Abstract
Photosystem I (PSI) is a large pigment–protein complex mediating light-driven charge separation and generating a highly negative redox potential, which is eventually utilized to produce organic matter. In plants and algae, PSI possesses outer antennae, termed light-harvesting complex I (LHCI), which increase the energy flux to the reaction center. The number of outer antennae for PSI in the green alga Chlamydomonas reinhardtii is known to be larger than that of land plants. However, their exact number and location remain to be elucidated. Here, applying a newly established sample purification procedure, we isolated a highly pure PSI–LHCI supercomplex containing all nine LHCA gene products under state 1 conditions. Single-particle cryo-EM revealed the 3D structure of this supercomplex at 6.9 Å resolution, in which the densities near the PsaF and PsaJ subunits were assigned to two layers of LHCI belts containing eight LHCIs, whereas the densities between the PsaG and PsaH subunits on the opposite side of the LHCI belt were assigned to two extra LHCIs. Using single-particle cryo-EM, we also determined the 2D projection map of the lhca2 mutant, which confirmed the assignment of LHCA2 and LHCA9 to the densities between PsaG and PsaH. Spectroscopic measurements of the PSI–LHCI supercomplex suggested that the bound LHCA2 and LHCA9 proteins have the ability to increase the light-harvesting energy for PSI. We conclude that the PSI in C. reinhardtii has a larger and more distinct outer-antenna organization and higher light-harvesting capability than that in land plants.
Keywords: photosynthesis, Chlamydomonas, light-harvesting complex (antenna complex), cryo-electron microscopy, single-particle analysis, green algae, LHCI, photosystem I, Chlamydomonas reinhardtii, cryo-electron microscopy (cryo-EM), photosynthesis, outer antenna, photosynthetic efficiency
Introduction
Life on Earth depends primarily on photosynthesis. In photosynthetic organisms, such as cyanobacteria, eukaryotic algae, and land plants, light energy from the sun is converted to chemical energy by two large membrane protein complexes, photosystem II and photosystem I (PSI),4 embedded in the thylakoid membranes. Using light energy, PSI catalyzes light-driven charge separation and transfers electrons from plastocyanin to ferredoxin (Fd) with nearly 100% quantum efficiency. The reducing equivalents generated downstream of PSI are finally utilized to produce organic matter, and thus, PSI is recognized as an indispensable protein complex on Earth.
The light-harvesting systems of PSI are diverse among photosynthetic organisms, such as cyanobacteria, eukaryotic algae, and land plants (1), although its core complex is highly conserved. In cyanobacteria, PSI exists as a trimer in which each monomeric complex shares light energy to achieve efficient light harvesting (2, 3). It was also reported that PSI in the filamentous cyanobacterium, Anabaena sp. PCC 7120, forms a tetramer and dimer instead of a trimer (4). Alternatively, green algae and land plant PSIs exist as monomers and do not have monomer–monomer cooperation between the core complexes. Instead, their PSI light-harvesting capacity is enhanced by acquiring an outer antenna system termed light-harvesting complex I (LHCI) in their thylakoid membranes (5).
The number of light-harvesting proteins (LHCA proteins) associated with PSI varies among the green plants. Six Lhca genes (Lhca1–6) have been identified in Arabidopsis thaliana (6), but only four of them are associated with the PSI core complex (7). The crystal structure of the plant PSI–LHCI supercomplex demonstrated that these four Lhca proteins, designated Lhca1–4, were associated with the side of the PsaF and PsaJ subunits (8–10). In contrast, it has been reported that the PSI core complex of the green alga, Chlamydomonas reinhardtii, associates with an antenna complex that is larger than that of the plant complex (11). Indeed, nine LHCA genes (LHCA1–9) have been identified, and all of them are expressed under normal growth conditions (12, 13). These LHCA proteins have been shown to coordinate the in vitro reconstitution of the chlorophylls (Chls) and carotenoids (14). Spectroscopic data also suggested that the C. reinhardtii PSI was able to harvest 40% more photons than the PSI of A. thaliana (15). Taken together, these results indicate that it is plausible that the C. reinhardtii PSI associates with a larger number of antenna proteins than the land plant PSI.
Although C. reinhardtii expresses nine LHCA gene products, the exact number of the LHCA proteins present in the PSI–LHCI supercomplex remains controversial. Gel electrophoresis and subsequent biochemical analyses led to varying estimates of 4–18 LHCA proteins per PSI core complex (11–13, 16–18). It has been reported that LHCA1 is the most abundant protein in LHCI based on Coomassie staining of the 2D electrophoresis gels (12, 16). Isotope dilution MS analysis showed that LHCA1, -4, and -7 were present at a ratio of 1:1, compared with the PSI core complex; LHCA2, -5, -6, -8, and -9 were present in substoichiometric amounts; and LHCA3 was present at a ratio of 2:1 compared with the PSI core complex (19). However, recent stoichiometry analysis using MS suggested that LHCA3, -5, -6, -7, and -8 were present at a ratio of 1:1 compared with the PSI core complex; LHCA2, -4, and -9 were present in substoichiometric amounts; and LHCA1 was present at a ratio of 2.4:1 compared with the PSI core complex (20). Ozawa et al. (21) recently reported that LHCA1 exists as approximately two copies in the PSI–LHCI supercomplex by uniform labeling of the PSI and LHCI subunits with 14C and subsequent separation of the polypeptides using three different SDS-PAGE systems. Furthermore, they determined the physical proximities of several LHCA and PSI subunits using a chemical cross-link method and proposed a 2D model of the PSI–LHC supercomplex involving 10 LHCAs (21). Considering the variety of stoichiometries previously reported for LHCA proteins, it is assumed that the LHCA proteins attached to the PSI core complex have not been consistent for all the experimental procedures. In addition, 2D gel electrophoresis showed some LHCA proteins at multiple positions due to processing and posttranslational modifications (16). These LHCA protein features make it difficult to determine the exact stoichiometry of the antenna proteins in the PSI–LHCI supercomplex of green algae.
Although the crystal structure of C. reinhardtii PSI–LHCI supercomplex is not available, the presence of a larger antenna complex than the land plant PSI–LHCI supercomplex has been supported by single-particle EM using negatively stained specimens. These results showed the presence of 6–14 LHCA subunits in the PSI–LHCI supercomplex (22–26). All of the models commonly proposed that the majority of LHCIs are located on one side of the PSI core complex. In addition, most of the models suggested that the LHCIs form two parallel concentric half-rings comprised of four to five LHCA proteins for each layer, designated the “LHCI belt” in the primary LHCI (22, 23, 25–27). In addition, it has been reported that the C. reinhardtii PSI–LHCI supercomplex has one to two additional LHCA proteins, possibly located on the PsaK and PsaH subunit side of the PSI core complex (22, 23, 26). Kargul et al. (24) reported that one LHCA protein is located at the PsaK and PsaH subunit side in the PSI core complex, whereas the other LHCA protein is located at the PsaG and PsaH subunit side, of the PSI core complex. Steinbeck et al. (26) reported that there were two LHCA proteins located between PsaG and PsaH. Because these EM observations were all performed at a relatively low resolution, the exact locations of the LHCA proteins in the C. reinhardtii PSI–LHCI supercomplex remain to be elucidated by careful structural analysis at higher resolution.
In this study, we purified a stable PSI–LHCI supercomplex from C. reinhardtii using a newly developed procedure, employing a combination of Fd affinity column chromatography and nonionic amphipol (NAPol) (28, 29). The biochemical properties of Fd, which exhibits high affinity for the PSI core complex with a specific protein–protein interaction, enables the efficient and high-purity purification of PSI, and NAPol maintains the integrity of the protein structure and function. Based on the results of biochemical stoichiometric analyses, single-particle EM analyses with ice-embedded and negatively stained specimens, and spectroscopic analysis, the structure of the C. reinhardtii PSI–LHCI supercomplex, which consists of 10 LHCA proteins (with nine LHCA protein types), was reconstructed at 6.9 Å, and the arrangement of all of the LHCI proteins was determined. Two of them were located at the side of the PsaG and PsaH subunits of the PSI core complex.
Results
Isolation of the PSI–LHCI supercomplex from C. reinhardtii
In this study, we applied a newly established method, using Fd affinity column chromatography and NAPol-derived complex stabilization, to purify the PSI–LHCI supercomplex from C. reinhardtii. Briefly, thylakoid membranes from the 137c WT strain were treated with 0.48% n-dodecyl-β-d-maltoside (β-DDM), and the solubilized protein complexes were subjected to sucrose density gradient (SDG) ultracentrifugation with 1 m betaine to stabilize the protein complexes. After centrifugation, the green band corresponding to the PSI–LHCI supercomplex (30) was collected and applied to an Fd affinity column as described under “Experimental procedures.” After the Fd column purification of the PSI–LHCI supercomplex, β-DDM was replaced with NAPol to further stabilize the supercomplex. To evaluate the purity of the PSI–LHCI supercomplex, we conducted SDS-PAGE analysis with Coomassie Brilliant Blue staining of the eluted sample, revealing that contamination from ATPases and LHCII was negligible (Fig. 1). In addition, the LHCA protein library was identified using MS (Table S1), confirming that the PSI–LHCI supercomplex was successfully purified with the PSI core complex and the nine LHCA proteins. Thus, we concluded that the method combining Fd affinity purification and NAPol stabilization is suitable for subsequent EM-based structural analysis.
Figure 1.

Purification of the PSI–LHCI supercomplexes from 137c WT. Shown is SDS-PAGE of the PSI–LHCI supercomplex purified only with a sucrose density gradient (before the Fd column) and with a combination of sucrose density gradient and Fd column affinity chromatography (after the Fd column). A total of 0.8 μg of Chl was loaded in each lane.
Supramolecular organization of the PSI–LHCI supercomplex in C. reinhardtii
To elucidate the exact number of LHCA proteins in the PSI–LHCI supercomplex and their supramolecular organization, single-particle cryo-EM was performed using the PSI–LHCI of the 137c WT strain. The highly purified and NAPol-stabilized PSI–LHCI supercomplex was suitable for single-particle cryo-EM analysis, in which the supercomplexes were uniformly dispersed and randomly oriented in amorphous ice with sufficient contrast (Fig. S1, A–C). The major subpopulation group comprising ∼70% of the sample preparation was the largest particle that contains 10 LHCA proteins. Because ∼30% of incomplete supercomplexes were included in the sample preparation, they were removed twice during the 3D classification procedure of the Relion single-particle analysis software (Fig. S1, D–F). An initial unmasked refinement resulted in an 11.9 Å map (Fig. S1E). A second round of 3D classification (Fig. S1F) improved the clarity of the nonpostprocessing map (Fig. S1G), which allowed successful 3D reconstruction of the PSI–LHCI supercomplex at 6.9 Å resolution (Fig. S1H and Movie S1) assessed by the “gold standard” Fourier shell correlation (FSC = 0.143) (Fig. S1, H and K). The local resolution assessment with ResMap (31) suggests that the structural resolution is higher for the PSI core complex than the peripheral LHCI complexes (Fig. S1I). The representative density and fitted models are shown in Fig. S2
The reliability of the cryo-EM map was evaluated by FSC for atomic models of the individual proteins and those of the proteins with the Chls (Fig. S1K, orange and blue lines, respectively). The FSC of the map for the sole protein models showed a resolution of 7.7 Å, but it was improved to 7.0 Å with respect to the proteins with the Chls. This suggests that the cryo-EM map in this study described the fine structure of the PSI–LHCI supercomplex from C. reinhardtii.
The final 2D class average and 3D density maps are shown in Figs. 2 and 3 and Fig. 4, respectively. In Figs. 2 and 3, the PSI core complex is viewed from the stromal side (stromal view). The PSI–LHCI supercomplex surrounded by NAPol in the stromal view had maximal dimensions of ∼230 × 200 Å, in which the dimensions of the protein complex were measured to be 200 × 170 Å when excluding the surrounding density caused by NAPol. This is the largest PSI–LHCI supercomplex size reported so far.
Figure 2.

Cryo-EM projection map of the PSI–LHCI supercomplex of 137c WT. White arrow, NAPol ring density. Scale bar, 5 nm.
Figure 3.

The 2D projection map of single-particle cryo-EM of the PSI–LHCI supercomplex. A, the 2D average of PSI–LHCI supercomplex from the 137c WT, where these transmembrane helices were used as a marker for the assignment of the orientation of the individual LHCA proteins. The white contours mark a border for each LHCA protein, and the red contours mark LHCA helix C. B, the 2D average of PSI–LHCI supercomplex from the lhca2 mutant. The crystal structure of the plant PSI–LHCI (PDB entry 5L8R) and the model structures of LHCA2 and LHCA9 were overlaid on the projection maps of the PSI–LHCI from the 137c WT (C) and the lhca2 mutant, highlighting the missing density with a dotted line (D). PsaA and PsaB are presented in blue; PsaC, PsaD, and PsaE are in yellow; PsaF and PsaJ are in lilac; PsaG is in cyan; PsaK is in red-purple; PsaI, PsaL, PsaH are in ocher; plant-LHCA1 is in green; plant-LHCA2 is in orange; plant-LHCA3 is in yellow orange; LHCA4 is in yellow green; LHCA9 is in magenta, and LHCA2 is in red. Images are the sums of 3814 (137c WT) and 3511 (lhca2 mutant) aligned projections. Scale bars, 5 nm. In A, the contrast of Fig. 2 has been increased to assist visualization; no contrast change was applied to B–D.
Figure 4.
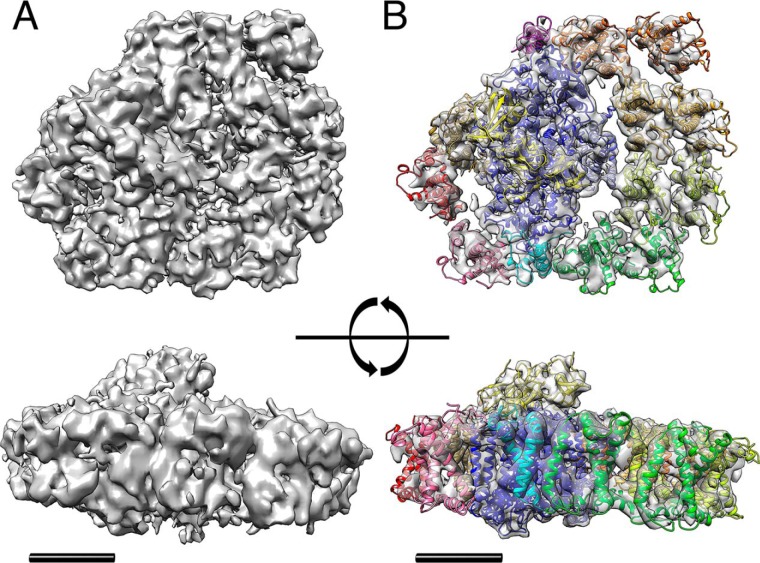
The 3D single-particle cryo-EM of the PSI–LHCI supercomplex. Surface representations of the PSI–LHCI supercomplex from 137c WT are shown. Densities are illustrated at the different contours (6σ (A) and 12σ (B)) with transmembrane helices rendered as a ribbon diagram using PDB structures (B). Top panels, view from the stromal side; bottom panels, view from the PsaG side (turned 90° along the x axis). PDB models and color codes are same as in Fig. 3. Scale bar, 5 nm.
In the 2D averages (Figs. 2 and 3), the NAPol density could be distinguished from the protein density; the boundaries of each LHCA protein were clear enough to fit the crystal structures derived from the land plant PSI–LHCI supercomplex (PDB code 5L8R). Because the primary structures of the PSI core are well-conserved among cyanobacteria, eukaryotic algae, and land plants (1), and primary structures of LHCA are also conserved between C. reinhardtii and land plants (17), we applied the crystal structure from land plants in this study. The projection map of the PSI core complex was fitted with the molecular models based on the correlation with specific features. For example, the typical quarter sector shape of the PSI core complex, and the extrinsic proteins, the PsaC, PsaD, and PsaE subunits, were clearly visible as strong densities compared with the uniform densities of the LHCI proteins (Figs. 2 and 3A). The PSI subunits from the crystal structure of PDB entry 5L8R matched well to the 3D cryo-EM map, except for the N-terminal domain with the helix-turn-helix of PsaF to which the plastocyanin binds. To fit this region into the density, we used the “fit in map” function in UCSF Chimera (32). As a result, the N-terminal of the PsaF subunit was located even closer to the PsaA subunit, which was located in the center of the PSI core, compared with the 5L8R model. The total correlation value was improved after the “fit in map” operation of the PsaF subunit from 0.7608 to 0.8230, implying that the fitted structures were more reliable.
The supercomplex of the 3D structure is viewed from the stromal side in the top panels of Fig. 4A. Their transmembrane helices were traceable in Fig. 4B. Horizontal views from the PsaG subunit side are shown in bottom panels of Fig. 4 (A and B). The 3D structure clearly indicates multiple protrusions of the PsaC, -D, and -E subunits on the stromal side and the single protrusion of the PsaF subunit on the luminal side, as reported previously (10). Based on these outstanding features, we were able to refit the molecular model of the PSI core complex into the density area without ambiguity.
The PsaF and PsaJ subunit-binding side of the PSI core complex was surrounded by a large area of relatively weak density (Figs. 2 and 3A). This plane area contained several typical features of the LHCI, including strong density dots (shown as red contour marks in Fig. 3B) corresponding to the LHCI helix C, which is perpendicular to the membrane, and weak density rods (shown as white contour marks in Fig. 3B), corresponding to the LHCI helices A and B, which are tilted, relative to the membrane, and cross each other. The size and features of these densities suggest that the eight LHCA proteins formed two layers of the LHCI belt with four LHCA proteins each, which is consistent with the reports of Ozawa et al. (21) and Steinbeck et al. (26). Based on these observations, we superimposed the eight crystal structures of LHCA proteins from the plant PSI–LHCI supercomplex (PDB code 5L8R) onto these densities (Fig. 3C).
We also found that there was extra density near the PsaG and PsaH subunits in the PSI–LHCI supercomplex (Figs. 2 and 3A). We presumed that this density corresponds to the LHCA2 and -9 proteins, because these two LHCI proteins had been localized at these positions using a chemical cross-link experiment (21). To confirm this possibility, we applied EM analysis to the PSI–LHCI supercomplexes purified from the lhca2 mutant (Fig. 3B and Fig. S3) (LMJ.RY0402.109691) (33). The structure of the PSI–LHCI supercomplex from the cc5325 WT, which is the parental strain of the lhca2 mutant, was also analyzed as a reference (Fig. S4). Prior to the structural analysis of the lhca2 mutant, stable isotope (15N)-labeled LC-MS/MS stoichiometry analysis was conducted to determine the LHCA composition in the PSI–LHCI supercomplex obtained from the lhca2 mutant. From the 14N:15N isotopomer values derived from the chromatograms, the relative abundance of each subunit from the PSI–LHCI supercomplex in the lhca2 mutant was determined (Fig. 5). Using these values, we compared the amount of LHCA proteins in the PSI–LHC supercomplex between these two strains. LHCA3, -7, and -8 existed almost equally in both strains, whereas LHCA1, -4, -5, and -6 in the lhca2 mutant were present at slightly larger amounts than in the WT strain with ratios ranging from 1.2 to 1.4. Remarkably, LHCA2 and -9 were present at a factor of less than 0.2, confirming that not only LHCA2, but also the association of LHCA9 with the PSI core complex, was severely affected in the lhca2 mutant. In the lhca2 mutant, the paromomycin-resistant cassette is inserted into an intron of the LHCA2 gene (intron 2). Therefore, it is possible that a full-length mRNA is transcribed, and the protein is synthesized at a very low level. The results of single-particle cryo-EM of the PSI–LHCI supercomplex purified from the lhca2 mutant demonstrated that no extra density was present near the PsaG and PsaH subunits (Fig. 3, B and D). Furthermore, the density of the PsaH subunit in this mutant was weak, possibly because PsaH became unstable in the absence of LHCA2 (Fig. 3B). Considering this cryo-EM observation together with the MS analysis shown in Fig. 5, we concluded that the extra density near the PsaG and PsaH subunits corresponded to the LHCA2 and -9 proteins.
Figure 5.
Relative abundance of the LHCA proteins and the PSI subunits in the lhca2 mutant. LHCA composition in the PSI–LHCI supercomplex from the lhca2 mutant was determined using stable isotope (15N)-labeled LC-MS/MS spectrometry. The relative amounts of each subunit for the PSI–LHCI supercomplex were determined based on the 14N:15N isotopomer values derived from the chromatograms between the 137c WT and the lhca2 mutant. Data shown are the mean ± S.E. (error bars) (n = 3).
To further determine the specific positions of LHCA2 and -9 proteins in the extra density, 3D structural information of the 137c WT PSI–LHCI supercomplex shown in Fig. 4 was analyzed. For the unknown molecular structures of LHCA2 and -9, we applied homology modeling using SWISS-MODEL (34) (Fig. S6) and fitted the models into the additional density of the 137c WT PSI–LHCI supercomplex. The alternative positions of LHCA2 and -9 in the two densities were evaluated using the correlation values between the models and the 3D densities calculated with the “fit in map” function in UCSF Chimera (32). The correlation values were 0.8767 and 0.8711, respectively, when the LHCA2 and -9 proteins were fitted into the densities along the edge from the PsaH to PsaG subunits (Fig. S7A and Table S2), whereas they were 0.8529 and 0.8544, respectively, when the LHCA2 and -9 proteins were assigned to the densities along the edge from the PsaH to the PsaG subunits in reverse order (Fig. S7B and Table S2). We applied other LHC models (LHCA1, LHCA3–8, LHCB4–5, LHCBM1–9) for calculating the “fit in map” to the densities. However, none of these alternative LHCs showed higher correlation than LHCA2 and LHCA9 (Table S2). Therefore, we tentatively concluded that LHCA2 and -9 are located along the edge from the PsaH to PsaG subunits in this order.
To obtain further insights into the molecular organization of the PSI–LHCI supercomplex, the 2D class averages of the PSI–LHCI supercomplexes from an lhca1 mutant (LMJ.RY0402.181250) were calculated from the negative-stain EM images (Fig. S5). The purified PSI–LHCI supercomplexes from the lhca1 mutant were separated by SDS-PAGE, but the LHCA1 band between the PsaD and PsaF bands was not detected (Fig. S3). Fig. S5 shows the top views projected from the stromal side, revealing that the PSI–LHCI supercomplex of the lhca1 mutant lost four LHCA subunits. Two of these subunits could be the LHCA2 and -9 proteins based on the results described above (labels by the red dotted circles in Fig. S5). The remaining two LHCA proteins were in the LHCI belts. One was located at the association site in the first layer of the LHCI belts in which the LHCA1 protein binds to the PSI core complex in land plants (10), and another was located near the PsaG subunit, but in the second layer of the LHCI belts (labels by the white dotted circles in Fig. S5).
Functional properties of the PSI–LHCI from C. reinhardtii
The absolute fluorescence emission spectra of the PSI–LHCIs purified from the cc5325 WT and lhca2 mutant strain at 77 K are shown in Fig. 6. The larger fluorescence emission of the lhca2 mutant PSI–LHCI supercomplex than that of the WT strain suggested that the energy-transfer efficiency from the antenna complex to the core complex was lower in the lhca2 mutant. The emission difference spectrum between the two PSI–LHCIs of the WT and lhca2 mutant showed emission maxima at 683 nm, implying that the energy-transfer efficiency from the “blue LHCA” with emission maxima at 682.5–683.5 nm (14) to the PSI core was hampered in the lhca2 mutant.
Figure 6.

Absolute fluorescence emission spectra at 77 K. Fluorescence emission spectra were obtained by excitation at 440 nm with an integrating sphere at 77 K with sample concentration of 3 μg of Chl/ml. a.u., arbitrary unit.
To estimate the energy transfer from the peripheral antenna complex to the PSI core complex, we analyzed the fluorescence kinetics at 685 and 710 nm, where the signals predominantly originate from the peripheral antenna complex and PSI core complex, respectively. The PSI–LHCI supercomplexes both from the cc5325 WT and from the lhca2 mutant showed a decay component at 685 nm and a rise component at 710 nm at 50–60 ps (Table 1 and Fig. S8), indicating that an energy transfer from the peripheral antenna complex to the PSI core complex occurred in this time domain. The amplitude of such an energy-transfer component in the lhca2 mutant (+0.806 at 685 nm and −0.704 at 710 nm, 60 ps) was smaller than in the cc5325 WT (+0.918 at 685 nm and −0.890 at 710 nm, 50 ps), showing that the energy-transfer efficiency from the peripheral antenna complex to the core complex in the lhca2 mutant was less efficient. These results suggest that LHCA2 and -9 function as light-harvesting antennas of the PSI–LHCI supercomplex in C. reinhardtii by binding to the PsaG and PsaH side.
Table 1.
Lifetime components and amplitudes of fluorescence kinetics at 685 and 710 nm
Fluorescence lifetime components were obtained using a convolution calculation. Positive and negative amplitudes represent decay and rise, respectively. Each amplitude represents the relative contribution of the lifetime component at the wavelength.
| Observed wavelength (nm) | Lifetime (amplitude) |
||||
|---|---|---|---|---|---|
| (ps) | (ps) | (ns) | (ns) | ||
| lhca2 mutant | 685 | 60 (0.806) | 280 (0.132) | 1.4 (0.047) | 6.4 (0.015) |
| 710 | 60 (−0.704) | 135 (0.665) | 0.61 (0.275) | 2.1 (0.060) | |
| cc5325 WT | 685 | 50 (0.918) | 290 (0.053) | 1.1 (0.012) | 4.5 (0.017) |
| 710 | 50 (−0.890) | 120 (0.648) | 0.57 (0.286) | 1.9 (0.066) | |
Discussion
In earlier studies, biochemical and EM analyses reported that green algae possess larger PSI antenna complexes than land plants (11, 13, 17, 21–23, 25, 27). However, both the number and location of the LHCA proteins in the PSI–LHCI supercomplex have been controversial. In this study, a cryo-EM analysis of the PSI–LHCI supercomplex from C. reinhardtii enabled the identification of 10 LHCA proteins around the PSI core complex with two layers of LHCI belts (each LHCI belt contains four LHCA proteins) on the PsaF and PsaJ side and two LHCA proteins on the PsaG and PsaH side. In total, we identified 10 LHCA proteins that were associated with the PSI core, although only nine of them were genetically encoded and confirmed by SDS-PAGE and MS (Fig. 1 and Table S1). This incongruity occurred because one LHCA protein was not present stoichiometrically with the other LHCA proteins. Recently, Ozawa et al. (21) determined the configuration of LHCA proteins within the PSI–LHCI supercomplex by a chemical cross-linking study and proposed that eight LHCA proteins bind to the PsaF side in two layers, and the other two LHCA proteins including LHCA2 and LHCA9 bind between PsaH and PsaG in this order. Our structural model agrees with their cross-linking results.
It has been reported that LHCA1 is the most abundant LHCA protein, as indicated by the evaluated spot volumes of individual LHCA proteins after staining 2D SDS-polyacrylamide gels with colloidal Coomassie Blue and subsequent LC-MS/MS analysis (12, 16). Recently, Marco et al. (20) and Ozawa et al. (21) reported that approximately two copies of the LHCA1 protein were present in the PSI–LHCI supercomplex in C. reinhardtii. Thus, we presumed that C. reinhardtii had two LHCA1 subunits per PSI core complex to establish a PSI–LHCI supercomplex with 10 LHCA proteins (Fig. 7).
Figure 7.

A schematic model of the PSI–LHCI supercomplex of C. reinhardtii. A, the PSI–LHCI supercomplex viewed from the stromal side. The LHCI belt at the PsaF (F, lilac) and PsaJ (J, lilac) side of the PSI core complex is composed of eight LHCA proteins (dark green oval) forming two layers. The LHCA2 and LHCA9 proteins exist between the PsaG (G, cyan) and PsaH (H, ocher), subunits of the PSI core complex. B, the PSI–LHCI supercomplex horizontally viewed from the LHCI belt side. Three extrinsic subunits, PsaC (C, yellow), PsaD (D, yellow), and PsaE (E, yellow), are located on the stromal side. The N-terminal region of PsaF subunit is projected to the luminal side. The color codes for PsaK (K) is red-purple. PsaM (M), which does not exist in C. reinhardtii, is shown by a dotted circle. Four of the LHCI subunits are assigned in the map as LHCA2 (2), LHCA9 (9), LHCA1a (1a), and LHCA1b (1b).
Consistent with previous studies using low-resolution models from negative-stain EM analysis, the cryo-EM analysis in this study revealed that the primary part of the LHCA proteins, designated the LHCI belts, was associated with the PsaF and PsaJ side of the PSI core complex (Figs. 3 and 4). These LHCA proteins formed a crescent shape, which is a common feature in the structures of the vascular plant PSI–LHCI supercomplexes (9, 10). However, the reported organization of the LHCI belt has not been consistent in C. reinhardtii in the literature. Kargul et al. (23) reported that the inner layer was composed of five LHCA proteins, and the outer layer was composed of four LHCA proteins (23, 25), whereas Drop et al. (24) reported that the inner layer comprised three LHCA proteins and the outer layer comprised five LHCA proteins. In this study, we visualized that the inner and outer layers each comprised four LHCA proteins (Figs. 3 and 4). The cryo-EM map density of the PsaK side of the outer layer was relatively weaker than the densities of the other LHCAs. In addition, this density was located slightly away from the primary complex. These results suggest that the LHCA protein located at this position may bind more loosely to the primary complex.
The PSI–LHCI supercomplex of the lhca1 mutant lost two LHCA proteins and the PsaG subunit, in addition to the LHCA1 protein (Fig. S5). As described above, two copies of the LHCA1 protein are likely to be present per PSI core complex. Together with the high homology of the LHCA1 proteins between land plants and green algae, we assigned two LHCA1 proteins beside the PsaG subunit of the PSI core complex. The first LHCA1 protein, designated LHCA1a, was assigned to the first layer, and the second LHCA1 protein, designated LHCA1b, was assigned to the location near the PsaG subunit of the PSI core complex, but in the second layer (Fig. 7). Recently, Ozawa et al. (21) reported that the LHCA1 protein could be cross-linked with the PsaG subunit and the LHCA9 protein. Thus, they assigned one LHCA1 protein in the first layer of the LHCI belt beside the PsaG subunit. They also detected two copies of the LHCA1 proteins in the single PSI–LHCI supercomplex, and the second LHCA1 protein was tentatively assigned to an open space beside the PsaG subunit in the second layer of the LHCI belt (21). Our EM results are consistent with these cross-linking results.
We identified two additional locations of the LHCA proteins between the PsaG and PsaH subunits on the opposite side of the LHCI belt. The presence of the additional LHC proteins in C. reinhardtii was suggested by two previous models. One model reported that 1–3 LHCA proteins were likely associated with the PsaH subunit of the PSI core complex (22, 23). Another model proposed that CP29, a minor monomeric antenna for photosystem II, could be associated with the PsaG and PsaH subunit side of the PSI core complex in the state 2 conditions (24). On the other hand, our research was done under state 1 conditions, and we could not detect CP29 in the MS. Based on the recent report, the LHCA2 and -9 proteins were cross-linked with the PsaH and PsaG subunits, respectively (21). Our investigation, based on the EM and MS studies using the PSI–LHCI supercomplex purified from the WT and lhca2 mutant, demonstrated that the subunits located between the PsaG and PsaH subunits were the LHCA2 and -9 proteins, not CP29.
The emission difference spectrum between the two PSI–LHCIs obtained from the WT and lhca2 mutant (Fig. 6) matched well with the reconstituted LHCA1, -3, and -7 proteins, reported previously, which were designated the “blue LHCA” group (14). These results indicated that a loss of the LHCA2 and -9 proteins in the lhca2 mutant might affect the stability of at least one of the blue LHCA group proteins. In addition, our negative stain EM analysis could not find the density of the LHCA2 and -9 proteins of the PSI–LHCI supercomplex in the lhca1 mutant (Fig. S5), suggesting that the LHCA1 and -9 proteins could indirectly interact and/or stabilize each other. Because the PsaG subunit is located between the LHCA9 and LHCA1a proteins in our model (Fig. 7), a loss of the LHCA9 protein in the lhca2 mutant may also affect the stability of the PsaG subunit and simultaneously cause the destabilization of the LHCA1a and -1b proteins.
A recent study showed that the red alga Cyanidioschyzon merolae (35) had additional antenna proteins at the side of the PsaG and PsaH subunits on the PSI core complex. In C. merolae, the peripheral antennas of the PSI–Lhcr supercomplex consist of five Lhcr proteins encoded by three genes, in which two copies of Lhcr1 and Lhcr2 and one copy of Lhcr3 are present (35). One of these, Lhcr1 and Lhcr2 were located at the side of the PsaG and PsaH subunits on the PSI core complex where the additional PsaM subunit is located between the PsaG and PsaH subunits. Pi et al. (35) suggested that the loss of the PsaM subunit in green algae and land plants might have caused the loss of additional LHCA proteins near the PsaM subunit, because the PsaM subunit seemed to provide a binding site for the additional Lhcr proteins in the red alga. However, in contrast to this hypothesis, the results in this study showed that the absence of the PsaM subunit did not prevent the association of additional antennas on this side of the PSI core complex in C. reinhardtii, suggesting that the PsaM subunit is not essential for the association of the peripheral antenna in green alga. Our results imply that LHCA2 and -9 could bind to PSI when their associations are stabilized by LHCA1a via PsaG. Therefore, we hypothesize that the PsaG subunit, instead of the PsaM subunit, can stabilize the association of the additional antennae beside the PsaG and PsaH subunits in C. reinhardtii.
Why are the additional antennae beside the PsaG and PsaH subunits conserved in green and red algae, but lost in land plants? The LHCA2 and -9 proteins are identified as C. reinhardtii-specific LHCA proteins (the C. reinhardtii LHCA2 is different from that of land plants), suggesting that these subunits may have a unique functional role in green algae. Recently, Steinbeck et al. (26) reported that LHCA2 and -9 have a role in blocking the binding of the cytochrome b6f dimer to the PSI–LHCI supercomplex. They also observed an increased cyclic electron flow (CEF) rate in the lhca2 mutant. Considering that the PSI–LHCI-cytochrome b6f had been known as a CEF supercomplex (36), they hypothesized that these LHCA proteins may function to control the CEF supercomplex assembly (26). The cryo-EM structure and spectroscopic results in this study indicate that both the LHCA2 and -9 proteins were able to increase the light-harvesting efficiency in the PSI–LHCI supercomplex, suggesting that these antennae facilitate light harvesting under weak light conditions, such as the aquatic environments in which most microalgae live. These LHCA proteins may have been abandoned in land plants, because their habitat provides adequate illumination, and thus they no longer require enhanced light-harvesting abilities.
PSI is thought to have evolved from the trimeric form to the monomeric form (1). In cyanobacteria, PSI forms a trimer to share the excitation energy between adjacent PSI protomers. In the case of green and red algae, PSI exists as a monomer, but it is still possible to take up excitation energy from the sides, which were considered to be boundary surfaces between the trimeric cyanobacterial monomers (3, 35). In this study, we found that the LHCA2 and -9 proteins associate with the side between the PsaG and PsaH subunits in the green alga C. reinhardtii. Pi et al. (35) reported that two Lhcr subunits associate between the PsaG and PsaL subunits in the red alga C. merolae. All of these antennae, including LHCA2, -9, and Lhcr, have been suggested to increase energy flux to the PSI core (this study) (35). These observations imply that the energy-transfer pathway that had developed in the trimeric interface of PSI in cyanobacteria (2) evolutionally diversified into the external antennae-dependent energy-transfer pathway using LHCA and Lhcr in green and red algae, respectively. Thus, the PSI monomer associated with the external antennae is allowed to perform advanced regulation of the energy flux toward the PSI core to successfully adapt to various light conditions.
Experimental procedures
C. reinhardtii strains and growth conditions
C. reinhardtii WT strains 137c, cc5325, and the lhca1 and lhca2 mutants were obtained from the Chlamydomonas Resource Center. The strains were grown photoautotrophically at 23 °C in a high-salt minimal medium (37) with 5% CO2 bubbling under continuous light (200 microeinsteins m−2 s−1) conditions. For the stoichiometry analysis, the WT cells were grown in Tris acetate medium (38) using 15N-labeled NH4Cl with air bubbling at 23 °C under continuous light (200 microeinsteins m−2 s−1) conditions.
Isolation of the thylakoid membranes
Thylakoid membranes were isolated from C. reinhardtii cells as reported previously (30) with the following modifications. The cells were resuspended in an isolation buffer containing 25 mm HEPES (pH 7.5), 10 mm MgCl2, 3 mm NaCl, and 0.33 m sucrose and disrupted twice using the BioNeb disruption system (Glas-Col, LLC).
Isolation of the PSI–LHCI supercomplexes
The thylakoid membranes were suspended in 25 mm HEPES buffer (pH 7.5) at 0.4 mg of Chl/ml and solubilized with 0.48% β-DDM for 15 min on ice. The samples were centrifuged at 10,000 × g for 1 min to remove the unsolubilized thylakoid membranes, and the supernatant was applied to a discontinuous SDG (0.1/0.4/0.7/1.0/1.3 m sucrose in a buffer containing 25 mm HEPES (pH 7.5), 1 m betaine, and 0.05% β-DDM) and subjected to ultracentrifugation at 90,000 × g on a P40ST rotor (Hitachi-koki, Tokyo, Japan) for 18 h at 4 °C. After SDG ultracentrifugation, the fractions containing the PSI–LHCI supercomplex were collected and applied to a PD10 column (GE Healthcare) equilibrated with 25 mm HEPES (pH 7.5) and 0.05% β-DDM for buffer exchange. The eluted PSI–LHCI supercomplex was loaded on a ferredoxin (Fd) affinity column as reported previously (3) with the following modifications. The sample-loaded column was washed using washing buffer (25 mm HEPES (pH 7.5) and 0.05% β-DDM) and eluted using 25 mm HEPES buffer (pH 7.5) containing 1 m betaine, 30 mm NaCl, and 0.05% β-DDM. The eluted PSI–LHCI was mixed with NAPol (nonionic amphipols, Anatrace, MI) in a 10:1 ratio with Chl for 30 min on ice to replace the β-DDM with NAPol. Extra β-DDM was removed by buffer exchange using a PD10 column equilibrated with 25 mm HEPES (pH 7.5) without β-DDM.
Preparation of the Fd column
For this step, 1.0 mg of CNBr-activated Sepharose 4B (GE Healthcare) was moistened with 1 mm HCl, and the resin was washed with 1 mm HCl. Subsequently, 8 mg of C. reinhardtii Fd-purified as reported previously (39) was resuspended in 100 mm NaHCO3 (pH 8.0) with 300 mm NaCl and gently stirred in the mixture at 4 °C overnight. The resin was washed with 50 mm Tris-HCl (pH 7.5) with 500 mm NaCl to block the remaining active sites on the resin.
SDS-PAGE analysis
The purified PSI–LHCI supercomplex corresponding to 0.8 μg of Chl was used for SDS-PAGE analysis as described previously (30).
Protein identification
Trypsin-digested peptides were separated using an EASY-nLC 1000 (Thermo Fisher Scientific). Digested peptides were desalted with a column (Acclaim PepMap 100 (75 μm × 2 cm nanoViper P/N 164946 Thermo Fisher Scientific). Then the column was switched with the separation column-packed nano-capillary column (NTCC-360/75-3-125 (C18, 0.075-mm inner diameter × 125 mm, particle diameter 3 μm) Nikkyo Technos Co. Ltd.). The mobile phases for peptide elution were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The flow rate was 300 nl/min and for the following gradient profiles: 0–30% B over 10 min, 30–80% B over 2 min. The LC was coupled Orbitrap Elite (Thermo Fisher Scientific) with the scan range m/z 350–2000. Peptide identification was performed using MASCOT (Matrix Science) and Proteome Discoverer software (Thermo Fisher Scientific). We used the database of nucleus-encoded genomes from JGI, Phytozome, and chloroplast-encoded genomes from NCBI. The ion score is a value of matching level between product ion peak and calculated fragment by MASCOT (Table S1).
Stoichiometry analysis
Stoichiometry analysis was performed as described previously (40).
Cryo-EM
Isolated PSI–LHCI supercomplexes were diluted to 50 μg of Chl/ml. For cryo-EM, 2.5 μl of the specimen suspension was applied to a R1.2/1.3 Mo Quantifoil grid (Quantifoil Micro Tools, Grosslöbichau, Germany) pretreated by glow discharge beforehand. The grid was blotted and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI Co.) with a setting of 95% humidity at 4 °C. The frozen sample grid was mounted on a Gatan 626 cryo-specimen holder (Gatan Inc.) and examined using a JEM-2200FS electron microscope (JEOL Inc., Tokyo, Japan) equipped with a field emission electron source at 200 kV and an in-column energy filter operated with a 20-eV energy slit. Micrographs were recorded using a DE20 direct-detector CMOS camera (Direct Electron LP) at a nominal magnification of ×30,000 or 1.992 Å/pixel at the sensor. A 2–4-μm underfocused condition was used. The total electron dose for each image was <20 e−/Å2 using a low-dose system. Individual frames were subjected to motion correction by a script provided by the manufacturer.
Single-particle analysis of cryo-EM
In total, 352 motion-corrected micrographs were imported into RELION 2.1 (41, 42), and the CTF calculation was conducted using CTFFIND4.1 (43) with the following parameters: Cs, 4.2 mm; acceleration voltage, 200 kV; amplitude contrast, 0.1; FFT box size, 2048; and resolution, 24 Å (minimum) and 4 Å (maximum). From these data, 62,486 particles were autopicked into 256 × 256 boxes. After 2D classification, 21,230 particles were subjected to an initial 3D classification into 10 models with a simple spherical mask of 290 Å. Model fitting and map visualization were performed using UCSF Chimera (32). The sizes of the cryo-EM images and volumes were measured using EMAN2 software (44). 3D models for LHCA2 and LHCA9 were created based on PDB entry 5L8R with SWISS-MODEL simulations (31). 3D classes were selected based on the presence of a PsaC/D/E subunit “peak” and density in the region between the PsaG and PsaH subunits. An initial 3D refinement of these combined classes resulted in an 11.9 Å map. Particles were re-extracted from the refined model coordinates, and a fresh round of 3D classification (without masking) into six classes was conducted. Of these, five classes were recombined for a total particle count of 10,835, and a further 3D refinement was performed. This final refinement was postprocessed with a 30 Å soft mask, resulting in a final reported resolution of 6.9 Å. The map resolution was assessed by the “gold standard” Fourier shell correlation (FSC = 0.143) criterion (45), whereas the map-to-model correlation was evaluated with the “half-bit” (FSC = 0.5) criterion. The procedures are summarized in Fig. S1.
Negative-stain EM
For negative-stain EM, the specimen suspension (5.0 μl) was applied to a carbon-coated copper grid pretreated by glow discharge beforehand. After removing the excess sample solution by blotting with filter paper (Whatman 1), the specimens were stained with a 2% (w/v) uranyl acetate solution for 30 s. The grids were dried in air after removing the staining solution using a piece of filter paper. The EM images were recorded as described above. The image pixel spacing was 5 Å on the camera. A 2–3-μm underfocused condition was selected to enhance the contrast of the EM images. The image analysis was performed in RELION (41, 42) as described above for the 2D classification.
Time-resolved fluorescence lifetime measurement
Isolated PSI–LHCI supercomplexes purified from the cc5325 WT and the lhca2 mutant were diluted to 3 μg of Chl/ml. Fluorescence kinetics were measured using a time-resolved single-photon counting system (SPC-630, Beker and Hickl, GmbH) as described (44). A picosecond pulse diode laser (PiL047X, Advanced Laser Diode Systems) was used to excite Chl at 459 nm with a 3-MHz repetition rate (≤0.1 nanojoule/pulse). Emission was detected with a 2.5-nm bandwidth. The lifetime components of fluorescence kinetics were obtained by a fitting with four-exponential components. Fluorescence lifetimes were determined using a convolution calculation (45).
Low-temperature fluorescence emission spectra
Fluorescence emission spectra were measured at 77 K using a FluoroMax 4 instrument (HORIBA Jobin-Yvon). The samples were excited at 440 nm, and the emission between 650 and 780 nm was monitored. Fluorescence intensities were normalized to the absolute fluorescence yield measured by an integrating sphere at 77 K.
Author contributions
J. M. and R. T. conceived the study and designed the experiments. The purification of PSI–LHCI was conducted by H. K.-K. and A. W. Spectroscopic measurement and analyses were conducted by S. A., M. Y., Y. U., and E. K. The EM measurement and analysis were performed by R. N. B.-S., C. S., and K. M. All of the authors contributed to writing and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the Chlamydomonas Mutant Library Group at Princeton University, the Carnegie Institution for Science, and the Chlamydomonas Resource Center at the University of Minnesota for providing the indexed Chlamydomonas insertional mutants. We thank C. Noda, T. Mori, and Y. Makino for technical help with this project.
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid 16H06553 (to R. T., S. A., and J. M.) and 17J40032 (to H. K.-K.) and by Advanced Bioimaging Support (ABiS) from Grant-in-Aid for Scientific Research on Innovative Areas 16H06280 (to K. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2, Figs. S1–S8, and Movie S1.
The 3D EM reconstruction of the C. reinhardtii PSI–LHCI supercomplex has been deposited in the EMDB with the accession number EMD-9700.
- PSI
- photosystem I
- Fd
- ferredoxin
- LHC
- light-harvesting complex
- Chl
- chlorophyll
- NAPol
- nonionic amphipol
- β-DDM
- n-dodecyl-β-d-maltoside
- FSC
- Fourier shell correlation
- PDB
- Protein Data Bank
- CEF
- cyclic electron flow.
References
- 1. Nelson N., and Ben-Shem A. (2005) The structure of photosystem I and evolution of photosynthesis. Bioessays 27, 914–922 10.1002/bies.20278 [DOI] [PubMed] [Google Scholar]
- 2. Baker D. R., Manocchi A. K., Lamicq M. L., Li M., Nguyen K., Sumner J. J., Bruce B. D., and Lundgren C. A. (2014) Comparative photoactivity and stability of isolated cyanobacterial monomeric and trimeric Photosystem I. J. Phys. Chem. B 118, 2703–2711 10.1021/jp407948p [DOI] [PubMed] [Google Scholar]
- 3. Kubota-Kawai H., Mutoh R., Shinmura K., Sétif P., Nowaczyk M. M., Rögner M., Ikegami T., Tanaka H., and Kurisu G. (2018) X-ray structure of an asymmetrical trimeric ferredoxin-photosystem I complex. Nat. Plants 4, 218–224 10.1038/s41477-018-0130-0 [DOI] [PubMed] [Google Scholar]
- 4. Watanabe M., Kubota H., Wada H., Narikawa R., and Ikeuchi M. (2011) Novel supercomplex organization of photosystem I in Anabaena and Cyanophora paradoxa. Plant Cell Physiol. 52, 162–168 10.1093/pcp/pcq183 [DOI] [PubMed] [Google Scholar]
- 5. Koziol A. G., Borza T., Ishida K., Keeling P., Lee R. W., and Durnford D. G. (2007) Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 143, 1802–1816 10.1104/pp.106.092536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240 10.1016/S1360-1385(99)01419-3 [DOI] [PubMed] [Google Scholar]
- 7. Scheller H. V., Jensen P. E., Haldrup A., Lunde C., and Knoetzel J. (2001) Role of subunits in eukaryotic Photosystem I. Biochim. Biophys. Acta 1507, 41–60 10.1016/S0005-2728(01)00196-7 [DOI] [PubMed] [Google Scholar]
- 8. Mazor Y., Borovikova A., and Nelson N. (2015) The structure of plant photosystem I super-complex at 2.8 Å resolution. Elife 4, e07433 10.7554/eLife.07433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qin X., Suga M., Kuang T., and Shen J. R. (2015) Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348, 989–995 10.1126/science.aab0214 [DOI] [PubMed] [Google Scholar]
- 10. Mazor Y., Borovikova A., Caspy I., and Nelson N. (2017) Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 3, 17014 10.1038/nplants.2017.14 [DOI] [PubMed] [Google Scholar]
- 11. Bassi R., Soen S. Y., Frank G., Zuber H., and Rochaix J. D. (1992) Characterization of chlorophyll-a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267, 25714–25721 [PubMed] [Google Scholar]
- 12. Stauber E. J., Fink A., Markert C., Kruse O., Johanningmeier U., and Hippler M. (2003) Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2, 978–994 10.1128/EC.2.5.978-994.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahashi Y., Yasui T. A., Stauber E. J., and Hippler M. (2004) Comparison of the subunit compositions of the PSI-LHCI supercomplex and the LHCI in the green alga Chlamydomonas reinhardtii. Biochemistry 43, 7816–7823 10.1021/bi035988z [DOI] [PubMed] [Google Scholar]
- 14. Mozzo M., Mantelli M., Passarini F., Caffarri S., Croce R., and Bassi R. (2010) Functional analysis of Photosystem I light-harvesting complexes (Lhca) gene products of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1797, 212–221 10.1016/j.bbabio.2009.10.005 [DOI] [PubMed] [Google Scholar]
- 15. Le Quiniou C., Tian L., Drop B., Wientjes E., van Stokkum I. H. M., van Oort B., and Croce R. (2015) PSI-LHCI of Chlamydomonas reinhardtii: increasing the absorption cross section without losing efficiency. Biochim. Biophys. Acta 1847, 458–467 10.1016/j.bbabio.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hippler M., Klein J., Fink A., Allinger T., and Hoerth P. (2001) Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28, 595–606 10.1046/j.1365-313X.2001.01175.x [DOI] [PubMed] [Google Scholar]
- 17. Tokutsu R., Teramoto H., Takahashi Y., Ono T. A., and Minagawa J. (2004) The light-harvesting complex of photosystem I in Chlamydomonas reinhardtii: protein composition, gene structures and phylogenic implications. Plant Cell Physiol. 45, 138–145 10.1093/pcp/pch013 [DOI] [PubMed] [Google Scholar]
- 18. Subramanyam R., Jolley C., Brune D. C., Fromme P., and Webber A. N. (2006) Characterization of a novel photosystem I-LHCI supercomplex isolated from Chlamydomonas reinhardtii under anaerobic (State II) conditions. FEBS Lett. 580, 233–238 10.1016/j.febslet.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 19. Stauber E. J., Busch A., Naumann B., Svatos A., and Hippler M. (2009) Proteotypic profiling of LHCI from Chlamydomonas reinhardtii provides new insights into structure and function of the complex. Proteomics 9, 398–408 10.1002/pmic.200700620 [DOI] [PubMed] [Google Scholar]
- 20. Marco P., Kozuleva M., Eilenberg H., Mazor Y., Gimeson P., Kanygin A., Redding K., Weiner I., and Yacoby I. (2018) Binding of ferredoxin to algal photosystem I involves a single binding site and is composed of two thermodynamically distinct events. Biochim. Biophys. Acta 1859, 234–243 10.1016/j.bbabio.2018.01.001 [DOI] [PubMed] [Google Scholar]
- 21. Ozawa S. I., Bald T., Onishi T., Xue H., Matsumura T., Kubo R., Takahashi H., Hippler M., and Takahashi Y. (2018) Configuration of ten light-harvesting chlorophyll a/b complex I subunits in Chlamydomonas reinhardtii photosystem I. Plant Physiol. 178, 583–595 10.1104/pp.18.00749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Germano M., Yakushevska A. E., Keegstra W., van Gorkom H. J., Dekker J. P., and Boekema E. J. (2002) Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525, 121–125 10.1016/S0014-5793(02)03100-9 [DOI] [PubMed] [Google Scholar]
- 23. Kargul J., Nield J., and Barber J. (2003) Three-dimensional reconstruction of a light-harvesting complex I-photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii: insights into light harvesting for PSI. J. Biol. Chem. 278, 16135–16141 10.1074/jbc.M300262200 [DOI] [PubMed] [Google Scholar]
- 24. Kargul J., Turkina M. V., Nield J., Benson S., Vener A. V., and Barber J. (2005) Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under State 2 conditions. FEBS J. 272, 4797–4806 10.1111/j.1742-4658.2005.04894.x [DOI] [PubMed] [Google Scholar]
- 25. Drop B., Webber-Birungi M., Fusetti F., Kouřil R., Redding K. E., Boekema E. J., and Croce R. (2011) Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J. Biol. Chem. 286, 44878–44887 10.1074/jbc.M111.301101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steinbeck J., Ross I. L., Rothnagel R., Gäbelein P., Schulze S., Giles N., Ali R., Drysdale R., Sierecki E., Gambin Y., Stahlberg H., Takahashi Y., Hippler M., and Hankamer B. (2018) Structure of a PSI-LHCI-cyt b6f supercomplex in Chlamydomonas reinhardtii promoting cyclic electron flow under anaerobic conditions. Proc. Natl. Acad. Sci. U.S.A. 115, 10517–10522 10.1073/pnas.1809973115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drop B., Yadav K. N. S., Boekema E. J., and Croce R. (2014) Consequences of state transitions on the structural and functional organization of photosystem I in the green alga Chlamydomonas reinhardtii. Plant J. 78, 181–191 10.1111/tpj.12459 [DOI] [PubMed] [Google Scholar]
- 28. Sharma K. S., Durand G., Gabel F., Bazzacco P., Le Bon C., Billon-Denis E., Catoire L. J., Popot J. L., Ebel C., and Pucci B. (2012) Non-ionic amphiphilic homopolymers: synthesis, solution properties, and biochemical validation. Langmuir 28, 4625–4639 10.1021/la205026r [DOI] [PubMed] [Google Scholar]
- 29. Bazzacco P., Billon-Denis E., Sharma K. S., Catoire L. J., Mary S., Le Bon C., Point E., Banères J. L., Durand G., Zito F., Pucci B., and Popot J. L. (2012) Nonionic homopolymeric amphipols: application to membrane protein folding, cell-free synthesis, and solution nuclear magnetic resonance. Biochemistry 51, 1416–1430 10.1021/bi201862v [DOI] [PubMed] [Google Scholar]
- 30. Tokutsu R., Kato N., Bui K. H., Ishikawa T., and Minagawa J. (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J. Biol. Chem. 287, 31574–31581 10.1074/jbc.M111.331991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kucukelbir A., Sigworth F. J., and Tagare H. D. (2014) Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 10.1038/nmeth.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goddard T. D., Huang C. C., and Ferrin T. E. (2007) Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 10.1016/j.jsb.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 33. Li X., Zhang R., Patena W., Gang S. S., Blum S. R., Ivanova N., Yue R., Robertson J. M., Lefebvre P. A., Fitz-Gibbon S. T., Grossman A. R., and Jonikas M. C. (2016) An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387 10.1105/tpc.15.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 35. Pi X., Tian L., Dai H. E., Qin X., Cheng L., Kuang T., Sui S. F., and Shen J. R. (2018) Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl. Acad. Sci. U.S.A. 115, 4423–4428 10.1073/pnas.1722482115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwai M., Takizawa K., Tokutsu R., Okamuro A., Takahashi Y., and Minagawa J. (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464, 1210–1213 10.1038/nature08885 [DOI] [PubMed] [Google Scholar]
- 37. Sueoka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 46, 83–91 10.1073/pnas.46.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gorman D. S., and Levine R. P. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mutoh R., Muraki N., Shinmura K., Kubota-Kawai H., Lee Y. H., Nowaczyk M. M., Rögner M., Hase T., Ikegami T., and Kurisu G. (2015) X-ray structure and nuclear magnetic resonance analysis of the interaction sites of the Ga-substituted cyanobacterial ferredoxin. Biochemistry 54, 6052–6061 10.1021/acs.biochem.5b00601 [DOI] [PubMed] [Google Scholar]
- 40. Oda Y., Huang K., Cross F. R., Cowburn D., and Chait B. T. (1999) Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 96, 6591–6596 10.1073/pnas.96.12.6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scheres S. H. W. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kimanius D., Forsberg B. O., Scheres S. H. W., and Lindahl E. (2016) Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 10.7554/eLife.18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rohou A., and Grigorieff N. (2015) CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akimoto S., Yokono M., Hamada F., Teshigahara A., Aikawa S., and Kondo A. (2012) Adaptation of light-harvesting systems of Arthrospira platensis to light conditions, probed by time-resolved fluorescence spectroscopy. Biochim. Biophys. Acta 1817, 1483–1489 10.1016/j.bbabio.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 45. O'Connor D. V., and Phillips D. (1984) Time-correlated Single Photon Counting, Academic Press, Inc., London [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.