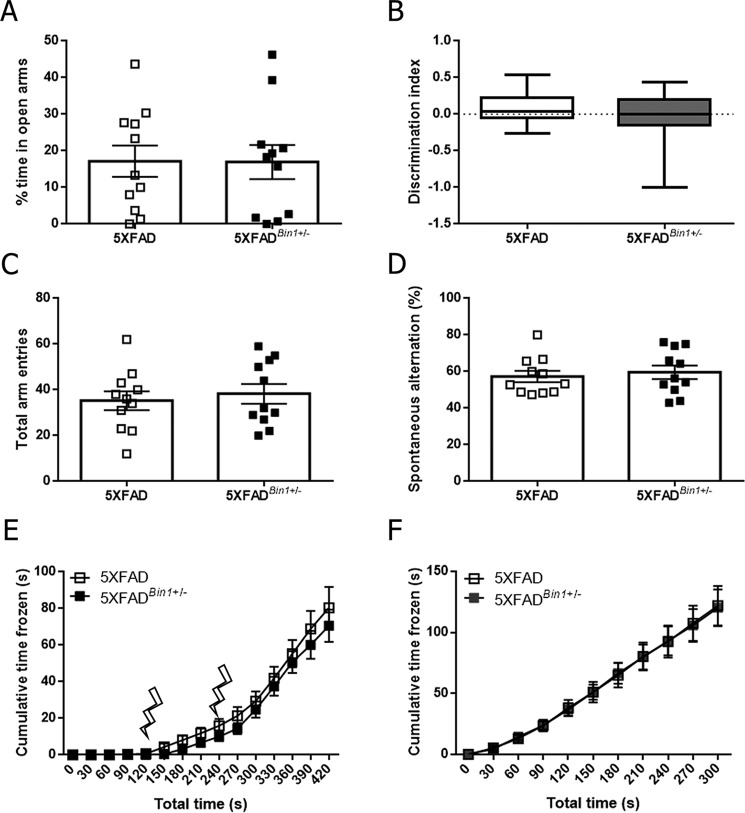
We performed behavioral tests in 5XFAD and 5XFADBin1+/−mice to determine whether the partial loss of BIN1 expression influenced the previously reported cognitive dysfunctions in the 5XFAD line in the absence of a change of cerebral amyloid burden. In the elevated-plus maze, designed to test anxiety phenotypes, no significant differences between the groups were observed in the percentage of time spent in open arms of the maze in female (Fig. 3A) or male mice (Fig. S3A). Novel object recognition, Y-maze, and contextual fear conditioning paradigms were used to test cognitive deficits in 5XFAD and 5XFADBin1+/− mice. No significant differences were observed in the discrimination index in female (Fig. 3B) or male cohorts (Fig. S3B). Similarly, in the Y-maze test, no significant difference in the number of arm entries or spontaneous alternation was observed in female (Fig. 3, C and D) or male mice (Fig. S3, C and D). In contextual fear conditioning, response to the initial shock was not significantly different between genotypes in female (Fig. 3E) or male (Fig. S3E) mice, nor was freezing behavior 24 h later upon reintroduction to the fear conditioning chamber (Fig. 3F and Fig. S3F). Thus, the partial loss of BIN1 expression in 5XFAD mice does not influence the anxiety or memory performance in behavior tests.
Figure 3.
Reduction in BIN1 expression does not alter behavioral deficits in the absence of changes in amyloid burden in female 5XFAD mice. A, the percentage of time in the open arms of the elevated-plus maze was quantified for 4-month-old 5XFAD and 5XFADBin1+/− mice. B, discrimination index score from novel object recognition for 5XFAD and 5XFADBin1+/− mice. Box plots show the median, maximum, and minimum values. C, the number of arm entries over the 8-min Y-maze trial. D, the number of spontaneous alternations in the Y-maze over the 8-min trial period. E, freezing behavior in female 5XFAD and 5XFADBin1+/− mice during day 1 of fear conditioning. Lightning bolts represent delivery of the shock stimulus. F, freezing behavior in female 5XFAD and 5XFADBin1+/− mice on day 2 of fear conditioning, 24 h after shock application (n = 11 for both genotypes in all analyses).
Neuronal conditional knockout of BIN1 does not alter presynaptic APP and BACE1 localization or endogenous Aβ generation
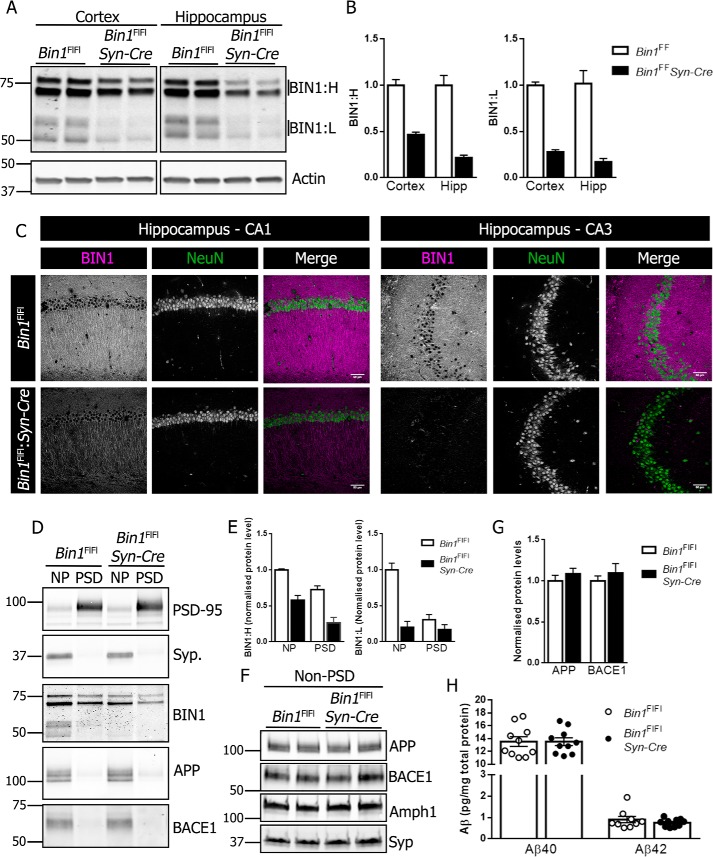
Previous reports showed a role for BIN1 in Aβ generation and BACE1 trafficking in primary neuronal cultures (26, 27). However, it was also reported that the knockdown or the overexpression of BIN1 in human neuroblastoma cells had no effect on APP processing (9). To test whether the select loss of BIN1 expression in neurons influences amyloidogenic processing of APP in vivo, we used the Synapsin1-Cre driver to generate neuronal Bin1 cKO mice (Bin1Fl/Fl:Syn-Cre). By crossing female Bin1Fl/Fl:Syn-Cre to male Bin1Fl/Fl mice, we avoided germline Bin1 deletion encountered in offspring generated with male Syn-Cre breeders (31). The cKO mice showed a significant reduction in BIN1 levels in the cortex and hippocampus as compared with littermate controls (Fig. 4A). Quantification showed greater than 50% reduction in both high and low molecular weight BIN1 isoforms (Fig. 4B). To visualize BIN1 expression, we performed immunohistochemical staining using BIN1 mAb 13463 in the absence of epitope retrieval of tissue sections, which preferentially labels neuronal BIN1 (Fig. S4). As expected, a marked diminution of neuronal BIN1 immunostaining could be observed in the hippocampus of Syn-Cre Bin1 cKO mice (Fig. 4C).
Figure 4.
Characterization of neuronal conditional Bin1 knockout. A, brain homogenates from the hippocampus and cortex of 4-month-old Bin1Fl/Fl and Bin1Fl/Fl:Syn-Cre were analyzed for BIN1 expression by immunoblotting. B, quantification of BIN1:H and BIN1:L in the cortex and hippocampus (Hipp) (n = 6 Bin1Fl/Fl and 4 Bin1Fl/Fl:Syn-Cre). C, immunohistochemical analysis of BIN1 and NeuN expression in the CA1 (left panel) and CA3 (right panel) of Bin1Fl/Fl and Bin1Fl/Fl:Syn-Cre mice. D, PSD and non-PSD (NP) membrane fractions were analyzed for PSD-95, synaptophysin, BIN1, APP, and BACE1 by immunoblotting. E, quantitative analysis of BIN1:H and BIN1:L in crude pre-and post-synaptic membranes (n = 4). F, non-PSD membranes were analyzed for APP, BACE1, Amph1, and synaptophysin by immunoblotting. G, quantitative analysis of APP and BACE1 levels in pre-synaptic fractions (n = 4). H, forebrain lysates were analyzed for steady-state levels of Aβ40 and Aβ42 using V-PLEX 4G8 immunoassay (Aβ40, n = 10 per genotype; Aβ42, n = 9 Bin1Fl/Fl and 10 Bin1Fl/Fl:Syn-Cre).
Synaptic activity has previously been shown to regulate Aβ production in vivo (35, 36), and both APP and BACE1 have been shown to localize at the synapse (24, 37, 38). BIN1, APP, and BACE1 were readily detectable in non–post-synaptic density (PSD) membrane fractions isolated from the mouse brain alongside enrichment of the synaptic marker synaptophysin (Fig. 4D). Levels of BIN1 in non-PSD and PSD fractions were significantly reduced in Bin1 cKO mice compared with controls (Fig. 4E). Non-PSD fractions were analyzed by immunobloting for APP, BACE1, synaptophysin, and Amph1 (Fig. 4F). The levels of APP and BACE1 remained unaltered in non-PSD fractions from cKO mice as compared with littermate controls (Fig. 4G). Finally, we ascertained whether the processing of endogenous APP was altered by the loss of neuronal BIN1 expression in the cKO mice. To this end, we performed ELISA on forebrain homogenates from 4-month-old Bin1Fl/Fl and Bin1Fl/Fl:Syn-Cre mice to measure the levels of endogenous Aβ40 or Aβ42. No significant differences were observed in the steady-state levels of Aβ40 or Aβ42 (Fig. 4H).
Discussion
In this study, we investigated a role for the Alzheimer's disease risk gene BIN1, in the development of amyloid pathology in a mouse model of Alzheimer's disease. Deletion of a single Bin1 allele resulted in the loss of 50% of endogenous BIN1 protein expression but did not alter BACE1 or APP levels and their localization or endogenous Aβ levels in the brain. In addition, 50% loss of BIN1 neither altered the steady-state Aβ levels and cerebral Aβ deposition nor influenced the behavioral outcomes in a widely used mouse model of Alzheimer's disease amyloid pathogenesis. Similar to the global reduction of BIN1 in heterozygous Bin1 KO mice, pan-neuronal conditional knockout of BIN1 resulted in 50% reduction of BIN1 levels in the cortex and hippocampus but did not alter BACE1 or APP levels in pre-synaptic fractions of mouse brains or endogenous Aβ levels. These results indicate the LOAD genetic risk associated with the BIN1 loci is unlikely mediated through a direct role in amyloid production in vivo.
GWAS over the past decade have advanced the field by the identification of several risk loci for LOAD. However, defining the molecular mechanisms through which GWAS genes contribute to LOAD risk remains a daunting task. Notably, two decades after the discovery that APOE is a major genetic risk factor, novel insights on how APOE modifies molecular and cellular phenotypes relevant to AD pathophysiology continue to emerge. Nevertheless, the investigations on APOE and, more recently LOAD-associated TREM2 mutations, have given invaluable insights into how genetic factors can contribute to disease risk and reveal potential pathways for disease modification or intervention (39). Despite its being the second most prevalent susceptibility locus for LOAD, little is known about the function of BIN1 in disease pathogenesis. Recent evidence implicates a reduction in BIN1 in the increased trans-synaptic transfer of extracellular tau in vitro through a mechanism involving endocytosis (40). A physical interaction between BIN1 and tau in neuronal cytosol has also been demonstrated (13, 41), although it is not clear whether this interaction in any way affects the endocytic function of BIN1.
Emerging evidence has also implicated BIN1 in the process of Aβ generation, namely through a role in endocytic trafficking of BACE1. The loss of Bin1 expression was shown in one study to increase BACE1 levels and Aβ production in neurons, with the subsequent analysis in immortalized cells lines implicating loss of endosomal trafficking and lysosomal degradation of BACE1 as the cause (26). Although we saw a small increase in BACE1 levels in the forebrain lysates of 5XFADBin1+/− male mice, this was not the case in female mice. Nevertheless, the loss of one Bin1 allele did not cause a measurable change in Aβ deposition in males or females. A second study showed a role for BIN1 in tubule scission and BACE1 exit from early endosomes, with a loss of BIN1 resulting in increased intracellular Aβ but no change in Aβ secretion (27). A lack of an effect of BIN1 knockdown or overexpression on APP processing and Aβ secretion was also reported in a study that used human neuroblastoma cells (9). In our study, we analyzed the pathology in 5XFAD mice at 4 months of age in both males and females to quantify pathogenesis before the brain gets overloaded with amyloid burden. It would be interesting to determine whether the loss of BIN1 expression influences BACE1 levels in older animals.
In addition to BIN1, other genes including SORL1, PICALM, and CD2AP have implicated endosomal dysfunction in AD. Indeed, studies have shown a role for PICALM in CME of γ-secretase and subsequent Aβ production (42) and SORLA in the intracellular trafficking and processing of APP (43). Although alterations in SORLA levels translated into changes in Aβ in both WT mice and mouse models of AD amyloidosis (44–46), our results using heterozygous KO and cKO models do not validate a similar translation from in vitro to in vivo data with the reduction of BIN1 expression. Nevertheless, the effect of BIN1 loss on intracellular Aβ accumulation identified by Ubelmann et al. (27) was rescued by the re-introduction of the neuronal BIN1 isoform but not the ubiquitous isoform, implicating potential isoform-specific differences in BIN1 function in the brain which may require further investigation in light of differential BIN1 isoform expression in AD (9–11). Moreover, given reports of cell-specific BIN1 isoform expression, more in-depth analysis of cell-autonomous roles for BIN1 in the context of amyloid deposition would also seem pertinent.
Mice with targeted deletion of both Bin1 alleles are not viable as the pups do not survive beyond a day or two after the birth (30). Our choice to use a global BIN1 heterozygous knockout, and a neuronal cKO circumvents the neonatal lethality of Bin1 KO mice and still models the reduction of neuronal BIN1 isoforms reported in patients with sporadic AD (9–11). The presence of the CLAP domain in longer brain-specific BIN1 isoforms predicts an important role in CME. Indeed, a role for BIN1 in endocytosis at the synapse has been proposed based on the documented role for amphiphysin 1 in synaptic vesicle recycling and dynamics (5, 8). However, previous studies did not reveal alterations in the density of synaptic vesicles in cultures of neurons from Bin1 knockout mice (30). BIN1 has also been identified among the 50 most highly expressed genes in oligodendrocytes and shows prominent expression in oligodendrocytes and white matter tracts in mouse and human brain tissue (11, 12, 47). White matter dysfunction is emerging as an important element of Alzheimer's disease pathogenesis (48), with evidence of a direct relationship between Aβ levels and white matter hyperintensities (49). Cause and effect relationship between Aβ and white matter dysfunction is yet to be fully understood, but evidence exists for white matter dysfunction in autosomal dominantly inherited forms of AD which precedes symptoms by 20 years (48). Demyelination of axons in the central nervous system is thought to leave them more vulnerable to Aβ toxicity. Interestingly, axonal damage in close proximity to amyloid deposits is a key feature of the 5XFAD mouse model (50). Furthermore, myelin deficits have been identified in the 5XFAD mouse model and some evidence exists for a role of myelin in amyloid deposition (51, 52). Despite high expression in oligodendrocytes, a definitive role for BIN1 in myelination is yet to be established, but our data indicate 50% loss of BIN1 expression does not recapitulate previously reported data for a role of myelin in amyloid deposition.
Despite an emerging body of data regarding the possible role of BIN1 in LOAD risk, a consensus on the mechanism through which its risk is conferred is yet to be reached. Our data do not support a role for BIN1 in Aβ generation in vivo despite evidence from some in vitro studies to suggest the opposite. Future investigation using Bin1 neuronal cKO in the 5XFAD model may provide additional information regarding a definitive role for BIN1 in neuronal amyloid production and deposition. Further understanding of the biological role of BIN1 in the brain, understanding the consequences of BIN1 SNPs on cellular BIN1 protein isoform abundance or subsequent function in select neuronal cell types remain essential pieces of information in understanding the role of BIN1 in LOAD disease progression.
Experimental procedures
Mice
Bin1Fl/Fl mice were generously provided by Dr. George C. Prendergast (Lankenau Institute for Medical Research) (53). Synapsin1-Cre line (Syn-Cre; stock no. 003966) was obtained from The Jackson Laboratory (Bar Harbor, ME). Bin1Fl/Fl:Syn-Cre and Bin1± mice were generated in-house and maintained in the C57BL/6 background. Male Bin1Fl/Fl:Syn-Cre mice were bred to female C57BL/6 mice to generate progeny with germline deletion of a single Bin1 allele. Bin1+/− mice were intercrossed, and pups sacrificed at postnatal day zero (P0) to confirm the generation of Bin1+/+, Bin1+/−, and Bin1−/− using PCR genotyping and immunoblot analyses. Bin1+/− mice were bred to 5XFAD animals (C57BL/6-SJL mixed background). All procedures related to animal care and treatment in this study have been approved by the Institutional Animal Care and Use Committee at the University of Chicago.
Antibodies
The following antibodies were used: mouse anti-Aβ mAb 3D6 (a kind gift from the late Dale Schenk, Elan Corporation PLC, South San Francisco, CA), anti–β-actin mAb (660099-1, Proteintech), anti–amphiphysin 1 mAb 8 (sc-21710, Santa Cruz Biotechnology), rabbit anti-APP C-terminal pAb CTM1 (54), pAb Y188 (ab32136, Abcam), rabbit anti-BACE1 mAb (EPR3956, Abcam), anti-BACE1 human IgG (a kind gift from Jasvinder Atwal, Genentech, South San Francisco, CA), rabbit anti-BIN1 pAb (14647-1, Proteintech), rabbit anti-BIN1 mAb EPR13463-25 (ab185950, Abcam), mouse anti-GFAP mAb (G3893, Sigma), mouse anti-NeuN mAb (MAB377, Millipore), mouse anti-PSD-95 monoclonal (P78352, NeuroMab, UC Davis), and mouse anti-synaptophysin mAb SVP38 (sc-12737, Santa Cruz Biotechnology).
Immunoblot analysis
Detergent lysates of mouse brain were prepared as described previously (55). The samples were resolved by SDS-PAGE on 4–20% Tris-glycine gels (Bio-Rad). APP-derived C-terminal fragments were resolved on NuPAGE 4–12% Bis-Tris protein gels (Thermo Fisher). Proteins were transferred to PVDF membranes and developed with IR680 anti-rabbit and IR800 anti-mouse secondary antibodies and imaged using the Odyssey IR Imaging System (LI-COR Biosciences). Densitometric analysis was performed using ImageJ.
Membrane fractionations
Post-synaptic and non–post-synaptic membrane fractions were purified from neuronal cKO mice using an adapted methodology (56). Briefly, forebrains were homogenized in 0.32 m sucrose and 10 mm HEPES, pH 7.4, and centrifuged at 800 × g for 10 min to pellet nuclei and large debris. The supernatant was centrifuged at 12,000 × g for 20 min to pellet a crude membrane fraction. The pellet was washed twice in wash buffer (4 mm HEPES and 1 mm EDTA, pH 7.4), resuspended in 20 mm HEPES, 100 mm NaCl, 0.5% Triton X-100, pH 7.2, and held on ice for 30 min. The suspension was centrifuged at 12,000 × g for 20 min to separate pre-synaptic proteins. The pellet was resuspended in 20 mm HEPES, 0.15 mm NaCl, 1% Triton X-100, 1% deoxycholic acid, and 1% SDS, pH 7.5, and held on ice for 1 h. The suspension was centrifuged at 800 × g for 10 min, and the supernatant saved as the post-synaptic fraction.
Histology and immunohistochemistry
For free-floating sections, hemibrains were processed as described previously and serially cut into 40-μm-thick sections (57). Sections were stained with primary antibodies described above and with secondary antibodies conjugated to Alexa Fluor 488, 555, or 647 (Life Technologies). Fibrillar amyloid deposits were stained using thioflavin S as described previously (58). Stained sections were imaged on a CRi Panoramic Scan Whole Slide Scanner (Cambridge Research and Instrumentation) using a 40× (0.95 N.A.) long-distance working Zeiss objective and Allied Vision Technologies Stingray F146C, 4.6-μm pixel size color camera. Confocal images were acquired on a Leica SP5 II STED-CW Super-resolution microscope with 20× (N.A. 0.8) and 40× (N.A. 0.8) objectives.
Image analysis
For amyloid burden quantification (thioflavin S and mAb 3D6), five to eight sections per mouse were analyzed using ImageJ as described previously (58).
Aβ ELISA
For endogenous Aβ ELISA, forebrain tissue from 4-month-old Bin1Fl/Fl, Bin1Fl/Fl:Syn-Cre, WT or Bin1+/− mice were homogenized in 10 volumes (w/v) of 1% SDS in PBS containing cOmplete™, EDTA-free Protease Inhibitor mixture (Sigma-Aldrich) and clarified by centrifugation. Aβ levels were measured using a V-PLEX Plus Aβ Peptide Panel 1 (4G8) Kit (Meso Scale Discovery). For human Aβ, brains from 4-month-old 5XFAD and 5XFADBin1+/− mice were homogenized in 20 volumes (w/v) of TBS containing protease inhibitor mixture and clarified by centrifugation at 100,000 × g for 1 h at 4 °C. The pellet was resuspended 700 μl of 70% formic acid and centrifuged at 100,000 × g for 1 h at 4 °C. The supernatant was neutralized with 9 volumes (v/v) of 1 m Tris-HCl, pH 11. TBS-soluble and formic acid–extracted Aβ was measured using a V-PLEX Plus Aβ Peptide Panel 1 (6E10) Kit (Meso Scale Discovery) according to the manufacturer's instructions. Samples were subjected to the bicinchoninic acid assay to determine protein concentration and Aβ concentrations adjusted to total protein concentrations.
Behavioral analyses
Elevated-plus maze
Mice were placed in the closed arm of an elevated-plus maze suspended 40 cm above the ground and allowed to explore freely for 8 min. The two opposite closed arms of the Greek cross-shaped maze had 15 cm high walls surrounding a 5-cm-wide platform and open arms had the same 5-cm-wide platform with no surrounding walls. Time spent in closed and open arms was monitored over the duration of the exploratory period.
Novel object recognition
Mice were introduced to the empty novel object recognition (NOR) arena for an 8-min session on day 1 of testing for habituation to the novel environment. After 24 h, mice were returned to the arena which contained two identical novel objects and allowed to explore freely for 8 min. After a further 24 h, mice were returned to the arena containing one familiar object from the previous day and one novel object. Time spent interacting with the novel and familiar objects was recorded and used to generate a discrimination index using the calculation (novel object(s) − familiar object(s))/(novel object(s) + familiar object(s)) (59).
Y-maze
4-month-old mice were allowed to freely explore the arms of a Y-maze for an 8-min session. The total number of arms entries (counted when all four limbs had entered the arm of the maze) and the sequence in which arms were entered were recorded. The percentage of spontaneous alternations was calculated and analyzed as described previously (58).
Contextual fear conditioning
Contextual fear conditioning was performed on 4-month-old mice in a Coulbourn HABITEST isolation cubicle with shocks applied using a Coulbourn Precision Animal Shocker (Harvard Apparatus) as described (58). Analysis of recordings from the video-based system was performed using FreezeFrame 4 software (Actimetrics) as described previously (58).
Statistical analysis
All statistical analyses were performed using GraphPad Prism Version 7 (GraphPad Software) with data represented as the mean ± S.E. unless described otherwise in the figure legend. Comparisons between two groups were performed using unpaired t tests. Cumulative time frozen in the contextual fear conditioning paradigm was analyzed by two-way analysis of variance (ANOVA).
Author contributions
R. J. A., P. D. R., and G. T. conceptualization; R. J. A., P. D. R., and P. N. data curation; R. J. A., P. D. R., P. N., R. C. R., and G. T. formal analysis; R. J. A., S. L. W., and G. T. supervision; R. J. A., P. D. R., and G. T. funding acquisition; R. J. A. validation; R. J. A., P. D. R., P. N., H. R. K., A. J. R., and T. G. investigation; R. J. A. visualization; R. J. A., P. D. R., S. V. K., and R. C. R. methodology; R. J. A. writing-original draft; R. J. A. and G. T. project administration; R. J. A., P. D. R., and G. T. writing-review and editing; S. V. K., L. L.-K., S. L. W., and G. T. resources.
Supplementary Material
Acknowledgments
We thank the University of Chicago Grossman Institute for Neuroscience for shared equipment support. Confocal imaging was performed at the Integrated Microscopy Core Facility at the University of Chicago (supported by grant S10OD010649).
This work was supported by National Institutes of Health Grants AG054223 and AG056061 (to G. T.), Cure Alzheimer's Fund (G. T. and S. L. W.), Alzheimer's Association (to P. D. R), Illinois Department of Public Health (to R. J. A.) and BrightFocus Foundation Fellowships (to P. D. R). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the United States government.
This article contains Figs. S1–S4.
- AD
- Alzheimer's disease
- Aβ
- β-amyloid
- APP
- amyloid precursor protein
- BACE1
- β-site APP cleaving enzyme 1
- BIN1
- bridging integrator 1
- cKO
- conditional KO
- CLAP
- clathrin and AP-2 adaptor complex binding
- CME
- clathrin-mediated endocytosis
- CTF
- C-terminal fragments
- GWAS
- genome-wide association study
- LOAD
- late-onset AD
- PSD
- post-synaptic density
- TBS
- Tris-buffered saline.
References
- 1. Musiek E. S., and Holtzman D. M. (2015) Three dimensions of the amyloid hypothesis: Time, space and “wingmen.” Nat. Neurosci. 18, 800–806 10.1038/nn.4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M. L., Pahwa J. S., Moskvina V., Dowzell K., Williams A., Jones N., Thomas C., Stretton A., Morgan A. R., Lovestone S., et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 41, 1088–1093 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naj A. C., Jun G., Beecham G. W., Wang L.-S., Vardarajan B. N., Buros J., Gallins P. J., Buxbaum J. D., Jarvik G. P., Crane P. K., Larson E. B., Bird T. D., Boeve B. F., Graff-Radford N. R., De Jager P. L., et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 43, 436–441 10.1038/ng.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seshadri S., Fitzpatrick A. L., Ikram M. A., DeStefano A. L., Gudnason V., Boada M., Bis J. C., Smith A. V., Carassquillo M. M., Lambert J. C., Harold D., Schrijvers E. M., Ramirez-Lorca R., Debette S., Longstreth W. T. Jr., et al. (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303, 1832–1840 10.1001/jama.2010.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prokic I., Cowling B. S., and Laporte J. (2014) Amphiphysin 2 (BIN1) in physiology and diseases. J. Mol. Med. 92, 453–463 10.1007/s00109-014-1138-1 [DOI] [PubMed] [Google Scholar]
- 6. Leprince C., Romero F., Cussac D., Vayssiere B., Berger R., Tavitian A., and Camonis J. H. (1997) A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J. Biol. Chem. 272, 15101–15105 10.1074/jbc.272.24.15101 [DOI] [PubMed] [Google Scholar]
- 7. Tan M.-S., Yu J.-T., and Tan L. (2013) Bridging integrator 1 (BIN1): Form, function, and Alzheimer's disease. Trends Mol. Med. 19, 594–603 10.1016/j.molmed.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 8. Wigge P., and McMahon H. T. (1998) The amphiphysin family of proteins and their role in endocytosis at the synapse. Trends Neurosci. 21, 339–344 10.1016/S0166-2236(98)01264-8 [DOI] [PubMed] [Google Scholar]
- 9. Glennon E. B., Whitehouse I. J., Miners J. S., Kehoe P. G., Love S., Kellett K. A., and Hooper N. M. (2013) BIN1 is decreased in sporadic but not familial Alzheimer's disease or in aging. PLoS One 8, e78806 10.1371/journal.pone.0078806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holler C. J., Davis P. R., Beckett T. L., Platt T. L., Webb R. L., Head E., and Murphy M. P. (2014) Bridging integrator 1 (BIN1) protein expression increases in the Alzheimer's disease brain and correlates with neurofibrillary tangle pathology. J. Alzheimer's Dis. 42, 1221–1227 10.3233/JAD-132450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Rossi P., Buggia-Prévot V., Clayton B. L., Vasquez J. B., van Sanford C., Andrew R. J., Lesnick R., Botté A., Deyts C., Salem S., Rao E., Rice R. C., Parent A., Kar S., Popko B., Pytel P., Estus S., and Thinakaran G. (2016) Predominant expression of Alzheimer's disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol. Neurodegener. 11, 59 10.1186/s13024-016-0124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams S. L., Tilton K., Kozubek J. A., Seshadri S., and Delalle I. (2016) Subcellular changes in bridging integrator 1 protein expression in the cerebral cortex during the progression of Alzheimer disease pathology. J. Neuropathol Exp. Neurol. 75, 779–790 10.1093/jnen/nlw056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chapuis J., Hansmannel F., Gistelinck M., Mounier A., Van Cauwenberghe C., Kolen K. V., Geller F., Sottejeau Y., Harold D., Dourlen P., Grenier-Boley B., Kamatani Y., Delepine B., Demiautte F., Zelenika D., et al. (2013) Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234 10.1038/mp.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Rossi P., Buggia-Prevot V., Andrew R., Krause S., Woo E., Nelson P., Pytel P., and Thinakaran G. (2017) BIN1 localization is distinct from Tau tangles in Alzheimer's disease. Matters 10.19185/matters.201611000018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Rossi P., Andrew R. J., Musial T. F., Buggia-Prevot V., Xu G., Ponnusamy M., Ly H., Krause S. V., Rice R. C., de l'Estoile V., Valin T., Salem S., Despa F., Borchelt D. R., Bindokas V. P., Nicholson D. A., and Thinakaran G. (2018) Aberrant accrual of BIN1 near Alzheimer's disease amyloid deposits in transgenic models. Brain Pathol. 10.1111/bpa.12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karch C. M., Jeng A. T., Nowotny P., Cady J., Cruchaga C., and Goate A. M. (2012) Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PLoS One 7, e50976 10.1371/journal.pone.0050976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cataldo A. M., Peterhoff C. M., Troncoso J. C., Gomez-Isla T., Hyman B. T., and Nixon R. A. (2000) Endocytic pathway abnormalities precede amyloid β deposition in sporadic Alzheimer's disease and Down syndrome. Am. J. Pathol. 157, 277–286 10.1016/S0002-9440(10)64538-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thinakaran G., and Koo E. H. (2008) Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 283, 29615–29619 10.1074/jbc.R800019200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajendran L., and Annaert W. (2012) Membrane trafficking pathways in Alzheimer's disease. Traffic 13, 759–770 10.1111/j.1600-0854.2012.01332.x [DOI] [PubMed] [Google Scholar]
- 20. Koo E. H., and Squazzo S. L. (1994) Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J. Biol. Chem. 269, 17386–17389 [PubMed] [Google Scholar]
- 21. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., et al. (1999) Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 10.1126/science.286.5440.735 [DOI] [PubMed] [Google Scholar]
- 22. Tampellini D., Rahman N., Gallo E. F., Huang Z., Dumont M., Capetillo-Zarate E., Ma T., Zheng R., Lu B., Nanus D. M., Lin M. T., and Gouras G. K. (2009) Synaptic activity reduces intraneuronal Aβ, promotes APP transport to synapses, and protects against Aβ-related synaptic alterations. J. Neurosci. 29, 9704–9713 10.1523/JNEUROSCI.2292-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buggia-Prévot V., Fernandez C. G., Udayar V., Vetrivel K. S., Elie A., Roseman J., Sasse V. A., Lefkow M., Meckler X., Bhattacharyya S., George M., Kar S., Bindokas V. P., Parent A. T., Rajendran L., Band H., Vassar R., and Thinakaran G. (2013) A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer's disease Aβ production. Cell Rep. 5, 1552–1563 10.1016/j.celrep.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buggia-Prévot V., Fernandez C. G., Riordan S., Vetrivel K. S., Roseman J., Waters J., Bindokas V. P., Vassar R., and Thinakaran G. (2014) Axonal BACE1 dynamics and targeting in hippocampal neurons: A role for Rab11 GTPase. Mol. Neurodegener. 9, 1 10.1186/1750-1326-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Das U., Scott D. A., Ganguly A., Koo E. H., Tang Y., and Roy S. (2013) Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron 79, 447–460 10.1016/j.neuron.2013.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyagawa T., Ebinuma I., Morohashi Y., Hori Y., Young Chang M., Hattori H., Maehara T., Yokoshima S., Fukuyama T., Tsuji S., Iwatsubo T., Prendergast G. C., and Tomita T. (2016) BIN1 regulates BACE1 intracellular trafficking and amyloid-β production. Hum. Mol. Genet. 25, 2948–2958 10.1093/hmg/ddw146 [DOI] [PubMed] [Google Scholar]
- 27. Ubelmann F., Burrinha T., Salavessa L., Gomes R., Ferreira C., Moreno N., and Guimas Almeida C. (2017) Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep. 18, 102–122 10.15252/embr.201642738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen D. V., Hanson J. E., and Sheng M. (2018) Microglia in Alzheimer's disease. J. Cell Biol. 217, 459–472 10.1083/jcb.201709069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Veeraraghavalu K., Zhang C., Zhang X., Tanzi R. E., and Sisodia S. S. (2014) Age-dependent, non-cell-autonomous deposition of amyloid from synthesis of beta-amyloid by cells other than excitatory neurons. J. Neurosci. 34, 3668–3673 10.1523/JNEUROSCI.5079-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muller A. J., Baker J. F., DuHadaway J. B., Ge K., Farmer G., Donover P. S., Meade R., Reid C., Grzanna R., Roach A. H., Shah N., Soler A. P., and Prendergast G. C. (2003) Targeted disruption of the murine Bin1/amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol. Cell Biol. 23, 4295–4306 10.1128/MCB.23.12.4295-4306.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rempe D., Vangeison G., Hamilton J., Li Y., Jepson M., and Federoff H. J. (2006) Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis 44, 44–49 10.1002/gene.20183 [DOI] [PubMed] [Google Scholar]
- 32. Hitt B., Riordan S. M., Kukreja L., Eimer W. A., Rajapaksha T. W., and Vassar R. (2012) β-Site amyloid precursor protein (APP)-cleaving enzyme 1 (BACE1)-deficient mice exhibit a close homolog of L1 (CHL1) loss-of-function phenotype involving axon guidance defects. J. Biol. Chem. 287, 38408–38425 10.1074/jbc.M112.415505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oakley H., Cole S. L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., Berry R., and Vassar R. (2006) Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadleir K. R., Eimer W. A., Cole S. L., and Vassar R. (2015) Aβ reduction in BACE1 heterozygous null 5XFAD mice is associated with transgenic APP level. Mol. Neurodegener. 10, 1 10.1186/1750-1326-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cirrito J. R., Kang J. E., Lee J., Stewart F. R., Verges D. K., Silverio L. M., Bu G., Mennerick S., and Holtzman D. M. (2008) Endocytosis is required for synaptic activity-dependent release of amyloid-β in vivo. Neuron 58, 42–51 10.1016/j.neuron.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cirrito J. R., Yamada K. A., Finn M. B., Sloviter R. S., Bales K. R., May P. C., Schoepp D. D., Paul S. M., Mennerick S., and Holtzman D. M. (2005) Synaptic activity regulates interstitial fluid amyloid-β levels in vivo. Neuron 48, 913–922 10.1016/j.neuron.2005.10.028 [DOI] [PubMed] [Google Scholar]
- 37. Wang P., Yang G., Mosier D. R., Chang P., Zaidi T., Gong Y. D., Zhao N. M., Dominguez B., Lee K. F., Gan W. B., and Zheng H. (2005) Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-like protein 2. J. Neurosci. 25, 1219–1225 10.1523/JNEUROSCI.4660-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilhelm B. G., Mandad S., Truckenbrodt S., Kröhnert K., Schäfer C., Rammner B., Koo S. J., Classen G. A., Krauss M., Haucke V., Urlaub H., and Rizzoli S. O. (2014) Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 10.1126/science.1252884 [DOI] [PubMed] [Google Scholar]
- 39. Shi Y., and Holtzman D. M. (2018) Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat. Rev. Immunol. 18, 759–772 10.1038/s41577-018-0051-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Calafate S., Flavin W., Verstreken P., and Moechars D. (2016) Loss of Bin1 promotes the propagation of Tau pathology. Cell Rep. 17, 931–940 10.1016/j.celrep.2016.09.063 [DOI] [PubMed] [Google Scholar]
- 41. Malki I., Cantrelle F. X., Sottejeau Y., Lippens G., Lambert J. C., and Landrieu I. (2017) Regulation of the interaction between the neuronal BIN1 isoform 1 and Tau proteins—role of the SH3 domain. FEBS J. 284, 3218–3229 10.1111/febs.14185 [DOI] [PubMed] [Google Scholar]
- 42. Kanatsu K., Morohashi Y., Suzuki M., Kuroda H., Watanabe T., Tomita T., and Iwatsubo T. (2014) Decreased CALM expression reduces Aβ42 to total Aβ ratio through clathrin-mediated endocytosis of γ-secretase. Nat. Commun. 5, 3386 10.1038/ncomms4386 [DOI] [PubMed] [Google Scholar]
- 43. Andersen O. M., Rudolph I. M., and Willnow T. E. (2016) Risk factor SORL1: From genetic association to functional validation in Alzheimer's disease. Acta Neuropathol. 132, 653–665 10.1007/s00401-016-1615-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Andersen O. M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C. A., Breiderhoff T., Jansen P., Wu X., Bales K. R., Cappai R., Masters C. L., Gliemann J., Mufson E. J., Hyman B. T., Paul S. M., Nykjaer A., and Willnow T. E. (2005) Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 10.1073/pnas.0503689102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dodson S. E., Andersen O. M., Karmali V., Fritz J. J., Cheng D., Peng J., Levey A. I., Willnow T. E., and Lah J. J. (2008) Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: Evidence for a proximal role in Alzheimer's disease. J. Neurosci. 28, 12877–12886 10.1523/JNEUROSCI.4582-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rohe M., Carlo A. S., Breyhan H., Sporbert A., Militz D., Schmidt V., Wozny C., Harmeier A., Erdmann B., Bales K. R., Wolf S., Kempermann G., Paul S. M., Schmitz D., Bayer T. A., Willnow T. E., and Andersen O. M. (2008) Sortilin-related receptor with A-type repeats (SORLA) affects the amyloid precursor protein-dependent stimulation of ERK signaling and adult neurogenesis. J. Biol. Chem. 283, 14826–14834 10.1074/jbc.M710574200 [DOI] [PubMed] [Google Scholar]
- 47. Dugas J. C., Tai Y. C., Speed T. P., Ngai J., and Barres B. A. (2006) Functional genomic analysis of oligodendrocyte differentiation. J. Neurosci. 26, 10967–10983 10.1523/JNEUROSCI.2572-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nasrabady S. E., Rizvi B., Goldman J. E., and Brickman A. M. (2018) White matter changes in Alzheimer's disease: A focus on myelin and oligodendrocytes. Acta Neuropathol. Commun. 6, 22 10.1186/s40478-018-0515-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee S., Viqar F., Zimmerman M. E., Narkhede A., Tosto G., Benzinger T. L., Marcus D. S., Fagan A. M., Goate A., Fox N. C., Cairns N. J., Holtzman D. M., Buckles V., Ghetti B., McDade E., et al. (2016) White matter hyperintensities are a core feature of Alzheimer's disease: Evidence from the dominantly inherited Alzheimer network. Ann. Neurol. 79, 929–939 10.1002/ana.24647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sadleir K. R., Kandalepas P. C., Buggia-Prevot V., Nicholson D. A., Thinakaran G., and Vassar R. (2016) Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, BACE1 elevation, and increased Aβ generation in Alzheimer's disease. Acta. Neuropathol. 132, 235–256 10.1007/s00401-016-1558-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gu L., Wu D., Tang X., Qi X., Li X., Bai F., Chen X., Ren Q., and Zhang Z. (2018) Myelin changes at the early stage of 5XFAD mice. Brain Res. Bull. 137, 285–293 10.1016/j.brainresbull.2017.12.013 [DOI] [PubMed] [Google Scholar]
- 52. Ou-Yang M. H., and Van Nostrand W. E. (2013) The absence of myelin basic protein promotes neuroinflammation and reduces amyloid β-protein accumulation in Tg-5xFAD mice. J. Neuroinflammation 10, 134 10.1186/1742-2094-10-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang M. Y., Boulden J., Katz J. B., Wang L., Meyer T. J., Soler A. P., Muller A. J., and Prendergast G. C. (2007) Bin1 ablation increases susceptibility to cancer during aging, particularly lung cancer. Cancer Res. 67, 7605–7612 10.1158/0008-5472.CAN-07-1100 [DOI] [PubMed] [Google Scholar]
- 54. Cheng H., Vetrivel K. S., Drisdel R. C., Meckler X., Gong P., Leem J. Y., Li T., Carter M., Chen Y., Nguyen P., Iwatsubo T., Tomita T., Wong P. C., Green W. N., Kounnas M. Z., and Thinakaran G. (2009) S-palmitoylation of γ-secretase subunits nicastrin and APH-1. J. Biol. Chem. 284, 1373–1384 10.1074/jbc.M806380200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thinakaran G., Borchelt D. R., Lee M. K., Slunt H. H., Spitzer L., Kim G., Ratovitsky T., Davenport F., Nordstedt C., Seeger M., Hardy J., Levey A. I., Gandy S. E., Jenkins N. A., Copeland N. G., Price D. L., and Sisodia S. S. (1996) Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17, 181–190 10.1016/S0896-6273(00)80291-3 [DOI] [PubMed] [Google Scholar]
- 56. De Rossi P., Harde E., Dupuis J. P., Martin L., Chounlamountri N., Bardin M., Watrin C., Benetollo C., Pernet-Gallay K., Luhmann H. J., Honnorat J., Malleret G., Groc L., Acker-Palmer A., Salin P. A., and Meissirel C. (2016) A critical role for VEGF and VEGFR2 in NMDA receptor synaptic function and fear-related behavior. Mol. Psychiatry 21, 1768–1780 10.1038/mp.2015.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gong P., Roseman J., Fernandez C. G., Vetrivel K. S., Bindokas V. P., Zitzow L. A., Kar S., Parent A. T., and Thinakaran G. (2011) Transgenic neuronal overexpression reveals that stringently regulated p23 expression is critical for coordinated movement in mice. Mol. Neurodegener. 6, 87 10.1186/1750-1326-6-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andrew R. J., Fernandez C. G., Stanley M., Jiang H., Nguyen P., Rice R. C., Buggia-Prévot V., De Rossi P., Vetrivel K. S., Lamb R., Argemi A., Allaert E. S., Rathbun E. M., Krause S. V., Wagner S. L., Parent A. T., Holtzman D. M., and Thinakaran G. (2017) Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 114, E9665–E9674 10.1073/pnas.1708568114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Giannoni P., Gaven F., de Bundel D., Baranger K., Marchetti-Gauthier E., Roman F. S., Valjent E., Marin P., Bockaert J., Rivera S., and Claeysen S. (2013) Early administration of RS 67333, a specific 5-HT4 receptor agonist, prevents amyloidogenesis and behavioral deficits in the 5XFAD mouse model of Alzheimer's disease. Front. Aging Neurosci. 5, 96 10.3389/fnagi.2013.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.