Abstract
Genetic ancestry is a critical co-factor to study phenotype-genotype associations using cohorts of human subjects. Most publicly available molecular datasets are, however, missing this information or only share self-reported race and ethnicity, representing a limitation to identify and repurpose datasets to investigate the contribution of ancestry to diseases and traits. We propose an analytical framework to enrich the metadata from publicly available cohorts with genetic ancestry information and a resulting diversity score at continental resolution, calculated directly from the data. We illustrate this framework using The Cancer Genome Atlas datasets searched through the DataMed Data Discovery Index. Data repositories and contributors can use this framework to provide genetic diversity measurements for controlled access datasets, minimizing the work involved in requesting a dataset that may ultimately prove inadequate for a researcher’s purpose. With the increasing global scale of human genetics research, studies on disease risk and susceptibility would benefit greatly from the adequate estimation and sharing of genetic diversity in publicly available datasets following a framework such as the one presented.
Keywords: genetics, diversity, ancestry, sharing
INTRODUCTION
To facilitate the identification and reuse of publicly available biomedical datasets, we have developed the DataMed (https://datamed.org), a search engine for indexed biomedical datasets.1 A large number of the datasets indexed and retrievable in DataMed are derived from human specimens (blood, cell lines, or tissues) and contain broad genetic information (genotypes, exome, genome or transcript sequences). Using established analysis frameworks, one can extract from the raw data useful metadata that is not necessarily collected or known from the investigators. These can include human leukocyte antigen haplotypes, telomere length, genetic admixture, tumor viral load or purity, or cell-line identity. We present here the framework to efficiently measure genetic diversity and global ancestry of DataMed-indexed cohorts and summarize and index the results through a diversity score (DS).
The role of race and ethnicity in disease etiology has been widely debated,2 but there are clear missed opportunities that continue to get ignored in the era of precision medicine.3,4 When accounting for race and ethnicity, studies generally rely on self-reporting. Self-reporting lacks accuracy to distinguish East Asian from South Asian,5 or when subjects have strong levels of genetic admixture (ie, they are related to 2 or more reference populations).6,7 The 1000 Genomes Project has identified variants in 26 reference populations that can be grouped into 5 continental superpopulations (European [EUR], African [AFR], East Asian [EAS], South Asian [SAS], and Admixed American [AMR]).8 From this reference dataset, one can estimate the level of admixture of any given individual using genotypes genome-wide, or preselected ancestry-informative markers.9,10 Admixture can then be used as a covariate in genetic studies, to account for population structure,11 or to identify signals specific to certain reference populations.12
The availability of a uniform, genetically-based ancestry estimation for all eligible human datasets indexed in DataMed would increase their usability, allowing the selection of diverse cohorts, preparing population specific meta-analyses, or simply monitoring diversity to identify understudied ancestry groups or, in contrast, highlight original cohorts of genetically diverse subjects. The DS can facilitate the identification and assembly of ancestry specific cohorts, and enable the monitoring of diversity in biomedical research datasets. Here, we present an analytical framework that uses continental admixture level estimates to calculate a cohort-wide genetic DS, apply it to 33 cohorts from the Cancer Genome Atlas (TCGA) dataset and benchmark its accuracy across diverse sources of genotypes such as genotyping arrays, exome, or transcriptome sequences.
MATERIALS AND METHODS
Data
We selected the TCGA13 cohort to implement the DS into DataMed. Indeed this cohort is large (N = 10 878), one of the most accessed cohorts in the database of Genotypes and Phenotypes (dbGAP) and contains self-reported race and ethnicity. In addition, the cohort can be split into 33 subcohorts corresponding to each cancer type, providing an opportunity to contrast the various collections. Finally, the vast majority of samples have multiple data types (genotypes, exomes, and transcriptomes) on which we can compare admixture estimation.
A total of 10 878 TCGA subjects (individuals) have been genotyped at ∼106 single nucleotide polymorphisms (SNPs). We called admixture from the 5 continental reference populations: EUR, AFR, EAS, SAS, and AMR. The data aggregated by cancer type can be used to query DataMed (Genomic Data Commons repository only) and is available online.14
Data access and preprocessing
The data specified below were retrieved through the National Cancer Institute Genomic Data Commons using the gdc-client application programming interface. We obtained the genotyping array data (germline blood DNA) in the birdseed format (the result of genotype calling by birdSuite),15 which were converted to the PLINK16 format (MAP and PED text files). To ensure the proper alleles were reported during the conversion, we established a relational database to decode the numeric genotype into alleles using information from the Affymetrix SNP Array 6.0 probe design and the corresponding dbSNP (v150) rsid. The RNA sequencing (RNA-Seq) reads (BAM files) from the patient tumors were used to call variants using the following steps: (1) duplicate reads removal (PICARD MarkDuplicates), (2) splitting of intron-spanning reads (GATK v3.8), and (3) variant calling (GATK v3.8 HaplotypeCaller). We called variants from the whole exome sequence (BAM files) from the blood using FreeBayes (v1.1.0).17 For both RNA-Seq and exome sequencing analysis, we restricted the variant calling to known SNP (dbSNP v150) located in the exons and coding sequence (CDS) regions of Gencode-v25,18 respectively. The variants were filtered (DP > 10 and GQ > 15), and then converted to PLINK format using vcftools. The analysis workflow is summarized in Supplementary Figure S1.
Admixture analysis
For each individual, the admixture fraction for the reference population was estimated using the iAdmix tool.10 In contrast to other supervised admixture estimation tools,9 which use individual genotypes for the reference populations, iAdmix uses allele frequencies to calculate, for each tested individual, the maximum likelihood estimates from each reference population. Relying directly on genome-wide allelic frequencies as reference dataset prevents the need for genotype imputation, increases the speed of the analysis and allows more flexibility by directly inferred from sequencing reads. The input data were individual genotypes (MAP and PED flat files in PLINK format), and the allele frequencies from the 1000 Genomes Project reference populations.8 The 1000 Genomes Project reference VCF file was based on the GRCh37 human genome build and contained allelic fractions calculated from 2504 individuals divided into 5 superpopulations: EUR, AFR, EAS, SAS, and AMR. To accommodate genotypes from different versions of the human genome reference, the SNP coordinates were converted to GRCh38 using liftOver (https://genome-store.ucsc.edu/). Notably, the reference SNPs from the 1000 Genomes Project reference population were not pruned for linkage disequilibrium to allow the use of the same reference across multiple assays with variable coverage breadth (genotyping arrays, exome, RNA-Seq). The output of iAdmix was a list of 5 admixture fractions, each with values ranging between 0 and 1, adding up to 1. These estimates correspond to maximum likelihood estimations through Broyden-Fletcher-Goldfarb and Shanno, a widely used, quasi-Newton optimization method.
Cumulative admixture fraction
The cumulative admixture fraction (CAF) was calculated as the overall fraction of admixture from the 5 reference populations after summing up individual admixture fractions across a given set of individuals:
where A(i, j) is the proportion of admixture from the reference population i for individual j and N the number of individuals in the cohort. Hence, the CAF reflects, at the cohort level, the fraction of total DNA from each reference population, rather than the fraction of individuals from a given dominant ancestry.
Diversity score
To calculate the DS of each cancer-specific cohort, we calculated the cumulative fraction of each reference population across all individuals in the cohort. We then computed the normalized entropy from the resulting 5-dimensional vector using R package entropy,19 as the empirical entropy divided by the maximal entropy for 5 dimensions.
where Ci is CAF for reference population i and Hmax=5*(0.2.ln(0.2))=−1.609438.
Data and code availability
The benchmarking study comparing admixture determination using genotyping vs exome vs transcriptome was conducted on 100 subjects specifically selected to have a sample that maximized diversity in self-reported race and ethnicity (Supplementary Table S1). The computational methods used to calculate admixture and DS can be found online.20 The cumulative admixture fraction and DS of each TCGA cancer type is available on figshare.14 The iAdmix tool is published10 and has been wrapped in a docker container, together with scripts to derive cohort-wide values.20
RESULTS
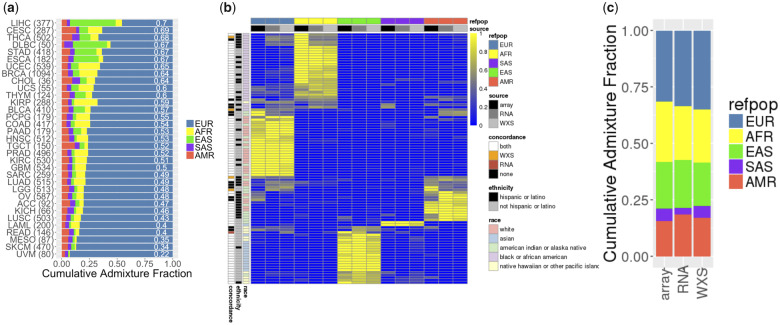
The dominant ancestry—representing more than 80% of admixture from one reference population—of each TCGA subject matched well the self-reported race and ethnicity: 76% White non-Hispanic were EUR dominant, 82% of Black were AFR dominant, and 89% of Asian were either SAS or EAS dominant. Similarly, 53% of subjects reported as Hispanic or Latino had at least 20% of AMR ancestry. We then determined the CAF for each cancer-specific cohort (Materials and Methods). While all cancer cohorts are predominantly EUR (Figure 1A) (46%-93%), the fraction of non-EUR varied: kidney renal cell carcinoma was the cohort with the highest AFR CAF (21%), while liver hepatocellular carcinoma had the highest EAS CAF (41%). While these differences may reflect the epidemiology of the disease, it is important to note that the TCGA cohort had significant ascertainment bias, including enrollment sites, tumors sizes, purity, and availability requirements. Finally, to summarize the overall diversity of each cohort, we used the CAF to compute a normalized DS: 0 for 1 reference population only, 1 for an even fraction of all 5 reference populations. Importantly, a maximum DS of 1 would reflect a perfectly diverse cohort in which all 5 continental ancestry contribute equally to its genetic background. The TCGA cohorts were ranked by decreasing diversity, revealing that the hepatocellular carcinoma dataset was the most diverse (DS = 0.7) and uveal melanoma the least diverse (DS = 0.22, Figure 1A). Both the DS and the minimal admixture estimate of a given reference population for each cohort are publicly available14 and can be used to query the DataMed index (Genomic Data Common repository only).
Figure 1.
Cumulative admixture in The Cancer Genome Atlas. (A) Cumulative admixture fraction of 33 cancer-specific cohorts, inferred from the 5 reference superpopulations. The cohorts are ranked by decreasing diversity score (white label). The number of samples in each cohort is indicated in parentheses. (B) Heatmap of the level of admixture estimated in 100 patients (rows) for 5 reference populations using genotypes derived from 3 sources (columns, genotyping array, transcriptome [RNA] or exome [WXS]). For each sample, concordance of the reference population with maximal admixture level is indicated using array derived admixture levels as reference. Self-reported race and ethnicity are indicated for each row. (C) Cumulative admixture fraction of a selected set of 100 diverse Cancer Genome Atlas subjects using genotypes from genotyping array, transcriptome (RNA), or exome (WXS). ACC: adrenocortical carcinoma; AFR: African; AMR: Admixed American; BLCA: bladder urothelial carcinoma; LGG: brain lower grade glioma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: cholangiocarcinoma; COAD: colon adenocarcinoma; DLBC: lymphoid neoplasm diffuse large B-cell lymphoma; EAS: East Asian; ESCA: esophageal carcinoma; EUR: European; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LAML: acute myeloid leukemia; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MESO: mesothelioma; OV: ovarian serous cystadenocarcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; SARC: sarcoma; SAS: South Asian; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THYM: thymoma; THCA: thyroid carcinoma; UCEC: uterine corpus endometrial carcinoma; UCS: uterine carcinosarcoma; UVM: uveal melanoma.
A subset of the subjects (n = 100) (Supplementary Table S1) representing diverse self-reported ancestry were further selected to evaluate the reproducibility of the approach using alternate sources of genotypes such as exome (germline DNA) or transcriptome (tumor RNA) sequencing. After variant calling and filtering (Materials and Methods), we identified a median of 21 327 and 838 usable variants in the exome and transcriptome of each subject, respectively. The populations corresponding to the maximum admixture level were consistent across for all 3 methods for 82 of 100 subjects (Figure 1B). The subjects with inconsistent results had higher admixture levels based on the genotyping array results (maximum admixture 0.89 ± 0.16 vs 0.72 ± 0.22). As a result, the CAF estimated from the exome or transcriptome variants were consistent with those of genotyping array (r = 0.97 for both) (Figure 1C) and all 3 DSs were similar: 0.93, 0.92, and 0.90 for genotyping, exome, and transcriptome, respectively.
DISCUSSION
A number of studies suggest important differences between genetic ancestry results and self-reported race and ethnicity.21–23 Such studies require investigators to determine admixture at the individual level, perhaps at ethnic or subcontinental resolution and to date, it is not possible to get such an accurate estimate before looking inside the dataset (ie, looking at the individual-level data) and calling genetic ancestry. The proposed DS is a low-resolution, cohort-wide summary of genetic ancestry according to the 5 super-populations. While DS may not be readily usable in an analysis, it represents a convenient and efficient metric to query and filter the growing number of biomedical datasets on the basis of their genetic diversity. Alternatively, more usable metrics could be derived from the distribution of continental admixture for each individuals such as the one presented in Figure 1B. Such information is however difficult to summarize, requiring an arbitrary threshold to establish the dominant ancestry or introducing an ambiguous “admixed” category not reflecting the ancestral populations. As such, the DS is a faithful summary of a cohort diversity, which can be used to efficiently sort and query a large number of biomedical datasets.
The choice of the reference populations can influence the accuracy of the admixture estimate and strategies exist to identify the optimal ancestral populations to use for a given cohort.24 The 5 superpopulations selected here are part of the 1000 Genomes Project, which guarantees their availability as a universal reference. Due to the history of human evolution and migration, it is theoretically impossible to identify a true reference population and the 5 superpopulations selected are expected to provide a reasonable approximation of continental ancestry that can be used to compare continental admixture between cohorts. However, it is important to acknowledge that they represent themselves different ancestries and admixtures levels (Spanish and Finnish in EUR, Puerto Rican and Peruvian in AMR).
To calculate the DS, the raw data access request had to be approved for ancestry analysis, a step that may not be permitted for certain cohorts or that is not necessarily scalable. However, the DS does not have to be generated by the DataMed curators, but could instead be computed by the data owners and shared as an additional piece of metadata that could be used downstream for cohort selection.
The admixture and DS generated are well applicable on a variety of broad molecular datasets. We demonstrated their validity from exome and transcriptome. To date, 176 × 103 and 201 × 103 human transcriptome (RNA-Seq) and exome datasets, respectively, are hosted by the National Center for Biotechnology Information Sequence Read Archive. Among those, 82% of transcriptomes and 12% of exomes are available without restriction, and likely none of them have associated genetic ancestry information. Beyond transcriptome or exomes, ancestry can also be called from chromatin immunoprecipitation sequencing datasets from human samples— more than 31 × 103 currently available in the National Center for Biotechnology Information Sequence Read Archive. A typical chromatin immunoprecipitation sequencing dataset may cover 106 bp from the human genome, harboring 1000 SNPs, the majority of which have been genotyped in the 1000 Genomes Project reference populations, representing a sufficient number to determine genetic admixture.
The same way the Gene Expression Omnibus has the ability to search and rank datasets based on differential expression of a specific gene, one can hope that future, innovative data sharing strategies will include as many of such data-derived features, like genetic admixture, generated in an automated, standardized way at the time of the deposition. The relative simplicity of calling admixture on molecular datasets may encourage more careful analytical design. While we know that germline variants may play a role in disease etiology or phenotypic differences, they are rarely considered in preclinical or clinical studies. Using admixture from known continental ancestry as a first-order surrogate for germline genetic differences, one could account for this important covariate and relate it to a population trait. In the past, preclinical studies based on a small number of cell lines or samples could not reasonably account for inherited genetic variation. Nowadays, preclinical studies are becoming larger and more systematic, such as the Cancer Cell Line Encyclopedia25 (N = 750 cell lines), but to our knowledge they still do not account for genetic ancestry. More recently, genetically diverse sets of lymphoblastoid cell lines26 or induced pluripotent stem cells27 have been made available for research, documenting the increasing interest in performing preclinical research in large sets of genetically diverse samples and cell lines. Similarly, while the individual-level data from clinical trials may not readily shared by the sponsors, the summary data related to demographics and ancestry, like the one we propose, could certainly pique the curiosity of researchers interested in health disparities associated with ancestry who may then want to collaborate with the investigators. The availability of genetic ancestry and diversity as a piece of metadata in the public datasets would therefore increase their visibility for inclusion in studies aimed at understanding the contribution of genetic ancestry to disease phenotypes. The DS featured in the DataMed index provides an optimal way for researchers to select the adequate datasets for this task, without the need to disclose individual-level data.
FUNDING
This work was supported by a grant from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (U24AI117966) (to LO-M).
AUTHOR CONTRIBUTORS
XX, JK, and OH performed the analysis; OH designed the study; and OH and LO-M wrote the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1. Ohno-Machado L, Sansone S-A, Alter G, et al. Finding useful data across multiple biomedical data repositories using DataMed. Nat Genet 2017; 49 (6): 816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shields AE, Fortun M, Hammonds EM, et al. The use of race variables in genetic studies of complex traits and the goal of reducing health disparities: a transdisciplinary perspective. Am Psychol 2005; 60 (1): 77–103. [DOI] [PubMed] [Google Scholar]
- 3. Rotimi C, Shriner D, Adeyemo A.. Genome science and health disparities: a growing success story? Genome Med 2013; 5 (7): 61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. West KM, Blacksher E, Burke W.. Genomics, health disparities, and missed opportunities for the nation’s research agenda. JAMA 2017; 317 (18): 1831–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mersha TB, Abebe T.. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum Genomics 2015; 9 (1): 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rishishwar L, Conley AB, Wigington CH, et al. Ancestry, admixture and fitness in Colombian genomes. Sci Rep 2015; 5: 12376.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sucheston LE, Bensen JT, Xu Z, et al. Genetic ancestry, self-reported race and ethnicity in African Americans and European Americans in the PCaP cohort. PLoS One 2012; 7 (3): e30950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alexander DH, Novembre J, Lange K.. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19 (9): 1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bansal V, Libiger O, Menozzi P, et al. Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics 2015; 16: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halder I, Shriver MD.. Measuring and using admixture to study the genetics of complex diseases. Hum Genomics 2003; 1 (1): 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fejerman L, Chen GK, Eng C, et al. Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Hum Mol Genet 2012; 21 (8): 1907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45 (10): 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harismendy O. DataMed-Admixture.TCGA.txt. figshare; 2017; 10.6084/m9.figshare.5695663.v1. [DOI] [Google Scholar]
- 15. Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008; 40 (10): 1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81 (3): 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garrison E, Marth G.. Haplotype-based variant detection from short-read sequencing. arXiv 2012; 9 http://arxiv.org/abs/1207.3907 [Google Scholar]
- 18. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res 2012; 22 (9): 1760–74. http://genome.cshlp.org/content/22/9/1760.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hausser J, Strimmer K.. Entropy inference and the James-Stein estimator, with application to nonlinear gene association networks. arXiv 2008. http://arxiv.org/abs/0811.3579 Accessed March 13, 2018. [Google Scholar]
- 20. Kim J, Harismendy O.. DataMed-Admixture Code Repository. GitHub; 2017. https://github.com/jihoonkim/DataMed-Admixture. [Google Scholar]
- 21. Spector SA, Brummel SS, Nievergelt CM, et al. Genetically determined ancestry is more informative than self-reported race in HIV-infected and -exposed children. Medicine (Baltimore) 2016; 95 (36): e4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith EN, Jepsen K, Arias AD, et al. Genetic ancestry of participants in the National Children’s Study. Genome Biol 2014; 15 (2): R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee YL, Teitelbaum S, Wolff MS, et al. Comparing genetic ancestry and self-reported race/ethnicity in a multiethnic population in New York City. J Genet 2010; 89 (4): 417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chimusa ER, Daya M, Moller M, et al. Determining ancestry proportions in complex admixture scenarios in South Africa using a novel proxy ancestry selection method. PLoS One 2013; 8 (9): e73971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. The International HapMap Consortium . A haplotype map of the human genome. Nature 2005; 437: 1299–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panopoulos AD, D’Antonio M, Benaglio P, et al. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Reports 2017; 8 (4): 1086–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The benchmarking study comparing admixture determination using genotyping vs exome vs transcriptome was conducted on 100 subjects specifically selected to have a sample that maximized diversity in self-reported race and ethnicity (Supplementary Table S1). The computational methods used to calculate admixture and DS can be found online.20 The cumulative admixture fraction and DS of each TCGA cancer type is available on figshare.14 The iAdmix tool is published10 and has been wrapped in a docker container, together with scripts to derive cohort-wide values.20