To the Editor:
Food allergy is a life-threatening disease that is common and increasing in prevalence, yet the factors leading to its development are poorly understood.1 Microbial composition has been associated with risk of food allergy,2 and integrative analysis of the human intestinal microbiome and metabolome could provide insights into mechanisms of microbial-associated pathogenic changes.3 Here, we performed a prospective, untargeted, integrative analysis of the intestinal bacterial microbiome and metabolome during infancy, testing associations with the development of clinical food allergy and sensitization to foods at age 3 years. Our goal was to identify microbial-associated metabolites that were associated with food allergy or sensitization.
For detailed methods, see this article’s Methods section in the Online Repository at www.jacionline.org. Subjects were offspring of participants in the Vitamin D Antenatal Asthma Reduction Trial (NCT00920621),4 a multicenter randomized controlled trial of vitamin D supplementation in pregnancy to prevent asthma in offspring. The study protocol was approved by the institutional review boards at each center and all participants provided written informed consent. Food allergy and sensitization at age 3 years were based on parental questionnaire responses and serum specific IgE testing. Stool samples were collected between age 3 and 6 months from 333 subjects. Microbiome composition analysis by bacterial 16S rRNA sequencing and metabolomic analysis with ultraperformance LC/MS-MS were performed on stool samples from 12 children with food allergy (see Table E1 in this article’s Online Repository at www.jacionline.org), 32 with food sensitization, and 37 controls.
TABLE E1.
Clinical history and serum specific IgE for food-allergic subjects
| Serum specific IgE (kU/L) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subject | Food | Reaction* | Age (mo) reported | Diagnosis method | Walnut | Peanut | Egg white | Milk | Wheat | Soybean |
| 1 | Sesame seed | Unknown | 24 | Blood and skin test | <0.10 | 1.00 | 5.42 | 0.67 | 0.42 | 0.63 |
| Egg | Unknown | 24 | ||||||||
| Other nut | Unknown | 24 | ||||||||
| Peanut | Rash | 24 | ||||||||
| 2 | Soy | Diarrhea, nausea, hives | 12 | Blood and skin test | <0.10 | 2.05 | 22.4 | 1.9 | 0.65 | 1.27 |
| Wheat | Diarrhea, nausea, hives | 12 | ||||||||
| Peanut | Diarrhea, nausea, hives | 12 | ||||||||
| Egg | Diarrhea, nausea, hives | 12 | ||||||||
| Other nut | Unknown | 15 | ||||||||
| Fish | Diarrhea, nausea, hives | 24 | ||||||||
| 3 | Milk | Hives, nausea, wheeze, worsened eczema | 12 | Blood test and skin test | 3.05 | 8.72 | 5.03 | 9.88 | 5.06 | 3.89 |
| Egg | Nausea, vomiting, wheeze, sneezing/runny nose, watery eyes, hives, worsened eczema, fatigue | 12 | ||||||||
| Peanut | Nausea | 12 | Blood test and skin test | |||||||
| Other fish | Hives, nausea, diarrhea, worsened eczema, facial swelling, fussiness | 12 | ||||||||
| Soy | Hives, fussiness, vaginal itching, nausea, diarrhea | 18 | ||||||||
| Shellfish | Facial swelling, hives, worsened eczema, fussiness, nausea, diarrhea | 18 | ||||||||
| Wheat | Hives, fussiness, vaginal itching, nausea | 21 | ||||||||
| 4 | Shellfish | Hives, worsened eczema, nausea | 12 | |||||||
| Other fish | Hives, worsened eczema, nausea | 12 | ||||||||
| Egg | Hives, wheeze, worsened eczema, nausea | 12 | ||||||||
| Hot dog | Hives | 15 | ||||||||
| Milk | Hives, worsened eczema, nausea | 24 | ||||||||
| Wheat | Hives | 24 | ||||||||
| Peanut | Hives, worsened eczema, nausea | 27 | ||||||||
| 5 | Egg | Nausea, hives, fatigue | 18 | Skin test | 0.66 | 1.32 | 1.59 | 4.24 | 0.78 | 2.28 |
| 6 | Peanut | Hives, wheezing, sneezing, red eyes | 24 | History | 12.7 | >100 | 38.5 | 26 | 29 | 96.3 |
| 7 | Other nut | Eyes swelled shut, itchy | 36 | Skin test | 0.72 | 2.07 | 4.41 | 8.51 | 1.95 | 0.96 |
| 8 | Unknown | Unknown | 15 | Blood test | 0.25 | >100 | 0.71 | <0.10 | 0.15 | 2.2 |
| Peanut | Hives | 24 | ||||||||
| 9 | Peanut | Hives on 1 body part only | 15 | Blood and skin test | 1.54 | 9.76 | 5.67 | 0.94 | 2.07 | 2.96 |
| Egg | Nausea, hives | 15 | ||||||||
| Pea | Hives on 1 body part only | 24 | ||||||||
| Lentil | Hives on 1 body part only | 24 | ||||||||
| Other nut | Unknown | 30 | ||||||||
| 10 | Peanut | Not asked† | 12 | Not asked† | <0.10 | 0.79 | 0.43 | 0.25 | 0.27 | <0.10 |
| Egg | Not asked† | 12 | ||||||||
| 11 | Egg | Not asked† | 12 | Not asked† | <0.10 | <0.10 | 0.62 | <0.10 | <0.10 | <0.10 |
| 12 | Egg | Hives on 1 body part only, nausea, cough | 27 | Blood test | <0.10 | 0.37 | 3.75 | 1.88 | 0.90 | 0.91 |
| Candy/sweets | Nausea | 36 | ||||||||
Unless it is specified that hives occurred on 1 body part only, hives occurred on at least 2 body parts.
A less detailed version of the questionnaire that did not ask about reaction type or method of food allergy diagnosis was used early in the study.
Subjects were well matched on baseline characteristics, with a few exceptions including age, solid food introduction at stool sample collection, and asthma/recurrent wheeze at age 3 years (see Table E2 in this article’s Online Repository at www.jacionline.org). Of several potential determinants of the intestinal microenvironment analyzed, only mode of delivery differed by phenotype, with a higher percentage of subjects born by Cesarean section among those with food allergy and a lower percentage among those with food sensitization. Accordingly, we adjusted for age in all analyses, performed sensitivity analyses of key results adjusting for other potential confounders, and tested for associations between mode of delivery and phenotype-associated microbiome and metabolome perturbations.
TABLE E2.
Baseline characteristics of children and upstream predictors of the intestinal microenvironment
| All subjects (n = 83) |
Subjects with iNKT-cell activity data (n = 57) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | All children (n = 81) | Food allergic (n = 12) | Food sensitive (n = 32) | Control (n = 37) | P value | Food allergic (n = 8) | Food sensitive (n = 22) | Control (n = 27) | P value |
| Sex, n (%) | .66 | .17 | |||||||
| Male | 41 (51) | 5 (42) | 18 (56) | 18 (49) | 2 (25) | 14 (64) | 13 (48) | ||
| Female | 40 (49) | 7 (58) | 14 (44) | 19 (51) | 6 (75) | 8 (36) | 14 (52) | ||
| Race/ethnicity, n (%) | .94 | .98 | |||||||
| Black, non-Hispanic | 34 (42) | 6 (50) | 13 (41) | 15 (41) | 4 (50) | 9 (41) | 10 (37) | ||
| White, non-Hispanic | 26 (32) | 4 (33) | 11 (34) | 11 (30) | 2 (25) | 6 (27) | 9 (33) | ||
| Hispanic | 21 (26) | 2 (17) | 8 (25) | 11 (30) | 2 (25) | 7 (32) | 8 (30) | ||
| Birth by Cesarean section, n (%) | 24 (30) | 6 (50) | 5 (16) | 13 (35) | .052 | 5 (63) | 2 (9) | 9 (33) | .01 |
| Gestational age (wk), mean ± SD | 39.1 ± 1.6 | 39.4 ± 1.7 | 38.9 ± 1.9 | 39.4 ± 1.4 | .48 | 39.3 ± 1.6 | 38.8 ± 2.2 | 39.4 ± 1.4 | .79 |
| Number of living children born to mother, mean ± SD | 0.9 ± 0.9 | 1.2 ± 0.9 | 1.0 ± 0.9 | 0.8 ± 0.9 | .40.16 | 1.3 ± 0.9 | 0.8 ± 0.8 | 0.8 ± 0.9 | .36 |
| Antibiotic exposure, n (%) | |||||||||
| Perinatal antibiotics | 35 (43) | 8 (67) | 11 (34) | 16 (43) | .16 | 5 (63) | 8 (36) | 11 (41) | .44 |
| By age 6 mo | 15 (19) | 2 (17) | 5 (16) | 8 (22) | .86 | 1 (13) | 3 (14) | 7 (26) | .59 |
| By age 3 y | 69 (85) | 11 (92) | 29 (91) | 29 (78) | .36 | 7 (87) | 21 (95) | 20 (74) | .10 |
| Pet dog in home, n (%) | |||||||||
| In first 6 mo of life | 22 (28) | 3 (33) | 6 (19) | 13 (36) | .31 | 2 (25) | 3 (14) | 10 (37) | .19 |
| Between age 6 and 36 mo | 32 (41) | 3 (33) | 13 (43) | 16 (43) | .52 | 2 (25) | 10 (50) | 11 (41) | .53 |
| Pet cat in home, n (%) | |||||||||
| In first 6 mo of life | 11 (14) | 2 (17) | 4 (13) | 5 (14) | .91 | 1 (13) | 2 (9) | 4 (15) | .86 |
| Between age 6 and 36 mo | 20 (25) | 6 (50) | 5 (17) | 9 (24) | .10 | 3 (38) | 2 (10) | 6 (22) | .22 |
| Daycare by age 3 y, n (%) | 43 (53) | 7 (58) | 15 (47) | 21 (57) | .72 | 4 (50) | 10 (45) | 14 (52) | .93 |
| Age at stool sample collection (mo), mean ± SD | 4.6 ± 1.1 | 5.0 ± 1.0 | 5.0 ± 1.0 | 4.1 ± 1.0 | <.01 | 5.0 ± 1.0 | 5.1 ± 1.0 | 4.2 ± 1.1 | .02 |
| Diet at stool sample collection, n (%) | |||||||||
| Breast-feeding | 34 (42) | 6 (50) | 13 (41) | 15 (41) | .91 | 4 (50) | 7 (32) | 12 (44) | .57 |
| Formula | 50 (62) | 7 (58) | 20 (63) | 23 (62) | 1 | 4 (50) | 16 (73) | 17 (63) | .51 |
| Solid foods | 33 (41) | 6 (50) | 19 (61) | 8 (22) | <.01 | 4 (50) | 12 (57) | 7 (26) | .07 |
| Asthma/recurrent wheeze by age 3 y, n (%) | 25 (31) | 8 (67) | 9 (28) | 8 (22) | .02 | 6 (75) | 7 (32) | 4 (15) | <.01 |
| VDAART treatment group, n (%) | 1 | 1 | |||||||
| 4400 IU/d vitamin D | 41 (51) | 6 (50) | 16 (50) | 19 (51) | 4 (50) | 11 (50) | 13 (48) | ||
| 400 IU/d vitamin D | 40 (49) | 6 (50) | 16 (50) | 18 (49) | 4 (50) | 11 (50) | 14 (52) | ||
| Study center, n (%) | .27 | .71 | |||||||
| Boston | 23 (28) | 4 (33) | 8 (25) | 11 (30) | 1 (13) | 2 (9) | 7 (26) | ||
| St Louis | 42 (52) | 7 (58) | 20 (63) | 15 (41) | 3 (38) | 7 (32) | 9 (33) | ||
| San Diego | 16 (20) | 1 (8) | 4 (13) | 11 (30) | 4 (50) | 13 (59) | 11 (41) | ||
| Maternal education, n (%) | .92 | .74 | |||||||
| Less than high school | 10 (12) | 1 (8) | 3 (9) | 6 (16) | 0 (0) | 3 (14) | 4 (15) | ||
| High school or technical school | 26 (32) | 5 (42) | 11 (34) | 10 (27) | 4 (50) | 8 (36) | 6 (22) | ||
| Some college | 10 (12) | 2 (17) | 4 (13) | 4 (11) | 1 (13) | 3 (14) | 3 (11) | ||
| College graduate or graduate school | 35 (43) | 4 (33) | 14 (44) | 17 (46) | 3 (38) | 8 (36) | 14 (52) | ||
| Household income (US $), n (%) | .57 | .55 | |||||||
| <30,000 | 29 (36) | 6 (50) | 12 (38) | 11 (30) | 5 (63) | 8 (36) | 8 (30) | ||
| 30,000–49,999 | 11 (14) | 1 (8) | 5 (16) | 5 (14) | 0 (0) | 4 (18) | 4 (15) | ||
| 50,000–74,999 | 9 (11) | 1 (8) | 3 (9) | 5 (14) | 1 (13) | 3 (14) | 4 (15) | ||
| 75,000–99,999 | 8 (10) | 1 (8) | 1 (3) | 6 (16) | 1 (13) | 0 (0) | 4 (15) | ||
| 100,000–149,999 | 10 (12) | 2 (17) | 2 (6) | 6 (16) | 1 (13) | 1 (5) | 4 (15) | ||
| At least 150,000 | 5 (6) | 0 (0) | 4 (13) | 1 (3) | 0 (0) | 2 (9) | 1 (4) | ||
| Refused to say or unknown | 9 (11) | 1 (8) | 5 (16) | 3 (8) | 0 (0) | 4 (18) | 2 (7) | ||
P value is for Kruskal-Wallis test for comparisons of gestational age, birth order, and age at stool sample collection. P value is for Fisher exact test for all other comparisons. Missing data: breast-feeding status for 1 subject, solid foods status for 1 subject, pet cat or dog in first 6 mo for 1 subject, and pet cat or dog between age 6 and 36 mo for 2 subjects. VDAART, Vitamin D Antenatal Asthma Reduction Trial.
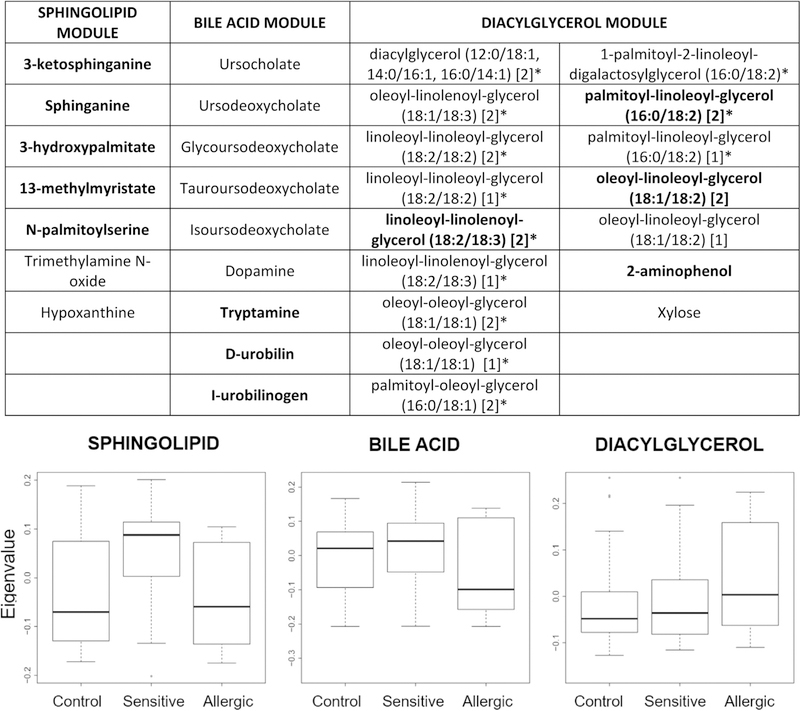
Logistic regression analyses revealed several individual metabolites that differed in relative abundance by food allergy/sensitization phenotype (see Table E3 in this article’s Online Repository at www.jacionline.org). Weighted gene coexpression network analysis identified 29 modules of highly correlated and likely functionally related metabolites. Eigenvalues of 3 modules were associated (P < .05) with food allergy or sensitization (Fig 1; see Table E4 in this article’s Online Repository at www.jacionline.org). We focused on a module that included several metabolites associated with de novo sphingolipid synthesis (sphinganine, 3-ketosphinganine, 3-hydroxypalmitate, N-palmitoylserine, 13-methylmyristate) that had significantly higher eigenvalues in subjects with food sensitization than in those with food allergy (P = .02) or controls (P = .02), and nonsignificantly higher eigenvalues in controls than in those with food allergy (P = .15). This pattern suggests that this module might be associated with protection from clinical food allergy, with the most pronounced protective effect among food-sensitized individuals.
TABLE E3.
Metabolites significantly different by phenotype
| Food-allergic (n = 12) vs control (n = 37, reference) |
Food-sensitive (n = 32) vs control (n = 37, reference) |
Food-allergic (n = 12) vs food-sensitive (n = 32, reference) |
||||||
|---|---|---|---|---|---|---|---|---|
| Metabolite | Odds ratio (95% CI) | P value | Metabolite | Odds ratio (95% CI) | P value | Metabolite | Odds ratio (95% CI) | P value |
| 7-Methylguanine | 0.29 (0.11–0.63) | .004 | 3-Ketosphinganine | 2.49 (1.37–4.89) | .004 | 3-Hydroxypalmitate | 0.23 (0.07–0.59) | .01 |
| Docosapentaenoate (n3 DPA; 22:5n3) | 0.24 (0.07–0.64) | .01 | Pimelate (heptanedioate) | 2.83 (1.38–6.29) | .01 | Ribonate | 0.32 (0.13–0.69) | .01 |
| 2-Aminophenol | 3.20 (1.40–8.68) | .01 | Pyridoxine (vitamin B6) | 3.08 (1.40–7.63) | .01 | 1-Palmitoyl-GPI (16:0) | 0.25 (0.08–0.63) | .01 |
| Bilirubin (E,Z or Z,E)* | 0.20 (0.05–0.62) | .01 | Cis-urocanate | 0.24 (0.07–0.66) | .01 | 1-Palmitoyl-GPS (16:0)* | 0.25 (0.08–0.65) | .01 |
| Histidine | 0.11 (0.01–0.53) | .01 | Diacetylchitobiose | 0.38 (0.16, -0.79) | .01 | I-urobilinogen | 0.42 (0.20–0.79) | .01 |
| N-formylmethionine | 0.28 (0.09–0.72) | .01 | N-acetylglutamate | 0.25 (0.07–0.73) | .02 | Sphinganine | 0.15 (0.03–0.56) | .01 |
| I-urobilinogen | 0.43 (0.20–0.82) | .02 | 5alpha-pregnan-3beta,20alpha-diol monosulfate (1) | 3.04 (1.31–8.77) | .02 | 2-Methylserine | 0.32 (0.11–0.75) | .02 |
| Dihomo-linolenate (20:3n3 or n6) | 0.25 (0.06–0.66) | .02 | N-carbamoylaspartate | 0.38 (0.15–0.81) | .02 | 13-Methylmyristate | 0.39 (0.17–0.82) | .02 |
| Serine | 0.11 (0.01–0.59) | .02 | N-acetylasparagine | 0.26 (0.08–0.78) | .02 | Dihomo-linolenate (20:3n3 or n6) | 0.24 (0.06–0.67) | .02 |
| 2-Methylserine | 0.20 (0.04–0.68) | .03 | Lysylleucine | 2.23 (1.15–4.69) | .02 | Glycylisoleucine | 0.25 (0.07–0.70) | .02 |
| 5,6-Dihydrothymine | 0.40 (0.16–0.86) | .03 | Vanillic alcohol sulfate | 0.42 (0.18–0.86) | .03 | D-urobilin | 0.40 (0.17–0.83) | .02 |
| 3-Hydroxypalmitate | 0.27 (0.07–0.79) | .03 | Docosahexenoylcarnitine (C22:6)* | 0.37 (0.14–0.83) | .03 | Docosapentaenoate (n3 DPA; 22:5n3) | 0.26 (0.07–0.74) | .02 |
| Phenethylamine | 0.32 (0.10–0.83) | .03 | Ursodeoxycholate sulfate (1) | 0.57 (0.34–0.92) | .03 | 7-Methylguanine | 0.47 (0.23–0.90) | .03 |
| D-urobilin | 0.42 (0.18–0.91) | .04 | 3-Methylglutarate/2-methylglutarate | 2.35 (1.14–5.35) | .03 | Sucrose | 0.37 (0.14–0.85) | .03 |
| Ursodeoxycholate sulfate (1) | 0.45 (0.20–0.90) | .04 | N-palmitoylserine | 1.87 (1.08–3.36) | .03 | 1-Palmitoyl-GPE (16:0) | 0.20 (0.04–0.73) | .03 |
| Tryptamine | 0.44 (0.19–0.90) | .04 | Malonylcarnitine | 0.39 (0.16–0.87) | .03 | 21-Hydroxypregnenolone disulfate | 0.31 (0.09–0.80) | .03 |
| 2,3-Dimethylsuccinate | 3.06 (1.09–9.72) | .04 | 13-Methylmyristate | 1.97 (1.08–3.80) | .03 | N-acetylglycine | 3.33 (1.21–11.18) | .03 |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) [1] | 2.56 (1.09–6.94) | .04 | Dopamine 3-O-sulfate | 0.45 (0.21–0.90) | .03 | Alpha-CEHC | 0.32 (0.10–0.840) | .03 |
| Palmitoyl-linoleoyl-glycerol (16:0/18:2) [1]* | 2.56 (1.08–7.07) | .04 | Glutaminylleucine | 2.58 (1.12–6.57) | .03 | Glycerophosphoserine* | 0.28 (0.08–0.82) | .03 |
| Tyrosine | 0.16 (0.02–0.86) | .04 | N2,N6-diacetyllysine | 0.41 (0.17–0.90) | .03 | Bilirubin (E,Z or Z,E)* | 0.32 (0.10–0.87) | .04 |
| Formiminoglutamate | 0.36 (0.11–0.90) | .04 | N-acetyl-aspartyl-glutamate (NAAG) | 0.41 (0.17–0.91) | .04 | Glycylvaline | 0.24 (0.05–0.87) | .04 |
| Linolenate (alpha or gamma; [18:3n3 or 6]) | 0.27 (0.06–0.86) | .04 | N-alpha-acetylornithine | 0.39 (0.15–0.91) | .04 | Linoleoyl ethanolamide | 0.42 (0.17–0.94) | .04 |
| Linoleoyl-linolenoyl-glycerol (18:2/18:3) [2]* | 2.39 (1.04–6.06) | .05 | Allo-threonine | 0.40 (0.15–0.90) | .04 | |||
| 3-Ureidoisobutyrate | 0.45 (0.19–0.95) | .05 | Homoarginine | 1.79 (1.05–3.20) | .04 | |||
| Lactobionate | 0.43 (0.18–0.93) | .04 | ||||||
| 2,3-Dimethylsuccinate | 2.28 (1.07–5.46) | .04 | ||||||
| Chiro-inositol | 2.07 (1.05–4.38) | .04 | ||||||
Only metabolites with significant (P < .05) associations in age-adjusted analyses are shown.
Compounds with annotations that have not been officially confirmed on the basis of a standard.
FIG 1.
Metabolite members of metabolite modules associated with food allergy or sensitization and box plots of module eigenvalues. Box plots summarize module eigenvalues for subjects with food allergy (n = 12), food sensitization (n = 32), and controls (n = 37). *Compounds with annotations that have not been officially confirmed on the basis of a standard. Bolded compounds were associated with phenotype in analyses of individual metabolites.
TABLE E4.
Association of metabolite module eigenvalues and sphingolipid module-associated OTUs with food allergy or sensitization, with sensitivity analyses including potential confounders as covariates
| Crude analysis |
Adjusted for age |
Adjusted for age and asthma/recurrent wheeze |
Adjusted for age and solid foods introduction status |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Module | Comparison | β (95% CI) | P value | β (95% CI) | P value | β (95% CI) | P value | β (95% CI) | P value |
| Sphingolipid | Food allergy (vs sensitization) | −8.3 (−15.6 to −1.8) | .02 | −8.3 (−15.7 to −1.8) | .02 | −8.8 (−16.9 to −1.9) | .02 | −8.5 (−16.4 to −1.9) | .02 |
| Food sensitization (vs control) | 7.2 (2.6 to 12.3) | <.01 | 6.1 (1.1 to 11.5) | .02 | 6.9 (1.8 to 12.7) | .01 | 6.1 (1.0 to 11.8) | .02 | |
| Food allergy (vs control) | −0.8 (−7.3 to 5.3) | .80 | −5.9 (−14.7 to 1.6) | .15 | −2.3 (−12.3 to 7.1) | .64 | −6.0 (−15.0 to 1.6) | .15 | |
| Bile acid | Food allergy (vs sensitization) | −6.2 (−13.1 to 0.1) | .06 | −6.3 (−13.4 to 0.0) | .06 | −6.1 (−13.3 to 0.4) | .08 | −6.4 (−13.9 to 0.1) | .07 |
| Food sensitization (vs control) | 3.3 (−1.2 to 8.2) | .16 | 1.2 (−4.0 to 6.4) | .66 | 0.9 (−4.4 to 6.3) | .73 | 0.6 (−4.7 to 6.1) | .82 | |
| Food allergy (vs control) | −2.6 (−8.5 to 3.0) | .36 | −7.0 (−14.7 to −0.4) | .05 | −6.8 (−15.4 to 0.7) | .09 | −7.1 (−14.9 to −0.4) | .05 | |
| Diacylglycerol | Food allergy (vs sensitization) | 2.9 (−2.5 to 8.3) | .29 | 3.0 (−2.5 to 8.6) | .28 | 2.7 (−3.2 to 8.7) | .36 | 2.9 (−2.6 to 8.6) | .30 |
| Food sensitization (vs control) | 1.5 (−2.9 to 6.1) | .51 | 2.5 (−2.3 to 7.5) | .32 | 2.6 (−2.2 to 7.8) | .30 | 2.2 (−2.7 to 7.4) | .37 | |
| Food allergy (vs control) | 5.1 (−0.8 to 11.2) | .09 | 7.4 (0.6 to 15.3) | .04 | 8.3 (0.8 to 17.2) | .04 | 7.5 (0.6 to 15.5) | .04 | |
| Taxa | Comparison | Log2 fold change | P value | Log2 fold change | P value | Log2 fold change | P value | Log2 fold change | P value |
| Bacteroides ID 182157 | Food allergy (vs sensitization) | −2.9 | .03 | −2.9 | .03 | −2.8 | .04 | −2.9 | .04 |
| Food sensitization (vs control) | 1.1 | .27 | 1.1 | .27 | 1.2 | .25 | 1.1 | .33 | |
| Food allergy (vs control) | −0.9 | .50 | −1.6 | .23 | −1.2 | .42 | −1.3 | .36 | |
| Bacteroides ID 184753 | Food allergy (vs sensitization) | −4.1 | <.01 | −5.2 | <.01 | −5.1 | <.01 | −5.2 | <.01 |
| Food sensitization (vs control) | 1.5 | .17 | 0.7 | .49 | 0.1 | .92 | 0.9 | .49 | |
| Food allergy (vs control) | −2.0 | .22 | −2.8 | .07 | −1.9 | .30 | −3.0 | .06 | |
| Bacteroides ID 188735 | Food allergy (vs sensitization) | −2.0 | .18 | −3.3 | .04 | −2.6 | .10 | −2.9 | .10 |
| Food sensitization (vs control) | 1.1 | .29 | 0.7 | .55 | 0.8 | .50 | 1.0 | .46 | |
| Food allergy (vs control) | −0.7 | .66 | −2.4 | .12 | −2.0 | .28 | −1.4 | .37 | |
| Bacteroides ID 577170 | Food allergy (vs sensitization) | −2.9 | .03 | −3.6 | .01 | −3.5 | .02 | −3.6 | .02 |
| Food sensitization (vs control) | 1.6 | .11 | 1.2 | .27 | 1.3 | .23 | 1.3 | .27 | |
| Food allergy (vs control) | −1.1 | .50 | −1.9 | .20 | −1.3 | .48 | −1.4 | .37 | |
| Bacteroides ID 3472078 | Food allergy (vs sensitization) | −0.9 | .53 | −3.3 | .04 | −3.1 | .06 | −3.2 | .07 |
| Food sensitization (vs control) | −0.1 | .92 | 1.9 | .09 | 2.1 | .09 | 1.9 | .16 | |
| Food allergy (vs control) | −0.8 | .63 | −1.4 | .36 | −1.1 | .55 | −1.4 | .40 | |
| Bacteroides ID 4354042 | Food allergy (vs sensitization) | −2.5 | .08 | −3.4 | .03 | −2.8 | .07 | −3.3 | .05 |
| Food sensitization (vs control) | 1.4 | .22 | 0.5 | .65 | 0.7 | .58 | 1.5 | .29 | |
| Food allergy (vs control) | −0.8 | .65 | −1.9 | .23 | −1.7 | .39 | −0.6 | .72 | |
Statistically significant associations (P < .05) are in boldface.
16S rRNA sequencing revealed 6 operational taxonomic units (OTUs), all of the genus Bacteroides, that were positively associated with the sphingolipid metabolite module and positively associated with food sensitization compared with food allergy (see Tables E4 and E5 in this article’s Online Repository at www.jacionline.org). Sphingolipids are produced by a minority of bacteria, including Bacteroides species.5 Mediation analysis showed that 95% of the association between having nonzero relative abundance of at least 1 of the 6 Bacteroides species OTUs and food sensitization was mediated by the sphingolipid module, with the proportion mediated ranging from 53% to 84% for individual sphingolipid metabolites (P value for indirect effect < .05 for all) (see Table E6 in this article’s Online Repository at www.jacionline.org). We investigated the possibility that Cesarean section could increase food allergy risk via reduced Bacteroides-associated sphingolipid intestinal abundances. In support of this hypothesis, Cesarean section was inversely associated with sphingolipid module eigenvalues (β = −0.09; P <.01) and with the 6 Bacteroides OTUs identified above (see Table E5 in Online Repository). Mediation analysis showed that 37% of the association between birth by Cesarean section and food sensitization was mediated by the sphingolipid module (P value for indirect effect = .02) (see Table E6 in Online Repository).
TABLE E5.
OTUs positively associated with sphingolipid metabolite module eigenvalues and with food sensitization compared with food allergy
| Association with sphingolipid module |
Association with food allergy (vs food sensitization) |
Association with food sensitization (vs control) |
Association with food allergy (vs control) |
Association with Cesarean section (vs vaginal delivery) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OTU ID | Taxa | Log2 fold change |
PBH
value |
Log2 fold change |
P
value |
Log2 fold change |
P
value |
Log2
fold change |
P
value |
Log2 fold change |
P
value |
| 577170 | Bacteroides unidentified species | 10.1 | .002 | −3.6 | .01 | 1.2 | .27 | −1.9 | .20 | −2.7 | .06 |
| 184753 | Bacteroides unidentified species | 9.2 | .01 | −5.2 | <.001 | 0.7 | .49 | −2.8 | .07 | −4.5 | .002 |
| 3472078 | Bacteroides unidentified species | 8.9 | .01 | −3.3 | .04 | 1.9 | .09 | −1.4 | .36 | −1.7 | .31 |
| 182157 | Bacteroides unidentified species | 7.8 | .02 | −2.9 | .03 | 1.1 | .27 | −1.6 | .23 | −2.9 | .02 |
| 188735 | Bacteroides unidentified species | 7.5 | .02 | −3.3 | .04 | 0.7 | .55 | −2.4 | .12 | −2.2 | .19 |
| 4354042 | Bacteroides unidentified species | 7.1 | .03 | −3.4 | .03 | 0.5 | .65 | −1.9 | .23 | −4.4 | .01 |
Negative binomial regression models of food allergy or sensitization phenotype were adjusted for age; models of mode of delivery were adjusted for age, sex, race/ethnicity, study center, maternal education, breast-feeding, and solid foods status.
P values denoting significant associations are in boldface. P values for associations with the sphingolipid module are adjusted for 420 comparisons (microbiome-wide analysis).
TABLE E6.
Causal mediation analyses of intestinal sphingolipids as mediators of the associations of intestinal Bacteroides and mode of delivery with food sensitization (n = 32) compared with food allergy or controls (n = 49)
| Average causal mediation effect |
Average direct effect |
|||||
|---|---|---|---|---|---|---|
| Association tested | Mediator | Estimate (95% CI) | P value | Estimate (95% CI) | P value | Proportion mediated (%) |
| Bacteroides* and food sensitization | Sphingolipid module eigenvalue | 0.15 (0.03 to 0.27) | .02 | −0.01 (−0.22 to 0.22) | .88 | 95 |
| 13-Methylmyristate | 0.13 (0.01 to 0.26) | .04 | 0.00 (−0.20 to 0.23) | .99 | 84 | |
| 3-Hydroxypalmitate | 0.12 (0.02 to 0.23) | .01 | 0.02 (−0.19 to 0.23) | .90 | 74 | |
| N-palmitoylserine | 0.12 (0.01 to 0.24) | .04 | 0.03 (−0.17 to 0.25) | .81 | 72 | |
| 3-Ketosphinganine | 0.11 (0.02 to 0.21) | .01 | 0.03 (−0.16 to 0.25) | .76 | 67 | |
| Sphinganine | 0.09 (0.01 to 0.19) | .02 | 0.05 (−0.15 to 0.26) | .61 | 53 | |
| Cesarean section delivery and food sensitization | Sphingolipid module eigenvalue | −0.08 (−0.18 to −0.01) | .02 | −0.13 (−0.34 to 0.09) | .22 | 37 |
| 13-Methylmyristate | −0.07 (−0.16 to 0.00) | .04 | −0.15 (−0.35 to 0.07) | .16 | 29 | |
| 3-Hydroxypalmitate | −0.05 (−0.13 to 0.01) | .09 | −0.16 (−0.36 to 0.06) | .16 | 22 | |
| N-palmitoylserine | −0.07 (−0.17 to 0.00) | .051 | −0.13 (−0.35 to 0.09) | .23 | 33 | |
| 3-Ketosphinganine | −0.11 (−0.22 to −0.02) | .01 | −0.11 (−0.33 to 0.12) | .34 | 49 | |
| Sphinganine | −0.03 (−0.10 to 0.02) | .23 | −0.18 (−0.37 to 0.05) | .11 | 27 | |
Bacteroides was analyzed as a dichotomous variable on the basis of presence or absence of at least 1 of the 6 Bacteroides OTUs associated with both the sphingolipid metabolite module and phenotype.
P values less than .05 are in boldface.
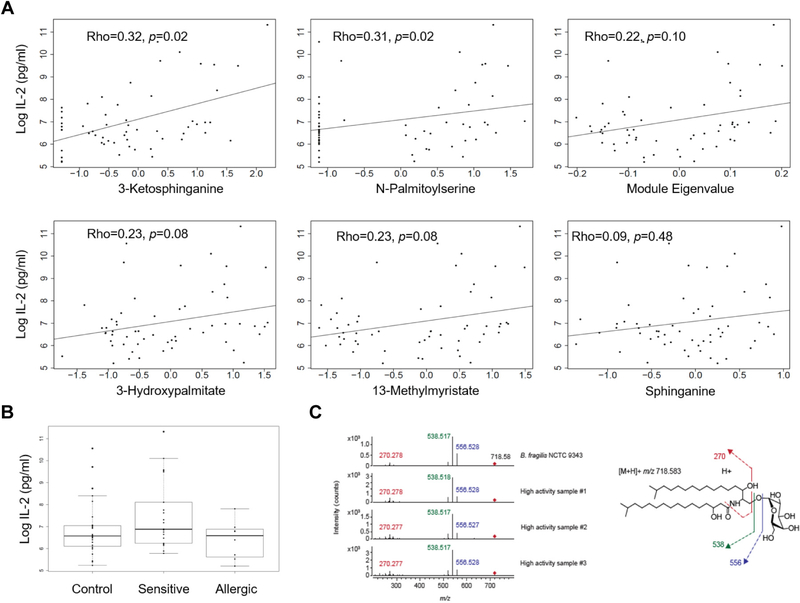
There is evidence that invariant natural killer T (iNKT)-cell number and cytokine production is perturbed in food allergy6 and that lipids recognized by iNKT cells include Bacteroides-derived glycosphingolipids.7 Accordingly, we tested the hypothesis that the sphingolipid metabolite module that we identified indicates the presence of a lipid antigen for iNKT cells. For 57 subjects with sufficient stool quantity (see Table E2 in Online Repository), we investigated whether fecal lipids could activate iNKT cells using a coculture assay with a T-cell hybridoma expressing an iNKT-cell T-cell receptor (DN32) and a macrophage cell line transfected with CD1d. This assay system detects lipid antigen, but is insensitive to innate pattern receptor agonists. Fecal lipid iNKT-cell activation was higher among subjects with food sensitization than among those with food allergy (P = .02) (Fig 2, B), recapitulating the pattern of association seen between the sphingolipid module and phenotype. All 5 sphingolipid members of the sphingolipid module were positively associated with iNKT-cell activation (Fig 2, A). iNKT-cell activation was strongly associated with B fragilis relative abundances (ρ = 0.49; P < .001). Finally, fecal lipid iNKT-cell activation was significantly lower among subjects born by Cesarean section than among subjects born by vaginal delivery (P = .049).
FIG 2.
A, Scatter plots of sphingolipid metabolite relative abundances vs iNKT-cell activation, as measured by IL-2 production. Spearman rho are displayed. B, Box plots of iNKT-cell activation comparing subjects with food allergy (n = 8), food sensitization (n = 22), and controls (n = 27). Fecal lipid iNKT-cell activation was higher in those with food sensitization compared with those with food allergy (t test P = .02 after log10-transformation of IL-2). C, Fragmentation of Bacteroides fragilis α-galactosylceramide. Left panel shows polar lipid extracts from B fragilis and 3 samples with high iNKT-cell activity. Two independent experiments were performed using high mass accuracy LC-MS-MS targeting the retention time of B fragilis α-galactosylceramide (m/z = 718.58). Right panel shows deduced fragmentation of B fragilis α-galactosylceramide (m/z = 718.58) based on the reported structure for this ion.7
Human B fragilis isolates produce α-galactosylceramide, a glycosphingolipid with activity at the iNKT-cell receptor.7 We quantified B fragilis–associated α-galactosylceramide in fecal lipids using high-performance liquid chromatography with quadrupole time-of-flight mass spectroscopy (Fig 2, C). The most abundant α-galactosylceramide molecular species (m/z = 718.58) was detectable in 12 (21%) of the 57 samples at the same retention time as in lipids extracted from B fragilis. B fragilis–associated α-galactosylceramide abundance was associated with sphingolipid module eigenvalues (ρ = 0.37; P = .005) and iNKT-cell activation (ρ = 0.55; P < .001). Comparison by phenotype was limited by the large proportion of subjects with no detectable α-galactosylceramide; however, α-galactosylceramide was present more frequently in fecal lipids of subjects with food sensitization (7 [32%] of 22) compared with controls (4 [15%] of 27) and was present in only 1 (13%) of 8 samples from subjects with food allergy. α-galactosylceramide was associated with B fragilis (ρ = 0.62; P < .001) and pooled Bacteroides species (ρ = 0.29; P = .03), but not with other Bacteroides species’ relative abundances. In contrast, 2 non–Bacteroides-derived control lipids, hexosylceramide (m/z = 828.69) and sphingomyelin (m/z = 703.58), were not associated with iNKT-cell activation, phenotype, or Bacteroides species. The high concordance among Bacteroides OTUs, B fragilis α-galactosylceramide ions, and iNKT-cell activation by fecal lipids suggest that together with other bioactive metabolites, α-galactosylceramide, likely produced by B fragilis, contributes to the observed differential iNKT-cell activation by clinical phenotype.
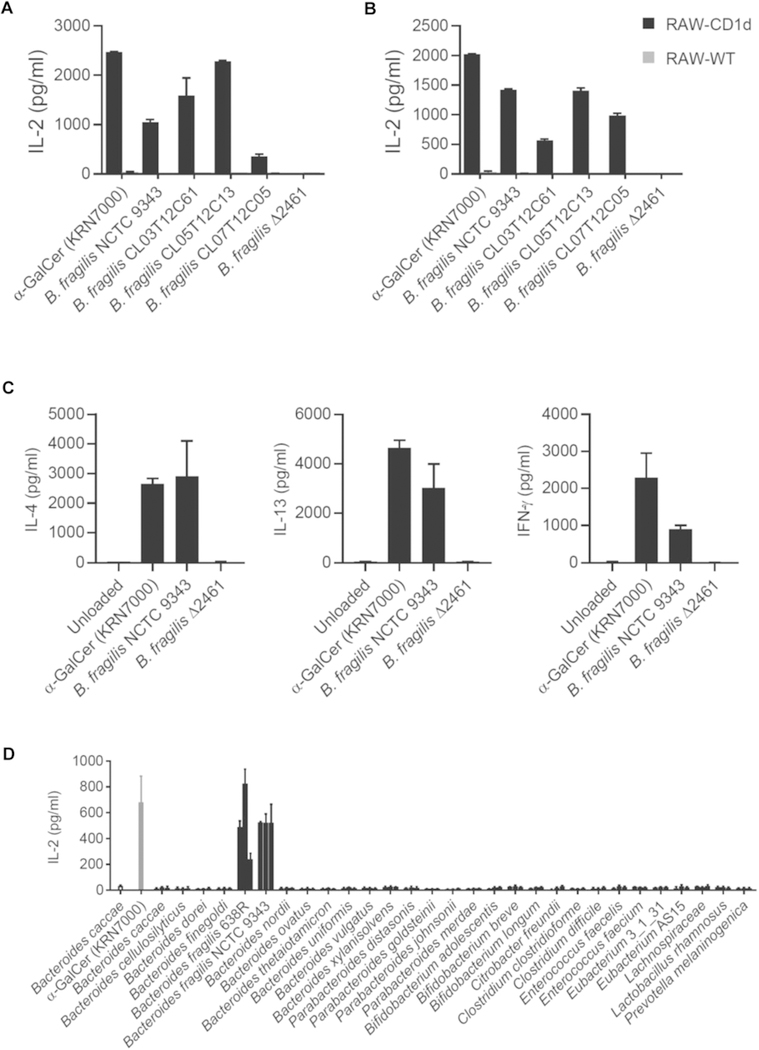
We tested several strains of B fragilis for the ability to activate iNKT cells. All B fragilis strains tested activated iNKT cells except a sphingolipid-deficient mutant (ΔBF2461, also known as BF9343_2380)7 (Fig E1, A and B). To confirm that iNKT-cell activation was the result of cognate interaction between the T-cell receptor and B fragilis lipids presented by CD1d, we performed a cell-free assay in which recombinant CD1d is loaded with lipids, fixed to solid phase, and tested for the ability to activate iNKT cells. In this assay, wild-type B fragilis lipids, but not lipids from the ΔBF2461 sphingolipid-deficient mutant, activated a primary iNKT-cell line to produce IL-4, IL-13, and IFN-γ (Fig E1, C). We next tested 29 human gut anaerobes8 from various genera for the ability to activate iNKT cells. Of the strains tested, only B fragilis showed activity (Fig E1, D). We concluded from these experiments that lipids produced by B fragilis activate iNKT cells, and that this bioactivity is neither shared by other common Bacteroides species, nor is it common among human gut anaerobes.
FIG E1.
B fragilis but not other species activate iNKT cells and produce α-galactosylceramides. Heat-killed whole bacteria (A) or polar lipid extracts (B) were added to cocultures of CD1d-transfected RAW246.7 cells and an iNKT-cell hybridoma (DN32). This assay system responds to lipid antigen, but not to microbial patterns. INKT-cell activation was assessed by IL-2 ELISA compared with α-galactosylceramide (α-GalCer KRN7000, 10 ng/mL), a prototypical iNKT cell lipid antigen. C, Lipids were loaded in recombinant, plate-bound CD1d before a primary mouse iNKT cell line was added. Cytokine production was measured by ELISA. Experiments were performed twice, and a representative experiment is shown. Error is SEM of 2 replicates. D, Twenty-nine human anerobic bacterial strains were tested for their ability to activate an iNKT-cell hybridoma (DN32) in coculture with a CD1d-transfected RAW246.7 macrophage cell line. Washed, heat-killed bacteria were added at 3 dilutions (left to right, 1:1, 1:5,1:25). α-Galactosylceramide (α-GalCer KRN7000) was added at 10 ng/mL as a positive control. iNKT-cell activation was assessed by IL-2 ELISA. Error bars indicate the SEM of 2 independent experiments.
This prospective and untargeted analysis of the infant intestinal microenvironment suggests that intestinal Bacteroides-derived sphingolipids, and particularly B fragilis–derived α-galactosylceramide and its effect on iNKT cells, could confer protection against food allergy among individuals predisposed to food sensitization. Additional Bacteroides-associated mechanisms may contribute to protection. The positive association between Cesarean section delivery and food allergy may be due in part to reduced Bacteroides abundance. Additional studies are needed to confirm our findings and discover additional mechanisms whereby the early-life intestinal microenvironment influences food allergy risk.
METHODS
Outcome ascertainment
Data used to ascertain food allergy and sensitization outcomes have been previously described.E1 Briefly, parents were asked to report on health care provider–diagnosed food allergy every 3 months from birth. In subjects who agreed to provide a blood sample at age 3 years, serum specific IgE was measured by ThermoFisher PIRL lab (Phadia Immunology Reference Laboratory, Portage, Mich) to food allergens (egg white, walnut, milk, peanut, soybean, and wheat). Food sensitization was defined by specific IgE concentration of 0.35 kU/L or more to at least 1 tested food allergen and no parental report of food allergy by age 3 years, though we could not confirm that subjects were eating and tolerating all tested foods. Food allergy was defined by parental report of allergy to at least 1 food and IgE level of 0.35 kU/L or more to the same food at age 3 years. No subjects had yet reported food allergy diagnoses at the time of stool sample collection. Control subjects had neither IgE level of 0.35 kU/L or more to any tested food nor parental report of food allergy diagnosis or reaction by age 3 years. Control and food-sensitized children were roughly matched to food-allergic children on sex and race/ethnicity.
Covariates
Additional characteristics were ascertained at study entry, birth, or via follow-up questionnaires and study visits. Asthma or recurrent wheeze was based on parental report of physician diagnosis of asthma or recurrent wheeze in the first 3 years of life as previously reported.4 Questionnaire responses were used to determine whether a child was ingesting breast milk and/or formula, and whether solid foods had been introduced at the time of stool sample collection as previously described.E1
Fecal sample collection and profiling
Stool collection and microbiome profiling methods have been described in detail previously.E2 DNA extraction was performed on the stool samples and sequencing of the bacterial 16S V3 to V5 hypervariable regions was performed by pyrosequencing (Roche 454 Titanium platform) at the Genome Center (TGI) at Washington University in St Louis, Mo. Filtering, trimming, and chimera checking were performed as previously described.E3,E4 Closed reference OTU classification was performed using QIIME.E5 Additional data processing was performed using Phyloseq (version 1.20.0).E6 All samples had total read counts of at least 1000. Of 1107 OTUs detected, those absent 5% of samples or more were excluded, leaving 420 OTUs for analysis.
Fecal metabolomic profiling was performed at Metabolon, Inc (Research Triangle Park, NC) using ultraperformance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS), as described earlier.E7 Of 887 identified metabolites, 148 xenobiotic metabolites and 38 metabolites with an interquartile range of 0 were excluded from analysis, leaving 701 metabolites. For each metabolite, missing values were replaced with half of the minimum value of that metabolite. Most metabolites were not normally distributed and all metabolite relative abundances were log10-normalized and pareto-scaled.
Identification of highly correlated metabolite modules
A network of highly correlated metabolites was constructed using the weighted gene correlation network analysis (WGCNA) R package (version 1.61)E8 using Spearman correlation coefficients and applying a minimum module size of 4 metabolites and a soft thresholding power of 8 (chosen by using the pickSoftThreshold function of the WGCNA R package to achieve a scale-free topology fitting index of >0.9). Eigenvalues summarizing relative abundances of metabolites of each metabolite module for each subject were used in subsequent analyses.
Statistical methods
Statistical analyses were conducted using R version 3.4.0 (R Foundation for Statistical Computing). Kruskal-Wallis and Fisher exact tests were used to test for differences in baseline characteristics by phenotype. Logistic regression analyses were used to determine the association between metabolites (first individual metabolites, then WGCNA-generated metabolite module eigenvalues) and binary phenotype variables. Adjusted analyses included only age as a covariate and for significant associations with metabolite module eigenvalues, sensitivity analyses were performed including age and individual potential confounders. All tests were 2-sided and the significance level was prespecified at a P value of less than .05.
Using Phyloseq (version 1.20.0) and DESeq2 (version 1.16.1),E6,E9 negative binomial regression models were used to analyze associations between intestinal microbial OTUs and phenotype-associated metabolite modules. For OTUs associated with metabolite modules, associations were also tested with phenotype in age-adjusted analyses. For OTUs associated with both metabolite modules and phenotype, sensitivity analyses were performed including age and individual potential confounders as covariates. Associations between mode of delivery and phenotype-associated microbes and metabolite modules were tested with adjusted negative binomial regression and multivariable linear regression.
Spearman correlation, ANOVA, and t tests were used to test for associations between log10-transformed IL-2 production in iNKT-cell activation assays and other variables. Spearman correlation was used to test for associations between α-galactosylceramide sphingolipid module eigenvalues, iNKT-cell activation, and Bacteroides species. Age-adjusted logistic regression analysis was used to test for associations between log-transformed α-galactosylceramide and phenotype.
Mediation analysis
Two series of mediation analyses were performed. The first estimated the direct association between phenotype- and sphingolipid-associated Bacteroides species OTUs and phenotype, and the indirect associations mediated through sphingolipid metabolites. The second estimated the direct association between mode of delivery and phenotype, and the indirect associations mediated through sphingolipid metabolites. Bacteroides was analyzed as a dichotomous variable on the basis of presence or absence of at least 1 of the 6 relevant Bacteroides OTUs. To ensure adequate sample size for adjusted regression, subjects with food sensitization (n = 32) were compared with all other subjects (n = 49). Sphingolipid metabolite module eigenvalues and individual sphingolipid relative abundances were tested as mediators. All analyses were adjusted for age at stool sample collection. The R package “mediation” was used and 95% CIs were based on quasi-Bayesian approximation with 2000 Monte-Carlo draws.E10,E11
In vitro iNKT-cell activity assay
In vitro iNKT-cell activity assays were performed using polar lipid extracts from fecal samples from all infants who had sufficient fecal sample volume. Polar lipids were extracted as previously described.E12,E13 Lipids were dried under nitrogen and sonicated in media immediately before assay. DN32 iNKT cell hybridoma cellsE14 (5 × 104) were cultured with 2.5 × 104 CD1d-transfected RAW-264.7 cellsE15 for 16 hours. The DN32 hybridoma expresses a uniform iNKT-cell T-cell receptor, and is robustly activated by known iNKT cell lipid antigens including α-galactosylceramide, α-glucosylceramide, and isoglobotrihexosylceramide. Plate-bound assays with primary iNKT cells were performed as described earlier.E16 α-Galactosylceramide KRN7000 (Avanti Polar Lipids) was used as a positive control. IL-2 for ELISA standards were from Peprotech (Rocky Hill, NJ). Two replicates were performed per sample and the average IL-2 per sample used for statistical analysis. This assay system was chosen because it is insensitive to innate activation mechanisms such as pattern receptor agonists, whereas primary iNKT cells would also respond indirectly to innate patterns. Activity was not observed when antigen-presenting cells lacking CD1d were used.
Mass spectroscopy analysis of fecal lipid fractions
HPLC-MS was performed on an Agilent 6520 Accurate-Mass Q-TOF using a normal-phase ternary gradient HPLC as previously described.E13 In the fecal lipid extracts, α-galactosylceramide m/z 718.58 was quantified from 4.5 to 5.5 minutes on the basis of the retention time and mass of lipids extracted from B fragilis NCTC 9343. Hexosylceramide m/z 828.69 was identified and sphingomyelin m/z 703.58 was quantified on the basis of mass and retention time of a standard. Ion abundance was quantified by centroid integration as area under the curve (MassHunter, Agilent, Santa Clara, Calif).
Bacterial strains
Bacteria were grown in supplemented basal mediumE17 under anerobic conditions to an OD of 0.5 to 1.5 at 600 nm for experiments. Where whole bacteria were used in culture with DN32 and RAW-246.7 cells, bacteria were pelleted at 1500g for 30 minutes, then heat killed at 65°C for 30 minutes. The top concentration of each bacteria added to assay (Fig E1, D) was 25 μL of culture (OD) of 0.5 to 1.0 at 600 nm in a 200-μL coculture. Strains included B fragilis 638R, B fragilis NCTC 9343B, B fragilis CL03T12C61, B fragilis CL05T12C13, B fragilis CL07T12C05, B xylanisolvens CL03T12C04, B caccae CL03T12C61, B cellulosilyticus CL02T12C19, B dorei CL02T12C06, B finegoldii CL09T03C10, B nordii CL02T12C05, Parabacteroides distasonis CL03T12C09, P goldsteinii CL02T12C30, P johnsonii CL02T12C29, and P merdae CL03T12C32.E17 B fragilis Δ2461 was generated from strain NCTC 9343B.7 Other strains included B ovatus NCTC 8483, B thetaiotamicron VPI-5482, B uniformis NCTC 8492, B vulgatus NCTC 8482, Bifidobacterium adolescentis L2–32 (HM-633), Bifidobacterium breve EX336960VC18 (HM-411), Bifidobacterium longum 44 (HM-845), Citrobacter freundii 4_7_47CFAA (HM-299), Clostridium clostridioforme (2_1_49FAA), Clostridium difficile NAP07 (HM-88), Enterococcus faecalis ERV103 (HM-934), Enterococcus faecium 503 (HM-952), Eubacterium sp.3_1_31 (HM-178), Eubacterium sp. AS15 (HM-766), Lachnospiraceae sp. 5_1_57AA (HM-157), Lactobacillus rhamnosus LMS2–1 (HM-106), and Prevotella melaninogenica (HM-49). Most of these bacterial strains were obtained from BEI Resources (Manassas, Va).
Acknowledgments
The Vitamin D Antenatal Asthma Reduction Trial was funded by the National Heart, Lung, and Blood Institute (grant no. U01HL091528). Additional funding came from the National Institutes of Health (NIH) (grant nos. R01HL108818 and 5T32AI007306–30). P.J.B. was supported by the NIH (grant no. AI102945) and generous support from the Goodfellow, Violin, and Karol families. P.J.B. and L.E.C. were supported by the Brigham and Women’s Hospital Innovation Evergreen Fund. T.-Y.C. and D.B.M. were supported by the NIH (grant no. R01AR048632).
Footnotes
Disclosure of potential conflict of interest: J. Lasky-Su is a consultant to Metabolon Inc. R. S. Zeiger participates on the Scientific Advisory Board of DBV Technologies, receives stock from DBV Technologies, and participates on the Data Safety Monitoring Committee of AIMMUNE. G. T. O’Connor is a coinvestigator on a grant from Janssen Pharmaceuticals to Boston University that funds a study of the pathogenesis of chronic obstructive pulmonary disease. L. B. Bacharier participates on the Data Safety Monitoring Board of DBV Technologies. A. Beigelman holds stock from DBV Technologies. S. Bunyavanich receives funding from the National Institutes of Health (grant no. R01AI118833). J. H. Savage is currently employed by Vertex Pharmaceuticals and has unvested stock in Vertex Pharmaceuticals, and her husband is employed by Shire and has unvested stock in Shire. S. T. Weiss is a paid consultant to UpToDate. A. A. Litonjua is a consultant to AstraZeneca, LP. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Yu W, Freeland DMH, Nadeau KC. Food allergy: immune mechanisms, diagnosis and immunotherapy. Nat Rev Immunol 2016;16:751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol 2014;80:2546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 2013;1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litonjua AA, Carey VJ, Laranjo N, Harshfield BJ, McElrath TF, O’Connor GT, et al. Effect of prenatal supplementation with vitamin D on asthma or recurrent wheezing in offspring by age 3 years: the VDAART randomized clinical trial. JAMA 2016;315:362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato M, Muto Y, Tanaka-Bandoh K, Watanabe K, Ueno K. Sphingolipid composition in Bacteroides species. Anaerobe 1995;1:135–9. [DOI] [PubMed] [Google Scholar]
- 6.Jyonouchi S, Abraham V, Orange JS, Spergel JM, Gober L, Dudek E, et al. Invariant natural killer T cells from children with versus without food allergy exhibit differential responsiveness to milk-derived sphingomyelin. J Allergy Clin Immunol 2011;128:102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, et al. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol 2013;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Turnbaugh PJ, Ley RE, Ley RE, Hamady M, Hamady M, et al. The human microbiome project. Nature 2007;449:804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.Savage JH, Lee-Sarwar KA, Sordillo J, Bunyavanich S, Zhou Y, O’Connor G A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018;73:145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E2.Sordillo JE, Zhou Y, MJ McGeachie, Ziniti J, Lange N, Laranjo N Factors influencing the infant gut microbiome at age 3–6 months: findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART). J Allergy Clin Immunol 2017;139:482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.HMP Consortium. A framework for human microbiome research. Nature 2012; 486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E4.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 2011;21:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E5.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E6.McMurdie PJ, Holmes S Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E7.Evans AM, Bridgewater B, Liu Q, Mitchell MW, Robinson RJ, Dai H, et al. Metabolomics: open access high resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 2014;4:1–7. [Google Scholar]
- E8.Langfelder P, Horvath S. WGCNA. an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E9.Love MI, Huber W, Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E10.Tingley D, Yamamoto T, Hirose K, Keele L, Imai K Mediation: R Package for Causal Mediation Analysis. J Stat Softw 2014;59:1–38.26917999 [Google Scholar]
- E11.Brennan PJ, Tatituri RV, Heiss C, Watts GF, Hsu FF, Veerapen N, et al. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci U S A 2014;111:13433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E12.Brennan P, Cheng TY, Pellicci DG, Watts GF, Veerapen N, Young DC, et al. Structural determination of lipid antigens captured at the CD1d-T-cell receptor interface. Proc Natl Acad Sci U S A 2017;114:8348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E13.Lantz O, Bendelac A An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–8− T cells in mice and humans. J Exp Med 1994;180:1097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E14.Muindi K, Cernadas M, Watts GF, Royle L, Neville DC, Dwek RA, et al. Activation state and intracellular trafficking contribute to the repertoire of endogenous glycosphingolipids presented by CD1d. Proc Natl Acad Sci U S A 2010;107:3052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E15.Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP, et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol 2011;12:1202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E16.Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun 1991;59:2075–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E17.Zitomersky NL, Coyne MJ, Comstock LE Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun 2011;79:2012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]