Abstract
A significant contributor to women’s cancer mortality worldwide is cervical cancer, which is caused by high-risk human papillomavirus (HR HPV). The two viral oncoproteins of HR HPV, E6 and E7, partner with host cell proteins to target oncogenic proteins and pathways. Previously, we have shown HR HPV type 16 E6 (16E6) interacts with the host protein NFX1-123 to target telomerase and cellular immortalization, requiring NFX1-123 to fully upregulate telomerase activity. We now report that NFX1-123 is highly expressed in primary cervical cancers. In vitro, cells expressing 16E6 and overexpressing NFX1-123 have extended active growth, decreased senescence marker staining, and more rapid cell cycling compared to 16E6 expressing cells with endogenous amounts of NFX1-123. These findings were associated with increased telomerase activity and augmented expression of its catalytic subunit, hTERT. In complement, HPV 16 positive cervical cancer cell lines with knocked down NFX1-123 had slowed growth and reduced hTERT over time. In cells that express HR HPV E6, greater expression of NFX1-123 can modify active cellular growth and augment hTERT expression and telomerase activity over time, potentially supporting the initiation and progression of HPV-associated cancers.
Keywords: telomerase, NFX1-123, HR HPV, HR E6, Growth
1. Introduction
High risk HPVs (HR HPVs) are the causative agent of cervical cancer, the fourth most common cancer in women, as well as other anogenital and head and neck cancers [1-8]. In total, HR HPV causes 5% of all cancers worldwide [3]. HPV-associated cancers universally express the viral oncogenes HR HPV E6 and E7, and these oncogenes activate pathways critical to cancer development. One of the most fundamental of these oncogenic pathways is cellular immortalization [9], driven by the enzyme telomerase.
Without telomerase, telomeric DNA is serially eroded and growth arrest is triggered once DNA becomes critically shortened [10, 11]. Telomerase is activated in all HPV-associated cancers, extending telomeric DNA and avoiding senescence, apoptosis, and cellular crisis [12-14]. Expression of the catalytic subunit of telomerase, hTERT, is rate-determining [15, 16]; HR HPV E6 activates its expression, thus driving cellular immortalization through telomerase [9, 17-21]. HR HPV E6 requires cellular proteins to regulate hTERT and telomerase activity, one of which is NFX1-123 [18, 22].
NFX1-123 binds HR HPV type 16 E6 (16E6) and is highly expressed in cervical cancer cell lines [22, 23]. With 16E6, NFX1-123 increases hTERT and telomerase through post-transcriptional stabilization of hTERT mRNA [18, 22]. Although it is known that NFX1-123 and 16E6 collaboratively increase hTERT and telomerase, what remains unexplored are the dynamics of NFX1-123 expression and its longitudinal effects on hTERT, telomerase, cellular growth, and longevity. Furthermore, although highly expressed in cervical cancer cell lines, the expression of NFX1-123 in the normal cervix or cervical cancers is unknown.
In the present study, we found that NFX1-123 was expressed in the cervix and highly expressed in HPV-positive cervical precancerous lesions and cancers. Expression of NFX1-123 to levels approaching cervical cancer led to extended active cellular growth, maintained cell cycling, and reduced senescence in 16E6 expressing human foreskin keratinoctyes (16E6 HFKs). In parallel with these increases in growth, hTERT expression and telomerase activity were greater in early passages and became further amplified over time. Knock down of NFX1-123 in the HPV 16 positive SiHa cervical cancer cell line resulted in slowed growth and reduced hTERT. These results indicate that NFX1-123 is markedly increased in cervical cancer, its greater expression in 16E6 expressing cells is associated with improved growth and telomerase activity, and that this association is maintained in HPV 16 positive cervical cancer cell lines.
2. Materials and Methods
2.1. Tissue culture.
Primary human foreskin keratinocytes (HFKs) were cultured as described previously [17]. Organotypic HFK raft cultures were grown as previously published in E-media containing 5% FBS using 6-well transwell inserts (Corning, NY) in place of wire mesh [24]. 293T and SiHa cells were grown in DMEM (Thermo Fisher Scientific, Grand Island, NY) containing 10% FBS and penicillin-streptomycin.
2.2. Histologic analysis.
Organotypic HFK raft cultures were formalin fixed and paraffin embedded (FFPE) following standard procedures. 32 normal cervical and 37 HPV 16+ cervical intraepithelial neoplasia (CIN) 2, CIN3, carcinoma in situ, and cervical cancer FFPE blocks were obtained from the University of Washington HPV Research Group Specimen Repository. They were deidentified and not considered human subjects by the Seattle Children’s Research Institute IRB. Sections were stained for NFX1-123 using a rabbit polyclonal anti-NFX1-123 antibody (1:1000 dilution) or an isotype control. To quantify NFX1-123 staining, three independent, blinded reviewers scored multiple 20x slide images. Scoring was based on staining intensity of the specimen: none (no stain), low (yellow), moderate (yellow-brown) or high (dark brown).
2.3. Plasmids.
FLAG-tagged NFX1-123WT cDNA (FN123), pBABE-puro 16E6 (16E6), LXSN vector control (LXSN), short hairpin 1 NFX1-123 (sh1), and scramble (scr) c-FUGW constructs have been described previously [18]. The short hairpin 2 NFX1-123 (sh2) c-FUGW construct was similarly cloned as sh1 and scr and designed specifically to the novel C terminus of NFX1-123. The sequence was BLAST searched at http://www.ensembl.org to confirm alignment with only NFX1-123. The short hairpin was made by annealing the following oligonucleotides: 5’ GATCCGCGTGAATAAGGGAAAGAATTTCAAGAGAATTCTTTCCCTTATTCACGTTTTTTGG 3’, and 5’ AATTCCAAAAAACGTGAATAAGGGAAAGAATTCTCTTGAAATTCTTTCCCTTATTCACGCG 3’, followed by ligation into the c-FUGW vector [25].
2.4. Retrovirus production and infection.
Retrovirus for FN123, 16E6, or LXSN was produced in 293T cells by a transient vesicular stomatitis virus G-pseudotyped virus (VSV-G) production protocol as previously described [26]. Lentivirus for NFX1-123 sh1, sh2, or scr control cFUGW constructs was produced as previously described [18]. Retrovirus transduction was confirmed with antibiotic resistance and lentivirus transduction by green fluorescent protein (GFP) expression. Typically over 95% of cells were transduced by lentivirus.
2.5. Growth assay in HFK cells.
Four biologically independent HFK cell lines were produced by transduction and selection with either empty vector LXSN or FN123, or by serial transduction and selection with 16E6 and then either LXSN or FN123. After transduction, 5×105 cells were plated onto three or more 10cm tissue culture (TC) dishes, fed every three days, and counted every four days. The total cell number of each plate was recorded, averaged, and divided by 5×105 to determine the number of population doublings achieved. After counting, 5×105 HFK cells were replated and the remaining cells were collected for experiments. HFK cell lines were grown until they no longer doubled within four days’ time, defined as the active growth phase.
2.6. Growth assay in SiHa cells.
SiHa cervical cancer cells were grown to 70-80% confluency, transduced overnight with NFX1-123 sh1, sh2 or scr lentivirus, and then recovered in fresh media. At 48 hours post-transduction (day 2), infection was confirmed by GFP expression, and cells were counted and plated into four 10cm dishes. At 72 hours post-transduction (day 3), cells were counted again, collected for experiments, and one plate was re-fed with media and kept in culture. This process was repeated again on days 5 and 6. The growth rate over 24 hours (doubling factor) for each cell line (scr, sh1, sh2) was calculated by the cell number on each plate on the day of collection divided by the cell number plated the previous day. At least three plates were included in each doubling factor calculation, for each cell line, at each timepoint. Statistical significance (p-values) was calculated using a one-way ANOVA with Bonferroni post hoc correction. The assays were repeated independently three times.
2.7. Western blot.
Whole cell lysates were prepared in WE16th lysis buffer as previously described [17], electrophoresed on a 4-12% gradient SDS-polyacrylamide gel (Thermo Fisher Scientific, Grand Island, NY), and transferred to an Immobilon-P or Immobilon-FL membrane (Millipore Sigma, Burlington, MA). Blots were probed with antibodies: GAPDH (ab8245) (1:100,000; Abcam, Cambridge, MA), and rabbit polyclonal anti-NFX1-123 or NFX1-91 (1:1,000). The rabbit polyclonal anti-NFX1-123 and NFX1-91 antibodies were a gift from Dr. Ann Roman.
2.8. Quantitative real-time PCR.
RNA was isolated using TRIzol reagent (Thermo Fisher Scientific, Grand Island, NY) as previously described [18]. Total RNA (1μg) was DNase treated and cDNA generated using random hexamer primers and SuperScript IV reverse transcriptase (Thermo Fisher Scientific, Grand Island, NY). Quantitative real-time PCR (qPCR) was performed using an ABI StepOne Plus system (Applied Biosystems, Foster City, CA). NFX1-123 primer sequences were 5’ CCACAGCTTCCCTCCCA 3’ (forward), and 5’ CCTGGACGTCAAAATAGTCAA 3’ (reverse). NFX1-91 primer sequences were 5’ TTACCCTCCAGTTCCCTGTG 3’ (forward), and 3’ CATGCGTGTGCAGGTATCTT 5’ (reverse). hTERT, 36B4, and FLAG-NFX1-123 primer sequences were described previously [18, 22]. Amplification was carried out in replicative triplicates using Power SYBR Green or Power UP SYBR Green master mix (Thermo Fisher Scientific, Grand Island, NY) [18]. Error bars represent 95% confidence intervals.
2.9. Telomeric repeat amplification protocol (TRAP) assay.
Telomerase activity was measured using a modified version of the qPCR-based TRAPeze RT kit (Millipore Sigma, Burlington, MA). HFKs were counted and lysed in CHAPS lysis buffer, using 200μL per 1×106 cells. To create a telomerase activity standard curve, 1×106 293T cells were lysed in 200μL of CHAPS lysis buffer and 1:10 dilutions prepared. qPCR was performed using an ABI StepOne Plus system (Applied Biosystems, Foster City, CA). Amplification was carried out in replicative triplicates and normalized to T1 16E6/LXSN. Error bars represent 95% confidence intervals.
2.10. Fluorescence-activated cell sorting (FACS) analysis.
Cells were synchronized by growing to confluency plus 24 more hours before release with staining at hour zero as previously described [23]. Analysis was performed using a LSR II flow cytometer (BD Biosciences, San Jose, CA) and FlowJo v10 software (Treestar, Ashland, OR).
2.11. Senescence Associated Beta Galactosidase (Beta-gal) Assay.
Beta-gal assays were completed using the Cellular Senescence Assay Kit (Millipore Sigma, Burlington, MA). Cells were plated onto 6-well plates at 5×104 cells per well, grown to 60-70% confluence, washed with PBS, and fixed for 15 minutes. X-gal detection solution was added to each well. Cells were incubated in the dark, overnight at 37°C. Images were acquired using a Keyence Imaging Scope (Keyence Corp., Itasca, IL) at 10x and 20x magnification. A minimum of ten fields were acquired, and total cell number, as well as the number of blue stained cells, were counted using the Keyence cell count program, with further analysis using ImageJ.
3. Results
3.1. High NFX1-123 expression in cervical cancer samples.
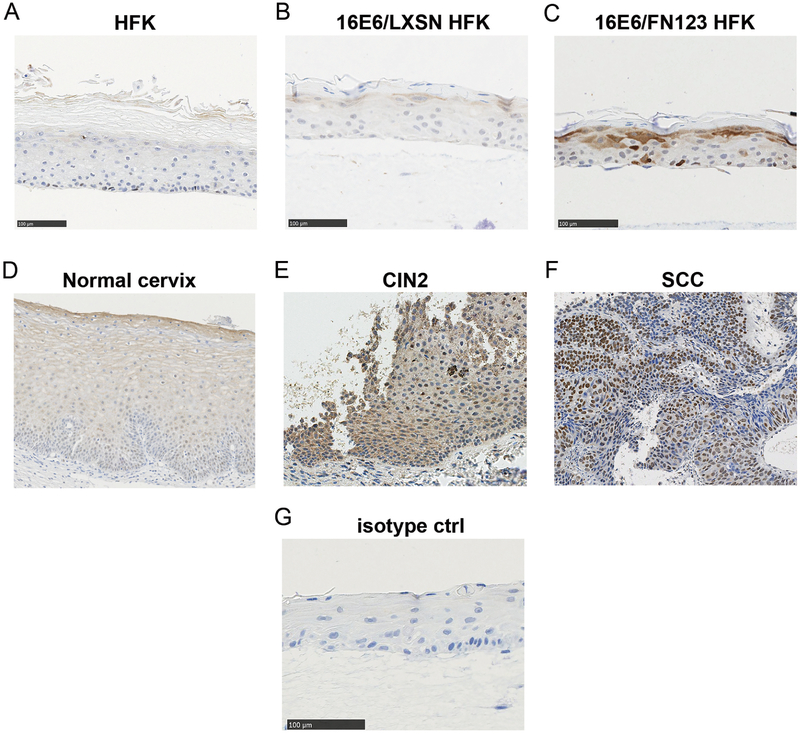
To study the effect of NFX1-123 gene expression on epithelial cells, a three-dimensional organotypic HFK raft culture system was used that mimics stratified squamous epithelium. NFX1-123 was detected in raft cultures in the upper, differentiated cells as well as sporadic basal layer cells (Figure 1A), unchanged by 16E6 co-expression (Figure 1B). With overexpression, greater NFX1-123 was seen both in the basal and upper differentiated cells (Figure 1C). In normal cervical specimens, NFX1-123 was seen throughout the epidermis, with greater expression in upper, differentiated cells that raft cultures mimicked (Figure 1D). In HPV 16 positive precancerous and cancer specimens, NFX1-123 protein was greatly increased: 11% stained with moderate (Figure 1E, CIN2) and 89% with high intensity (Figure 1F, SCC).
Figure 1. NFX1-123 expression in three dimensional raft cultures, cervical tissues, cervical precancerous lesions, and cervical cancer samples.
(A) HFKs (B) 16E6/LXSN and (C) 16E6/FN123 HFKs were grown in raft cultures and stained for NFX1-123 protein. Overexpression of NFX1-123 in 16E6/FN123 HFKs had increased staining in basal and differentiating keratinocytes. (D) NFX1-123 stained throughout the normal cervical epithelium, with greater expression seen in upper differentiating keratinocytes. (E) HPV 16 positive cervical intraepithelial neoplasia 2 (CIN2) stained moderately for NFX1-123. (F) HPV 16 positive squamous cell carcinoma (SCC) stained highly for NFX1-123. (G) Rabbit IgG isotype control.
3.2. NFX1-123 increased total population doublings and length of active growth in 16E6 HFKs but not HFKs alone.
NFX1-123 is highly expressed in cervical cancer cell lines [23] and primary cervical cancers (Figure 1). Therefore, it was important to determine if increased NFX1-123 expression in HFKs would affect growth patterns and longevity, and whether changes were dependent on HR HPV E6 co-expression. To quantify growth rate changes, we developed an objective definition of active cellular growth: a doubling in cell number within four days’ time at minimum. To identify trends separate from the primary cell background, studies were repeated in four biologically independent cell lines.
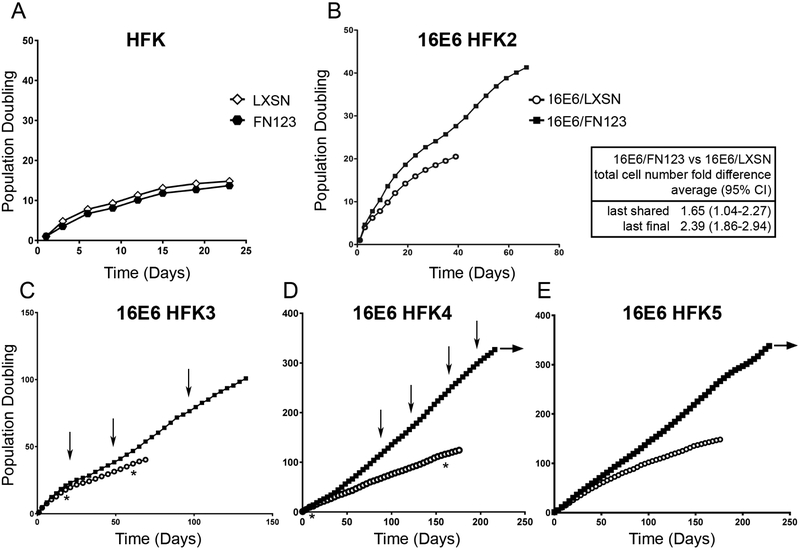
First, the effect of increased NFX1-123 on active cellular growth in non-16E6 HFKs was studied. HFKs transduced with FLAG-tagged NFX1-123 (FN123) had no difference in overall growth, total cell counts, or cumulative population doublings when compared to HFKs transduced with an empty vector control (LXSN) (Figure 2A). Next, to explore the role of 16E6 in combination with greater NFX1-123 expression, HFK cell lines were serially transduced with 16E6 and then with either FN123 (16E6/FN123) or LXSN vector (16E6/LXSN). Confirming effects of 16E6 from previous studies, HFKs with 16E6 and endogenous levels of NFX1-123 had a longer period of active growth than HFKs without 16E6 [20]. Across all four cell lines, 16E6 HFKs with greater NFX1-123 (16E6/FN123) achieved more population doublings during active growth at both their last shared timepoint with 16E6/LXSN HFKs and their last final timepoint (Figure 2B-E closed squares versus open circles). Although there were variations in total population doublings and length of time in active growth across the four HFK cell lines, 16E6/FN123 HFKs consistently had longer active growth periods and more population doublings than their isogenic controls (16E6/LXSN HFKs). In fact, after more than 200 days in culture 16E6/FN123 HFK3 and HFK4 cells continue to be actively growing (Figure 2D and E, horizontal arrow). These results indicate that 16E6 HFKs overexpressing NFX1-123 had more robust and longer active growth periods compared to their matched control cells. Subsequent studies shown were conducted in three or all four 16E6 HFK lines, with results shown for two (HFK3 and HFK4) that bracket the shorter and longer growth ranges, respectively, observed across the cell lines.
Figure 2. NFX1-123 overexpression enhanced active growth and population doublings in 16E6 HFKs.
5×105 cells were plated and grown in culture until they no longer doubled within four days. Cumulative population doublings (PD) were calculated. (A) HFKs transduced with FLAG-tagged NFX1-123 (FN123, black hexagon) or an empty vector (LXSN, open diamond) grew for 14 PDs. (B-E) HFKs (HFK2-5) were transduced with 16E6 and then FN123 (16E6/FN123, black square) or LXSN (16E6/LXSN, open circle). Across 4 independent cell lines, 16E6/FN123 HFKs grew for more PDs and total cell numbers than matched 16E6/LXSN HFKs (average difference and 95% CI shown). Horizontal arrows indicate continued active growth of 16E6/FN123 HFKs in culture. Vertical arrows note T1-T4 timepoints for Figures 3 and 5. Asterisks note early and late shared timepoints for Figure 4.
3.3. NFX1-123 overexpression was sustained in 16E6/FN123 HFKs.
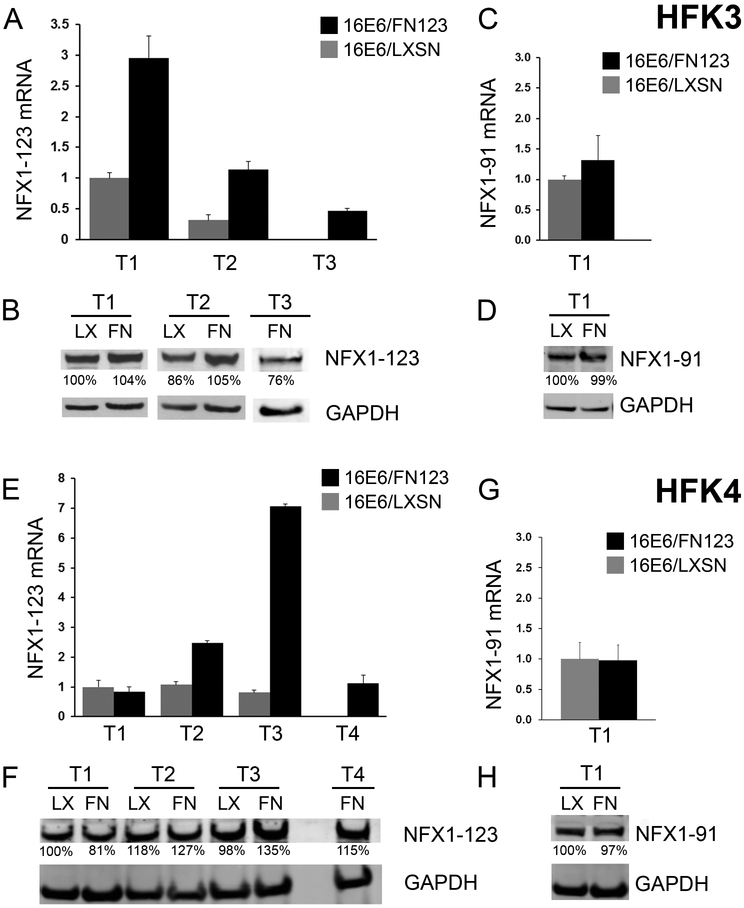
These growth studies spanned an extended period in culture, so it was important to evaluate if NFX1-123 expression changed during this time. NFX1-123 expression was quantified serially in all 16E6/LXSN and 16E6/FN123 HFK cell lines (Figure 3). Timepoints shown include two or three matching shared timepoints, as well as one timepoint for 16E6/FN123 past active growth for 16E6/LXSN (Figure 2, vertical arrows). Over time, total NFX1-123 mRNA (Figure 3A and E) remained greater in 16E6/FN123 HFKs when compared to matching 16E6/LXSN HFK timepoints, with FLAG-tagged NFX1-123 mRNA stably detected at each 16E6/FN123 HFK time point (data not shown). NFX1-123 protein amounts also increased over time in 16E6/FN123 HFKs relative to matching 16E6/LXSN HFK timepoints (Figure 3B and F).
Figure 3. NFX1-123 overexpression was sustained in 16E6/FN123 HFKs.
(A-D) HFK3; T1 and T2 are matched shared timepoints for 16E6/LXSN and 16E6/FN123. T3 is one timepoint for 16E6/FN123 past the end of active growth for 16E6/LXSN (noted as vertical arrows in Figure 2C). (E-H) HFK4; T1-3 are matched shared timepoints for 16E6/LXSN and 16E6/FN123. T4 is one timepoint for 16E6/FN123 past the end of active growth for 16E6/LXSN (noted as vertical arrows in Figure 2D). (A) Mean expression of NFX1-123 mRNA in HFK3 at T1-T3, relative to 16E6/LXSN HFK3 at T1, was quantified. (B) NFX1-123 protein was increased in 16E6/FN123 HFK3 (FN) relative to the 16E6/LXSN HFK3 (LX). (C) Mean expression of NFX1-91 mRNA, relative to 16E6/LXSN HFK3 at T1, was quantified. (D) NFX1-91 protein was equivalent in 16E6/FN123 HFK3 compared to 16E6/LXSN. (E) Mean expression of NFX1-123 mRNA in HFK4 at T1-T4, relative to 16E6/LXSN HFK4 at T1, was quantified. (F) NFX1-123 protein was moderately increased in 16E6/FN123 HFK4 (FN) relative to the 16E6/LXSN HFK4 (LX). (G) Mean expression of NFX1-91 mRNA, relative to 16E6/LXSN HFK4 at T1, was quantified. (H) NFX1-91 protein was equivalent in 16E6/FN123 HFK4 compared to 16E6/LXSN. (A, C, E, and G: all qPCRs were normalized to the housekeeping gene 36B4, and all error bars represent 95% confidence intervals from the technical replicates shown (n = 3). B, D, F and H: GAPDH used as a loading control.)
The endogenous NFX1 gene expresses two splice variant isoforms in keratinocytes, NFX1-123 and NFX1-91 [17, 18, 23, 27]. The mRNA and protein expression of NFX1-91 was unaffected by overexpression of FLAG-tagged NFX1-123 (Figure 3C, D, G, and H).
3.4. 16E6/FN123 HFKs had reduced senescence marker expression during long-term active growth.
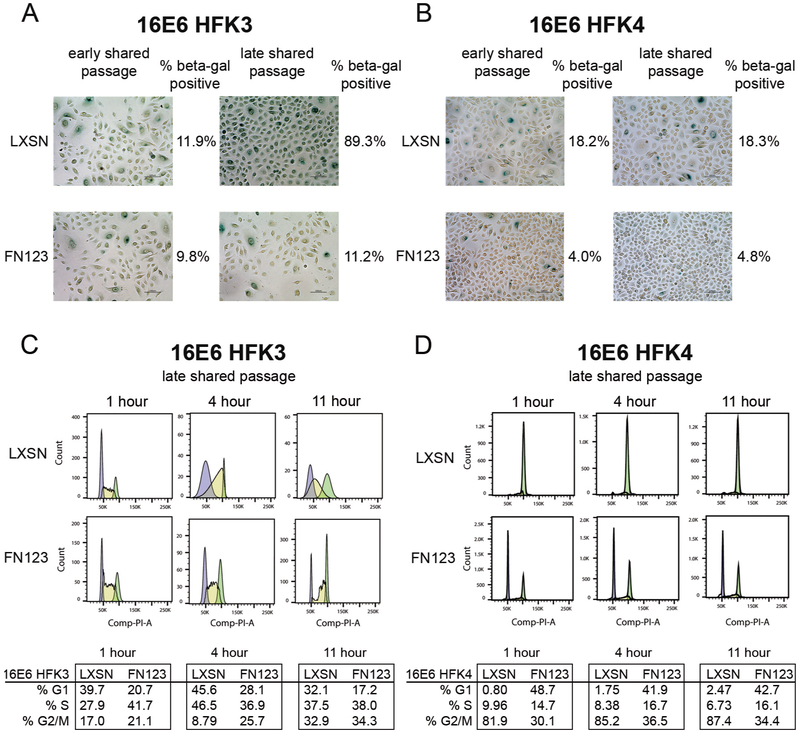
The greater cell numbers that 16E6/FN123 HFKs consistently achieved, as compared to 16E6/LXSN HFKs, could be due to differences in cellular senescence initiation, cell cycle changes, or both. To quantify differences in cellular senescence associated with NFX1-123 amounts, cells were stained for senescence-associated beta-galactosidase (beta-gal) at matching timepoints (Figure 2, asterisks). At each shared timepoint, 16E6/LXSN HFKs had a greater percentage of cells with positive staining for beta-gal compared to 16E6/FN123 HFKs (Figure 4). In the shorter growing HFK3 cell line, 89% of 16E6/LXSN HFKs stained positive at the latest shared timepoint whereas 16E6/FN123 HFKs maintained a low positivity of 11% (Figure 4A). For the longer growing HFK4 cell line, 16E6/LXSN HFK4 had a greater percent of cell staining for beta-gal at both early and late shared passages than 16E6/FN123 HFK4, although they overall did not have as dramatic an increase in beta-gal staining as HFK3. This suggests that the biologic background of 16E6 HFKs can affect the pace of senescence initiation, but in each case greater NFX1-123 was linked with reduced senescence associated beta-gal staining.
Figure 4. 16E6/FN123 HFKs at shared timepoints had reduced senescent marker expression and maintained cell cycling compared to 16E6/LXSN HFKs.
(A) 16E6/LXSN and 16E6/FN123 HFK3 (A) or HFK4 (B) cells were stained for senescence associated beta-galactosidase (beta-gal) at early and late shared passages (noted as asterisks in Figure 2C and D). (A) At an early shared passage, HFK3 cells were equivalently stained (11.9% vs 9.8%). At a late shared passage, 16E6/LXSN HFK3 had increased staining (89.3% vs 11.2%). (B) At an early shared passage, 16E6/LXSN HFK3 cells had increased staining (18.2% vs 4.0%). At a late shared passage, this was maintained (18.3% vs 4.8%). (C and D) 16E6/LXSN and 16E6/FN123 HFK3 and HFK4 cells were synchronized by density arrest and then released. HFK incorporation of BrdU and propidium iodide were quantified at 1, 4, and 11 hours after release. (C) 16E6/FN123 HFK3 had more than 1½ times as many cells in S phase at 1 hour compared to 16E6/LXSN HFK3, and 16E6/FN123 HFK3 advanced through the cell cycle more rapidly than 16E6/LXSN HFKs at 4 and 11 hours. (D) 16E6/LXSN HFK3 remained in G2/M at 1, 4, and 11 hours after release whereas 16E6/FN123 HFK4 maintained a typical cycling pattern.
3.5. 16E6/FN123 HFKs cycled faster during long-term cell culture.
To quantify the rate of cell cycle progression in 16E6/LXSN and 16E6/FN123 HFKs, the cell cycle profiles of density arrested and released cells were determined using BrdU incorporation and FACS analysis. In both HFK3 and HFK4 lines, 16E6/LXSN HFKs had slowed progression through the cell cycle compared to matched 16E6/FN123 HFKs (Figure 4C and D). For the shorter growing HFK3 cell line, 16E6/FN123 HFK3 quickly entered S phase after release (41.7% at one hour) whereas most 16E6/LXSN HFK3 did not begin to enter S phase until four hours post-release. By 11 hours, 16E6/LXSN HFK3 had progressed to G2/M while most 16E6/FN123 HFK3 had entered G1 and S phases (Figure 4C). For the longer growing HFK4 line, 16E6/LXSN HFK4 remained in G2/M even at 11 hours post-release while 16E6/FN123 HFK4 maintained a typical cycling pattern (Figure 4D). Therefore, 16E6/LXSN cells had both increased senescence and slowed of cell cycle progression in late active phase growth passages compared to 16E6/FN123 cells.
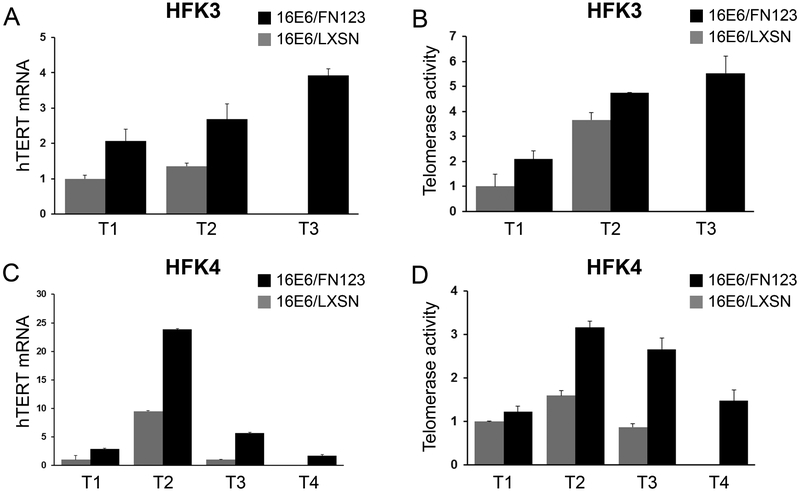
3.6. 16E6/FN123 HFKs had greater hTERT expression and telomerase activity that was amplified over time.
In the short term, NFX1-123 overexpression in 16E6 HFKs increases hTERT expression and telomerase activity [18, 22]. Over the long term, increased hTERT expression improves growth and longevity in tissue culture [20, 28, 29]. For these reasons, the expression of hTERT was quantified over time in 16E6/LXSN and 16E6/FN123 HFKs. At the early shared timepoint (T1), 16E6/FN123 HFK3 had two fold greater hTERT mRNA (Figure 5A) similar to our previously published findings [18, 22]. Interestingly, hTERT expression in 16E6/FN123 HFK3 rose over time (Figure 5A – black bars). For HFK4, hTERT was three fold greater in 16E6/FN123 cells at T1 (Figure 5C – black bars) and at T2, 16E6/FN123 HFK4 had nearly 25 times the hTERT amount seen in 16E6/LXSN at T1. Although this did decline at later passages, 16E6/FN123 HFKs always had greater hTERT expression relative to 16E6/LXSN at every shared time point.
Figure 5. hTERT expression and telomerase activity was increased and rose further over time in 16E6/FN123 HFKs.
Relative levels of hTERT mRNA and telomerase activity were quantified, and values shown were the mean fold change relative to 16E6/LXSN HFKs at T1 (noted as vertical arrows in Figure 2C and D). (A) 16E6/FN123 HFK3 (black bars) had 2 to 4 fold greater hTERT. (B) 16E6/FN123 HFK3 (black bars) had 2 to 6 fold greater telomerase activity. (C) 16E6/FN123 HFK4 (black bars) had 2 to 25 fold greater hTERT. (D) 16E6/FN123 HFK4 (black bars) had 1 1/3 to 3 1/2 fold greater telomerase activity. (A and C: hTERT qPCR was normalized to the mRNA levels of the housekeeping gene 36B4. A-D: error bars represent 95% confidence intervals from the technical replicates shown (n = 3).)
Telomerase activity was assessed in these cell lines as well. 16E6/FN123 HFK3 and HFK4 had increased telomerase activity when compared to 16E6/LXSN HFKs at early shared timepoints (Figure 5B and D). Like hTERT, telomerase activity rose over time in 16E6/FN123 HFKs and remained increased relative to 16E6/LXSN HFKs during long-term culture. Although hTERT and telomerase increases did not occur in synchrony with any relative differences in NFX1-123, 16E6 HFKs with greater NFX1-123 over time had higher hTERT levels and telomerase activity.
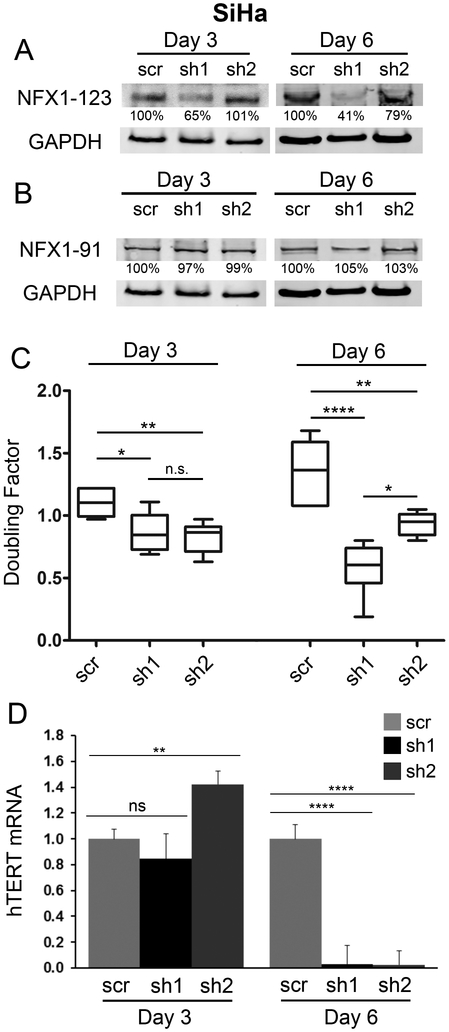
3.7. Knock down of NFX1-123 in SiHa cells slowed growth and decreased levels of hTERT.
NFX1-123 is highly expressed in cervical cancer cell lines [23] and in primary cervical cancers (Figure 1), indicating that increased expression of NFX1-123 may be selected for over the course of oncogenic progression, or that greater expression may promote subsequent development of cancers. To study the role of NFX1-123 in cellular growth and hTERT expression in the context of cancer, NFX1-123 was knocked down in HPV 16 positive SiHa cells. Cells were transduced with two different short hairpin RNAs (sh1 or sh2) targeting NFX1-123, or a scrambled short hairpin RNA (scr) control. NFX1-123 protein was reduced by sh1 within three days, but at six days post transduction both sh1 and sh2 decreased NFX1-123 (38% and 80%, respectively), when compared to the scramble (scr) control (Figure 6A). NFX1-91 was also measured to assure no off-target effect of the short hairpin constructs (Figure 6B).
Figure 6. Knock down of NFX1-123 in SiHa cells resulted in a slowed growth rate and decreased hTERT.
SiHa cells were transduced with either lentivirus containing a short hairpin targeting NFX1-123 (sh1 or sh2) or containing scrambled short hairpin control (scr). (A) NFX1-123 protein levels in sh1 and sh2 cells were decreased at day 6 compared to scr cells at day 3 and day 6 post-transduction. (B) NFX1-91 protein remained constant after short hairpin transduction. A and B: GAPDH used as a loading control (C) For each cell line (scr, sh1, sh2), at both day 3 and day 6, scr cells achieved more growth and had a higher doubling factor than either sh1 or sh2. (D) hTERT mRNA was quantified and values shown were the mean fold change normalized to scr SiHa cells. sh1 and sh2 cells had decreased hTERT compared to scr cells at day 6. hTERT qPCR was normalized to the mRNA levels of the housekeeping gene 36B4, and error bars represent 95% confidence intervals from the technical replicates shown (n = 3).) Doubling factor and hTERT p values: * ≤ 0.05, ** ≤ 0.01, **** ≤ 0.0001
By day 6, there was reduced SiHa cell doubling when compared to scr SiHa cells (Figure 6C), paralleling NFX1-123 protein expression. Additionally, there was a dramatic 97% decrease in hTERT mRNA in SiHa cells even with only a 20% decrease in NFX1-123 (Figure 6D).
4. Discussion
NFX1-123 has been shown to be highly expressed in cervical cancer cell lines and greater NFX1-123 leads to increased hTERT in 16E6 expressing HFKs [23, 30]. Now, we demonstrate that NFX1-123 is normally expressed in the cervix and highly expressed in primary cervical precancers and cancers. Additionally, 16E6 HFKs that begin with greater NFX1-123 have extended periods of active growth in culture, completing more population doublings at every parallel timepoint, and achieving at least twice the total number of cells in culture. This is associated with increased hTERT and telomerase activity, faster cell cycling, and a relative reduction in senescence. The effect of NFX1-123 on growth and hTERT is also seen in HPV 16 positive SiHa cells, where although growth regulation is likely more complex in a cervical cancer cell line, even transient knock down of NFX1-123 led to pronounced hTERT reduction and a slowing of growth.
NFX1-123 expression was measured across long-term cultures to evaluate whether it shifted over time. This measurement was important for two reasons. First, long-term tissue culture leads to cellular stress [28, 31], and in other systems NFX1 gene expression is important for growth maintenance during stress [32, 33]. Therefore, the expression of endogenous NFX1-123 may have changed over time during long-term culture. However, we found 16E6/LXSN HFKs maintained stable NFX1-123 protein levels. Second, assuring that NFX1-123 overexpression was maintained throughout the study was fundamental; this was especially important as knock down of NFX1-123 is quickly surmounted in tissue culture making reductions only transient (unpublished data). We found NFX1-123 overexpression was consistently increased overall in FN123 cells, although as evident in Figure 3, the NFX1-123 overexpression construct transduced in 16E6 HFKs led to only moderate increases in NFX1-123. This overexpression of NFX1-123 in 16E6/FN123 HFKs was well within the range seen in cervical cancer cell lines [23], and it remained increased relative to 16E6/LXSN HFKs throughout long-term cell culture.
16E6 HFKs that were transduced with NFX1-123 had greater hTERT and telomerase activity when compared to 16E6 HFKs with endogenous levels of NFX1-123. Interestingly over time, these greater hTERT levels were further augmented – paralleling increased active growth and population doublings in 16E6/FN123 HFKs. Long-term studies of HR HPV expressing HFKs have demonstrated increasing hTERT and telomerase activity [29], and our data in 16E6/LXSN HFKs support that (Figure 5, gray bars). Intriguingly, a recent study noted that 16E6 was not the only determinant of hTERT expression over time [34]. Now, we have demonstrated that over time, tonically increased NFX1-123 is a determinant of augmented hTERT expression and telomerase activity in 16E6 HFKs (Figure 5, black bars). Additionally, regardless of their relative synchrony in increased expression levels, NFX1-123, hTERT, and telomerase collaborate to impact active growth.
Telomerase is universally expressed in HPV-associated cervical cancers [35, 36], and upregulation of hTERT is associated with worse clinical grade of cervical intraepithelial neoplasia and cervical dysplasia [36-38]. As a regulator of hTERT in the context of 16E6 expression, NFX1-123 appears to be an important factor in cervical cancer development. Because NFX1-123 is highly expressed in cervical cancer cell lines and primary cervical cancers, understanding the associative or causal role it plays in supporting and maintaining HPV-associated malignancies will help define factors driving transformation from HR HPV-infected keratinocytes to cancer.
Highlights.
NFX1-123 is highly expressed in cervical cancers
NFX1-123 and HPV 16E6 extend active cellular growth
NFX1-123 remains required for telomerase activity in cervical cancer cell lines
HPV 16E6 cells with increased NFX1-123 have greater hTERT and telomerase over time
Acknowledgements
This study was supported by NIH R01 CA 172742 to R.A.K and NIH T32 AI083203 to J.L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing Interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors contributed to the design and execution of the work, the interpretation of results, the drafting of the manuscript. They give their final approval and agree to be accountable for the work herein.
Reference List
- [1].Forman D, de MC, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J, Bray F, Plummer M, Franceschi S, Global burden of human papillomavirus and related diseases, Vaccine., 30 (2012) F12–F23. [DOI] [PubMed] [Google Scholar]
- [2].Daling JR, Madeleine MM, Schwartz SM, Shera KA, Carter JJ, McKnight B, Porter PL, Galloway DA, McDougall JK, Tamimi H, A population-based study of squamous cell vaginal cancer: HPV and cofactors, Gynecol.Oncol, 84 (2002) 263–270. [DOI] [PubMed] [Google Scholar]
- [3].de Martel C, Plummer M, Vignat J, Franceschi S, Worldwide burden of cancer attributable to HPV by site, country and HPV type, International journal of cancer, 141 (2017) 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chaturvedi AK, Beyond cervical cancer: burden of other HPV-related cancers among men and women, J.Adolesc.Health., 46 (2010) S20–S26. [DOI] [PubMed] [Google Scholar]
- [5].Munoz N, Castellsague X, de Gonzalez AB, Gissmann L, Chapter 1: HPV in the etiology of human cancer, Vaccine., 24 (2006) S1–S10. [DOI] [PubMed] [Google Scholar]
- [6].Gross G, Pfister H, Role of human papillomavirus in penile cancer, penile intraepithelial squamous cell neoplasias and in genital warts, Med.Microbiol.Immunol, 193 (2004) 35–44. [DOI] [PubMed] [Google Scholar]
- [7].Daling JR, Madeleine MM, Johnson LG, Schwartz SM, Shera KA, Wurscher MA, Carter JJ, Porter PL, Galloway DA, McDougall JK, Krieger JN, Penile cancer: importance of circumcision, human papillomavirus and smoking in in situ and invasive disease, Int.J.Cancer., 116 (2005) 606–616. [DOI] [PubMed] [Google Scholar]
- [8].Chaturvedi AK, Engels EA, Anderson WF, Gillison ML, Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States, J.Clin.Oncol, 26 (2008) 612–619. [DOI] [PubMed] [Google Scholar]
- [9].Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ, Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells, Nature, 396 (1998) 84–88. [DOI] [PubMed] [Google Scholar]
- [10].Hayflick L, The limited in vitro lifetime of human diploid cell strains, Exp.Cell Res, 37 (1965) 614–636. [DOI] [PubMed] [Google Scholar]
- [11].Hayflick L, Moorhead PS, The serial cultivation of human diploid cell strains, Exp.Cell Res, 25 (1961) 585–621. [DOI] [PubMed] [Google Scholar]
- [12].Plug-DeMaggio AW, Sundsvold T, Wurscher MA, Koop JI, Klingelhutz AJ, McDougall JK, Telomere erosion and chromosomal instability in cells expressing the HPV oncogene 16E6, Oncogene., 23 (2004) 3561–3571. [DOI] [PubMed] [Google Scholar]
- [13].Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE, Extension of life-span by introduction of telomerase into normal human cells, Science., 279 (1998) 349–352. [DOI] [PubMed] [Google Scholar]
- [14].Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, Wang ZQ, Kestler HA, d'Adda di FF, Gunes C, Rudolph KL, Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion, EMBO J., 34 (2015) 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Counter CM, Meyerson M, Eaton EN, Ellisen LW, Caddle SD, Haber DA, Weinberg RA, Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase, Oncogene., 16 (1998) 1217–1222. [DOI] [PubMed] [Google Scholar]
- [16].Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR, Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts, Cancer Res., 58 (1998) 4168–4172. [PubMed] [Google Scholar]
- [17].Gewin L, Myers H, Kiyono T, Galloway DA, Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex, Genes Dev., 18 (2004) 2269–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Katzenellenbogen RA, Egelkrout EM, Vliet-Gregg P, Gewin LC, Gafken PR, Galloway DA, NFX1-123 and Poly(A) Binding Proteins Synergistically Augment Activation of Telomerase in Human Papillomavirus Type 16E6 Expressing Cells, J.Virol, 81 (2007) 3786–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA, Cytoplasmic poly(A) binding proteins regulate telomerase activity and cell growth in human papillomavirus type 16 E6-expressing keratinocytes, J.Virol, 84 (2010) 12934–12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Klingelhutz AJ, Foster SA, McDougall JK, Telomerase activation by the E6 gene product of human papillomavirus type 16, Nature, 380 (1996) 79–82. [DOI] [PubMed] [Google Scholar]
- [21].Veldman T, Horikawa I, Barrett JC, Schlegel R, Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein, J Virol, 75 (2001) 4467–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA, NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes, J.Virol, 83 (2009) 6446–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA, Human papillomavirus 16E6 and NFX1-123 potentiate notch signaling and differentiation without activating cellular arrest, Virology, 478 (2015) 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee D, Norby K, Hayes M, Chiu YF, Sugden B, Lambert PF, Using Organotypic Epithelial Tissue Culture to Study the Human Papillomavirus Life Cycle, Current protocols in microbiology, 41 (2016) 14b. 18.11–14b. 18.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D, Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors, Science., 295 (2002) 868–872. [DOI] [PubMed] [Google Scholar]
- [26].Bartz SR, Vodicka MA, Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein, Methods., 12 (1997) 337–342. [DOI] [PubMed] [Google Scholar]
- [27].Xu M, Luo W, Elzi DJ, Grandori C, Galloway DA, NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes, Mol.Cell Biol, 28 (2008) 4819–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Smith LL, Coller HA, Roberts JM, Telomerase modulates expression of growth-controlling genes and enhances cell proliferation, Nat.Cell Biol, 5 (2003) 474–479. [DOI] [PubMed] [Google Scholar]
- [29].Schutze DM, Snijders PJ, Bosch L, Kramer D, Meijer CJ, Steenbergen RD, Differential in vitro immortalization capacity of eleven, probable high-risk human papillomavirus types, J.Virol, 88 (2014) 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA, NFX1-123 and Human Papillomavirus 16E6 Increase Notch Expression in Keratinocytes, J.Virol, 87 (2013) 13741–13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Indran IR, Hande MP, Pervaiz S, hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells, Cancer Res, 71 (2011) 266–276. [DOI] [PubMed] [Google Scholar]
- [32].Lisso J, Altmann T, Mussig C, The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress, FEBS Lett, 580 (2006) 4851–4856. [DOI] [PubMed] [Google Scholar]
- [33].Mussig C, Schroder F, Usadel B, Lisso J, Structure and putative function of NFX1-like proteins in plants, Plant Biol.(Stuttg). 12 (2010) 381–394. [DOI] [PubMed] [Google Scholar]
- [34].Baege AC, Berger A, Schlegel R, Veldman T, Schlegel R, Cervical epithelial cells transduced with the papillomavirus E6/E7 oncogenes maintain stable levels of oncoprotein expression but exhibit progressive, major increases in hTERT gene expression and telomerase activity, Am.J Pathol, 160 (2002) 1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shay JW, Bacchetti S, A survey of telomerase activity in human cancer, Eur.J.Cancer., 33 (1997) 787–791. [DOI] [PubMed] [Google Scholar]
- [36].Snijders PJ, van DM, Walboomers JM, Steenbergen RD, Risse EK, Helmerhorst TJ, Verheijen RH, Meijer CJ, Telomerase activity exclusively in cervical carcinomas and a subset of cervical intraepithelial neoplasia grade III lesions: strong association with elevated messenger RNA levels of its catalytic subunit and high-risk human papillomavirus DNA, Cancer Res., 58 (1998) 3812–3818. [PubMed] [Google Scholar]
- [37].Branca M, Giorgi C, Ciotti M, Santini D, Di BL, Costa S, Benedetto A, Bonifacio D, Di BP, Paba P, Accardi L, Mariani L, Ruutu M, Syrjanen S, Favalli C, Syrjanen K, Upregulation of telomerase (hTERT) is related to the grade of cervical intraepithelial neoplasia, but is not an independent predictor of high-risk human papillomavirus, virus persistence, or disease outcome in cervical cancer, Diagn.Cytopathol, 34 (2006) 739–748. [DOI] [PubMed] [Google Scholar]
- [38].Kailash U, Soundararajan CC, Lakshmy R, Arora R, Vivekanandhan S, Das BC, Telomerase activity as an adjunct to high-risk human papillomavirus types 16 and 18 and cytology screening in cervical cancer, Br.J.Cancer., 95 (2006) 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]