Abstract
Sensitive assays of biochemical specificity, affinity, and capacity are valuable both for basic research and drug discovery. We created fluorescent sensors that monitor high-affinity binding reactions and used them to study iron acquisition by ESKAPE bacteria, which are frequently responsible for antibiotic-resistant infections. By introducing site-directed Cys residues in bacterial iron transporters and modifying them with maleimide fluorophores, we generated living cells or purified proteins that bind but do not transport target compounds. These constructs sensitively detected ligand concentrations in solution, enabling accurate, real-time spectroscopic analysis of membrane transport by other cells. We assessed the efficacy of these “fluorescent decoy” (FD) sensors by characterizing active iron transport in the ESKAPE bacteria. The FD sensors monitored uptake of both ferric siderophores and hemin by the pathogens. An FD sensor for a particular ligand was universally effective in observing the uptake of that compound by all organisms we tested. We adapted the FD sensors to microtiter format, where they allow high-throughput screens for chemicals that block iron uptake, without genetic manipulations of the virulent target organisms. Hence, screening assays with FD sensors facilitate studies of mechanistic biochemistry, as well as discovery of chemicals that inhibit prokaryotic membrane transport. With appropriate design, FD sensors are potentially applicable to any pro- or eukaryotic high-affinity ligand transport process.
Keywords: membrane transport, fluorescence, ligand-binding protein, siderophore, pathogenesis, iron, spectroscopy, heme, ESKAPE pathogen
Introduction
Sensitive assays of biochemical specificity, affinity, and capacity are valuable in many experimental settings, including biochemical measurements of living cells, and the discovery of new therapeutic compounds. At present, a need exists for new antibiotics against multidrug-resistant bacteria because without them treatment options for many infections will dwindle (1). Gram-negative strains cause about two-thirds of the mortality from bacteria in United States hospitals, and up to 20% of these strains are resistant to all antibiotics (2). The World Health Organization identified the Gram-negative ESKAPE5 pathogens Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. as well as carbapenem-resistant strains of K. pneumoniae and Escherichia coli as critical targets for drug discovery (3). Compounds that inhibited bacterial cell envelope processes were once the most potent antibiotics, but now their efficacy has diminished as a result of bacterial adaptation (4). Gram-negative bacterial antibiotic resistance largely originates from cell envelope physiology: the outer membrane (OM) excludes large or hydrophobic compounds (5), periplasmic enzymes degrade or inactivate chemicals that breach the OM barrier, and inner membrane (IM) pumps expel hydrophobic molecules, including antibiotics (6). Similar processes in Gram-positive bacteria heighten the need for new treatments of bacterial infections.
Nevertheless, bacterial pathogens face nutritional obstacles in human and animal hosts that we may exploit against them. Iron, for example, is a cofactor in many biochemical pathways, including catabolism, DNA synthesis, and bioenergetics (7). Iron deprivation retards bacterial growth, whereas iron availability promotes it (8). Animals sequester iron in transferrin, lactoferrin, and ferritin to minimize bacterial infections (9). However, microbial siderophores (10–12) capture the metal from host proteins (13), and ferric siderophores then enter bacterial cells through high-affinity acquisition systems (14). The balance of these competing processes influences the outcome of infection, so bacterial iron uptake systems are potential targets for antibiotics.
Toward that end, we studied the ferric enterobactin (FeEnt) uptake system (14) of Gram-negative bacteria. FeEnt enters through the OM protein FepA, which selectively binds and transports it. The uptake reaction requires the additional cell envelope protein TonB (15), so FepA and other OM proteins in this superfamily are called “TonB-dependent transporters” (TBDTs) (16). TonB action, which is proposed to transmit the potential energy of the electrochemical proton gradient across the IM to TBDT in the OM (17), underlies the active intake of bound iron complexes (18). TonB is ubiquitous in Gram-negative bacteria and a determinant of their pathogenesis (12, 14, 19). Iron acquisition through TBDT contributes to the invasiveness of the Gram-negative ESKAPE pathogens (20), the determination of their ultimate localization in human and animal tissues (21, 22), and the overall outcome of their infections (9, 12, 14).
We previously designed fluorescence methods that monitored high-affinity ligand binding and uptake (23, 24). By genetically engineering FepA and then labeling it with fluorescein maleimide (FM; Fig. S1), we created a sensor that reflected [FeEnt] in the environment. When FeEnt bound to FepA-FM, it quenched its fluorescence. Quenching originated from binding-induced conformational motion in the external loops of FepA, which subjected attached fluorophores to collisions with other residue side chains or elements of protein structure or increased interactions with the aqueous environment (23, 24). When cellular uptake depleted FeEnt from solution, fluorescence rebounded. Consequently, FepA-FM tracked the binding and transport of FeEnt by living bacteria.
In this study, we created both species-specific and generic fluorescence spectroscopic assays (Figs. 1 and S1) that measure ligand uptake. Fluorescently labeled, binding-competent, but transport-defective cells and fluorescently labeled purified proteins were key innovations in these experiments. Such genetically engineered cells or proteins comprise “fluorescent decoy” (FD) sensors, which report ligand concentrations in solution and monitor transport of the same ligand by other cells. We illustrate these applications by observations of iron transport activity through Gram-negative bacterial TBDTs and Gram-positive bacterial NEAT domain–dependent hemin (Hn) uptake systems. Both high-affinity pathways function at nanomolar concentrations, and both contribute to bacterial colonization of eukaryotic hosts (10, 12). We adapted these fluorescence tests to a microtiter high-throughput screening format to enable discovery of inhibitors that retard iron acquisition and thereby prevent bacterial growth and pathogenesis.
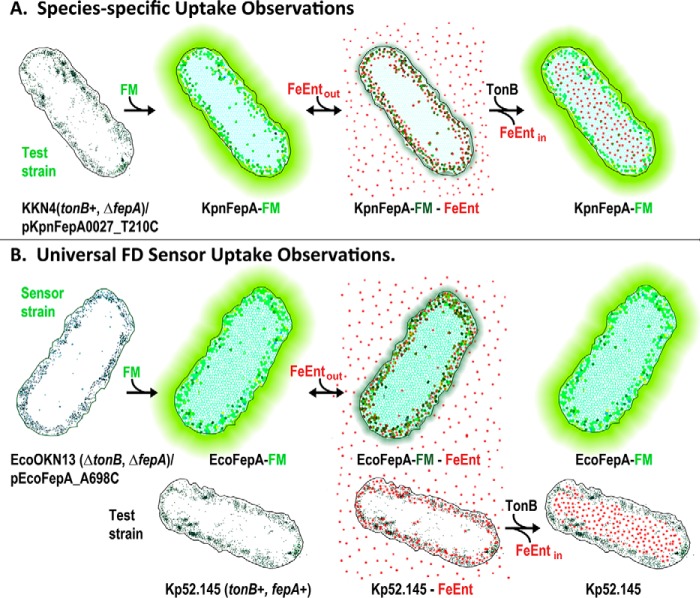
Figure 1.
Fluorescence observations of membrane transport. A, species-specific assay design. The target organism (e.g. K. pneumoniae) contains a Cys substitution for a surface-exposed residue in a TonB-dependent transporter (KpnFepA_T210C) labeled with FM. Fluorescence emissions depict FeEnt binding and TonB-dependent FeEnt transport in K. pneumoniae. B, universal FD sensor assay design. The fluorescence of a TonB-deficient E. coli “sensor” strain OKN13 (ΔtonB, ΔfepA)/pEcoFepA_A698C-FM reflects TonB-dependent FeEnt transport by a second test strain (e.g. K. pneumoniae strain Kp52.145) in the same solution.
Results
Universal fluorescence sensor of FeEnt acquisition
We originally devised species- or transporter-specific fluorescence assays of FeEnt uptake for FepA of E. coli (23) and A. baumannii (25); the same approach was effective for FepA of K. pneumoniae (Fig. 2). We cloned and genetically engineered KpnfepA of strain Kp52.145 to enable site-directed fluoresceination in its native cellular (i.e. K. pneumoniae) environment and in E. coli. These studies compared the acquisition of FeEnt by FepA orthologs in different bacteria. As for FepA of E. coli (23) and A. baumannii (25), FM exclusively labeled KpnFepA in living cells (Figs. S1 and S2; see Table S1 for strains, plasmids, and abbreviations), and the fluoresceinated OM protein detected and quantified its ligand, FeEnt. Binding of FeEnt to KpnFepA-FM quenched its fluorescence, but as the transporter's activity depleted FeEnt from solution, fluorescence intensity rebounded (Fig. 2A). Thus, by the species-specific approach, EcoFepA-FM (23), AbaFepA-FM (25), and KpnFepA-FM (Fig. 2A) all detected FeEnt binding and reflected its transport. Each of these transporters requires TonB action for FeEnt uptake: its absence or inhibition prevented fluorescence recovery. By engineering the individual transporters of specific bacteria, we observed, monitored, and characterized their TonB-dependent iron uptake processes.
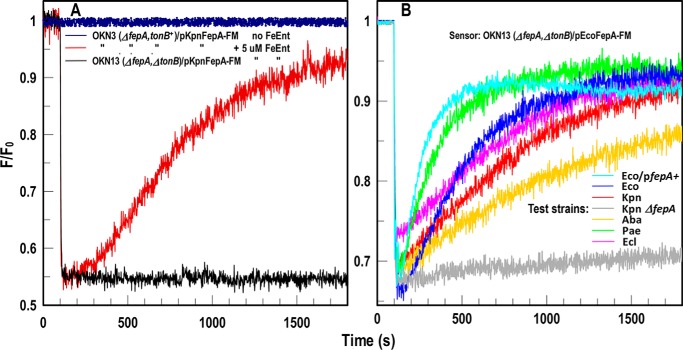
Figure 2.
FD sensor analysis of FeEnt acquisition by ESKAPE pathogens. A, species-specific fluorescence assay of FeEnt uptake. KpnFepA-FM. E. coli OKN3 (ΔfepA) expressing KpnFepA-FM was exposed to FeEnt in PBS + 0.2% glucose. The tracings show emissions of living cells in the absence (dark blue) or presence (dark red or black) of FeEnt. In tonB+ cells (dark red), FeEnt binding to KpnFepA-FM (at 100 s) quenched emissions, but subsequent uptake depleted FeEnt from solution, restoring fluorescence. In TonB-deficient E. coli OKN13 (ΔfepA, ΔtonB)/pKpnFepA-FM; black), which cannot transport FeEnt, quenching occurred but not recovery. B, Universal fluorescence assay of FeEnt uptake using sensor strain OKN13/pEcoFepA-FM. FeEnt binding at 100 s quenched the fluorescence of the FD sensor strain E. coli OKN13/pEcoFepA-FM. It cannot transport FeEnt, so subsequent recovery did not occur. However, the fluorescence of the E. coli strain reflected FeEnt uptake by other bacteria in the same solution: as they decreased [FeEnt] by cellular transport, the intensity of OKN13/pEcoFepA-FM returned to initial levels (as described in Fig. 1B). The assay monitored FeEnt uptake by E. coli expressing chromosomal (Eco; cyan) or plasmid-mediated (Eco/pfepA+; blue) FepA, and ESKAPE species K. pneumoniae (Kpn; red), A. baumannii (Aba; gold), P. aeruginosa (Pae; green), and E. cloacae (Ecl; magenta). FepA-deficient K. pneumoniae (KKN4; black) does not transport FeEnt and did not recover. Hence, OKN13/pEcoFepA-FM was a universal sensor of [FeEnt] that monitored FeEnt acquisition by all the bacteria. The panels show the results of single representative experiments that were performed in triplicate, from which we calculated and plotted the mean values. Standard deviations of the mean measurements (data not shown) were 1.7–2.7%.
The species-specific method was sensitive and accurate, but genetic and biochemical manipulations of ESKAPE organisms and other pathogens are technically challenging and potentially hazardous. We discovered a modification that extends the scope of the assay to allow observations of FeEnt uptake by other organisms, including clinical isolates, without genetically engineering them. TonB-deficient Gram-negative cells adsorb ferric siderophores and other metal complexes (e.g. vitamin B12 (15, 16)) but cannot transport them. Thus, FeEnt bound to E. coli OKN13 (ΔtonB, ΔfepA)/pEcoFepA-FM, quenching its fluorescence, but the strain's TonB deficiency prevented FeEnt internalization (Figs. 1B and 2B). The placement of EcoFepA-FM in OKN13 transformed the cell into an FD sensor, whose emissions inversely related to [FeEnt]. The transport-defective, physiologically inert, fluorescent ΔtonB bacteria detected the presence and concentration of FeEnt. When this strain cohabited an environment with other bacteria, it reflected their uptake of FeEnt. Solution levels that saturated EcoFepA-FM (Kd = 0.2 nm (24, 26)) quenched its fluorescence, but if the ambient [FeEnt] decreased from its uptake by other cells, then fluorescence rebounded. The upshot is that the ΔtonB FD sensor cells monitored FeEnt transport by wild isolates of K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter cloacae (Figs. 1B and 2B). The FD sensor observed FeEnt uptake by all the organisms we tested, reflecting TonB action in the pathogens.
Affinity determinations by spectroscopic analyses
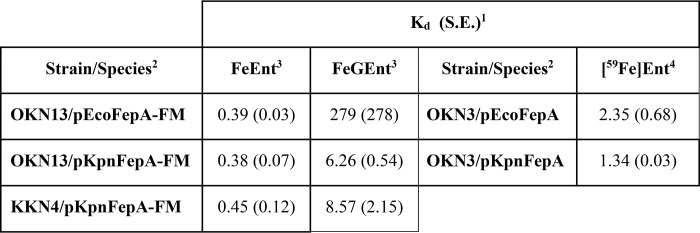
The analytical capabilities of the fluorescence spectroscopic determinations stood out when we compared the recognition of FeEnt and its glucosylated form, FeGEnt (also known as ferric salmochelin (27)), by EcoFepA and KpnFepA. The ability of Gram-negative pathogens, including K. pneumoniae, to glucosylate enterobactin and transport FeGEnt enhances their invasiveness (21), in part because glucosylation impairs recognition of the iron complex by lipocalins (28, 29). Additionally, whereas E. coli K12 contains a single chromosomal fepA locus that encodes its FeEnt transporter, pathogenic K. pneumoniae contain at least three fepA orthologs (chromosomal loci 1658 and 4984 and plasmid PII locus 0027). IroN of Salmonella enterica serovar Typhimurium has a role in FeGEnt uptake (27); its closest ortholog in K. pneumoniae is plasmid-mediated fepA0027. After bioinformatics analyses and structural modeling of KpnFepA0027, we cloned its structural gene and engineered the Cys substitution T210C (Fig. S1), which is nominally equivalent to T216C in EcoFepA (24). To define its recognition specificity, we expressed and FM-labeled KpnFepA0027_T210C (KpnFepA-FM) in E. coli OKN13 (ΔtonB, ΔfepA) and in K. pneumoniae KKN4 (ΔentB, ΔfepA0027, ΔfepA1658, ΔfepA4984) relative to EcoFepA-FM in OKN13. The comparison of FeEnt and FeGEnt adsorption to KpnFepA-FM and EcoFepA-FM in different bacteria revealed several things. First, KpnFepA was fluoresceinated to 3-fold higher levels in E. coli (Fig. 3B) than in its native K. pneumoniae (Fig. 3C). The better labeling of KpnFepA in the E. coli sensor strain made it more responsive to quenching, resulting in more accurate measurements (smaller variations in the mean extent of quenching). Second, both KpnFepA-FM and EcoFepA-FM preferred FeEnt (Kd ≈ 0.38 nm) over FeGEnt (Kd = 6–280 nm; Fig. 3 and Table 1). These data recapitulate previously published affinities and specificities of EcoFepA for FeEnt (26, 30). FeGEnt adsorbed to KpnFepA with 20-fold lower affinity (Kd ≈ 6.3 nm) than FeEnt, and FeGEnt barely bound to EcoFepA at all (Fig. 3A), so weakly that it was problematical to make an accurate measurement (Table 1). Despite its 3-fold lower specific fluoresceination in K. pneumoniae, KpnFepA-FM showed the same preferences and affinities in that environment (Fig. 3, B and C). Lastly, we compared the spectroscopic tests with conventional radioisotopic binding measurements. The latter approach with [59Fe]Ent yielded comparable affinities of EcoFepA and KpnFepA for FeEnt (Fig. 3E and Table 1), but the former fluorescence methods were more sensitive, with equivalent or better reproducibility and more accurate statistical fits (Table 1). Overall, the experiment showed that individual, fluorescently labeled transporters (i.e. the species-specific approach) accurately depicted the concentration dependence of ligand binding, with best sensitivity in the E. coli cell envelope and less but acceptable accuracy in the more complex cell envelopes of the infectious bacteria.
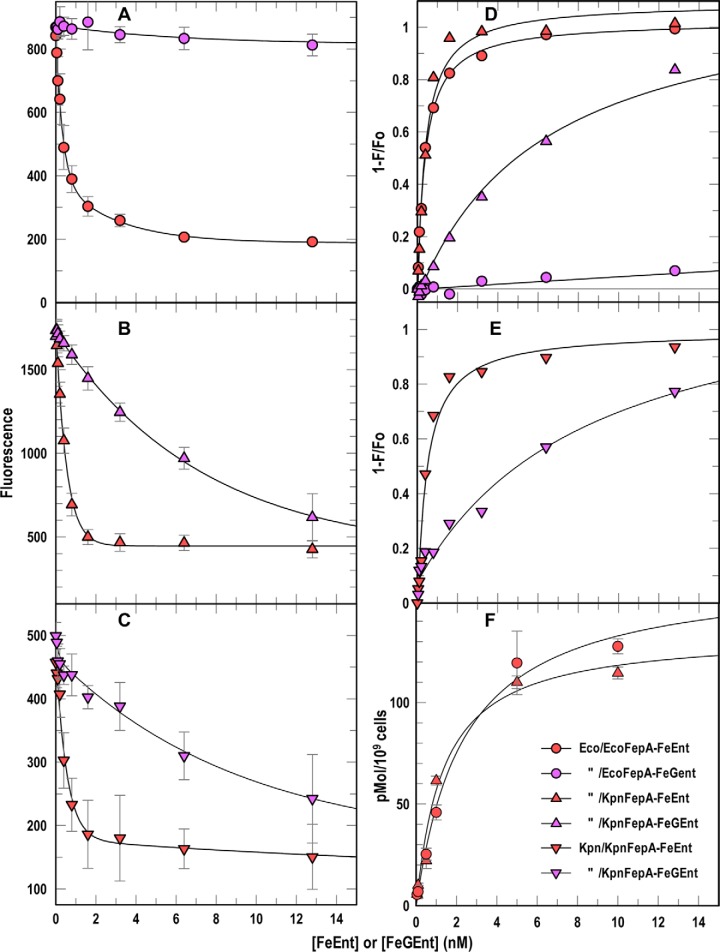
Figure 3.
Binding affinities from spectroscopic assays. A–E, we added FeEnt (red symbols) or FeGEnt (purple symbols) to E. coli OKN13/pEcoFepA-FM (A and D, circles) or OKN13/pKpnFepA-FM (B and D, triangles) or K. pneumoniae KKN4/pKpnFepA-FM (C and E, inverted triangles). A–C, quenching data from this species-specific test quantified binding of the iron complexes. Nonlinear fits of 1 − F/F0 (D and E) using the one-site (with background) equation of Grafit 6.02 revealed the affinities (Kd) of the binding reactions (Table 1). F, [59Fe]Ent binding measurements. We added varying concentrations of [59Fe]Ent to OKN3/pEcoFepA (circles) or OKN3/pKpnFepA (triangles), measured adsorption of the ferric siderophore by filter-binding assays (26), and obtained nonlinear fits to the one-site (with background) equation of Grafit 6.02, which gave the affinities (Kd) of the binding reactions (Table 1). In each panel the error bars represent S.D. of triplicate measurements at each concentration tested.
Table 1.
Affinities of EcoFepA and KpnFepA0027 for ferric catecholates
1 Kd values were calculated by the one-site (with background) equation of Grafit 6.02.
2 Bacteria were grown in iron-deficient minimal MOPS medium to 5 × 108 cells/ml. For fluorescence studies, they were modified with FM and washed with and resuspended in PBS.
3 Binding determinations were conducted in a 10 ml volume in an Olis Clarity spectrofluorometer by adding varying concentrations of purified FeEnt or FeGEnt to bacteria at 1.25 × 107 cells/ml and observing quenching. The excitation/emission maxima were 488/520 nm.
4 [59Fe]Ent binding determinations were conducted in 10 ml of ice-cold PBS (26), with cells chilled on ice.
Transport rates from spectroscopic analyses
The rate of fluorescence recovery in the spectroscopic system correlates to the rate of ligand depletion by bacterial transport (23), and comparisons of test strains in universal FD uptake assays reiterated this point. The various ESKAPE bacteria conferred fluorescence recovery at different rates (Fig. 2B). Because we tested the individual bacteria at equal cell concentrations, their rates were directly comparable with one another. However, absolute calculations of rate parameters (i.e. Vmax and kcat) from FD recovery time courses are complicated by other factors besides the kinetic attributes of the transporters, including their expression levels and, in this system, the expression of TonB. Consequently, FD assays are best suited to comparative rate measurements among the test strains, from either the slope of the recovery curve or the duration of quenching after initiation of uptake (Fig. 4). In the former case, when uptake of FeEnt reverts fluorescence to a level of 50%, the EcoFepA-FM sensor is half-saturated, so [FeEnt] = Kd (0.38 nm). The slope of the time course at this point reveals the uptake rate of the test strain at [FeEnt] = 0.38 nm. In the latter case, the test strain's transport rate determines the elapsed time from addition of FeEnt to half-maximal quenching (i.e. half-saturation). For example, at 107 cells/ml, K. pneumoniae Kp52.145 required 520 s to deplete 5 nm FeEnt to 0.38 nm (i.e. half-saturation), which translates into a mean uptake rate over the first 520 s of 53 pmol/109 cells/min. The hypothetical mean uptake rate over this range of saturation (14.3Km → Km) is ∼ 0.8 Vmax, suggesting that for K. pneumoniae Kp52.145, Vmax ≈ 53 pmol/109 cells/min ÷ 0.8 ≈ 66 pmol/109 cells/min. This value approximates Vmax for chromosomally expressed FepA in E. coli (50–100 pmol/min/109 cells (31)). We conducted this prototypic assay with K. pneumoniae in the presence of an E. coli EcoFepA-FM FD sensor, but the test similarly functioned for all of the Gram-negative bacteria (Fig. 2). FD assays are conceptually universal in that an appropriately configured sensor will function in many biological systems, with few exceptions (see “Discussion”), to monitor changes in the concentration of its target ligand.
Figure 4.
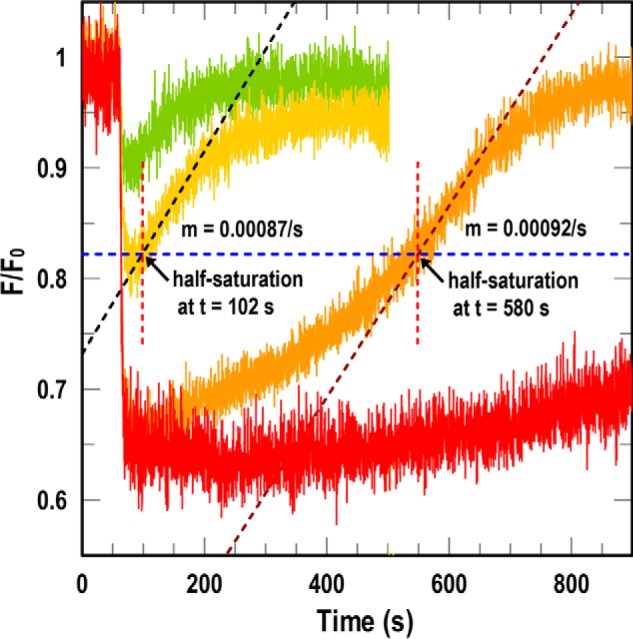
Uptake rates from FD assays. At t = 60 s we added varying concentrations of FeEnt ((0.5 (green), 1 (yellow), 5 (orange), and 10 nm (red)) to sensor strain OKN13/pEcoFepA-FM in the presence of test strain K. pneumoniae isolate Kp52.145. 10 nm FeEnt gave maximal quenching. For 1 and 5 nm FeEnt, at half-saturation (blue dashed line) we determined the slope of the time course (black dashed line) and the elapsed time (dashed red line). Either parameter estimated the transport rate of the test strain (see text).
Universal FD sensor of ferrichrome (Fc) acquisition
The FD sensor approach was relevant to the uptake of other iron complexes by E. coli and other bacteria. We engineered mutation D396C in EcoFhuA, the OM Fc transporter (32), and modified it with FM (Fig. S1B). EcoFhuA-FM measured Fc uptake in tonB+ E. coli (Fig. 5A), but in the TonB-deficient strain OKN1, EcoFhuA-FM was an FD sensor that detected Fc uptake by any of the carbapenem-resistant/ESKAPE bacteria (Fig. 5B). As seen for FeEnt–EcoFepA-FM, binding of Fc to EcoFhuA-FM quenched its emissions, and depletion of Fc from solution by microbial transport restored fluorescence intensity. This additional test of a second TonB-dependent transport reaction by the ESKAPE species, without the need to genetically modify them, showed the breadth of the FD concept: an appropriately labeled TBDT in a ΔtonB host became a transport-deficient FD sensor cell that reflected the concentration of its ligand in solution. This approach potentially encompasses the diverse repertoire of microbial ferric siderophores (33) and their membrane protein receptors. It is also adaptable to other metal complexes that microbes utilize and nonmetal, high-affinity ligand–receptor pairs.
Figure 5.
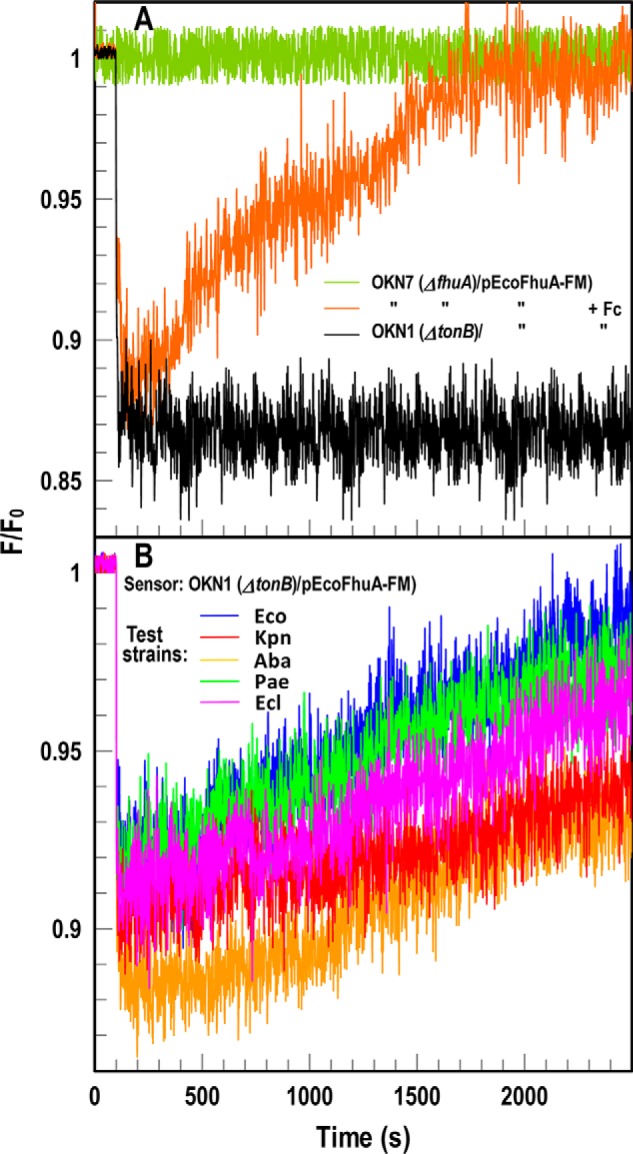
FD sensor analysis of Fc acquisition by ESKAPE pathogens. A, species-specific fluorescence assay of Fc uptake. OKN7 (ΔfhuA)/pEcoFhuA-FM reflects Fc binding and transport (orange). In OKN1 (ΔtonB; black) the same construct became an inert sensor of [Fc]: binding of Fc quenched fluorescence, and the absence of transport prevented recovery to initial levels. B, universal assay of Fc uptake. We incubated the sensor strain with 1.5 × 107 cells of Gram-negative ESKAPE pathogens. Colored tracings denote the same strains as in Fig. 2. In each case, the pathogens acquired Fc, causing fluorescence recovery of the sensor strain. Uptake of Fc occurred at a slower rate than FeEnt (30), resulting in slower fluorescence recovery. The panels show the results of single representative experiments that we performed in triplicate, from which we calculated and plotted the mean values. Standard deviations of the mean measurements (data not shown) were 2.5–4%.
Universal FD sensor of heme uptake
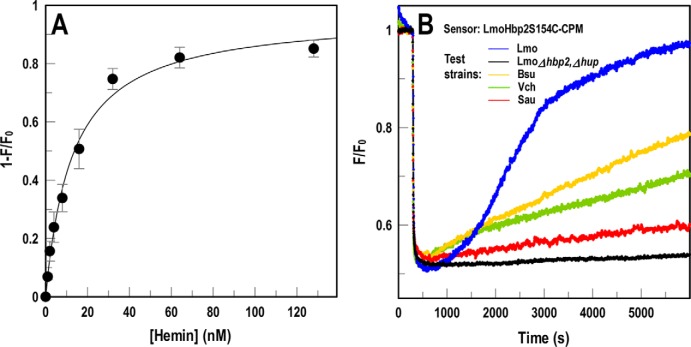
Using a slightly modified method, the FD sensor technology described a third relevant microbial transport system: Hn uptake by Gram-positive bacteria. Listeria monocytogenes, Staphylococcus aureus, Streptococcus pyogenes, Bacillus anthracis, and other Gram-positive bacteria produce NEAT domain–containing Hn-binding proteins, which either anchor to peptidoglycan by the actions of sortases A (34) and B (35) or pass to the external environment where they act as hemophores (36). Some Gram-negative species like Serratia marcescens also secrete hemophores (e.g. HasA (37)), and others (e.g. Vibrio cholerae (38)) directly transport the iron porphyrin. We engineered the listerial NEAT-domain protein Hbp2 to create an FD sensor of Hn binding and uptake (Figs. S1C and S3). Introduction of Cys in NEAT1 of LmoHbp2 allowed its modification by extrinsic fluorophores, transforming the purified binding protein into a sensitive sensor whose emissions inversely related to [Hn]. LmoHbp2_S154C-CPM (LmoHbp2-CPM) detected Hn in solution (Fig. 6A) with an affinity (Kd = 12 ± 3.1 nm) that was consistent with isocalorimetric (Kd = 25–48 nm (39)) and radioisotopic (Kd = 12 nm (36)) data. The sensor also revealed the ability of other bacteria to transport Hn (Fig. 6B) at different rates. Overall, the bacterial cells OKN13/pEcoFepA-FM and OKN1/pEcoFhuA-FM and the purified protein LmoHbp2-CPM functioned as universal sensors of extracellular iron in different forms: FeEnt, Fc, and hemin, respectively.
Figure 6.
FD sensor assay of hemin uptake. We substituted Cys for residue Ser-154 in NEAT1 of LmoHbp2, expressed and purified the binding protein, and covalently modified it with CPM. A, concentration dependence of hemin binding to LmoHbp2-CPM. Using purified LmoHbp2-CPM, plots of 1 − F/F0 versus [hemin], analyzed by nonlinear fit to the one-site (with background) equation of Grafit 6.02, produced a saturation curve with Kd = 12 ± 3.1 nm. The error bars represent S.D. of triplicate measurements at each concentration tested. B, Hbp2-CPM detects and quantifies bacterial hemin transport. When we mixed Hbp2-CPM with heterologous bacteria in solution, the assay reflected their hemin uptake. Addition of hemin at 300 s quenched LmoHbp2-CPM emissions, but fluorescence rebounded as bacteria (L. monocytogenes (Lmo; blue), Bacillus subtilis (Bsu; gold), S. aureus (Sau; red), and V. cholerae (Vch; green)) transported the porphyrin and depleted it from solution. Conversely, hemin transport–deficient cells (EGDe Δhbp2, Δhup; black) did not elicit fluorescence recovery. The panel shows the results of a single representative experiment that was performed in triplicate, from which we calculated and plotted the mean values. Standard deviations of the mean measurements (data not shown) were 2–2.9%.
Adaptation of FD sensors to fluorescence high-throughput screening (FLHTS)
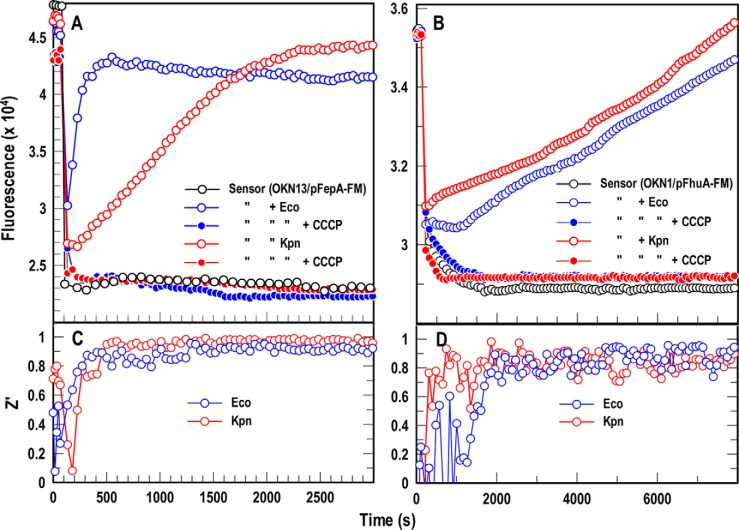
Compounds that block iron acquisition by inhibiting TonB action in Gram-negative bacteria or by reducing Hn uptake into Gram-positive cells may combat bacterial proliferation in human and animal hosts. We previously used the species-specific spectroscopic test in microtiter format to screen chemical libraries for inhibitors of E. coli TonB. Consequently, we transposed the FD sensor assays to microtiter format for FLHTS. However, despite the fact that all the Gram-negative ESKAPE bacteria utilized FeEnt (Fig. 2), the adaptation of this method to screen against them faced obstacles. First, bacterial pathogens often manifest reduced cell envelope permeability that lowers antibiotic susceptibility. Compounds that penetrate E. coli may not enter ESKAPE organisms. Second, protein orthologs in different bacteria typically manifest cross-species sequence divergence, so compounds that inhibit EcoTonB may not similarly impair TonB in Gram-negative ESKAPE bacteria. For these reasons it is preferable to directly screen chemical libraries against the target ESKAPE pathogens. The species-specific approach is adaptable to ESKAPE bacteria for FLHTS (as seen for A. baumannii (25)) and K. pneumoniae (Fig. 2), but more extensive lipopolysaccharides and capsular polysaccharides in clinical isolates reduce the efficiency of modifying their OM proteins with fluorophores (e.g. Kp52.145 in Fig. 3). FD sensor assays with fluorescent cells or proteins circumvent this problem because they allow FLHTS against the pathogens (with adequate Z′ of 0.8–1.0; Fig. 7) without the need to fluorescently modify them. The E. coli FD sensor cells in microtiter wells followed the TonB-dependent uptake of both FeEnt and Fc by K. pneumoniae. The sensors minimized the experimental manipulations of the pathogen while allowing spectroscopic observations of its physiology that were sensitive to varying concentrations of both the ligands and the bacterial cells (Fig. S4).
Figure 7.
A and B, FD HTS assay for Gram-negative bacterial TonB-dependent FeEnt (A) and Fc (B) transport. We used a ΔtonB host harboring either pEcoFepA-FM (A) or pEcoFhuA-FM (B), suspended in microtiter plates, to create universal FD sensors of FeEnt or Fc uptake, respectively. The inert sensors effectively monitored active iron transport in 96-well microtiter plates by both E. coli (blue) and K. pneumoniae (red). The proton ionophore CCCP (23) abrogated iron uptake, so fluorescence did not recover in the presence of these compounds. C and D, statistical comparisons of these positive and negative controls yielded Z′ factors approaching 1.0 within 10 min in the case of FeEnt (C) and 35 min in the case of Fc (D). The panels show the results of single representative experiments that were performed in triplicate, from which we calculated and plotted the mean values. Standard deviations of the mean measurements (data not shown) were 1.9–4.1%.
Discussion
We considered several attributes of the FD constructs in making the designation “universal.” (i) Sensors for FeEnt, Fc, or heme had the ability to detect and quantify the uptake of their target ligand by any Gram-positive or -negative bacterium we tested. This unrestricted scope alone strongly supported the term universal, but FD sensors had additional universalities. (ii) The technical concept was applicable to virtually any high-affinity ligand whether ferric siderophores, other metal complexes, or other molecules. (iii) FD sensors were straightforward to create from any cloned, high-affinity binding protein as we showed for EcoFepA, KpnFepA, EcoFhuA, and LmoHbp2. These fluorescently labeled, Cys-mutant proteins detected and discriminated FeEnt, FeGEnt, Fc, and hemin. Hence, it is conceivable to create sensors for any iron complex of interest or other high-affinity ligand. (iv) The site-directed Cys-fluorophore labeling approach succeeded with any maleimide fluorophore we tried: FM, 7-diethylamino-3-(4′-maleimidylphenyl)-4-methyl coumarin (CPM), and in previous work Alexa Fluors 546, 555, 647 and 680, which differ in mass, structure, and spectral properties. (v) Because the sensors are expressed from plasmids, the engineered BSL-1 sensor strains that host them may be centrally stored and distributed to any interested researcher. (vi) FD sensors may achieve many biochemical and microbiological purposes: determination of the presence and concentrations of siderophores in natural and host environments; identification of the siderophores elaborated or the ferric siderophores transported by an organism; surveys of microbiological families, genera, or species to determine which members use a certain metal complex; and measurements of the specificities, affinities, and kinetic attributes of a transport system for a single ligand or a family of ligands. All these applications have the inherent analytical power to discriminate ligands in the nanomolar concentration range.
A technology that quantitatively monitors (i) any microorganism for uptake of (ii) any specific ligand using (iii) any cloned, high-affinity binding protein modified by (iv) any maleimide fluorophore with (v) central distribution to any investigator for applications that span (vi) a variety of microbiological and biochemical purposes has a claim to the term universal. Exceptions or clarifications may arise to this designation, but these may be recognized and stipulated.
The general applicability and technical attributes of FD sensors will aid antibiotic discovery. An HTS plan must include secondary screens and controls to exclude false positives. Fluorescence assays, for example, are susceptible to quenching artifacts that mimic inhibition. FLHTS against TonB (25) was complicated by the involved secondary screening needed to eliminate nonspecific quenchers and other irrelevant compounds. However, FD technology enables new fluorescence tests, in microtiter format, that measure the uptake of relevant compounds or identify artifacts. The FD sensor of Fc uptake is one such assay, which may validate or refute the TonB specificity of primary hits from FeEnt uptake screens. FD sensors of other relevant ligands (vitamin B12 and ferric aerobactin) are within reach, and each additional test of TonB-dependent transport focuses the experiments on the most valid candidates. FD sensors in FLHTS format permit more rapid screening, allowing expansion of screening programs to larger chemical libraries.
We found that FM modified KpnFepA to higher levels in E. coli than in a clinical isolate of K. pneumoniae, which was a technical advantage. This higher intensity, which likely derived from better fluoresceination in the unencapsulated, rough laboratory strain, led to better sensitivity to quenching during ligand binding. Consequently, the FD sensor yielded more accurate measurements in E. coli than in K. pneumoniae.
From this work and our experience with binding determinations (24, 26, 30), fluorescence studies are faster, simpler, more sensitive, more reproducible, and less hazardous than radioisotopic experiments. Optimum measurements of [59Fe]Ent–FepA affinity require a large assay volume (∼25 ml), to avoid substrate depletion at low concentrations, and [59Fe] with high specific radioactivity. The short half-life of the isotope (44.6 days) results in rapid loss of sensitivity. Fluorescence binding assays are not confounded by such problems because of the high intensity and environmental sensitivity of selected fluorophores (e.g. FM). Our fluorescent sensors reported higher affinities (Kd = 0.4 nm; Table 1) than the [59Fe]Ent–EcoFepA binding studies (Kd = 1–2 nm). The spectroscopic tests were also most consistent with previous determinations (Kd = 0.2–0.5 nm (26, 30)).
FD sensor assays are not undermined by adventitious iron or by siderophores secreted by test bacteria. We supplied chelated iron, as FeEnt, Fc, or Hn, for transport by the target cells. Neither extraneous Fe2+ nor Fe3+ interfere because FepA, FhuA, and Hbp2 have no affinity for unchelated iron. FD sensor cells carry mutations that block siderophore production. Aposiderophores from the test cells (e.g. enterobactin, aerobactin, acinetobactin, etc.) may bind adventitious iron in the assay buffers, but we curtail this problem by washing cells before analysis to remove any associated siderophores. The relatively short FD assay duration (<30 min) minimizes siderophore production by the test bacteria, which generally requires extended growth in iron-deficient media for secretion of substantial amounts. The FD assay buffer (PBS + 0.2% glucose) contains little adventitious iron and no metabolites, so it constrains biosynthesis by the test bacteria.
It is worthwhile to stipulate the limitations of the fluorescence approaches in determination of membrane transport parameters. Light absorption/scatter by the bacterial cells, which obscures emitted light from fluorophores, is an obstacle to accurate measurements. Dilute bacterial suspensions (i.e. low turbidity) produce the best data. The assay requires high-level, specific labeling of the cells or binding protein, resulting in bright emission from dilute cells (i.e. ∼107/ml) or protein. FD sensor assays do not directly yield cellular binding capacities or absolute transport rates (e.g. Vmax and kcat), both of which are available from radioisotopic studies (24, 26, 30). The FD sensor approach may fail for certain transport processes. The Tbp-mediated iron uptake systems of pathogenic Neisseria, which do not deplete their external transferrin ligand (40), illustrate this point. Although a neisserial TbpB-FM sensor that discriminates between holo- and apotransferrin (e.g.) is feasible, this objective requires further research and development.
The overall advantages of the FD sensor technology include high sensitivity, the technical sophistication of fluorescence instrumentation, and facile miniaturization/automation. In the absence of excessive turbidity, the strategic selection of maleimide fluorophores confers the potential to quantify ligands in numerous environments, including solutions of blood, serum, and eukaryotic cells. Appropriate adaptations may enable observations of bacterial IM or mammalian plasma membrane uptake reactions. If applied to eukaryotic phenomena like cancer, neurobiology, or metabolism, FD sensors will measure molecular concentrations, facilitate in vivo thermodynamic or kinetic characterizations of biochemical reactions, and empower identification of clinically relevant therapeutic agents.
Experimental procedures
Bacteria, plasmids, genetic engineering, and cell growth
E. coli strains were descended from BN1071 (entA (8)): OKN1 (ΔtonB), OKN3 (ΔfepA), OKN7 (ΔfhuA), and OKN13 (ΔtonB, ΔfepA) (41). ESKAPE strains included K. pneumoniae Kp52.145 (42) (courtesy of Regis Tournebiz, Institut Pasteur), A. baumannii ATCC 17978, P. aeruginosa PA01 (courtesy of Stephen Lory, Harvard University), and E. cloacae (43). K. pneumoniae KKN4 originated from Kp52.145 by sequential site-directed deletions of entB and three annotated kpnfepA structural genes, chromosomal loci 1658 and 4984 and plasmid (pII) locus 0027. The resultant mutant strain did not synthesize or secrete enterobactin or transport ferric catecholate iron complexes. However, it was an effective host for plasmids carrying fepA alleles.
Cys-mutant proteins
To generate Cys substitution mutants of FepA and FhuA we used QuikChange mutagenesis (Agilent) of the WT genes on pITS23 (41) and pITS11 (30), respectively. Both plasmids were derived from the low-copy plasmid pHSG575 (44). pEcoFepA_A698C is a pITS23 derivative with the engineered substitution A698C, pITS11 hosts WT ecofhuA (30), and pEcoFhuA_D396C is a pITS11 derivative with the engineered mutation D396C (this study). pKpnFepA_T210C is a pHSG575 derivative that encodes KpnFepA (allele 0027) under the control of its native promoter, with the engineered substitution T210C (this study). For structural predictions of KpnFepA, which has 82% identity to EcoFepA, we relied on the guideline that >25% sequence identity strongly predicts an overall identical protein fold (45, 46). To determine labeling targets in KpnFepA, we performed a ClustalW alignment against EcoFepA (Protein Data Bank (PDB) code 1FEP (47)) and used the Modeller function of UCSF Chimera to predict its tertiary structure, including surface loops. Cells expressing T210C, located in L2, were quantitatively modified by FM (Fig. S2).
To generate Cys mutants in LmoHbp2, we cloned the hbp2 structural gene (lmo2185) in pAT28 (48), introduced Cys substitutions at positions of interest by QuikChange mutagenesis, and verified the mutations by DNA sequencing. We transformed the pAT28 derivative pLmoHbp2_S154C into L. monocytogenes EGDeΔhbp2 (36), grew the strain to late exponential phase in iron-deficient MOPS-L (36) media, and purified the secreted Cys-mutant protein from iron-deficient bacterial cultures (49). After removing the cells by centrifugation, we precipitated secreted Hbp2_S154C from the supernatant with 0.6 m neutralized TCA, collected the precipitate at 10,000 × g for 30 min, washed the precipitated protein with 70% acetone, and then washed it with and resuspended it in 50 mm NaHPO4, pH 6.7.
Fluorescence modifications of Cys-mutant proteins
We inoculated OKN3, OKN11, OKN13, and KKN4, harboring plasmids and expressing Cys-mutant FepA proteins, from frozen stocks into LB broth, grew them overnight, and subcultured them at 1% into iron-deficient MOPS minimal medium with shaking (200 rpm) at 37 °C for 6–7 h until late exponential phase. We labeled Cys-mutant FepA proteins in these strains with 5 μm FM in 50 mm NaHPO4, pH 6.7, for 15 min at 37 °C.
We labeled LmoHbp2_S154C with 5 μm CPM in 50 mm NaHPO4, pH 6.7, for 30 min at 37 °C; precipitated the reaction mixture with 70% acetone; and collected LmoHbp2-CPM by centrifugation at 10,000 × g for 20 min. We chromatographically purified LmoHbp2-CPM over Sephacryl S-300HR in PBS (Fig. S3) and consolidated fractions 42–47 for spectroscopic studies. In all experiments with Hbp2 and Hn, we took precautions to maximize the solubility of the porphyrin (36, 39) by using fresh solutions of Hn formulated in DMSO and dispensed to physiological concentrations in alkaline buffers.
Fluorescence spectroscopy
We observed fluorophore-labeled cells in an SLM-Aminco 8100 fluorescence spectrometer, upgraded with an Olis operating system and software (Olis SpectralWorks, Olis Inc., Bogart, GA), to control its shutters, polarizers, and data collection. We also utilized an Olis Clarity fluorescence spectrometer for fluorescence assays of ferric catecholate binding. For species-specific ferric siderophore transport assays of Gram-negative bacteria we added an iron complex to 2 × 107 labeled cells in a quartz cuvette (final volume, 2 ml) with stirring at 37 °C and monitored the time course of fluorescence emissions at 520 nm. For FD assays, we incubated 107 sensor cells and 1.5 × 107 cells of the test organism (E. coli MG1655, K. pneumoniae Kp52.145 or KKN4, A. baumannii 17978, P. aeruginosa PA01, or E. cloacae) together in 2 ml of PBS + 0.4% glucose at 37 °C in a quartz cuvette. After addition of the ferric siderophore, we monitored the time course of fluorescence emissions at 520 nm for 15–30 min with stirring. FD assays in microtiter plates (23) contained 4 × 106 FD sensor cells and 3 × 106 cells of test organisms (E. coli MG1655 or K. pneumoniae Kp52.145) in 200 μl of PBS + 0.4% glucose at 37 °C. We monitored quenching and recovery of fluorescence by ferric siderophores added to a final concentration of 5 nm in the presence or absence of CCCP.
For fluorescence spectroscopic determinations of Gram-positive bacterial heme acquisition we used the FD sensor LmoHbp2_S154C-CPM. We determined its affinity for Hn by titrations of 30 nm LmoHbp2_S154C-CPM with 1–255 nm hemin while monitoring fluorescence in an SLM-Olis fluorimeter at excitation/emission wavelengths of 390/480 nm and plotted the ensuing fluorescence quenching with Grafit 6.012 (Erithacus Ltd., Middlesex, UK). For measurements of Hn uptake by heterologous bacteria we suspended the LmoHbp2_S154C-CPM sensor protein at 30 nm in PBS + 0.2% glucose in a 2-ml cuvette, added Hn to 15 nm after 300 s, and then added iron-starved bacterial cells at 600 s to a final concentration of 5 × 107/ml, and monitored fluorescence for 2 h.
When collecting fluorescence spectroscopic data of bacterial transport processes, we performed each experiment at least three times and repeated each individual time course or condition within an experiment in triplicate. We calculated the mean values and standard deviations of the triplicate measurements and plotted the resulting data with Grafit 6.02.
Author contributions
S. C., Y. S., A. K., S. M. N., and P. E. K. data curation; S. C., Y. S., and P. E. K. formal analysis; S. C., Y. S., A. K., S. M. N., and P. E. K. validation; S. C., Y. S., A. K., A. M., T. Y., B. N., S. M. N., and P. E. K. investigation; S. C., Y. S., A. K., A. M., T. Y., S. M. N., and P. E. K. methodology; Y. S., S. M. N., and P. E. K. conceptualization; T. Y. and P. E. K. resources; S. M. N. and P. E. K. supervision; S. M. N. and P. E. K. funding acquisition; S. M. N. and P. E. K. project administration; S. M. N. and P. E. K. writing-review and editing; P. E. K. visualization; P. E. K. writing-original draft.
Supplementary Material
Acknowledgments
We thank Stephen Lory, Regis Tournebiz, and Luis Actis for providing bacterial strains and Jesse Aaron and Teng Leong Chew at the Advanced Imaging Center of Howard Hughes Medical Institute Janelia Farms for assistance with microscopic imaging of bacteria.
This work was supported by NIAID, National Institutes of Health Award R21AI115187 (to P. E. K. and S. M. N.) and a Kansas IDeA Network of Biomedical Research Excellence (K-INBRE; National Institute of General Medical Sciences of the National Institutes of Health Grant P20GM103418) postdoctoral research award (to S. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Table S1.
- ESKAPE
- E. faecalis, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, Enterobacter spp.
- FD
- fluorescent decoy
- OM
- outer membrane
- IM
- inner membrane
- FeEnt
- ferric enterobactin
- TBDT
- TonB-dependent transporter
- FM
- fluorescein maleimide
- Hn
- hemin
- FeGEnt
- glucosylated form of FeEnt
- Fc
- ferrichrome
- CPM
- 7-diethylamino-3-(4′-maleimidylphenyl)-4-methyl coumarin
- HTS
- high-throughput screening
- FLHTS
- fluorescence high-throughput screening
- Eco
- E. coli
- Lmo
- L. monocytogenes
- Kpn
- K. pneumoniae
- Aba
- A. baumannii
- CCCP
- carbonyl cyanide p-chlorophenylhydrazone
- NEAT
- NEAr Transporter.
References
- 1. Khan S. N., and Khan A. U. (2016) Breaking the spell: combating multidrug resistant 'superbugs'. Front. Microbiol. 7, 174 10.3389/fmicb.2016.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elemam A., Rahimian J., and Mandell W. (2009) Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clin. Infect. Dis. 49, 271–274 10.1086/600042 [DOI] [PubMed] [Google Scholar]
- 3. Tacconelli E., and Magrini N. (2017) Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics, World Health Organization, Geneva [Google Scholar]
- 4. Fisher J. F., and Mobashery S. (2016) β-Lactam resistance mechanisms: Gram-positive bacteria and Mycobacterium tuberculosis. Cold Spring Harb. Perspect. Med. 6, a025221 10.1101/cshperspect.a025221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu E. W., McDermott G., Zgurskaya H. I., Nikaido H., and Koshland D. E. Jr. (2003) Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300, 976–980 10.1126/science.1083137 [DOI] [PubMed] [Google Scholar]
- 7. Rouault T. A., and Maio N. (2017) Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 292, 12744–12753 10.1074/jbc.R117.789537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klebba P. E., McIntosh M. A., and Neilands J. B. (1982) Kinetics of biosynthesis of iron-regulated membrane proteins in Escherichia coli. J. Bacteriol. 149, 880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parrow N. L., Fleming R. E., and Minnick M. F. (2013) Sequestration and scavenging of iron in infection. Infect. Immun. 81, 3503–3514 10.1128/IAI.00602-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hammer N. D., and Skaar E. P. (2011) Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65, 129–147 10.1146/annurev-micro-090110-102851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neilands J. B. (1952) A crystalline organo-iron pigment from a rust fungus (Ustilago sphaerogena). J. Am. Chem. Soc. 74, 4846–4847 10.1021/ja01139a033 [DOI] [Google Scholar]
- 12. Tsolis R. M., Bäumler A. J., Heffron F., and Stojiljkovic I. (1996) Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64, 4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Konopka K., Bindereif A., and Neilands J. B. (1982) Aerobactin-mediated utilization of transferrin iron. Biochemistry 21, 6503–6508 10.1021/bi00268a028 [DOI] [PubMed] [Google Scholar]
- 14. Nagy T. A., Moreland S. M., Andrews-Polymenis H., and Detweiler C. S. (2013) The ferric enterobactin transporter Fep is required for persistent Salmonella enterica serovar Typhimurium infection. Infect. Immun. 81, 4063–4070 10.1128/IAI.00412-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noinaj N., Guillier M., Barnard T. J., and Buchanan S. K. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 10.1146/annurev.micro.112408.134247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schauer K., Rodionov D. A., and de Reuse H. (2008) New substrates for TonB-dependent transport: do we only see the 'tip of the iceberg'? Trends Biochem. Sci. 33, 330–338 10.1016/j.tibs.2008.04.012 [DOI] [PubMed] [Google Scholar]
- 17. Jordan L. D., Zhou Y., Smallwood C. R., Lill Y., Ritchie K., Yip W. T., Newton S. M., and Klebba P. E. (2013) Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc. Natl. Acad. Sci. U.S.A. 110, 11553–11558 10.1073/pnas.1304243110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klebba P. E. (2016) ROSET model of TonB action in Gram-negative bacterial iron acquisition. J. Bacteriol. 198, 1013–1021 10.1128/JB.00823-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pi H., Jones S. A., Mercer L. E., Meador J. P., Caughron J. E., Jordan L., Newton S. M., Conway T., and Klebba P. E. (2012) Role of catecholate siderophores in Gram-negative bacterial colonization of the mouse gut. PLoS One 7, e50020 10.1371/journal.pone.0050020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams P. H. (1979) Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect. Immun. 26, 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bachman M. A., Lenio S., Schmidt L., Oyler J. E., and Weiser J. N. (2012) Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. MBio 3, e00224–11 10.1128/mBio.00224-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russo T. A., Shon A. S., Beanan J. M., Olson R., MacDonald U., Pomakov A. O., and Visitacion M. P. (2011) Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One 6, e26734 10.1371/journal.pone.0026734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanson M., Jordan L. D., Shipelskiy Y., Newton S. M., and Klebba P. E. (2016) High-throughput screening assay for inhibitors of TonB-dependent iron transport. J. Biomol. Screen. 21, 316–322 10.1177/1087057115613788 [DOI] [PubMed] [Google Scholar]
- 24. Smallwood C. R., Jordan L., Trinh V., Schuerch D. W., Gala A., Hanson M., Hanson M., Shipelskiy Y., Majumdar A., Newton S. M., and Klebba P. E. (2014) Concerted loop motion triggers induced fit of FepA to ferric enterobactin. J. Gen. Physiol. 144, 71–80 10.1085/jgp.201311159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nairn B. L., Eliasson O. S., Hyder D. R., Long N. J., Majumdar A., Chakravorty S., McDonald P., Roy A., Newton S. M., and Klebba P. E. (2017) Fluorescence high-throughput screening for inhibitors of TonB action. J. Bacteriol. 199, e00889–16 10.1128/JB.00889-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newton S. M., Trinh V., Pi H., and Klebba P. E. (2010) Direct measurements of the outer membrane stage of ferric enterobactin transport: postuptake binding. J. Biol. Chem. 285, 17488–17497 10.1074/jbc.M109.100206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams P. H., Rabsch W., Methner U., Voigt W., Tschäpe H., and Reissbrodt R. (2006) Catecholate receptor proteins in Salmonella enterica: role in virulence and implications for vaccine development. Vaccine 24, 3840–3844 10.1016/j.vaccine.2005.07.020 [DOI] [PubMed] [Google Scholar]
- 28. Smith K. D. (2007) Iron metabolism at the host pathogen interface: lipocalin 2 and the pathogen-associated iroA gene cluster. Int. J. Biochem. Cell Biol. 39, 1776–1780 10.1016/j.biocel.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Valdebenito M., Müller S. I., and Hantke K. (2007) Special conditions allow binding of the siderophore salmochelin to siderocalin (NGAL-lipocalin). FEMS Microbiol. Lett. 277, 182–187 10.1111/j.1574-6968.2007.00956.x [DOI] [PubMed] [Google Scholar]
- 30. Scott D. C., Cao Z., Qi Z., Bauler M., Igo J. D., Newton S. M., and Klebba P. E. (2001) Exchangeability of N termini in the ligand-gated porins of Escherichia coli. J. Biol. Chem. 276, 13025–13033 10.1074/jbc.M011282200 [DOI] [PubMed] [Google Scholar]
- 31. Thulasiraman P., Newton S. M., Xu J., Raymond K. N., Mai C., Hall A., Montague M. A., and Klebba P. E. (1998) Selectivity of ferric enterobactin binding and cooperativity of transport in Gram-negative bacteria. J. Bacteriol. 180, 6689–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pawelek P. D., Croteau N., Ng-Thow-Hing C., Khursigara C. M., Moiseeva N., Allaire M., and Coulton J. W. (2006) Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312, 1399–1402 10.1126/science.1128057 [DOI] [PubMed] [Google Scholar]
- 33. Hider R. C., and Kong X. (2010) Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657 10.1039/b906679a [DOI] [PubMed] [Google Scholar]
- 34. Mazmanian S. K., Liu G., Ton-That H., and Schneewind O. (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 10.1126/science.285.5428.760 [DOI] [PubMed] [Google Scholar]
- 35. Bierne H., Garandeau C., Pucciarelli M. G., Sabet C., Newton S., Garcia-del Portillo F., Cossart P., and Charbit A. (2004) Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186, 1972–1982 10.1128/JB.186.7.1972-1982.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao Q., Jiang X., Moore K. J., Shao Y., Pi H., Dubail I., Charbit A., Newton S. M., and Klebba P. E. (2011) Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol. Microbiol. 80, 1581–1597 10.1111/j.1365-2958.2011.07667.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Létoffé S., Deniau C., Wolff N., Dassa E., Delepelaire P., Lecroisey A., and Wandersman C. (2001) Haemophore-mediated bacterial haem transport: evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol. Microbiol. 41, 439–450 10.1046/j.1365-2958.2001.02530.x [DOI] [PubMed] [Google Scholar]
- 38. Henderson D. P., and Payne S. M. (1993) Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7, 461–469 10.1111/j.1365-2958.1993.tb01137.x [DOI] [PubMed] [Google Scholar]
- 39. Malmirchegini G. R., Sjodt M., Shnitkind S., Sawaya M. R., Rosinski J., Newton S. M., Klebba P. E., and Clubb R. T. (2014) Novel mechanism of hemin capture by Hbp2, the hemoglobin-binding hemophore from Listeria monocytogenes. J. Biol. Chem. 289, 34886–34899 10.1074/jbc.M114.583013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostberg K. L., DeRocco A. J., Mistry S. D., Dickinson M. K., and Cornelissen C. N. (2013) Conserved regions of gonococcal TbpB are critical for surface exposure and transferrin iron utilization. Infect. Immun. 81, 3442–3450 10.1128/IAI.00280-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma L., Kaserer W., Annamalai R., Scott D. C., Jin B., Jiang X., Xiao Q., Maymani H., Massis L. M., Ferreira L. C., Newton S. M., and Klebba P. E. (2007) Evidence of ball-and-chain transport of ferric enterobactin through FepA. J. Biol. Chem. 282, 397–406 10.1074/jbc.M605333200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lery L. M., Frangeul L., Tomas A., Passet V., Almeida A. S., Bialek-Davenet S., Barbe V., Bengoechea J. A., Sansonetti P., Brisse S., and Tournebize R. (2014) Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 12, 41 10.1186/1741-7007-12-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rutz J. M., Abdullah T., Singh S. P., Kalve V. I., and Klebba P. E. (1991) Evolution of the ferric enterobactin receptor in Gram-negative bacteria. J. Bacteriol. 173, 5964–5974 10.1128/jb.173.19.5964-5974.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hashimoto-Gotoh T., Kume A., Masahashi W., Takeshita S., and Fukuda A. (1986) Improved vector, pHSG664, for direct streptomycin-resistance selection: cDNA cloning with G:C-tailing procedure and subcloning of double-digest DNA fragments. Gene 41, 125–128 10.1016/0378-1119(86)90275-1 [DOI] [PubMed] [Google Scholar]
- 45. Ginalski K. (2006) Comparative modeling for protein structure prediction. Curr. Opin. Struct. Biol. 16, 172–177 10.1016/j.sbi.2006.02.003 [DOI] [PubMed] [Google Scholar]
- 46. Stokes-Rees I., and Sliz P. (2010) Protein structure determination by exhaustive search of Protein Data Bank derived databases. Proc. Natl. Acad. Sci. U.S.A. 107, 21476–21481 10.1073/pnas.1012095107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buchanan S. K., Smith B. S., Venkatramani L., Xia D., Esser L., Palnitkar M., Chakraborty R., van der Helm D., and Deisenhofer J. (1999) Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6, 56–63 10.1038/4931 [DOI] [PubMed] [Google Scholar]
- 48. Trieu-Cuot P., Carlier C., Poyart-Salmeron C., and Courvalin P. (1990) A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 18, 4296 10.1093/nar/18.14.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Newton S. M., Klebba P. E., Raynaud C., Shao Y., Jiang X., Dubail I., Archer C., Frehel C., and Charbit A. (2005) The svpA-srtB locus of Listeria monocytogenes: Fur-mediated iron regulation and effect on virulence. Mol. Microbiol. 55, 927–940 10.1111/j.1365-2958.2004.04436.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.