Abstract
In the 1950s the myxoma virus was released into European rabbit populations in Australia and Europe, decimating populations and resulting in the rapid evolution of resistance. We investigated the genetic basis of resistance by comparing the exomes of rabbits collected before and after the pandemic. We found a strong pattern of parallel evolution, with selection on standing genetic variation favouring the same alleles in Australia, France and the United Kingdom. Many of these changes occurred in immunity-related genes, supporting a polygenic basis of resistance. We experimentally validated the role of several genes in viral replication and showed that selection acting on an interferon protein has increased its antiviral effect.
The emergence of new infectious diseases can result in intense selective pressures on populations and cause rapid evolutionary change in both host and parasite. While pathogens must adapt to a new ecology and cellular environment, hosts can rapidly evolve resistance in a continuous arms-race. One of the most emblematic examples of this coevolutionary process arose when the wild European rabbit (Oryctolagus cuniculus) populations in Australia and Europe were exposed for the first time to the myxoma virus (MYXV; genus Leporipoxvirus, family Poxviridae). MYXV circulates naturally in American cottontail rabbits (Sylvilagus spp.) where it causes benign skin tumours, but in European rabbits it causes the highly lethal systemic disease myxomatosis (1).
Rabbits were initially introduced into Australia by European settlers, resulting in extensive ecological and economic damage (2). In an attempt to control the rabbit populations, MYXV was released in 1950 in Australia, after which it spread rapidly across the country, causing massive population reductions (1). In 1952 it was released in France and in 1953 it reached the United Kingdom (UK) with similar outcomes (2). In a series of classic experiments, the evolution of the virus and rabbits was tracked, and in all three countries substantial declines in case fatality rates in wild rabbits were observed both due to evolution of less virulent viral phenotypes and increased resistance in rabbit populations (3–5). Considered “one of the greatest natural experiments in evolution”, these observations ultimately became a textbook example of host-parasite coevolution (2).
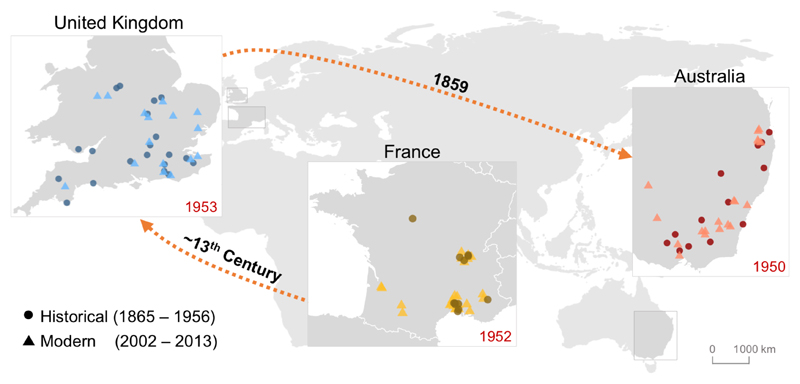
Sixty-nine years have passed since myxomatosis was first released in Australia and today the virus continues to evolve in an ongoing arms-race against the rabbit immune system (6, 7). Despite much research on the genetics of MYXV, little is known about the genetic basis of resistance to myxomatosis. The intense selective pressure exerted by the disease in rabbits and the parallel phenotypic evolution across multiple populations, provides an exceptional framework to study the evolution of resistance to a highly lethal pandemic in natural host populations. To understand the genetic basis of these changes we focused on three countries where genetic resistance independently emerged: Australia, France and the UK. For each one, we compared modern rabbits with historical specimens from museums that were collected before or soon after the release of the virus (1856-1956; Figs. 1 and S1, File S1).
Fig. 1. Rabbit origins and sampling locations.
Historical (circles) and modern (triangles) sampling locations. Dates in red inside the maps show the date of the first myxomatosis outbreak in the respective countries. Orange dashed arrows and dates reflect historical and archaeological records of the colonisation of European rabbits from France to the United Kingdom and Australia.
Colonisation route of rabbits
To obtain genome-wide polymorphism data we used oligonucleotide probes to capture 32.10Mb (1.17%) of the rabbit genome that includes the exome (19,293 genes), the mitochondrial genome, and three genomic regions that contain the Major Histocompatibility Complex region (MHC) encompassing 1.75 Mb. We sequenced 152 rabbits from Australia (historical: n=17, modern: n=26), France (historical: n=29, modern: n=26) and the UK (historical: n=29, modern: n=25). The mean sequence coverage on-target per individual after filtering was 33X. After filtering, the number of bi-allelic SNPs was 757,333.
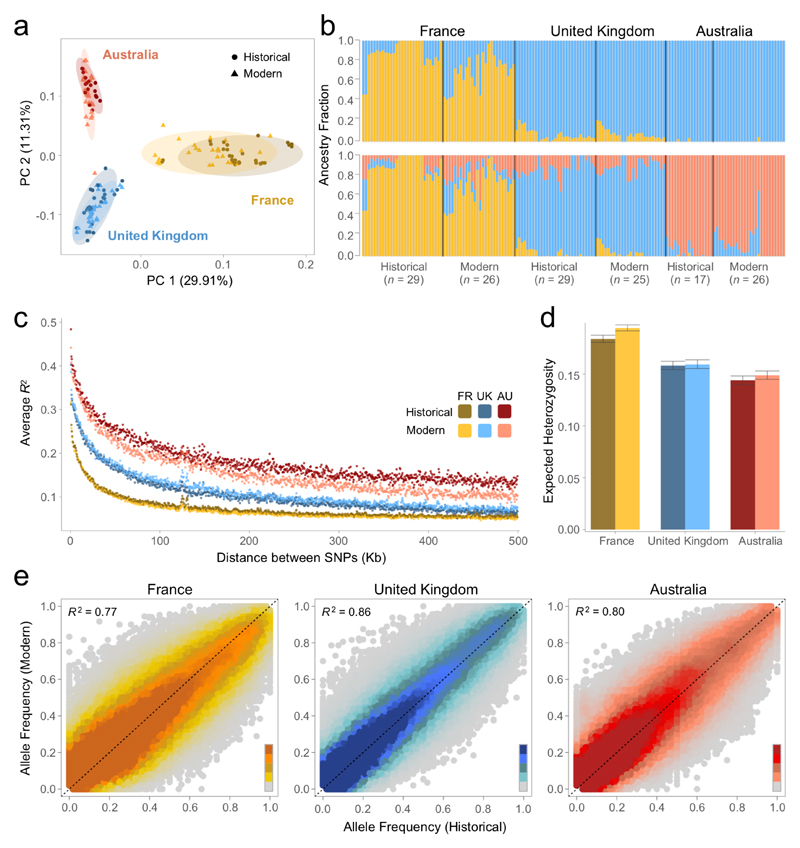
Historical records support the introduction of rabbits to the UK mainland from continental Europe by the 13th century (8) and most Australian rabbits are thought to be derived from an introduction in 1859 from the UK (9) (Fig. 1). Our genetic data reflects these historical records. Both Principal Components Analysis (PCA; Fig. 2a) and structure analyses (Fig. 2b; K=3) reveal three clusters composed of individuals from the same country. The sequential colonisation from France to the UK and then Australia is reflected in the levels of genetic differentiation which are greatest between France and Australia, and lowest between UK and Australia (Table S1). This pattern is repeated with the Australian populations clustering together with the UK in the structure analysis (Fig. 2b; K=2). Population bottlenecks during colonisation can reduce genetic diversity and increase linkage disequilibrium (LD). Both aspects are reflected in the successive increases in LD (Fig. 2c) and decreases in heterozygosity (Fig. 2d) as rabbits moved from continental Europe to the UK and then Australia.
Fig. 2. Genetic structure and diversity in historical and modern populations of France, United Kingdom and Australia.
(a) Principal components analysis (PCA). Ellipses show 95% confidence intervals. (b) Ancestry fractions inferred with Ohana structure analysis for K=2 (top) and K=3 (bottom). Each bar shows the inferred ancestry fraction for an individual. Black lines between bars separate populations. Labels above bars identify country and labels below bars identify temporal set and sample size. Individuals are ordered geographically within each population. (c) Decay of linkage disequilibrium for each population. Each dot represents the averaged pairwise R2 values between pairs of SNPs in non-overlapping 500bp windows. Colours represent different populations. (d) Expected heterozygosity for each population. The bars are the mean across chromosome arms, and error bars are 95% bootstrap confidence intervals from resampling chromosome-arms. Colours represent different populations. (e) The correlation between the frequency of the alternative allele in historical and modern populations for France, the UK and Australia. Colours reflects the relative density of points according to the scale on the bottom right of each plot, from darker (more density) to lighter (less density).
Genetic variation in historical and modern populations
To detect changes in allele frequency due to natural selection, it is important that the genome-wide allele frequencies in our modern and historical samples are similar. Although changes in allele frequency are an expected signal of selection, artefactual variation generated by DNA degradation can generate similar signals between modern and historical DNA. To mitigate this, we sequenced samples to a high coverage, corrected for the effects of DNA damage patterns, and used a stringent set of filters at read and variant level. Genome-wide differences in allele frequency between our modern and historical samples could also arise due to population substructure. To minimise this effect, we sampled modern rabbits from locations near to where the historical specimens had been sampled (Figs. 1 and S1).
We consistently found that historical and modern populations from the same country exhibit similar patterns of genetic structure and diversity. In the PCA, the 95% confidence ellipses for historical and modern samples from the same country are interspersed (Fig. 2a). In the structure analysis, the patterns again reflect geography rather than the time of sampling (Fig. 2b, K=3), and increasing the number of ancestral populations (K) reveals finer population substructure instead of a split between the different time points (Fig. S2). More generally, across all SNPs the allele frequencies of historical and modern populations are highly correlated in the three countries (Fig. 2e).
The collapse of populations due to myxomatosis has not increased levels of LD or decreased genetic diversity as both these parameters are similar between historical and modern populations (Figs. 2c and 2d). This is to be expected as the effective population size of rabbits is ~106 (10), which is large for a vertebrate. Therefore, even if an ~99% reduction in population size (as seen in some populations immediately after the first epidemic (6)), had been sustained for the subsequent 65 generations, the expected reduction in heterozygosity would have only been 0.35% (per generation reduction heterozygosity = 1-1/(2Ne)). Similarly, bottlenecks of this size are expected to have little effect on LD (11). Together, these results demonstrate that historical and modern samples are drawn from genetically similar populations. This strong correlation in allele frequencies between the two time points within each country is conducive to the identification of unusual shifts in allele frequency resulting from natural selection.
Parallel changes in allele frequency
With a 99.8% case fatality rate of the originally released strain of MYXV, the myxomatosis pandemic imposed intense selection on rabbits, resulting in rapid and parallel evolution of increased resistance in exposed populations (6). To investigate whether parallel genetic changes occurred in Australia, France and the UK, we calculated FST between the historical and modern samples for each country and identified the 1000 SNPs that show the highest FST. By intersecting these three lists we found that more alleles have changed in frequency across two or three countries than expected by chance (Fig. 3a; File S2). Furthermore, considering the 92 SNPs in the intersections of Fig. 3a, in 87 cases it was the same allele that had increased in frequency in the countries involved. It is particularly striking that SNPs that were among the top 1000 most differentiated in any two populations tend to show elevated FST in the third population (Fig. 3b). These results demonstrate the occurrence of parallel changes in allele frequency across the three populations.
Fig. 3. Parallel changes in allele frequency across three countries.
(a) Venn diagram showing the overlap of the 1000 SNPs with the highest changes in allele frequency between modern and historical samples (FST) in France, the UK, and Australia. Numbers in black show the observed number of SNPs and numbers in red show the expected overlap after 1000 random permutations of modern and historical samples within each country. (b) Scaled histogram of the FST values in the three countries. Bars with dark colours reflect SNPs that are in the top 1000 in both of the other two countries. Bars with light colours are SNPs that are in the top 1000 of only one of the other countries. Grey bars are all the remaining SNPs. (c) Genome-wide selection scan based on allele frequency changes after the introduction of myxomatosis. (supplementary methods, Equation 5; the strength of selection in each population is allowed to vary independently) (d) Selection scan testing whether selection has acted in all three populations (positive values) or just one population (negative values; supplementary methods, Equation 6). (c and d) Y-axis shows likelihood ratio statistic of each model. Orange dotted line shows genome-wide 95% significance threshold from permuting sample collection dates within each country. Shaded boxes show SNPs located in the highlighted genes. Different shades of blue show chromosomes. (e) Mean of the posterior distribution of the derived allele frequency as a function of time for the IFN-α21A and FCRL3 loci from the Bayesian selection analysis (additive model). 95% credible intervals are shaded. Triangles across the bottom represent years of samples (only samples post-1940 are shown). Dotted lines show dates of first reports of MYXV and RHDV. List of the top 1000 SNPs for all figures is available in Files S2, S3, and S4.
To locate the putative targets of selection in the genome, we searched for SNPs that experienced large changes in allele frequency since the release of the MYXV. We accounted for the effects of population structure by scanning for outliers where the difference in allele frequency between modern and historical populations was larger than expected given the covariance matrix describing the joint allelic frequencies among populations inferred from the structure analysis (Fig. 2b, K=3). In each population, we assumed selection started in the year the virus was introduced. For each SNP, we then compared the likelihood of a model where the change in allele frequency followed that predicted from the genome-wide amount of genetic drift across all historical and modern samples to a model that allowed additional changes in allele frequency through time (Fig. S3). Combining data across the three populations and allowing the strength of selection to vary independently in the three countries, we identified 193 SNPs in 98 genes and 7 intergenic regions that experienced significant changes in allele frequency (genome-wide significance p<0.05; Fig. 3c; File S3. The results were similar when we assumed the same strength of in all countries (Fig. S4; File S3).
To explicitly test for parallel evolution and identify variants that significantly changed in frequency in all three populations, we compared the likelihood of our previous model, that assumed the equal selection across the three populations, to a model where we allowed the alleles to change in frequency due to selection in one population only. We identified 94 SNPs that had a significant signature of parallel selection (genome-wide significance p<0.05; Fig. 3d, positive values).
It is common to find considerable standing genetic variation in susceptibility to infection within populations (12, 13), which may allow populations to rapidly evolve resistance when they encounter novel pathogens (14). This can be explicitly tested with our experimental design that incorporated historical samples. We found that in the large majority of cases, the SNPs under selection were also present in at least one of our historical populations (93% and 84% of the SNPs with a genome-wide P<0.05 in Figs. 3c and 3d, respectively). Therefore, parallel evolution has occurred because natural selection has been acting on standing genetic variation that was present over 800 years ago in continental Europe, and was carried with rabbits to the UK and Australia. The existence of this variation likely explains the rapid development of resistance to myxomatosis observed almost immediately after the first outbreaks.
Population-specific evolution
Despite the common selection pressure imposed by myxomatosis, the populations of France, UK and Australia will have experienced their unique selection pressures, as resistance was evolving in a different ecological and genetic background. We can identify SNPs that have experienced population-specific changes in allele frequency from our previous analysis when the model of selection in one population is preferred over the model of selection in all three populations (negative values in Fig. 3d).
To quantify the proportion of SNPs under parallel or population-specific selection we used a Bayesian approach. First, we analysed data from each population independently to identify variants under selection (Fig. S5, File S4; genome-wide p<0.05). This minimises the inherent bias towards detecting SNPs under parallel selection that occurs when the all populations are combined (Figs. 3c and 3d). We then returned to the combined dataset, and for each of the SNPs that were significant in the single population analysis we calculated a Bayes factor to compare models of population-specific versus parallel selection (Fig. S5). For each gene we retained only the most significant SNP (n=43, File S5). We found evidence that 20 SNPs were under population-specific and 20 were under parallel selection (for 3 SNPs we could not distinguish the models), implying that a large component of the recent selection in the three populations has occurred on a common set of variants.
Due to population bottlenecks as rabbits colonised new areas (Figs. 2c and 2d), alleles selected in one population may be rare or missing in other populations. This means that population-specific adaptation could result not just from differences in selection pressures but also due to differences in the variants available for selection to act on. To test this, we examined the frequency of the 20 alleles under population-specific selection in our historical samples. There were no consistent differences in the ancestral allele frequencies between the populations where we detected the effects of selection and populations where we did not (Table S2). Therefore, we can conclude that population-specific changes in allele frequency result from population-specific selection pressures, perhaps due to differences in ecology, genetic background or independent paths of co-evolution with MYXV (15).
Changes in the strength of selection
The proportion of rabbits killed by myxomatosis has fallen since the 1950s, due to declines in the virulence of the virus and increases in resistance (6). Therefore, the strength of selection on variants that confer resistance to MYXV is expected to have declined. However, in 1984 a new lethal viral pathogen was identified in rabbits, the rabbit haemorrhagic disease virus (RHDV; genus Lagovirus, family Caliciviridae) (16). RHDV, which has a similar case-fatality rate to myxomatosis, was first found in domestic rabbits in China, from where it spread to continental Europe in 1986, the UK in 1992 and Australia in 1995 (2, 17, 18). Like myxomatosis, RHDV caused high mortality across the two continents, which could have contributed to the observed changes in allele frequency.
To evaluate the role of RHDV in our selection signatures and understand how the strength of selection has changed through time, we obtained 70 rabbit samples that were collected before or soon after RHDV between 1985 and 1996, in the UK and Australia (Fig. 3e: File S1). We selected four SNPs that had experienced significant changes in allele frequency since the 1950s and that were located in or near genes with known immune functions (CD96, FCRL3, IFN-α21A and MHC Class I), and genotyped them in these samples by Sanger sequencing. Combining these genotypes with data from our exome sequences, we used a Bayesian approach (19) to estimate the selection coefficient acting on these SNPs. We allowed two phases of selection. The first from when MYXV was released to the appearance of RHDV, and the second from the appearance of RHDV to the present day (Fig. 3e; note the date when the strength of selection changes is fixed in the model).
We found that the strength of selection on FCRL3 and IFN-α21A has declined through time in both the UK and Australia (Fig. 3e, File S6). This is consistent with selection being driven by MYXV, whose case fatality rate has decreased since its release, and does not support a role for RHDV. Intriguingly, the data suggests that these variants have been negatively selected in recent decades (Fig. 3e), which is predicted if they have negative pleiotropic costs on other fitness components. For CD96 and MHC, there was no significant change in the strength of selection through time (File S6). Nonetheless, both the genes in the UK and CD96 in Australia have a significant signal of positive selection before the release of RHDV.
We observed large changes in allele frequency across the two time periods (Fig. 3e). Averaging across populations during the first phase of selection, we estimate the per year selection coefficient was 0.16 for IFN-α21A, 0.13 for FCRL3, 0.08 for CD96 and 0.07 for MHC (File S6). These analyses assume the alleles were additive, but estimates of the timing and strength of selection remained similar if we assumed recessive or dominant mode of inheritance.
Selection on the immune system
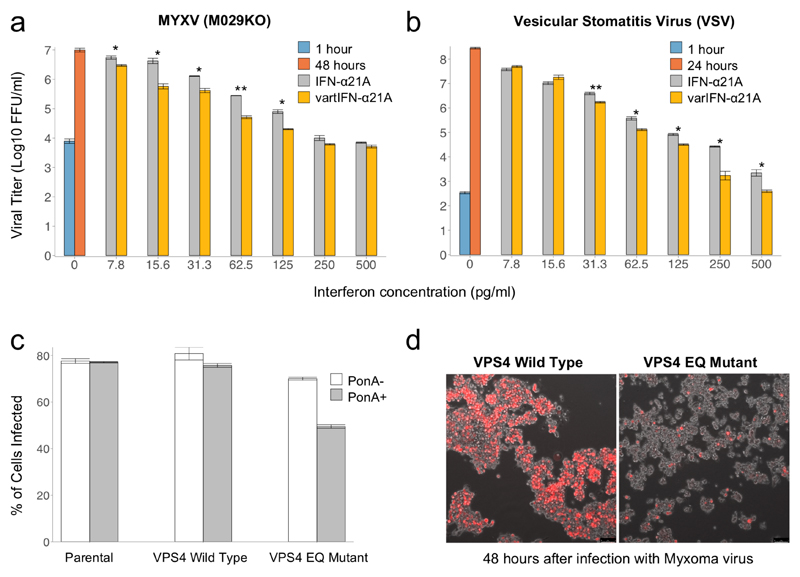
The innate immune response provides the first line of defence against viruses. It is activated by the secretion of cytokines including interferon-α (IFN-α), which binds to receptors on uninfected neighbouring cells and induces an antiviral state. To circumvent this, MYXV encodes potent inhibitors of the interferon response, including the double-stranded RNA binding protein M029 (20, 21). Among the most significant increases in frequency in our dataset, there are three non-synonymous variants that segregate as a haplotype in the interferon-α21A gene (IFN-α21A; Fig. 3c). To evaluate the role of these SNPs in the antiviral activity of IFN-α21A, we synthesised the two corresponding protein isoforms and tested their antiviral effect by measuring MYXV viral replication in a rabbit cell line. Neither isoform of IFN-α21A affected the replication of the wild-type MYXV (Lausanne strain) (Fig. S6a), however both reduced significantly the replication of an attenuated strain of MYXV (M029 mutant) (22) (Figs. 4a and S6b). Moreover, we found that the isoform of IFN-α21A (varIFN-α21A) that had been favoured by selection more strongly inhibited the replication of M029 MYXV. This indicates that natural selection has increased the potency of the interferon response in modern rabbit populations that have co-evolved with MYXV. The selected allele also had an antiviral effect on vesicular stomatitis virus (VSV), an RNA virus (Fig. 4b). This suggests a general increase in the protein’s antiviral activity and it may be that selection by MYXV has increased resistance to other viruses, including RHDV. While this effect was not apparent with wild-type MYXV, this could be a limitation of the cell culture-based experiment, since in vivo innate immune responses involve the coordinated action of multiple cytokines across many tissues. In such circumstances, it is possible that the isoform of IFN-α21A that has been favoured by selection may contribute to attenuating wild-type MYXV infection.
Fig. 4. The effect of IFN-α21A and VPS4 on viral titres.
(a and b) wild-type (IFN-α21A, grey bars) and variant (varIFN-α21A, yellow bars) IFN-α21A were added at different concentrations to cell culture before infection with (a) MYXV-M029KO and (b) vesicular stomatitis virus (VSV). Viral titre was measured 1-hour post-infection (blue bars) and 24/48 hours post infection (orange bars). Error bars show standard error of the mean. Statistical significance between wild and mutated interferon treatments was inferred with a Student's t-test across three replicate assays (* P<0.05; ** P<0.01). (c) HEK293 cell lines stably expressing the wild-type isoform of human VPS4 (wild-type) or a dominant-negative VPS4 (EQ Mutant) under the control of ponasterone A (PonA). The HEK293 non-transfected cell line (Parental) was included as an additional control. Cells were either untreated (PonA-) or pre-treated (PonA+) with 1 µM PonA for 20-24 hours and then infected with wild-type MYXV expressing a red fluorescent protein (vMyx-tdTomato) at a multiplicity of infection (MOI) 10. The percentage of infected cells (tdTomato+) was assessed by flow cytometry. Error bars show standard error of the mean. (d) Fluorescence microscope images of VPS4 wild-type and VPS4 EQ mutant HEK293 cells pre-treated with PonA (20-24 hours), 48 hours post-infection with vMyx-tdTomato (MOI 10). The live cell images were taken using an inverted fluorescence microscope at 10x magnification. FFU, focus forming unit.
We found a strong population-specific signal of selection on a non-synonymous variant in CD200-R, which is the receptor for the negative regulator of innate immune responses CD200. The selected allele was not observed in any of the historical populations, but it increased to a frequency of 56% in the modern French population (Fig. 3d). The selected residue is part of the CD200 binding interface (23) (Fig. S7). Mutation of this site in the human protein does not prevent CD200 binding (24), but it may affect binding affinity in a quantitative way. Therefore, any effects of this variant may occur via modulation of the rabbit CD200:CD200R interaction.
Alongside the innate immune response, hosts mount an adaptive antiviral immune response mediated by MHC class I proteins. Polymorphisms in the peptide-binding region of MHC proteins affect the repertoire of peptides they can present and are frequently associated with variation in susceptibility to infectious disease (25). We found multiple SNPs in the MHC region that have experienced positive selection across the three populations (Fig. 3c). However, the top hits in this region were located in an intergenic region of 35.7Kb between two MHC class I genes (grey shaded box; Fig. 3c). We failed to sequence the regions encoding the peptide-binding regions of these proteins for most individuals (either the sequence capture or read mapping failed). Therefore, the variants that have increased in frequency in the intergenic region may be in LD with variants in the protein coding sequence that affect susceptibility to MYXV.
Other immunity genes that exhibit parallel changes in frequency include four non-synonymous variants in FCRL3 (Fc Receptor-Like 3), a cell surface receptor that inhibits the activation of B cells (26). Three highly significant non-synonymous variants are in CD96 which is a transmembrane receptor of T and NK cells involved in modulating immune responses (27, 28). The four SNPs with the highest likelihood in the entire dataset were all intronic variants present in the MFSD1, which encodes a transmembrane protein in the major facilitator superfamily. Several paralogs of this gene have been identified in a genome-wide RNAi screen for genes affecting replication of vaccinia virus, a poxvirus related to MYXV (29). Using RNAi, we found that this gene also modulated MYXV titres in rabbit cells (Fig. S6e).
Selection on proviral genes
Resistance to viruses can evolve not only through improvement of antiviral defences, but also through changes in host proviral proteins that viruses hijack for their own benefit. Among our significant associations was a SNP in the 3’ prime UTR of VPS4 (Fig. 3c). This gene has no known role in the replication cycle of poxviruses, but is required for the envelopment of herpes simplex virus (HSV) in the cytoplasm (30). To evaluate the role of VPS4 in MYXV replication we took advantage of a human cell line expressing a dominant-negative form of the VPS4 protein (30). By comparing the effect of MYXV in wild-type and VPS4 dominant-negative human cell lines, we found that the latter strongly inhibited MYXV replication (Figs. 4c, 4d, S6c and S6d). In contrast to HSV, the effect at a high multiplicity of infection suggests VPS4 may affect earlier replication steps in MYXV. Therefore, the VPS4 protein is proviral, and it is possible that selection by MYXV may have altered its expression in the modern rabbit populations.
The proteasome is required both for poxvirus uncoating after the cell entry (31) and the processing of antigenic peptides, and three variants in the gene PSMG3 (pac3, proteasome assembly chaperone 3) increased 83% in frequency in France, with smaller changes in other populations. Using Sanger sequencing we found that these SNPs were associated with a 7-base pair insertion in the first exon and a 50-base pair deletion spanning the first intron and second exon. However, since genome annotation of this gene is incomplete the importance of these changes is unclear.
Conclusions
The myxomatosis pandemic caused massive mortality in rabbit populations, leading to the evolution of genetic resistance to the disease in Australia, France and the UK. We have found that over the last 60 years, the convergent phenotype of viral resistance in these populations was also accompanied by parallel genetic changes. This was a consequence of natural selection acting on standing genetic variation that was present in the ancestral rabbit populations in continental Europe and was retained in the subsequent colonisation process of the UK and Australia. The presence of this variation likely explains the rapid development of resistance to myxomatosis observed in rabbit populations almost immediately after the first outbreaks and may frequently be critical to allow populations to respond to novel pathogens. This is in striking contrast to the evolution of the virus where parallel changes in MYXV virulence do not have a common genetic basis (15).
Despite rapid phenotypic evolution, only 1% of the alleles favoured by selection have reached fixation in any of the modern populations (Fig. 3c, 3 out of 193 SNPs). Together with our estimates of selection coefficients and the moderate antiviral effect of the three interferon SNPs, this suggests genetic resistance to myxomatosis is a polygenic trait resulting from the cumulative effect of multiple alleles shifting in frequency across the genome, as opposed to a few major-effect changes to the immune response. Such adaptation is likely to result in a gradual ‘quantitative’ increase in resistance. When resistance reduces the within–host replication rate of the virus rather than completely preventing infection, selection will favour increases in viral virulence (32). Consistent with this prediction, it has recently been observed in wild populations of rabbits that the decline in virulence seen in the years immediately after the virus was released has been reversed and highly virulent viral genotypes have emerged (7).
The evolution of resistance to MYXV is associated with enhanced innate antiviral immunity (6). Homologs of CD96, FCRL, CD200-R and IFN-α all play a regulatory role in the innate immune response, including effects on lymphocyte proliferation and activation (21, 26, 27, 33). The increased virulence seen in recent MYXV isolates is associated with the virus becoming extremely immunosuppressive, causing the loss of cellular inflammatory responses, lymphocytes and neutrophils (7). Therefore, viral evolution may have found ways to counter the effect of many of the genetic adaptations that we have observed. In conclusion, our results reveal how standing genetic variation in the immune system allowed populations to rapidly evolve resistance to a novel and highly virulent pathogen and describe the molecular and genetic basis of an iconic example of natural selection.
Supplementary Material
Acknowledgements
We thank the Australian Museum, American Museum of Natural History, Booth Museum of Natural History, Natural History Museum (London), Museum of Comparative Zoology (Harvard University), Musée des Confluences, Muséum National d'Histoire Naturelle (Paris), Museum Victoria, Queensland Museum, Museum of Zoology (University of Michigan), Smithsonian Institution National Museum of Natural History and all the curators and museum technicians who generously sampled and provided historical samples. Marie-Dominique Wandhammer from Musée Zoologique de la Ville de Strasbourg that helped to track historical French samples. The British Association for Shooting and Conservation (BASC), Amanda Holroyd, Simon Whitehead and all volunteers that contributed with modern rabbit samples. Peter Kerr provided rabbit samples from NSW (Australia) from before RHDV. Peter Elsworth and Will Dobbie contributed Queensland post-RHDV samples. Colin Crump provided human cells lines expressing VPS4. Rute Fonseca provided valuable advice concerning ancient DNA bioinformatics.
Funding: This work was funded by grants from the Programa Operacional Potencial Humano–Quadro de Referência Estratégica Nacional funds from the European Social Fund and Portuguese Ministério da Ciência, Tecnologia e Ensino Superior to M.C. (IF/00283/2014/CP1256/CT0012), to P.J.E. (IF/00376/2015) and to J.M.A. (SFRH/BD/72381/2010). J.M.A. was supported by a travel grant from the Middleton Fund (Cambridge) to undertake work in the Centre of GeoGenetics (Copenhagen), AM was supported by the European Research Council (grant 647787-LocalAdaptation). FJ was supported by the European Research Council (grant 281668). LL was supported by the European Research Council grant (339941-ADAPT). McFadden Lab is supported by National Institute of Health (NIH) grant R01 AI080607. S.C.G. holds a Sir Henry Dale Fellowship, co-funded by the Wellcome Trust and the Royal Society (098406/Z/12/Z).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: None declared.
Author contributions: J.M.A, M.C., P.E., N.F., and F.M.J. designed and led the study. J.M.A, P.F.C., N.W., S.A., J.P.D and M.T.P.G. generated sequencing data. J.M.A., T.S., D.J.B, S.J.F, S.M., W.J.P., G.Q., A.K.S. did fieldwork and extracted DNA. A.L.M., M.M.R, S.C.G., L.B. and G.M., performed the experiments in cell culture. J.M.A., M.C., J.Y.C., L.L., A.E., A.M., F.G.V., R.N., and F.M.J. analysed the data. J.M.A., M.C. and F.M.J. wrote the paper with input from the other authors. All authors approved the manuscript before submission.
Data and materials availability: Original sequence data are available in the Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) under BioProject PRJNA393806 (SRP118358). The variant calls are available in the Cambridge Data Repository (https://doi.org/10.17863/CAM.35707). French modern samples are available from S.M. under a material agreement with Office national de la chasse et de la faune sauvage.
References and Notes
- 1.Fenner F, Ratcliffe FN. Myxomatosis. Cambridge University Press; Cambridge ; New York: 1965. [Google Scholar]
- 2.Fenner F, Fantini B. Biological Control of Vertebrate Pests: The History of Myxomatosis - an Experiment in Evolution. CABI publishing; New York, NY, USA: 1999. [Google Scholar]
- 3.Ross J, Sanders MF. The development of genetic resistance to myxomatosis in wild rabbits in Britain. J Hyg (Lond) 1984;92:255–261. doi: 10.1017/s0022172400064494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenner F, Ross J. In: The European rabbit: The History and Biology of a Successful Colonizer. Thompson HV, King CM, editors. Oxford University Press; Oxford ; New York: 1994. pp. 205–239. [Google Scholar]
- 5.Marshall ID, Fenner F. Studies in the epidemiology of infectious myxomatosis of rabbits. V. Changes in the innate resistance of Australian wild rabbits exposed to myxomatosis. J Hyg (Lond) 1958;56:288–302. doi: 10.1017/s0022172400037773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerr PJ. Myxomatosis in Australia and Europe: a model for emerging infectious diseases. Antiviral Res. 2012;93:387–415. doi: 10.1016/j.antiviral.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Kerr PJ, et al. Next step in the ongoing arms race between myxoma virus and wild rabbits in Australia is a novel disease phenotype. Proc Natl Acad Sci USA. 2017;114:9397–9402. doi: 10.1073/pnas.1710336114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veale EM. The Rabbit in England. Agric Hist Rev. 1957;5:85–90. [Google Scholar]
- 9.Myers K, Parer I, Wood P, Cooke BD. In: The European rabbit: The History and Biology of a Successful Colonizer. Thompson HV, King CM, editors. Oxford University Press; Oxford ; New York: 1994. pp. 108–157. [Google Scholar]
- 10.Carneiro M, Ferrand N, Nachman MW. Recombination and speciation: loci near centromeres are more differentiated than loci near telomeres between subspecies of the European rabbit (Oryctolagus cuniculus) Genetics. 2009;181:593–606. doi: 10.1534/genetics.108.096826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 12.Hill AVS. Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Phil Trans R Soc B. 2012;367:840–849. doi: 10.1098/rstb.2011.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magwire MM, et al. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 2012;8:e1003057. doi: 10.1371/journal.pgen.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens JC, et al. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet. 1998;62:1507–1515. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr PJ, et al. Evolutionary history and attenuation of myxoma virus on two continents. PLoS Pathog. 2012;8:e1002950. doi: 10.1371/journal.ppat.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrantes J, van der Loo W, Le Pendu J, Esteves PJ. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res. 2012;43:12. doi: 10.1186/1297-9716-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller HE, Chasey D, Lucas MH, Gibbens JC. Rabbit haemorrhagic disease in the United Kingdom. Vet Rec. 1993;133:611–613. [PubMed] [Google Scholar]
- 18.Morisse JP, Le Gall G, Boilletot E. Hepatitis of viral origin in Leporidae: introduction and aetiological hypotheses. Rev Sci Tech. 1991;10:269–310. [PubMed] [Google Scholar]
- 19.Loog L, et al. Inferring Allele Frequency Trajectories from Ancient DNA Indicates That Selection on a Chicken Gene Coincided with Changes in Medieval Husbandry Practices. Mol Biol Evol. 2017;34:1981–1990. doi: 10.1093/molbev/msx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith GL, Symons JA, Alcamí A. Poxviruses: interfering with interferon. Semin Immunol. 1998;8:409–418. [Google Scholar]
- 21.Upton C, Mossman K, McFadden G. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 22.Rahman MM, Liu J, Chan WM, Rothenburg S, McFadden G. Myxoma virus protein M029 is a dual function immunomodulator that inhibits PKR and also conscripts RHA/DHX9 to promote expanded host tropism and viral replication. PLoS Pathog. 2013;9:e1003465. doi: 10.1371/journal.ppat.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatherley D, Lea SM, Johnson S, Barclay AN. Structures of CD200/CD200 receptor family and implications for topology, regulation, and evolution. Structure. 2013;21:820–832. doi: 10.1016/j.str.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatherley D, Barclay AN. The CD200 and CD200 receptor cell surface proteins interact through their N-terminal immunoglobulin-like domains. Eur J Immunol. 2004;34:1688–1694. doi: 10.1002/eji.200425080. [DOI] [PubMed] [Google Scholar]
- 25.Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- 26.Kochi Y, et al. FCRL3, an autoimmune susceptibility gene, has inhibitory potential on B-cell receptor-mediated signaling. J Immunol. 2009;183:5502–5510. doi: 10.4049/jimmunol.0901982. [DOI] [PubMed] [Google Scholar]
- 27.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs A, Colonna M. The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance. Semin Cancer Biol. 2006;16:359–366. doi: 10.1016/j.semcancer.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Sivan G, et al. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc Natl Acad Sci USA. 2013;110:3519–3524. doi: 10.1073/pnas.1300708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crump CM, Yates C, Minson T. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol. 2007;81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt FI, et al. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. 2013;4:464–476. doi: 10.1016/j.celrep.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Gandon S, Michalakis Y. Evolution of parasite virulence against qualitative or quantitative host resistance. Proc Biol Sci. 2000;267:985–990. doi: 10.1098/rspb.2000.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stack G, et al. CD200 receptor restriction of myeloid cell responses antagonizes antiviral immunity and facilitates cytomegalovirus persistence within mucosal tissue. PLoS Pathog. 2015;11:e1004641. doi: 10.1371/journal.ppat.1004641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. available at http://www.R-project.org/ [Google Scholar]
- 35.Becker RA, Wilks AR, Brownrigg R, Minka TP. Maps: draw geographical maps. R package version 2.3-9. 2010 [Google Scholar]
- 36.Brownrigg R. Mapdata: Extra Map Databases, R Package Version 2.2-5. 2013 [Google Scholar]
- 37.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 38.Wales N, et al. New insights on single-stranded versus double-stranded DNA library preparation for ancient DNA. BioTechniques. 2015;59:368–371. doi: 10.2144/000114364. [DOI] [PubMed] [Google Scholar]
- 39.Carneiro M, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345:1074–1079. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 available at http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc.
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.H IM, Quinlan Aaron R. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Briggs AW, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 49.Schwalb B, et al. Package “LSD”. 2015 [Google Scholar]
- 50.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng JY, Mailund T, Nielsen R. Fast admixture analysis and population tree estimation for SNP and NGS data. Bioinformatics. 2017;33:2148–2155. doi: 10.1093/bioinformatics/btx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. 2014:1–2. [Google Scholar]
- 54.Soetaert K, Petzoldt T, Setzer RW. Solving differential equations in R: package deSolve. Journal of Statistical Software. 2010 [Google Scholar]
- 55.Rahman MM, McFadden G. Myxoma Virus dsRNA Binding Protein M029 Inhibits the Type I IFN-Induced Antiviral State in a Highly Species-Specific Fashion. Viruses. 2017;9 doi: 10.3390/v9020027. [DOI] [Google Scholar]
- 56.Liu J, et al. Myxoma virus expressing interleukin-15 fails to cause lethal myxomatosis in European rabbits. J Virol. 2009;83:5933–5938. doi: 10.1128/JVI.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zemp FJ, et al. Treating brain tumor-initiating cells using a combination of myxoma virus and rapamycin. Neuro-oncology. 2013;15:904–920. doi: 10.1093/neuonc/not035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bond CS, Schüttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.