Abstract
The O-linked N-acetylglucosamine (O-GlcNAc) modification is an essential component in cell regulation. A single pair of human enzymes conducts this modification dynamically on a broad variety of proteins: O-GlcNAc transferase (OGT) adds the GlcNAc residue and O-GlcNAcase (OGA) hydrolyzes it. This modification is dysregulated in many diseases, but its exact role on particular substrates remains unclear. In addition, no apparent sequence motif was found in the modified proteins and the factors controlling the substrate specificity of OGT and OGA are largely unknown. In this concept, we will discuss recent developments of chemical and biochemical methods toward addressing the challenge of OGT and OGA substrate recognition. We hope the new concept and knowledge from these studies will promote research in this area to advance understanding of O-GlcNAc regulation in health and disease.
Keywords: glycosylation, O-GlcNAc, substrate recognition, microarrays, chemical probes
Graphical Abstract
Introduction
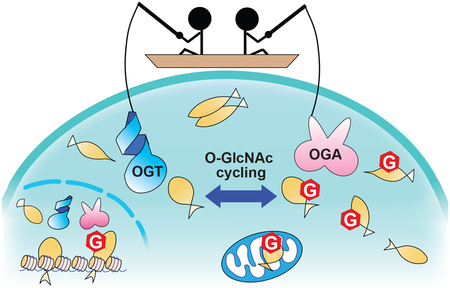
O-GlcNAcylation is an essential and reversible post-translational modification in metazoans in which N-acetylglucosamine (GlcNAc) is appended to serine and threonine residues of over 1,000 proteins (Figure 1).[1–5] Dynamic O-GlcNAc cycling regulates a wide variety of cellular functions, including transcription, translation, and signal transduction.[1,2] Importantly, many proteins are aberrantly O-GlcNAcylated in diseases such as cancer,[3,4] diabetes,[5,6] and Alzheimer’s disease.[7,8] Only one enzyme, O-GlcNAc transferase (OGT), is known to add this modification to nucleocytoplasmic and mitochondrial proteins,[9,10] while one other, O-GlcNAcase (OGA), removes this modification (Figure 1).[11,12] Recent advances in quantitative mass spectrometry studies have revealed that the O-GlcNAc level of certain proteins changes in response to environmental stimuli and in different disease states.[13–23] Therefore, understanding what mechanisms regulate the enzyme activity and substrate specificity of OGT and OGA will help elucidate the biological functions of this modification in health and disease.
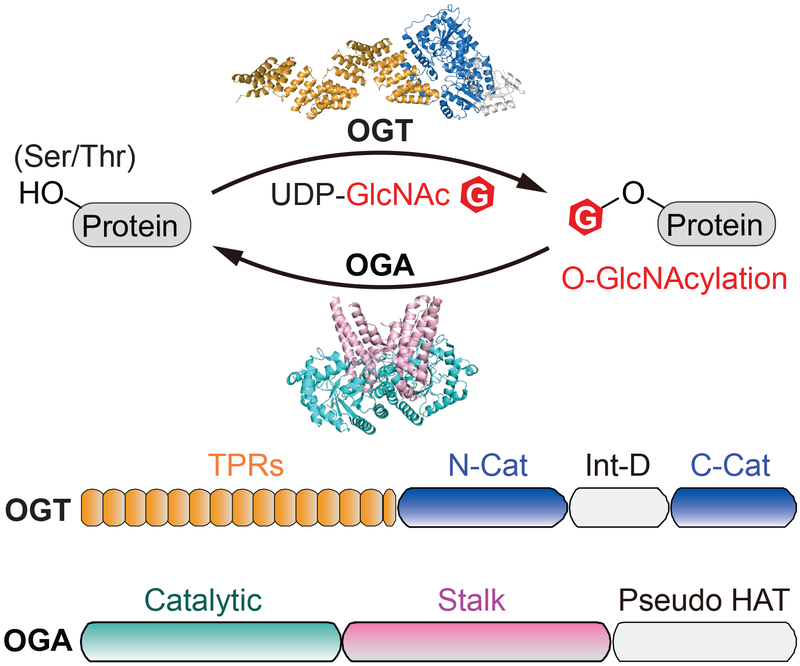
Figure 1.
Overview of O-GlcNAcylation and its cycling enzymes. Top panel: schematic of reversible O-GlcNAcylation. O-GlcNAc cycling enzymes, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), are represented by the full-length ncOGT model (generated from PDB 3PE4 and 1W3B) and a truncated human OGA structure (OGAcryst from PDB: 5TKE). Bottom panel: the schematic of domain architecture of ncOGT and OGA. OGT is comprised of a tetratricopeptide repeat domain (TPR, orange), catalytic domain (N-Cat and C-Cat, blue), and intervening domain (Int-D, light gray). OGA is comprised of a catalytic domain (Catalytic, cyan), stalk domain (Stalk, pink), and pseudo-histone acetyltransferase domain (Pseudo HAT, light gray).
OGT is a member of the GT-B glycosyltransferase family.[24] It is composed of an N-terminal tetratricopeptide repeat (TPR) region and a C-terminal catalytic region that is split by an intervening domain (Figure 1).[10,25,26] The TPR region folds into a super alpha helix and has been suggested to play an important role in protein interactions.[27,28] Furthermore, mutations in the TPR region have been implicated in intellectual disability.[29–31] Three isoforms of human OGT have been reported,[32,33] which differ only in the number of TPR repeats: 13.5 for ncOGT (the most abundant isoform in cytoplasm and nucleus), 9 for mOGT (mitochondria), and 2.5 for a short isoform, sOGT. However, the physiological importance of the two shorter isoforms remain mostly unknown.[34–36] Several studies reported the requirement of the extended TPR region for binding and glycosylating protein- but not peptide-substrates.[37–40] While the full-length ncOGT is not yet amenable to crystallization, structural studies of OGT4.5 (an N-terminal truncated OGT construct containing 4.5 TPR repeats) in complexes with sugar donor and acceptor peptide substrates have revealed key features in substrate recognition: a) the active site-bound sugar donor forms extensive interactions with the acceptor peptide near the catalytic pocket, and b) a series of asparagine residues lining the inside of the TPR play an important role in binding to the backbones of peptide substrates.[26,41–43] To better understand the substrate specificity of OGT, future research will be needed to illustrate the binding modes of this essential enzyme with protein substrates as well as other interacting partners.
OGA is a multi-domain protein, with an N-terminal catalytic domain sharing significant sequence homology to glycoside hydrolase 84 (GH84) family of enzymes, a stalk domain, and a C-terminal domain showing sequence homology to histone acetyltransferase (pseudo-HAT domain) (Figure 1).[12,44] However, the pseudo-HAT domain seems to lack residues necessary for binding acetyl coenzyme A, and its functions, including any in substrate binding, are not yet determined.[45,46] OGA substrate recognition is also a topic of intense interest; the extent to which OGA recognizes protein substrates beyond the GlcNAc residue has been a long-standing question.[47,48] As a major breakthrough, structures of truncated human OGA containing the catalytic and stalk domains have recently been solved.[49–51] Intriguingly, a potential substrate-binding cleft created by the dimerized OGA protein was discovered. In support of this, the structures of OGA in complex with each of five distinct glycopeptides demonstrate that all the peptide substrates are bound in the substrate-binding cleft.[49,52] The interactions of GlcNAc in the OGA catalytic pocket are highly conserved, while the peptides are bound in a bidirectional yet generally similar V-shaped conformation. Notably, some peptides are engaged in side-chain specific interactions with OGA residues on the substrate-binding cleft, and mutagenesis analysis confirmed the importance of some of these interactions. It is expected that future research to elucidate how OGA interacts with protein substrates beyond the catalytic domain will provide a more complete understanding on the molecular basis of OGA-substrate recognition.
Besides the progress made with structural studies, traditional approaches such as immunoprecipitation and yeast two-hybrid screening have also been applied to investigate the interactions between OGT and its binding partners.[53–57] However, these methods may not be optimal for studying O-GlcNAcylation due to its dynamic nature, transient interactions, and large number of substrates. Other complementary methods have therefore been introduced and employed to start addressing these challenges, which will be critical to discover how these enzymes’ activities and specificities are altered in diseases. Here, we will highlight recently reported chemical and biochemical approaches that have not been widely reviewed. Some of these approaches have only started to be developed, while others are already being applied to investigate OGT- and OGA-substrate recognition. Methods to simply detect O-GlcNAcylation or measure O-GlcNAc stoichiometry will not be emphasized here, as they have been extensively reviewed elsewhere.[15,58–62] We hope this concept paper will spur efforts to make new discoveries in this exciting field.
Microarrays to elucidate substrate preferences of OGT and its variants
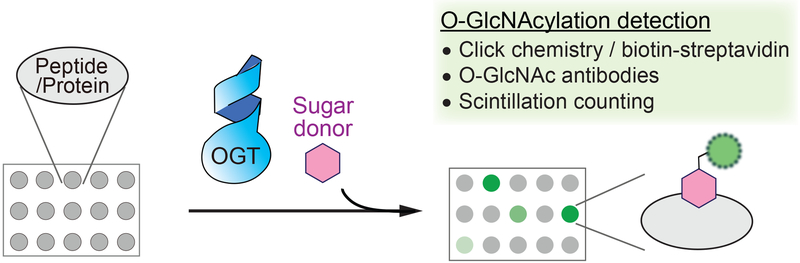
One of the popular approaches for studying OGT substrate preferences has been microarrays, which can be performed with peptides or purified proteins (Figure 2). The first peptide microarray applied to OGT examined the preference for different types of residues surrounding the glycosylation site.[63] It utilized a panel of biotinylated peptides derived from α-crystallin, a known OGT substrate, where each peptide had one amino acid changed relative to the parent peptide (AIPVSREEK). Following incubation with OGT and UDP-GlcNAz, an azido analogue of UDP-GlcNAc, the glycosylated peptides were labeled with phosphine-FLAG for detection in an azido-ELISA assay. They found that OGT had variable tolerance for amino acid substitutions at different sites. For instance, mutations at the +3 position (Glu) relative to the serine glycosylation site produced generally moderate reductions in O-GlcNAcylation, while changing the +2 subsite (Glu) increased glycosylation. In contrast, mutations at the −2 (Pro), −1 (Val), and +1 (Arg) positions led to dramatic reductions in modification. These results suggested that OGT possesses some preferences for residues flanking the glycosylation site, but the extent to which this applies to other peptide substrates remained to be examined. Another recent study utilized radiolabeled UDP-[3H]GlcNAc for peptide O-GlcNAcylation. Following quantification on a scintillation counter, 70 out of 720 peptides were significantly glycosylated, implying that OGT has a substantial level of substrate specificity dictated by the peptide-binding site alone.[43] Peptide microarrays were later used to examine the substrate specificity of different OGT isoforms. Using RL2 (an anti-O-GlcNAc antibody) for O-GlcNAc detection, one study found that a number of peptides were glycosylated at different levels by each of the three OGT isoforms, indicating differential substrate specificities.[64] Besides the applications to study OGT, a microarray with synthetic glycopeptides was also developed for OGA,[65] which may be used in the future to investigate the substrate preferences of this hydrolase.
Figure 2.
Microarray strategy to study OGT substrate preferences. A large number of immobilized peptides or proteins on the chip are incubated with OGT and UDP-GlcNAc (or a sugar donor analogue). Several methods can be used for detection of O-GlcNAcylation, including click chemistry of azido sugar with alkyne-linked biotin and detection by fluorescent streptavidin, anti-O-GlcNAc antibodies, and scintillation counting.
In addition to peptide microarrays, a protein microarray method for OGT has also been established. One commercially available microarray contains about 8,000 purified human proteins and the O-GlcNAcylation of these proteins was detected by CTD110.6, another anti-O-GlcNAc antibody.[66] This protein microarray method identified 230 possible OGT protein substrates in humans, most of which were not previously known. To further improve detection sensitivity and specificity, the same group applied UDP-GlcNAz and detected O-GlcNAzylated proteins with cyclooctyne-linked biotin and fluorescent streptavidin.[67] Using this assay to evaluate the activity of WT OGT and its mutant, they provided direct evidence that an asparagine ladder within the TPR of OGT is important for glycosylation of a large majority of proteins on the microarray.
There are a number of notable features of microarrays. They can test up to thousands of substrates at a time quantitatively and present simplified substrate identification. Peptide microarrays allow quick follow-up experiments with both mutated and modified peptide sequences. Protein microarrays are advantageous in their ability to test all the purified proteins at similar concentrations, which may otherwise be present in cells at highly variable levels or only expressed in certain cell types or conditions. This technique also offers a more accurate assessment of which proteins OGT prefers to glycosylate. However, microarrays have some limitations. These include potential interference with OGT binding due to attachment of peptides or proteins to a solid surface, high cost, low reproducibility, and that it is only applicable in vitro. Endogenous glycosylation or other modifications may already be present on proteins in microarrays as well. Pre-treatment of arrays with bacterial OGA and lambda phosphatase could be an option to reduce the interference from endogenous O-GlcNAcylation and phosphorylation.[68] Nevertheless, microarray is an efficient approach to evaluate O-GlcNAc cycling enzymes and their variants against a large number of substrates in high throughput.
Chemical probes to study OGT substrate recognition
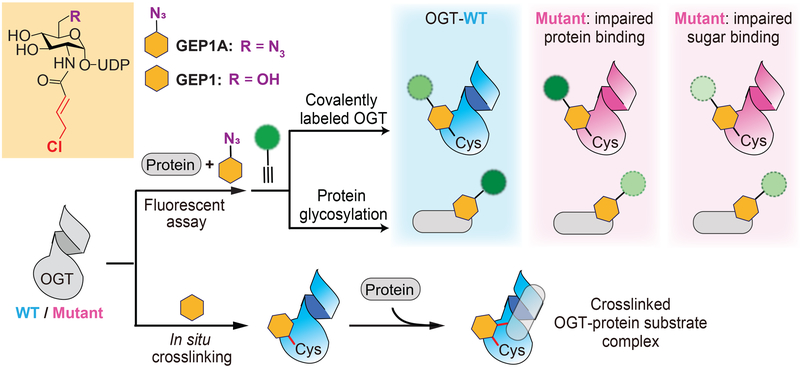
One of the challenges in studying OGT substrate recognition is that the binding of protein substrate is dependent on prior UDP-GlcNAc binding,[26] and there is a lack of strategies to efficiently distinguish them. A UDP-GlcNAc analogue called GlcNAc Electrophilic Probe 1 (GEP1) was recently reported to address this limitation. In combination with structure-guided mutagenesis, this new probe can rapidly assess the role of specific OGT residues in binding of sugar donor compared to acceptor substrate (Figure 3).[69] The GEP1 probe contains an allyl chloride electrophile extended from the N-acetyl group of UDP-GlcNAc. In the presence of protein substrates, this probe can be used as a regular sugar donor to conduct glycosylation by OGT. In the absence of acceptor substrates, this probe can efficiently label an OGT active site residue C917 to generate a covalent modification on OGT. The binding ability of OGT toward sugar and protein substrate can be evaluated by varied levels of probe modified protein and OGT. This can be reported using click-chemistry conjugation of an alkyne fluorophore with the 6’ azido group on a modified GEP1 probe (named GEP1A) in a fluorescence assay (Figure 3). The assay was applied to identify a couple of important asparagine residues in the OGT TPR region for protein substrate binding. Orthogonal radiolabeled kinetic experiments further confirmed the data of this assay. In agreement with the results from other methods (e.g., protein microarrays), these findings suggest that the asparagine residues on the inner surface of TPR domain are a generic binding region contributing to OGT interactions with different acceptor substrates. The results of GEP1A fluorescence assay also suggest that the importance of particular TPR residues could be dependent on the protein substrates, thus conferring modest substrate specificity to OGT. Taken together, this study demonstrated that the GEP1A fluorescence assay can be applied to rapidly screen the binding ability of OGT variants with protein substrates, accelerating the discovery of crucial residues for substrate recognition. Moreover, these experiments highlighted a notable feature of GEP1A assay, complementary to the conventional activity-based assays: even when no glycosylation is detected, GEP1A fluorescence assay can still potentially discriminate the impact of the mutated OGT residues on sugar binding compared to protein substrate binding.
Figure 3.
Application of GlcNAc Electrophilic Probes (GEPs) to characterize OGT-protein substrate recognition. Top: In the presence of protein substrate, GEP1A can label both OGT and its protein substrate, which can be readily detected by click chemistry-based fluorescence assay. The relative level of modified proteins between OGT-WT and its mutants (derived from structure-guided mutagenesis) reflects the altered OGT ability of protein binding compared to sugar binding. Bottom: Preincubation of GEP1 enables OGT crosslinking with various protein substrates in situ.
The GEP1 probe can also potentially stabilize OGT-protein substrate complexes to facilitate investigation of their interactions. Following a brief pre-incubation with OGT, GEP1 was able to in situ crosslink OGT with over 100 proteins in cell lysates (Figure 3).[69] Most of these crosslinked proteins were reported to be O-GlcNAcylated, and dozens of possible new OGT substrate proteins were also identified. The crosslinking assay of GEP1 can facilitate identification of novel OGT substrates and may be optimized for characterizing the binding mode of particular substrates with OGT. It should be noted that this probe is not currently applicable in cells, and the electrophiles and assay conditions may require optimization for different substrates. Another chemical probe with potential to be adapted for this type of research is Ac4GlcNDAz.[70,71] This GlcNAc analogue is modified with the photoactivatable crosslinking group diazirine and is resistant to hydrolysis by WT OGA. It has been used to identify O-GlcNAcylated protein binding partners, and because OGT and OGA can be O-GlcNAcylated, this probe may be used to study their interactions as well. Overall, small molecule probes are versatile since they can be modified with different functional groups to tune their reactivity or to be applied in entirely new strategies. Consequently, chemical probes have great potential to overcome challenges in the field, including those that have not yet been applied to O-GlcNAc cycling enzymes (such as unnatural amino acids).[72] By improving the specificity, sensitivity, and efficiency of the labeling and detection methods, chemical probes are promising tools for cellular or in vivo studies.
Engineered OGA for enriching glycoprotein substrates and binding partners
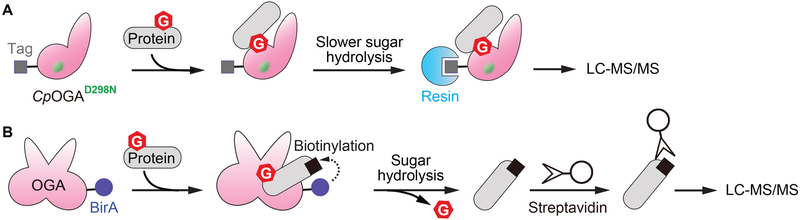
Due to the lack of a conserved motif in OGA substrates, approaches to engineer OGA for identifying interacting proteins have recently been developed. One method utilized a catalytically impaired bacterial OGA mutant (CpOGAD298N) that binds to O-GlcNAcylated proteins without quickly hydrolyzing the sugar, and thus can be used for enrichment of O-GlcNAcylated proteins (Figure 4A).[73] Despite the unclear function of bacterial OGA orthologs, affinity purification of this mutant followed by mass spectrometry analysis led to identification of over 2,000 enriched proteins from Drosophila embryo lysates, and 43 of them were mapped with O-GlcNAc sites.[74] Genetic interaction experiments were then applied to validate two novel O-GlcNAc substrates along with their physiological roles. This study revealed the potential of using OGA mutant to not only investigate OGA interaction with particular substrates in vitro or in cells, but also examine the OGA-regulated O-GlcNAc repertoire under different conditions.
Figure 4.
Engineered OGA to enrich substrates and binding partners. A) Catalytically impaired bacterial OGA (CpOGAD298N) binds to substrate without quickly hydrolyzing the sugar. The interacting substrates can then be enriched by affinity purification of CpOGAD298N and identified by LC-MS/MS. B) The expression of an OGA-BirA fusion protein in cells results in proximity-dependent biotinylation of OGA-bound proteins, including OGA substrates. The enriched biotinylated proteins can then be detected by LC-MS/MS.
The second approach applied the proximity-dependent biotinylation assay (BioID), wherein the protein of interest is expressed as a fusion with biotin ligase (BirA), which biotinylates proteins within about 10 nm (Figure 4B).[75] Biotin capture can then be applied to enrich the interacting proteins. One study combined this with stable isotopic labeling of amino acids in cell culture (SILAC) to quantitatively analyze the proteins enriched by OGA.[76] They identified 90 proteins that were potential interacting partners of OGA and validated that one of them, fatty acid synthase, reduces OGA activity upon binding. Some considerations should be taken into account for OGA BioID methods. For example, exogenous expression of OGA may cause unintended signaling that alters O-GlcNAc homeostasis, and fused proteins may also interfere with binding (the BioID study expressed two different fusion proteins, one each with BirA on either terminus of OGA). Biotin ligation may also not be efficient for transient OGA-substrate interactors, as overnight incubation was required. Of note, CpOGAD298N and OGA-BirA methods could also identify protein interactors that are not direct substrates of human OGA, thus control experiments are needed to discriminate non-substrate binders. Despite that, these approaches can be potentially applied in different cellular conditions and will be valuable to illustrate the potential of protein engineering in studying OGA protein interactions.
Imaging techniques to investigate OGT substrate- and localization-specific activity in cells
Since the O-GlcNAcylation in cells is dynamic, it is essential to learn how OGT and OGA interact with different substrates in a temporal and spatial manner. Methods that can be applied to answer this question, particularly in live cells, will greatly extend our knowledge about the protein substrate recognition of O-GlcNAc cycling enzymes. A promising strategy to accomplish this is fluorescence imaging, which offers notable advantages over other approaches, including that it can account for protein expression level and cellular localization. Several efforts have utilized genetically encoded fluorescent proteins to detect O-GlcNAcylation on specific substrates. One such approach expressed a construct containing two fluorescent proteins linked by an OGT peptide substrate and an O-GlcNAc binding lectin.[77,78] Glycosylation of the substrate peptide brought the two fluorescent proteins into proximity, which could be detected by changes in their fӧrster resonance energy transfer (FRET). This method was utilized to monitor the spatiotemporal changes of OGT activity in cells upon serum stimulation. A consideration is that the excess acceptor fluorophore in regular FRET experiments usually produces high background signal and reduces detection sensitivity, which may not be suitable for certain substrates with low expression or O-GlcNAcylation level.
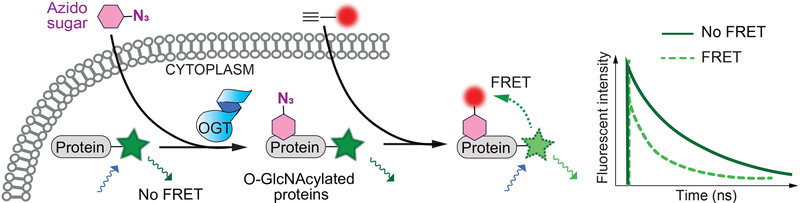
An alternative detection method, called fluorescence lifetime imaging microscopy (FLIM-FRET), was applied in an effort to increase detection sensitivity. One such study incorporated Ac4GalNAz (a cell-permeable acetylated GalNAc analogue), which can be metabolically converted to UDP-GlcNAz and used as a sugar donor by OGT in cells (Figure 5).[79,80] The OGT protein substrate of interest was fused with a fluorescent protein (FRET donor). After the substrate is O-GlcNAzylated, it can be conjugated with a fluorophore-linked alkyne FRET acceptor. Stronger FRET between the protein and small molecule fluorophore renders a faster decrease of donor fluorescence, which can sensitively report the O-GlcNAcylation on the particular substrate. This technique overcomes the issue of high background signal produced by excess acceptor fluorophore. The researchers applied this strategy to study O-GlcNAcylation on Tau and β-catenin proteins, which are implicated in neurodegenerative diseases and cancer, respectively.[81,82] In a similar study, O-GlcNAcylation on several proteins was imaged using cyclopropene-tetrazine click chemistry, which avoids the use of potentially toxic copper catalyst and is more specific than cyclooctyne-azide click chemistry.[83] FLIM-FRET imaging of O-GlcNAcylation allows real-time analysis of different OGT variants in specific conditions and compartments of live cells, an advantage not available with many other methodologies. The O-GlcNAcylation observed in the FRET approaches could, however, differ from that in unaltered cells due to the need for exogenous expression and fusion to a fluorophore of the substrate protein. Moreover, differential O-GlcNAcylation could result from an equilibrium of OGT and OGA activities. A recent study also reported nonspecific reactions of cysteine residues with acetylated GalNAc/GlcNAc analogues that have been widely used for metabolic labeling of O-GlcNAc in cells.[23] Finally, imaging may be difficult with proteins that are not highly glycosylated, and distance limitation between the fluorophores may restrict the application of FRET on certain large substrate proteins. Other strategies such as SNAP-tag, which exploits an alkyltransferase (O6-alkylguanine-DNA-alkyltransferase) to label fusion proteins with a fluorophore, may also be adapted to investigate OGT substrate preferences in cells.[84]
Figure 5.
Fluorescence lifetime imaging microscopy-fӧrster resonance energy transfer (FLIM-FRET) to monitor substrate-specific OGT activity in live cells. An OGT substrate fused with fluorescent protein can be metabolically labeled by azido sugar in cells. Click-chemistry conjugation of glycosylated protein to an acceptor fluorophore leads to proximity-induced FRET between donor and acceptor on the glycosylated substrate. The reduced fluorescence lifetime of the donor can report the O-GlcNAcylation on the protein substrate.
Conclusions
In the past few years, different types of strategies have been introduced and applied to investigate the substrate recognition of O-GlcNAc cycling enzymes. While current knowledge on this topic was mainly derived from structural studies, the new complementary chemical and biochemical strategies mentioned here will be valuable for characterizing OGT and OGA protein substrate interactions under different regulatory conditions in vitro and in cells. These studies, in addition to published structural data, allow us to propose general concepts for substrate recognition of OGT. Although there is no strict sequence motif specifically targeted by OGT, amino acid preferences close to the glycosylated residue and extended substrate binding conformation appear to be important. A series of asparagine residues lining the concave surface of the TPR domain are also likely to provide additional binding affinity and substrate selectivity through interactions with other regions of the protein substrates. Compared to OGT, there is even less known about OGA substrate recognition. As with OGT, most discoveries to date are derived from X-ray crystallography, which revealed that OGA can bind substrates beyond the GlcNAc moiety. However, little is known about the extent of these interactions, and few assays like the ones described above have been applied to OGA. Thus, many questions remain for the substrate recognition of O-GlcNAc cycling enzymes, including:
What is the molecular basis of TPR region contribution to the substrate selectivity of OGT?
Is the pseudo-HAT domain important for substrate recognition or protein binding to OGA?
How much does protein sequence or conformational preference direct OGT or OGA to different substrates?
Whether recruiter proteins guide OGT or OGA to substrates and if so under what conditions?
What differences in substrate recognition exist among the isoforms of O-GlcNAc cycling enzymes?
New approaches will still be needed to address the questions above. Future studies of OGT and OGA substrate recognition will likely expand our understanding of cell regulation and promote development of novel molecular tools to modulate O-GlcNAc cycling on particular substrates for biomedical applications.
Acknowledgements
This work was supported by NIH grants R01 GM121718 and R01 GM126300.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Bond MR, Hanover JA, J. Cell Biol. 2015, 208, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hardiville S, Hart GW, Cell Metab. 2014, 20, 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferrer CM, Sodi VL, Reginato MJ, J. Mol. Biol. 2016, 428, 3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Singh JP, Zhang K, Wu J, Yang X, Cancer Lett. 2015, 356, 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ma J, Hart GW, Expert Rev. Proteomics 2013, 10, 365–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Qin CX, Sleaby R, Davidoff AJ, Bell JR, De Blasio MJ, Delbridge LM, Chatham JC, Ritchie RH, Pharmacol. Res. 2017, 116, 45–56. [DOI] [PubMed] [Google Scholar]
- [7].Yuzwa SA, Vocadlo DJ, Chem. Soc. Rev. 2014, 43, 6839–6858. [DOI] [PubMed] [Google Scholar]
- [8].Skorobogatko YV, Deuso J, Adolf-Bryfogle J, Nowak MG, Gong Y, Lippa CF, Vosseller K, Amino Acids 2011, 40, 765–779. [DOI] [PubMed] [Google Scholar]
- [9].Haltiwanger RS, Blomberg MA, Hart GW, J. Biol. Chem. 1992, 267, 9005–9013. [PubMed] [Google Scholar]
- [10].Lubas WA, Frank DW, Krause M, Hanover JA, J. Biol. Chem. 1997, 272, 9316–9324. [DOI] [PubMed] [Google Scholar]
- [11].Dong DL, Hart GW, J. Biol. Chem. 1994, 269, 19321–19330. [PubMed] [Google Scholar]
- [12].Gao Y, Wells L, Comer FI, Parker GJ, Hart GW, J. Biol. Chem. 2001, 276, 9838–9845. [DOI] [PubMed] [Google Scholar]
- [13].Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, Pitteri SJ, Mol. Cell. Proteomics 2018, 17, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qin K, Zhu Y, Qin W, Gao J, Shao X, Wang Y, Zhou W, Wang C, Chen X, ACS Chem. Biol. 2018, 13, 1983–1989. [DOI] [PubMed] [Google Scholar]
- [15].Rexach JE, Clark PM, Hsieh-Wilson LC, Nat. Chem. Biol. 2008, 4, 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang S, Yang F, Petyuk VA, Shukla AK, Monroe ME, Gritsenko MA, Rodland KD, Smith RD, Qian W-J, Gong C-X, et al. , J. Pathol. 2017, 243, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC, Nat. Chem. Biol. 2012, 8, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC, Nat. Chem. Biol. 2007, 3, 339–348. [DOI] [PubMed] [Google Scholar]
- [19].Wang X, Yuan ZF, Fan J, Karch KR, Ball LE, Denu JM, Garcia BA, Mol. Cell. Proteomics 2016, 15, 2462–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao P, Viner R, Teo CF, Boons G-J, Horn D, Wells L, J. Proteome Res. 2011, 10, 4088–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van ADMF, Agnew B, Kuster B, J. Proteome Res. 2013, 12, 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Myers SA, Daou S, Affar EB, Burlingame A, Proteomics 2013, 13, 982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Qin W, Qin K, Fan X, Peng L, Hong W, Zhu Y, Lv P, Du Y, Huang R, Han M, et al. , Angew. Chem. Int. Ed. 2018, 57, 1817–1820. [DOI] [PubMed] [Google Scholar]
- [24].Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B, Nucleic Acids Res. 2009, 37, D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kreppel LK, Blomberg MA, Hart GW, J. Biol. Chem. 1997, 272, 9308–9315. [DOI] [PubMed] [Google Scholar]
- [26].Lazarus MB, Nam YS, Jiang J, Sliz P, Walker S, Nature 2011, 469, 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E, Nat. Struct. Mol. Biol. 2004, 11, 1001–1007. [DOI] [PubMed] [Google Scholar]
- [28].Rafie K, Raimi O, Ferenbach AT, Borodkin VS, van ADMF, Kapuria V, Open Biol. 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vaidyanathan K, Niranjan T, Selvan N, Teo CF, May M, Patel S, Weatherly B, Skinner C, Opitz J, Carey J, et al. , J. Biol. Chem. 2017, 292, 8948–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gundogdu M, Llabres S, Gorelik A, Ferenbach AT, Zachariae U, van Aalten DMF, Cell Chem. Biol. 2018, 25, 513–518.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Selvan N, George S, Serajee FJ, Shaw M, Hobson L, Kalscheuer V, Prasad N, Levy SE, Taylor J, Aftimos S, et al. , J. Biol. Chem. 2018, 293, 10810–10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, Love DC, Arch. Biochem. Biophys. 2003, 409, 287–297. [DOI] [PubMed] [Google Scholar]
- [33].Levine ZG, Walker S, Annu. Rev. Biochem. 2016, 85, 631–657. [DOI] [PubMed] [Google Scholar]
- [34].Trapannone R, Mariappa D, Ferenbach AT, van Aalten DMF, Biochem. J. 2016, 473, 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lazarus BD, Love DC, Hanover JA, Glycobiology 2006, 16, 415–421. [DOI] [PubMed] [Google Scholar]
- [36].Shin S-H, Love DC, Hanover JA, Amino Acids 2011, 40, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lubas WA, Hanover JA, J. Biol. Chem. 2000, 275, 10983–10988. [DOI] [PubMed] [Google Scholar]
- [38].Iyer SPN, Hart GW, J. Biol. Chem. 2003, 278, 24608–24616. [DOI] [PubMed] [Google Scholar]
- [39].Yang X, Zhang F, Kudlow JE, Cell 2002, 110, 69–80. [DOI] [PubMed] [Google Scholar]
- [40].Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J, J. Biol. Chem. 2014, 289, 5986–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lazarus MB, Jiang J, Gloster TM, Zandberg WF, Whitworth GE, Vocadlo DJ, Walker S, Nat. Chem. Biol. 2012, 8, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schimpl M, Zheng X, Borodkin VS, Blair DE, Ferenbach AT, Schuettelkopf AW, Navratilova I, Aristotelous T, Albarbarawi O, Robinson DA, et al. , Nat. Chem. Biol. 2012, 8, 969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, Schuttelkopf AW, Albarbarawi O, van Aalten DMF, Nat. Struct. Mol. Biol. 2015, 22, 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE, J. Biol. Chem. 2004, 279, 53665–53673. [DOI] [PubMed] [Google Scholar]
- [45].Rao FV, Schuttelkopf AW, Dorfmueller HC, Ferenbach AT, Navratilova I, van ADMF, Open Biol. 2013, 3, 130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].He Y, Roth C, Turkenburg JP, Davies GJ, Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schimpl M, Schuettelkopf AW, Borodkin VS, van Aalten DMF, Biochem. J. 2010, 432, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shen DL, Gloster TM, Yuzwa SA, Vocadlo DJ, J. Biol. Chem. 2012, 287, 15395–15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li B, Li H, Lu L, Jiang J, Nat. Struct. Mol. Biol. 2017, 24, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Elsen NL, Patel SB, Ford RE, Hall DL, Hess F, Kandula H, Kornienko M, Reid J, Selnick H, Shipman JM, et al. , Nat. Chem. Biol. 2017, 13, 613–615. [DOI] [PubMed] [Google Scholar]
- [51].Roth C, Chan S, Offen WA, Hemsworth GR, Willems LI, King DT, Varghese V, Britton R, Vocadlo DJ, Davies GJ, Nat. Chem. Biol. 2017, 13, 610–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li B, Li H, Hu C-W, Jiang J, Nat. Commun. 2017, 8, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW, J. Biol. Chem. 2008, 283, 33935–33941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Olszewski N, West C, Sassi S, Hartweck L, Biochim. Biophys. Acta Gen. Subj. 2010, 1800, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tseng T-S, Salome PA, McClung CR, Olszewski NE, Plant Cell 2004, 16, 1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Robertson M, Plant Physiol. 2004, 136, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng T-S, Kieffer M, Eshed Y, Olszewski N, Weiss D, Plant Cell 2012, 24, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ma J, Hart GW, Clin. Proteomics 2014, 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cecioni S, Vocadlo DJ, Curr. Opin. Chem. Biol. 2013, 17, 719–728. [DOI] [PubMed] [Google Scholar]
- [60].Worth M, Li H, Jiang J, ACS Chem. Biol. 2017, 12, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Thompson JW, Sorum AW, Hsieh-Wilson LC, Biochemistry 2018, 57, 4010–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Thompson JW, Griffin ME, Hsieh-Wilson LC, Methods Enzymol. 2018, 598, 101–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Leavy TM, Bertozzi CR, Bioorg. Med. Chem. Lett. 2007, 17, 3851–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shi J, Sharif S, Ruijtenbeek R, Pieters RJ, PLoS One 2016, 11, e0151085/1–e0151085/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sharif S, Shi J, Bourakba M, Ruijtenbeek R, Pieters RJ, Anal. Biochem. 2017, 532, 12–18. [DOI] [PubMed] [Google Scholar]
- [66].Ortiz-Meoz RF, Merbl Y, Kirschner MW, Walker S, J. Am. Chem. Soc. 2014, 136, 4845–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Levine ZG, Fan C, Melicher MS, Orman M, Benjamin T, Walker S, J. Am. Chem. Soc. 2018, 140, 3510–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DMF, Cantrell DA, Nat. Immunol. 2016, 17, 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hu C-W, Worth M, Fan D, Li B, Li H, Lu L, Zhong X, Lin Z, Wei L, Ge Y, et al. , Nat. Chem. Biol. 2017, 13, 1267–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ, Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rodriguez AC, Yu SH, Li B, Zegzouti H, Kohler J, J. Biol. Chem. 2015, 290, 22638–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Neumann-Staubitz P, Neumann H, Curr. Opin. Struct. Biol. 2016, 38, 119–128. [DOI] [PubMed] [Google Scholar]
- [73].Mariappa D, Selvan N, Borodkin VS, Alonso J, Ferenbach AT, Shepherd C, Navratilova IH, van Aalten DMF, Biochem. J. 2015, 470, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Selvan N, Williamson R, Mariappa D, Campbell DG, Gourlay R, Ferenbach AT, Aristotelous T, Hopkins-Navratilova I, Trost M, van Aalten DMF, Nat. Chem. Biol. 2017, 13, 882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Roux KJ, Kim DI, Raida M, Burke B, J. Cell Biol. 2012, 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Groves JA, Maduka AO, O’Meally RN, Cole RN, Zachara NE, Maduka AO, O’Meally RN, Cole RN, J. Biol. Chem. 2017, 292, 6493–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Carrillo LD, Krishnamoorthy L, Mahal LK, J. Am. Chem. Soc. 2006, 128, 14768–14769. [DOI] [PubMed] [Google Scholar]
- [78].Carrillo LD, Froemming JA, Mahal LK, J. Biol. Chem. 2011, 286, 6650–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lin W, Gao L, Chen X, ChemBioChem 2015, 16, 2571–2575. [DOI] [PubMed] [Google Scholar]
- [81].Ballatore C, Lee VM-Y, Trojanowski JQ, Nat. Rev. Neurosci. 2007, 8, 663–672. [DOI] [PubMed] [Google Scholar]
- [82].Duchartre Y, Kim Y-M, Kahn M, Crit. Rev. Oncol. Hematol. 2016, 99, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Doll F, Buntz A, Spaete AK, Schart VF, Timper A, Schrimpf W, Hauck CR, Zumbusch A, Wittmann V, Angew. Chem. Int. Ed. 2016, 55, 2262–2266. [DOI] [PubMed] [Google Scholar]
- [84].Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K, Nat. Biotechnol. 2003, 21, 86–89. [DOI] [PubMed] [Google Scholar]