Abstract
Genomic features are used as biomarkers of sensitivity to kinase inhibitors used widely to treat human cancer, but effective patient stratification based on these principles remains limited in impact. Insofar as kinase inhibitors interfere with signaling dynamics, and, in turn, signaling dynamics affects inhibitor responses, we investigated associations in this study between cell-specific dynamic signaling pathways and drug sensitivity. Specifically, we measured 14 phosphoproteins under 43 different perturbed conditions (combinations of 5 stimuli and 7 inhibitors) in 14 colorectal cancer cell lines, building cell line-specific dynamic logic models of underlying signaling networks. Model parameters representing pathway dynamics were used as features to predict sensitivity to a panel of 27 drugs. Specific parameters of signaling dynamics correlated strongly with drug sensitivity for 14 of the drugs, 9 of which had no genomic biomarker. Following one of these associations, we validated a drug combination predicted to overcome resistance to MEK inhibitors by co-blockade of GSK3, which was not found based on associations with genomic data. These results suggest that, in order to better understand cancer resistance and move toward personalized medicine, it is essential to consider signaling network dynamics that can not be inferred from static genotypes.
Keywords: cancer resistance, combinatorial therapy, signaling networks, computational models, biomarkers
Introduction
Patient response to anticancer therapies is extremely variable and understanding the reasons for this variability is a major challenge in cancer research. One approach to address this problem is to identify biomarkers that correlate with therapy response. However, except for a few examples, no efficient biomarkers are available (1). The problem of finding biomarkers has become even more important with the advent of targeted cancer therapies that are designed to affect specific molecular changes that drive the cancer, providing a larger number of treatment options. However, biomarkers that can be used to stratify patients for targeted drugs also remain largely elusive (2).
The most common approaches for patient stratification are currently based on genomic biomarkers, typically consisting of either copy number alteration or mutation of specific genes. The popularity of those biomarkers has been favored by the advancements of sequencing technologies and their subsequent decrease in cost. While for some drugs they have shown strong potential (3–5), for many other cases no efficient genomic marker for patient stratification exists, and for those where markers exist, they have rather low power due to the complex nature of cancer. Furthermore, their actual clinical significance for precision oncology is questionable (1).
Many targeted therapies inhibit specific signaling molecules, mostly kinases, and elicit a response that can affect and rewire large parts of the signaling network. Therefore, investigating the dynamics of signaling pathways can help to unveil new therapeutic strategies and biomarkers (6,7). Understanding how drugs affect the signaling network as a whole, is particularly important as signaling dynamic differs not only between tumors of different tissue, but also between tumors of the same tissue. In fact, for different cancer cell-types, the dynamics of signaling networks have been shown to differ strongly between different cells of the same tissue of origin (8–11).
Furthermore, models parameterized and experimentally calibrated using a specific cell line have been used to define pathway dynamic biomarkers of therapeutic outcome, such as drug sensitivity (12) or patient survival (13), based on model simulations in breast cancer and neuroblastoma, respectively.
The understanding of mechanisms of cellular response (network structure and dynamic behavior) can also be useful to tackle the development of compensatory signaling mechanisms of drug resistance (14), which is a recurrent problem for targeted therapies and is challenging to predict. Signal transduction is carried out by complex dynamic networks, and mutations in oncogenes or tumor suppressors can drastically change the properties of the network. This complex wiring confers robustness to the cells that often allows them to escape single-agent-targeted treatments (15,16). Based on this idea, modeling of signaling pathways has been recently used to suggest combinatorial targeted therapies to effectively block multiple molecular pathways (8,17–20): by integration of prior pathway knowledge with experimental observations cell line-specific models were built, which could be used to simulate and thus prioritize possible combinatorial perturbation experiments.
In this paper, we investigated to what extent dynamic interactions between different signaling pathways play a role in characterizing the specific cellular responses to drugs and how this can be used to suggest targeted combinatorial therapies. Furthermore we assessed how these dynamic features of signaling fare against static genomic traits as markers of drug response. We did so by characterizing cell-type-specific models for a panel of 14 different colorectal cancer (CRC) cell lines that are integrated with a large-scale drug screening (21). We found associations with model parameters for 14 of the 27 drugs targeting our pathways of interests, for 9 of which there were no genomic biomarkers. These associations were used to define pathway dynamic biomarkers and to propose combinatorial therapies.
Materials and methods
Cell lines
All cell lines used in this study (CAR1, CCK81, COLO-320-HSR, DIFI, HCT116, HT115, HT29, SK-CO-1, SNU-C5, SNU-C2B, SW1116, SW1463, SW620, SW837) are mycoplasma negative. To exclude cross-contaminated or synonymous lines, a panel of 92 SNPs was profiled for each cell line (Sequenom, San Diego, CA) and a pair-wise comparison score calculated for in-house identity checking across a panel of 1000 cell lines (21). We confirmed the identity of cancer cell line set against those provided by the repositories, where possible, using a panel of 16 STRs (AmpFLSTR Identifiler KIT, ABI), which includes the 9 currently used by most of the cell line repositories (ATCC, Riken, JCRB and DSMZ). STR or SNP data are available at COSMIC database (http://cancer.sanger.ac.uk/cell_lines). All cell lines are sourced from collaborators or repositories; they have been used for COSMIC cell line project and GDSC project (21) and have been stored in the lab as cryopreserved aliquots in liquid N2 for 7 years. A single cryovial is thawed for use and propagated for a maximum of 3 months before being discarded.
Experimental setup of perturbation screen
Cell lines were cultured in either RPMI or DMEM/F12 medium with 10% FBS and were incubated at 37°C and 5% CO2. Before perturbation commenced cells were starved overnight in serum free medium. At 90 minutes before lysis the cells were treated with inhibitors (or solvent control DMSO) and at 30 minutes before lysis cells were stimulated with ligand (or solvent control H2O). This procedure was conducted for all single inhibitor and combined inhibitor-ligand perturbations. The following inhibitors were used: MEKi AZD6244 (4µM), PI3Ki AZD6482 (10µM), mTORi AZD8055 (2µM), TBK1i BX-795 (10µM), IKKi BMS-345541 (25µM), BRAFi PLX4720 (5µM) and TAK1i 5Z-7-Oxozeanol (5µM). Concentrations were selected to inhibit target while minimizing off-target activity. We used the following ligands: EGF (25ng/ml), HGF (50ng/ml), IGF1 (10ng/ml), TGFb (5ng/ml) and TNFa (10ng/ml). After treatment and incubation, lysates were collected and analyzed with the Bio-Plex Protein Array system (Bio-Rad, Hercules, CA) as described in (8) using magnetic beads specific for AKTS473, c-JunS63, EGFRY1068, ERK1/2T202,Y204/T185,Y187, GSK3A/BS21/S9, IkBaS32,S36, JNKT183,Y185, MEK1S217,S221, mTORS2448, p38T180,Y182, PI3KY458, RPS6S235,S236, p90RSKS380 and SMAD2S465,S467. The beads and detection antibodies were diluted 1:3. For data acquisition, the Bio-Plex Manager software and R package lxb was used.
Data preprocessing
For each cell line, data were processed separately for each of the 14 measured phosphoproteins. The value of the control (unperturbed condition) was estimated as the median value of the 4 replicates and log2 fold changes with respect to the control were computed for each of the 42 perturbed conditions (data in Supplementary Table S1). As required by our logic formalism, data were linearly scaled between 0 and 1, with 0.5 corresponding to the basal (control) condition.
Definition of logic ordinary differential equations
The logic ordinary differential equation formalism (22) is based on ordinary differential equations derived from logic models using a continuous update function Bi for each species (i.e. node in the network) xi, which can assume continuous values {0,1}. Differential equation for species i is defined as follow:
Where τi is the life-time of species i: τi =0 corresponds to a node not responsive to activation by upstream nodes and higher values of τi represent faster response, interpretable as more functional species. x1,i, x2,i, …, xN,i are the N regulators of xi and each regulation is defined by a sigmoidal transfer function f(xj,i) (Supplementary Fig. S1), where parameter kj,i characterizes the strength of the regulation of species i dependent on species j (i.e. edge j → i): kj,i=0 means no regulation, while higher values represent stronger interactions.
Optimization of parameters τi and kj,i was performed using a modified version of the R package CNORode (add-on to CellNOptR (23)) and the optimization toolbox MEIGO (24). In order to improve parameter estimates, we redefined the objective function (i.e. sum of the squared difference between model predictions and measured values, QLS) introducing an L1 penalty term on the parameters (L1 norm is the sum of the absolute values of the parameters, P). The balance between prioritizing good fit (low QLS) or sparse model (low P) is regulated by a tunable parameter λ, with new objective function:
Additionally, the objective function includes a penalty to promote that the simulated values at t=30 min (corresponding to the measured time point) are at steady state in order to match the experimental assumptions. The regularization was performed sequentially first on τi (to remove unresponsive nodes), and then, fixing τi, on kj,i (to induce edge sparsity). At each step the term λ was tuned to guarantee a good compromise between goodness of fit and model sparsity (Supplementary Fig. S2 - S3). Finally, bootstrap distribution was obtained for parameters kj,i by repeating the optimization on resampled experimental data with replacement 150 times. More details are in Supplementary Methods, and code for modified version of the CNORode package at https://github.com/saezlab/CNORode2017.
Elastic net
In order to select only robust features, Elastic net (25) training was iterated M times (M is the total number of observations), each time leaving out one observation (sampling without replacement). For each iteration, leave-one-out cross-validation was applied to tune Elastic Net hyperparameters (using caret R package). Median values were computed for the weights of the features across the M iterations, in order to select only features appearing in at least half of the iterations.
Statistical Analysis
ANOVA was used to define genomic markers of drug sensitivity using the GDSC tools (http://gdsctools.readthedocs.io/). Microsatellite instability was used as covariate and threshold on minimum size of the positive and negative population for each feature set to 3. The threshold for significance of the p-value was set to 0.05, but no correction for multiple hypothesis testing was applied.
Kruskal-Wallis rank sum test (one-way analysis of variance on ranks) was used to test if estimated parameters (from bootstrap) derive from the same distribution for all 14 cell lines (null hypothesis rejected if different for at least one group). Effect size w was computed as where χ2 is the statistics from the test and N is the number of observations. Effect size >0.5 is considered large. P-values were Bonferroni corrected to compensate for multiple comparisons, and threshold set to 0.05. Wilcoxon rank sum test was then used for post hoc pairwise test on parameters passing Kruskal-Wallis rank sum test, and effect size where Z is the statistics from the test and N is the number of observations. Effect size >0.5 is considered large. P-values were Bonferroni corrected and threshold was set to 0.05. Rank type test were preferred over parametric tests because they are highly robust against non-normality.
Drug combination sensitivity experiments
Experiments were performed using MEKi Trametinib (0.005 μM) as anchor drug, testing its effect on our 14 colorectal cancer cell lines alone and in combination with 5 concentrations (from 10 μM with 4-fold dilutions: 0.039 μM, 0.156 μM, 0.625 μM, 2.5 μM, 10 μM) of two GSKi (SB 216763 and CHIR-99021). For available replicates median value was computed. Dose-response curves were normalized between 0 and 100 with 100 corresponding to the negative control (cells with no treatment) and 0 to the positive control (cells treated with Staurosporine 2 μM). Same DMSO volume was maintained for all experiments (negative and positive controls, single drug and drug combination). Cell number was measured after 3 days of constant drug exposure using CellTiter-Glo reagent as described by the manufacturer (Promega).
Results
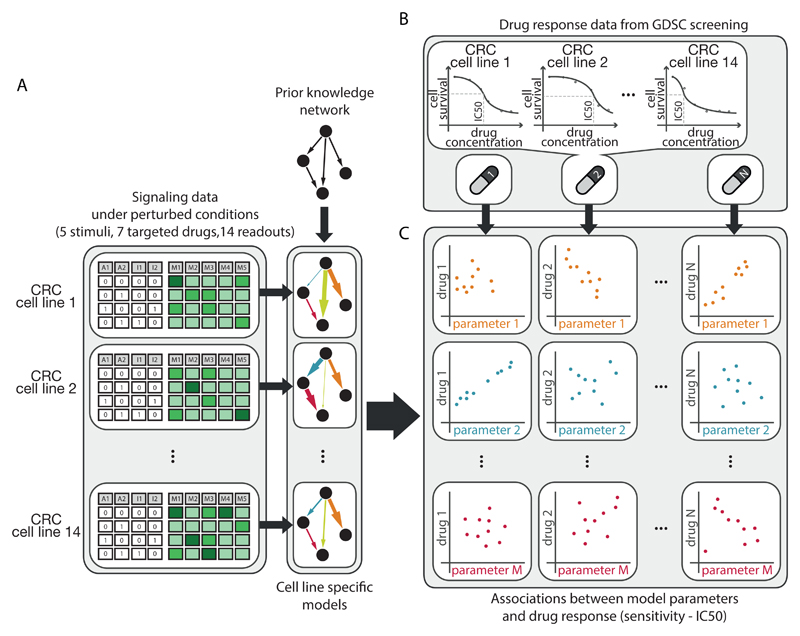
To identify biomarkers of drug response from dynamic logic models, we proceeded as follows (Fig. 1). First, for 14 colorectal cancer (CRC) cell lines, we systematically obtained antibody-based signaling data on how key molecules respond to perturbations in the signaling network. By combining this data with a literature-derived prior knowledge network, we generated cell line-specific computational models for each of the 14 cell lines. Each model consists of logic ordinary differential equations (22) representing the dynamics of the signaling network, which was parameterized and refined by fitting it to the experimental data using CellNOpt (23) (Fig. 1A). Second, we retrieved drug sensitivity data for these 14 cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) (21), filtering for drugs targeting kinases acting either inside the modeled network or as direct neighbors (Fig. 1B). Third, we investigated correlations between model parameters and drug response to select strong associations using Elastic Net regression (25) (Fig. 1C). All steps will be detailed in the following sections.
Fig. 1.
Schematic of the approach used to define biomarkers of drug response from pathway models. A. Specific predictive models for 14 colorectal cancer (CRC) cell lines were built from perturbation data and prior information about network structure (PKN). B. Drug sensitivity data for the same 14 CRC cell lines were retrieved from the Genomics of Drug Sensitivity in Cancer (GDSC) panel in response to 27 different drugs targeting the PKN or first neighbor. C. Elastic Net was used to select strong associations between model parameters and drug sensitivity.
Systematic perturbation data for signaling in a large colon cancer cell line panel
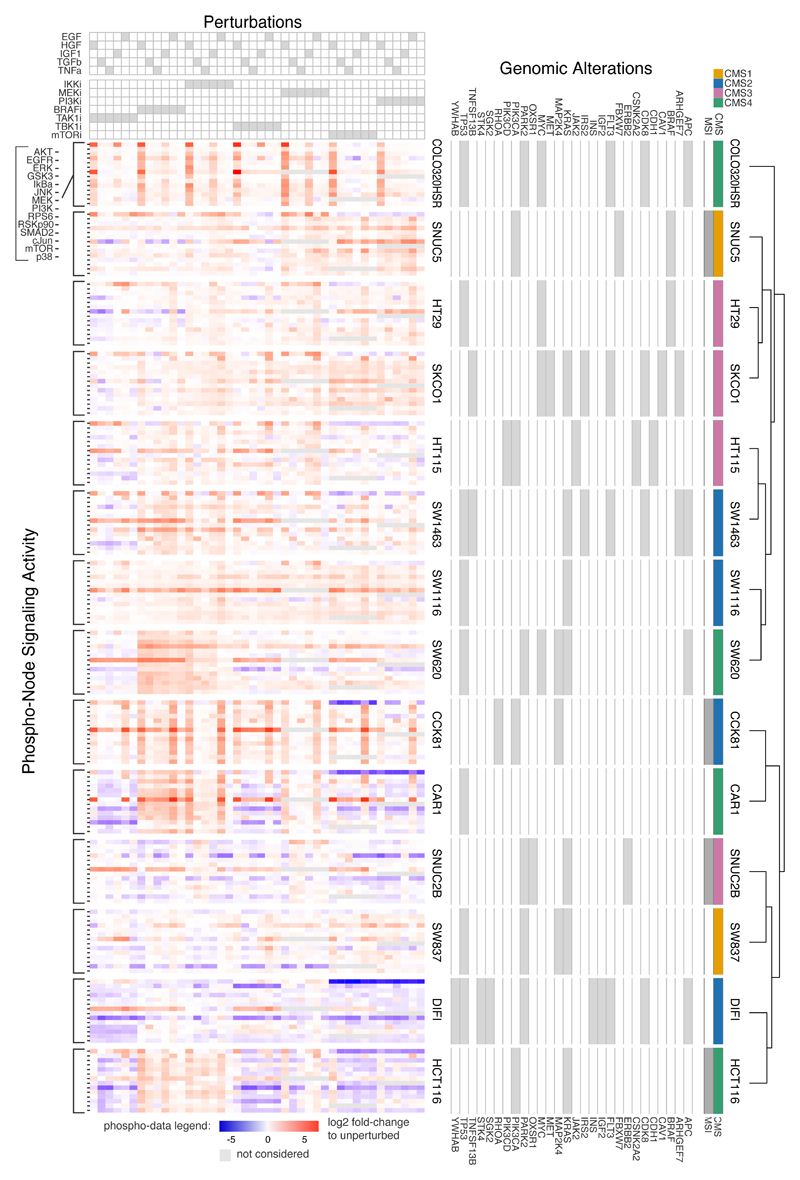
To generate a solid dataset to build cell line specific signaling models and characterize signaling heterogeneity in colon cancer, we performed a large-scale signaling perturbation screening in a panel of 14 genetically heterogeneous colorectal cancer (CRC) cell lines. We chose the cell lines to reflect the heterogeneity of colon cancer (26), covering typical recurrent mutations in CRC (such as KRAS, TP53, APC, BRAF). Four cell lines had Microsatellite Instabilities (MSI), and our cell lines covered all classes of the consensus molecular subtype (CMS) described for CRC patients (27) and computed for CRC cell lines (28), see Fig. 2.
Fig. 2.
Large-scale experimental perturbation dataset. Data for 14 colorectal cancer cell lines in response to 43 different perturbed conditions with 14 measured phosphoproteins. Genomic alterations affecting investigated pathways are also shown along with molecular subtypes. Cell lines are clustered based on phosphorylation profile across all perturbations.
Our perturbation data consists of changes in 14 phosphoproteins measured after the signalling network was systematically perturbed in 43 different scenarios. Perturbations consist of combinations of 7 small-molecule inhibitors (targeting the kinases IKK, MEK, PI3K, BRAF, TAK1, TBK1, mTOR) and 5 ligands stimulating receptors (EGF, HGF, IGF1, TGFb, TNFa), see details in Materials and Methods. Altogether we obtained 8428 different experimental data points. When visualizing the perturbation-response profiles (Fig. 2), we noticed that they differ strongly between cell lines. For example, several cell lines showed strong attenuation of AKT signaling upon mTOR and PI3K inhibition (blue areas in the top right of each cell line, such as for CAR1, HCT116 and DIFI), while others did not (such as SW837 and SW620). Notably, these differences were not correlated with consensus subtype or mutation status of e.g. PIK3CA. Cluster analyses of the perturbation response profiles also unveil no clear correlation between genotype data or consensus subtype and perturbation response data, highlighting the idea that the state and responsiveness of the signaling is not fully determined by mutations, and that the obtained data set provides information that is not contained in genomic data.
We next aimed to build computational network models from the perturbation-response signaling data, in order to get more insights into the underlying changes in the signaling networks that lead to these heterogeneous responses.
Compiling a signaling network as a basis for the computational model
As a basis to model the signaling network, we compiled a comprehensive prior-knowledge network (PKN, shown in Supplementary Fig. S4) including our measured and perturbed pathways and their connections, using literature and OmniPath (29), a curated ensemble of multiple pathway resources. Overall, the network had 49 nodes (signaling molecules) and 86 edges (regulatory interactions) and could be interpreted as a logic model. As the PKN contained many nodes that were neither measured nor perturbed, we compressed it as described in (30) to reduce model complexity without affecting logic consistency, while maintaining interactions between measured or perturbed nodes. The model definition considered that PLX4720, a selective BRAF inhibitor, inhibits mutated BRAF (V600E), but induces paradoxical activation in wild type BRAF cells (31).
Using the final network, we created a mathematical model that represents the dynamics of the signaling network using logic ordinary differential equations (ODEs) (22). For each node in the network, this model has one parameter τi representing the responsiveness of node i, and for each interaction, there is one parameter kj,i describing the strength of the edge from node j to node i (see Materials and Methods). Note that this model represents a rather generic signaling network, and it is likely that not all interactions are functional in a specific cell line.
Strategy for model optimization: an in silico example
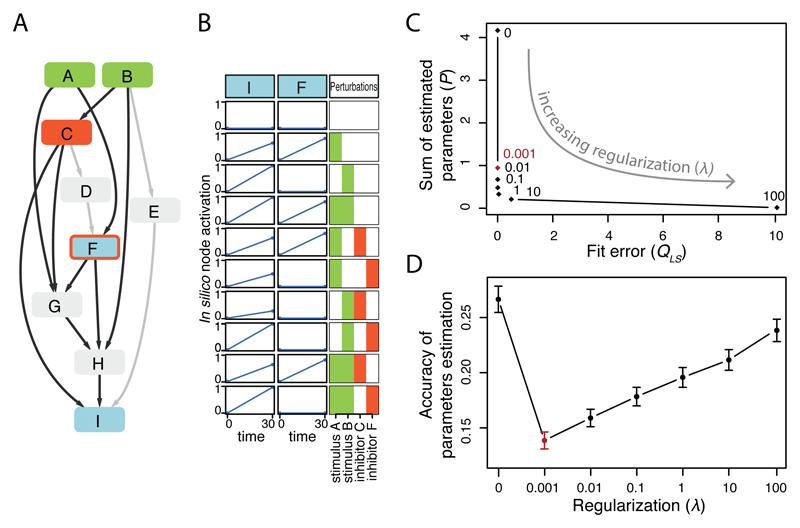
After having established a generic computational model for signaling dynamics, we next aimed to adjust the parameters for each of the 14 cell lines so that we obtain 14 cell line-specific models. A major challenge when attempting to parameterize such large-scale networks is to avoid overfitting, i.e. to estimate too many parameters from insufficient data. Building on (23,32) we therefore developed a regularization scheme that favors sparser networks (i.e. networks that contain fewer edges and nodes). This is done by adding to the objective function the sum of the absolute parameter values multiplied by a parameter (λ) (as described in Materials and Methods and Supplementary Material). This way, interactions or responses of nodes that are not supported by the data will be removed and the network will be pruned. The approach is illustrated and characterized in Fig. 3 using an in silico model, where the prior knowledge network (PKN) includes 9 nodes and 14 edges. To simulate that some interactions are present in a specific cell line, while others might not, we chose 10 edges (shown in black in Fig. 3A) to be present, and 4 to be absent to generate in silico data for 2 readouts (nodes shown in blue in Fig. 3A) recorded after perturbation with 2 stimuli (green nodes) and 2 inhibitors (targeting red nodes). In silico data contained in total 10 different perturbation scenarios and were generated for 10 random sets of parameters (one example shown in Fig. 3B).
Fig. 3.
In silico example to illustrate modeling approach. A. In silico general network (PKN) with edges used for data simulation highlighted in black. B. One (out of 10) example of simulated perturbation data used to illustrate L1 regularization. C. Goodness of fit (QLS, i.e. sum of squared residuals) versus model sparsity (P, i.e. sum of estimated parameters) for different values of tunable regularization term λ, (λ=0, 0.001, 0.01, 0.1, 10, 100) resulting in an L-shaped curve (each dot is the mean value obtained from the 10 in silico datasets). D. Mean accuracy (error bars represent standard deviation) of the estimated parameters (sum of the square of the difference between estimated and true parameters) as function of λ. Optimal λ is depicted in red.
When we did not include a penalty for complexity (corresponding to λ=0) the fit of the optimized models to the data was perfect (Fig. 3C with λ =0), however, parameters could not be well estimated due to model redundancy and low identifiability (Fig. 3D). However, when we increase the penalty for the node parameters τi using the regularization parameter λ (see Materials and Methods), we tend to have worse fit to the data (higher fit error QLS) and sparser models (lower sum of estimated parameters P), as shown in Fig. 3C. Good values of λ are those on the elbow of the L-shaped curve in Fig. 3C, corresponding to the best compromise between good fit and sparse model. Interestingly, these values (especially λ=0.001 which represents the most conservative choice in terms of regularization) also correspond to the best accuracy in estimating the parameters in Fig. 3D. Looking at the estimated parameters we could verify that for λ=0.001 the parameters τD and τE, corresponding to the unconnected nodes D and E, were indeed set to zero thanks to the regularization term in the objective function.
Generation of cell line-specific models
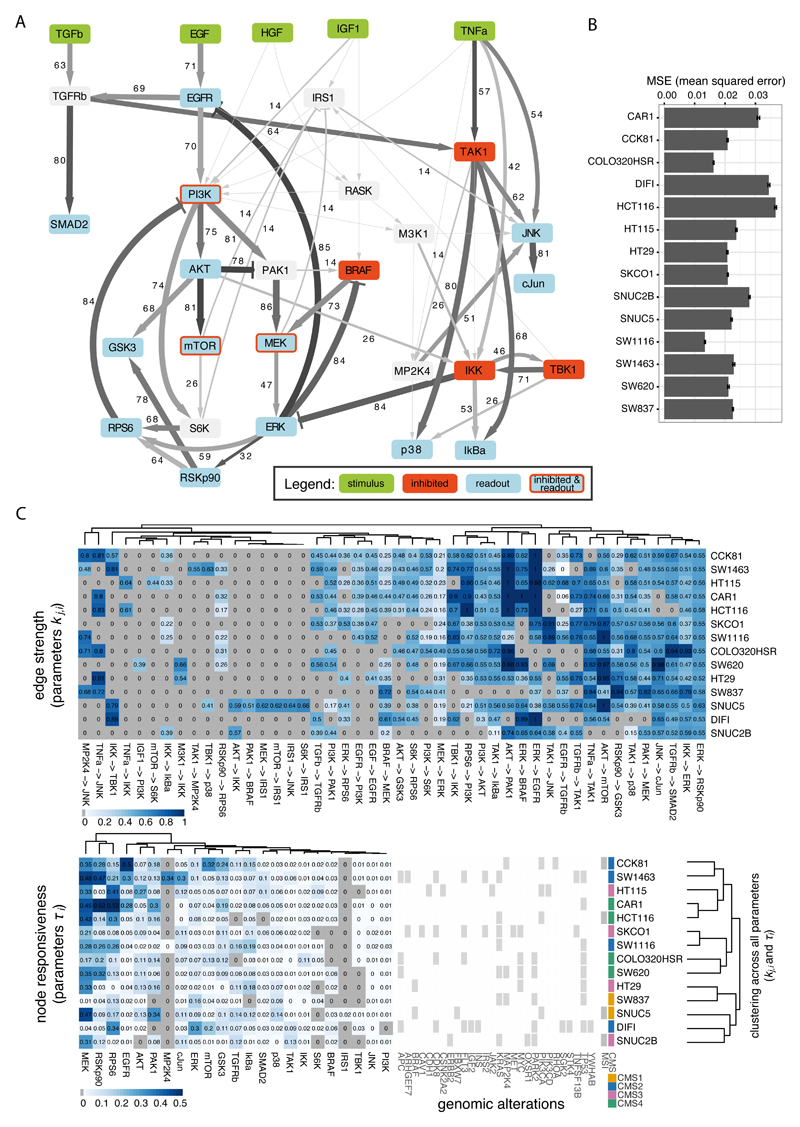
Having established a modeling framework that can handle the complexity of our data and network (Fig. 4A), we generated cell line-specific models for the 14 cell lines, performing also bootstrapping to obtain confidence intervals for the edge parameters kj,i (see details in Materials and Methods and Supplementary Methods). The resulting models were comparably sparse across cell lines, with a percentage of values set to zero ranging between 12-28% for node parameters τi and 51-73% for edge parameters kj,i. The optimized models showed good fit to the experimental data for all cell lines, with mean squared error (MSE) ranging between 0.013 and 0.036 (see Fig. 4B). Estimated median values for the edge parameters kj,i and node parameter values τi are shown in Fig. 4C and reported in Supplementary Table S2 for all cell lines. Cluster analysis of the model parameters re-emphasized that cell lines with similar genetic alterations are not associated with specific states of their signaling networks represented by the parameters (Fig. 4C).
Fig. 4.
Results of cell type specific model optimization. A. Compressed PKN with edge width and values representing the level of heterogeneity of the corresponding model parameter among cell lines (percentage of pairwise tests between cell lines for which null hypothesis of equal distribution was rejected) and edge color representing median value of the corresponding parameter across all cell lines. B. Mean squared error (MSE) for each cell line, error bars represent standard deviation derived from bootstrap. C. Estimated parameters kj,i and τi, representing edges strength and nodes responsiveness respectively, which are different from zero in at least one cell line. Clustering on the right is based on all estimated model parameters and does not correlate well with genomic alterations.
Heterogeneity of the cell line specific models
To further quantify how much the signaling networks differ between cell lines, we statistically investigated the heterogeneity of the estimated edge parameters kj,i. 46 of the 63 parameters showed a difference in at least one cell line (Kruskal-Wallis rank sum test, adjusted p-value < 0.05, effect size > 0.5, see Materials and Methods). Notably, all other 17 parameters were zero for all cell lines, indicating signaling interactions that are not functional in any of the colon cancer cell lines.
As an attempt to quantify heterogeneity, we tested how often each of the 46 parameters that show variability between cell lines, differs between pairs of cell lines (Wilcoxon rank sum post hoc test for all 91 combinations of 14 cell lines, adjusted p-value < 0.05 and effect size > 0.5, see Materials and Methods). The resulting percentages of pairwise differences are visualized as line width and numbers in Fig. 4A, while edge color intensity maps its mean value across cell lines. It is interesting to see that the major signaling routes are among the most variable between cells (PI3K/AKT and BRAF/MEK/ERK), however, also other often overlooked interactions such as feedbacks from RPS6 to PI3K or ERK to BRAF have high variability. The highly variable interaction between IKK and ERK may reflect variability of off-target effects of the chosen IKK inhibitor, as shown previously (8). Importantly for further analysis, we noted that only 34 of 46 edge parameters kj,i were different from zero in at least three cell lines, and decided to concentrate our further analysis on these highly heterogeneous parameters.
When investigating the parameters for the nodes, τi, only 1 out of 25 parameters was zero across all cell lines and 21 parameters were different from zero in at least three cell lines. Among these, 12 were defined as highly heterogeneous, showing reasonable variability across cell lines (standard deviation > 0.04). From our analysis, we conclude that cell lines are extremely heterogeneous and hence it is important to characterize each cell line with a specific signaling model.
Association of model parameters with drug response data
We next investigated if the highly heterogeneous model parameters can be associated with the efficacy of drugs. We reasoned that since many kinase inhibitors target the pathways that show highly variable parameters, these parameters might in turn determine the efficiency of these drugs. Drug sensitivity of our 14 colorectal cancer cell lines has been characterized for hundreds of drugs as part of the Genomics of Drug Sensitivity in Cancer (GDSC) project (21). From that data set, we selected those drugs that target molecules within our network, or molecules that are directly interacting with our network (based on Omnipath (29)). We excluded drugs with low variability in the response across our 14 cell lines (less than three sensitive and three resistant cell lines based on the classification used in (21)) and those for which sensitivity data were missing for more than three of our 14 cell lines. For the remaining 27 drugs, we investigated if the sensitivity of our cell lines (as measured by the IC50) is correlated with any of the model parameters (using cross-validated Elastic Net, details in Materials and Methods). Out of all 1242 potential correlations, we found in total 146 significant and robust parameter-drug associations. These associations covered 14 of our 27 drugs (Supplementary Fig. S5 - S6). We then performed a similar analysis to obtain associations between drugs and genomic alterations (functional mutations and copy number alterations) acting on PKN nodes or first neighbor (using ANOVA as described in (21), without correction for multiple hypothesis testing due to too large number of comparisons), we obtained associations for 12 of our 27 drugs, five of which also showed associations with model parameters (significant associations are reported in Supplementary Fig. S7).
Interpretation and validation of the dynamic biomarkers
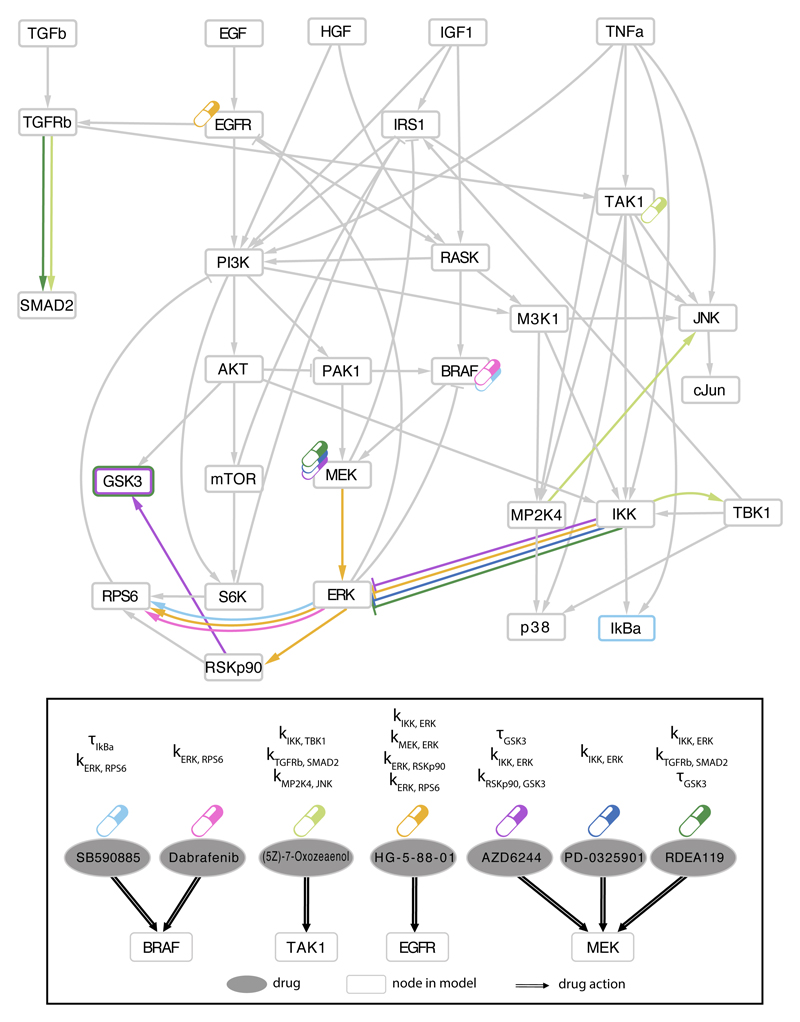
We then focused on the 9 drugs whose efficacy was not significantly associated with mutations or copy number alterations but instead with pathway biomarkers, to explore how our approach provides insights in otherwise unexplained drug efficacy. Top associations are illustrated in Fig. 5, visualized on the network (PKN) to show, for each drug, both the target(s) (indicated with pills) and the associated pathway parameter(s) (colored edges).
Fig. 5.
Associations between model parameters and drug sensitivity. Among the top 25 parameter-drug associations resulting from Elastic Net selection, those involving drugs with no genetic biomarkers are schematically mapped to the signaling pathways. Drug targets for the 7 drugs listed as ellipses in the bottom panel, are shown in the compressed PKN (upper panel) using the corresponding colored drug symbols. Corresponding model parameter associations for each drug are shown in the PKN using the same color code to mark edges corresponding to parameters kj,i (strength of edge from j to i) and nodes border for corresponding responsiveness parameters τi (node i).
For example, multiple parameters related with MEK-ERK pathway showed association with EGFR signaling inhibitor (HG-5-88-01) and with BRAF inhibitors (SB590885 and Dabrafenib). Interestingly, many recent studies reported synergistic effects when combining MEK inhibitors with EGFR inhibitors (8,33,34) or with BRAF inhibitors (8,35,36) in different cancer types. In particular, focusing on colorectal cancer, combination of EGFR and MEK inhibitors was suggested to overcome resistance to MEK inhibitors in BRAF and KRAS mutants (8,37) and it was suggested in (34) as potential therapy for CRC patients who become refractory to anti-EGFR therapies. A significant increase in response was also shown on a phase I study for CRC patients treated with a combination of three drugs targeting MEK, BRAF and EGFR respectively, in BRAF V600E mutants (38) and is currently in Phase I/II study (NCT01750918 in clinicaltrials.gov). These examples underline the potential of our approach in suggesting targeted combinatorial therapies.
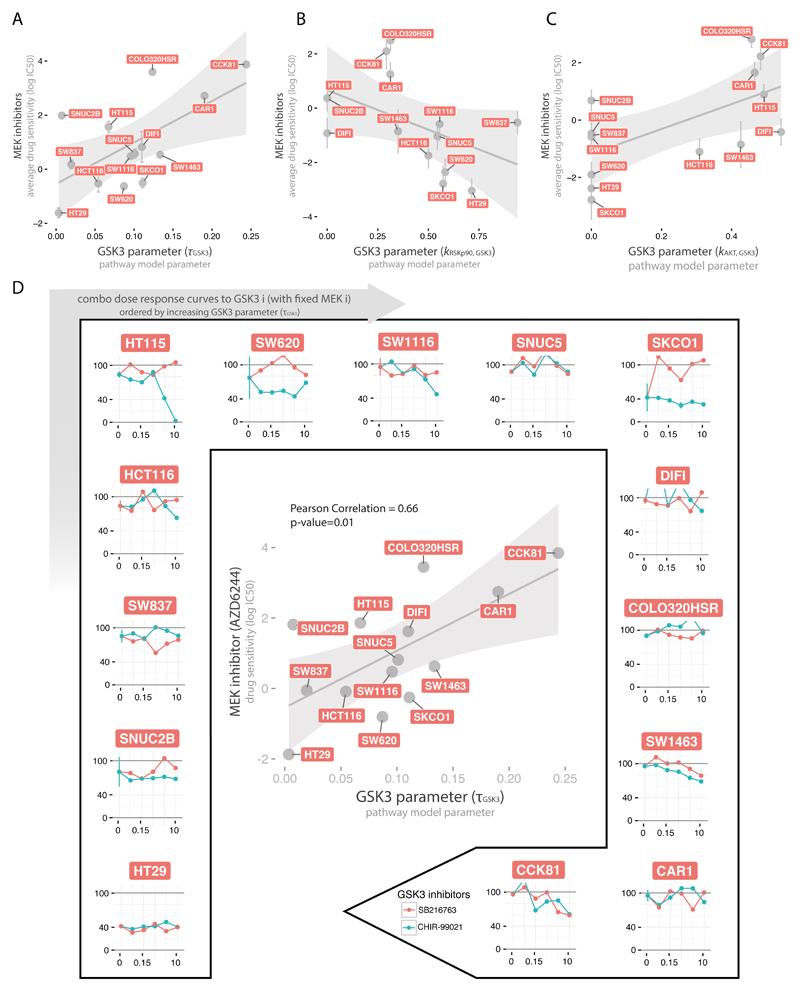
Another interesting example is the association of different MEK inhibitors (AZD6244, RDEA119, Trametinib, PD-0325901, CI-1040) with multiple model parameters related to GSK3 (τGSK3, kRSKp90,GSK3, kAKT,GSK3), as shown in Fig. 6A-C, with each parameter associated with at least two of the inhibitors. The top association among these, between AZD6244 and τGSK3, is shown in the center of Fig. 6D. Our interpretation of these associations is that inhibition of GSK3 would improve the sensitivity for cell lines resistant to MEK inhibitors (i.e. high IC50) and with functional GSK3 (i.e. high τGSK3). On the contrary, cell lines with nonfunctional GSK3 (i.e. low τGSK3) should not be affected by GSK3 inhibitors. In order to experimentally test this hypothesis, we combined a MEK inhibitor (Trametinib) with two GSK3 inhibitors (SB216763 and CHIR-99021) at increasing concentrations (Materials and Methods) as shown in the external plots in Fig. 6D. As expected, cell lines with nonfunctional GSK3 do not show an increased sensitivity when treated in combination with any of the two tested GSK3 inhibitors, regardless of their response to MEK inhibitors (e.g. HT29 and SNUC2B for a sensitive and a more resistant case, respectively). On the contrary, cell lines with more functional GSK3 tend to show an improved sensitivity under the combinatorial treatment (see HT115, SW1116, SW1463 and CCK81) even at the low drug concentrations tested.
Fig. 6.
Association between MEK inhibitors and GSK3 model parameters. A-C. Scatterplots of the three GSK3 related model parameter (τGSK3, kRSKp90,GSK3, kAKT,GSK3 in panel A, B and C respectively) versus the mean sensitivity of the respective associated MEK inhibitors (at least two for each parameter), with error bars representing the standard error. D. In the center, scatterplot of the strongest association between MEK inhibitor (AZD6244) and GSK3 node parameter (τGSK3). In the external plots, dose-response curves when combining MEK inhibitor (Trametinib) (at fixed concentration 0.005 μM) with two GSK3 inhibitors (SB216763 and CHIR-99021) at increasing concentrations (0, 0.039, 0.156, 0.625, 2.5, 10 μM). Plots for the 14 CRC cell lines are ordered clockwise according to their τGSK3 parameter value.
Discussion
It is broadly accepted that alterations in signaling pathways largely determine the efficacy of kinase inhibitors used in the clinic. Yet, in only few examples we have a clear understanding of how changes in signaling networks shape drug response. In this study we systematically investigated how the dynamics of signaling pathways can determine the efficacy of targeted drug treatments and found several examples where quantitative differences in signaling networks are associated with drug sensitivity that cannot be explained by genomic data. Additionally, we show that this information can be used to guide combinatorial therapies.
To uncover cell-specific changes in signaling, we constructed mechanistic models of signal transduction for 14 colorectal cancer cell lines based changes in phosphorylation in key signaling molecules upon perturbations. We built our models based on differential equations to capture the continuous aspects of signal strength and time-dynamics, but using a logic formalism that kept the model simple and with easily interpretable parameters. We could then study how these model parameters, which determine the dynamic behavior of the pathway, are related with the cellular sensitivity to cancer drugs. We found 146 strong associations between model parameters and drug sensitivity, including drugs without genetic markers. This suggests that differential wiring of these pathways is related to the efficacy of the drugs, and these differences could hint at alternative markers.
A key aspect of our study is that we capture and catalogue the heterogeneity of signaling dynamics (which is described by the model parameters) for a large panel of colorectal cancer cell lines. It uses a unique dataset, consisting of 14 phospho-proteins measured under 43 differently perturbed conditions, which allowed us to generate and characterize cell line-specific models for 14 cell lines. In our models trained on this data, we found that about half of the model parameters are highly heterogeneous in our panel of cell lines and that this heterogeneity cannot be explained by genomic alterations alone. Since these findings were obtained with a rather focused set of antibody-based assays, we expect that many more associations can be found by a broader characterization. Mass-spectrometry based approaches, while still more costly and laborious than antibody-based methods, are under active development and likely to enable such an analysis in the near future (39).
Associations between model parameters and drug sensitivity were used to define interesting points of intervention to overcome resistance to specific drugs in specific cell lines, thus paving the way for personalized combinatorial therapies. Some of the combinatorial therapies suggested using this approach, like the combination of MEK inhibitors with EGFR and/or BRAF inhibitors, are supported by literature (as reported in the Results) and are currently in clinical trials. We found a rather surprising association between GSK3-related parameters and the efficacy of MEK inhibitors, which suggests that GSK3 inhibition would sensitize cells for drugs targeting MEK, which we validated experimentally. This interaction could not be unveiled using genomic data, but required information about the dynamics of signaling. Interestingly, the inhibition of GSK3 showed promising anticancer effect in recent studies on cancer cell lines (40,41) and its potential interaction with the MEK-ERK pathway has also been suggested in this context (41,42) but, as far as we know, no proven synergistic effect had previously been reported.
Our study clearly highlights the influence of inter-tumor heterogeneity in signaling networks on drug response; however, each colorectal tumor itself is also a very heterogeneous system. The tumor cells form hierarchies with stem cells and more differentiated cells, giving rise to a mix of different interacting cell types, including fibroblasts and immune cells (43,44). As each of these cell types has different signaling capabilities they will likely respond differently to the targeted drugs, and in turn the drugs will influence the heterocellular composition of the tumors. Therefore it will be very important to study signaling response in these heterogeneous cells in future, such as by single cell technologies, to fully appreciate the complexity of the system in vivo.
Our findings underscore the value of studying the dynamics of signaling pathways to better understand tumor phenotypes and to exploit this knowledge to suggest new therapeutic strategies (45). Such an approach is fundamentally different from the currently common characterization of various ‘omics’ layers at the basal level and it can be exploited to anticipate the dynamic adaptation mechanisms underlying drug resistance (17). Accordingly, we were able to find various pathway dynamic biomarkers for drugs with no genetic biomarkers. Hence, we believe that a perturbation-based strategy, even if restricted to fewer genetic backgrounds and only monitoring selected proteins, can provide a complementary strategy for biomarker discovery in cancer and beyond.
Supplementary Material
Acknowledgments
We thank Markus Morkel and members of the Saez laboratory for useful feedback, especially Luz Garcia-Alonso for help on mutations and Attila Gabor for fruitful discussion on model definition and David Henriques for support with CNORode. We thank Howard Lightfoot for help and assistance with drug combination data.
Financial support: N. Blüthgen acknowledges funding by the Berlin Institute of Health (BIH) and by the Federal Ministry of Research and Education (BMBF), grants OncoPath and ColoSys. F. Eduati thanks European Molecular Biology Laboratory Interdisciplinary Post-Docs (EMBL EIPOD) and Marie Curie Actions (COFUND) for funding.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Prasad V. Perspective: The precision-oncology illusion. Nature. 2016;537:S63. doi: 10.1038/537S63a. [DOI] [PubMed] [Google Scholar]
- 2.de Gramont A, Watson S, Ellis LM, Rodón J, Tabernero J, de Gramont A, et al. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol. 2015;12:197–212. doi: 10.1038/nrclinonc.2014.202. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 4.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Kirouac DC, Onsum MD. Using network biology to bridge pharmacokinetics and pharmacodynamics in oncology. CPT Pharmacometrics Syst Pharmacol. 2013;2:e71. doi: 10.1038/psp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niepel M, Hafner M, Pace EA, Chung M, Chai DH, Zhou L, et al. Profiles of Basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci Signal. 2013;6:ra84. doi: 10.1126/scisignal.2004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klinger B, Sieber A, Fritsche-Guenther R, Witzel F, Berry L, Schumacher D, et al. Network quantification of EGFR signaling unveils potential for targeted combination therapy. Mol Syst Biol. 2013;9:673. doi: 10.1038/msb.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saez-Rodriguez J, Alexopoulos LG, Zhang M, Morris MK, Lauffenburger DA, Sorger PK. Comparing signaling networks between normal and transformed hepatocytes using discrete logical models. Cancer Res. 2011;71:5400–11. doi: 10.1158/0008-5472.CAN-10-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkle R, Steiert B, Salopiata F, Depner S, Raue A, Iwamoto N, et al. Identification of Cell Type-Specific Differences in Erythropoietin Receptor Signaling in Primary Erythroid and Lung Cancer Cells. PLoS Comput Biol. 2016;12:e1005049. doi: 10.1371/journal.pcbi.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill SM, Nesser NK, Johnson-Camacho K, Jeffress M, Johnson A, Boniface C, et al. Context Specificity in Causal Signaling Networks Revealed by Phosphoprotein Profiling. Cell Syst. 2016 doi: 10.1016/j.cels.2016.11.013. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirouac DC, Du JY, Lahdenranta J, Overland R, Yarar D, Paragas V, et al. Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Sci Signal. 2013;6:ra68. doi: 10.1126/scisignal.2004008. [DOI] [PubMed] [Google Scholar]
- 13.Fey D, Halasz M, Dreidax D, Kennedy SP, Hastings JF, Rauch N, et al. Signaling pathway models as biomarkers: Patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal. 2015;8:ra130. doi: 10.1126/scisignal.aab0990. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–66. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 15.Fritsche-Guenther R, Witzel F, Sieber A, Herr R, Schmidt N, Braun S, et al. Mol Syst Biol. Vol. 7. EMBO Press; 2011. Strong negative feedback from Erk to Raf confers robustness to MAPK signalling; p. 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Müller U, et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell. 2012;22:796–811. doi: 10.1016/j.ccr.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Korkut A, Wang W, Demir E, Aksoy BA, Jing X, Molinelli EJ, et al. Perturbation biology nominates upstream-downstream drug combinations in RAF inhibitor resistant melanoma cells. Elife. 2015:4. doi: 10.7554/eLife.04640. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flobak Å, Baudot A, Remy E, Thommesen L, Thieffry D, Kuiper M, et al. Discovery of Drug Synergies in Gastric Cancer Cells Predicted by Logical Modeling. PLoS Comput Biol. 2015;11:e1004426. doi: 10.1371/journal.pcbi.1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverbush D, Grosskurth S, Wang D, Powell F, Gottgens B, Dry J, et al. Cell-specific computational modeling of the PIM pathway in acute myeloid leukemia. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-16-1578. [Internet] [DOI] [PubMed] [Google Scholar]
- 20.Guex N, Crespo I, Bron S, Ifticene-Treboux A, Hull EF-V, Kharoubi S, et al. Angiogenic Activity of Breast Cancer Patients’ Monocytes Reverted by Combined Use of Systems Modeling and Experimental Approaches. PLoS Comput Biol. 2015;11:e1004050. doi: 10.1371/journal.pcbi.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016 doi: 10.1016/j.cell.2016.06.017. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittmann DM, Krumsiek J, Saez-Rodriguez J, Lauffenburger DA, Klamt S, Theis FJ. Transforming Boolean models to continuous models: methodology and application to T-cell receptor signaling. BMC Syst Biol. 2009;3:98. doi: 10.1186/1752-0509-3-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terfve C, Cokelaer T, Henriques D, MacNamara A, Goncalves E, Morris MK, et al. CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Syst Biol. 2012;6:133. doi: 10.1186/1752-0509-6-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egea JA, Henriques D, Cokelaer T, Villaverde AF, MacNamara A, Danciu D-P, et al. MEIGO: an open-source software suite based on metaheuristics for global optimization in systems biology and bioinformatics. BMC Bioinformatics. 2014;15:136. doi: 10.1186/1471-2105-15-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou H, Hastie T. J R Stat Soc Series B Stat Methodol. Vol. 67. Blackwell Publishing Ltd; 2005. Regularization and variable selection via the elastic net; pp. 301–20. [Google Scholar]
- 26.Fessler E, Medema JP. Colorectal Cancer Subtypes: Developmental Origin and Microenvironmental Regulation. Trends Cancer Res. 2016 Sep;2:505–18. doi: 10.1016/j.trecan.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roumeliotis TI, Williams SP, Goncalves E, Zamanzad Ghavidel F, Aben N, Michaut M, et al. Genomic determinants of protein abundance variation in colorectal cancer cells. bioRxiv. 2016 doi: 10.1016/j.celrep.2017.08.010. [Internet] biorxiv.org; Available from: http://biorxiv.org/content/early/2016/12/09/092767.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Türei D, Korcsmáros T, Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods. 2016;13:966–7. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- 30.Saez-Rodriguez J, Alexopoulos LG, Epperlein J, Samaga R, Lauffenburger DA, Klamt S, et al. Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol Syst Biol. 2009;5:331. doi: 10.1038/msb.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriques D, Rocha M, Saez-Rodriguez J, Banga JR. Reverse engineering of logic-based differential equation models using a mixed-integer dynamic optimization approach. Bioinformatics. 2015;31:2999–3007. doi: 10.1093/bioinformatics/btv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov. 2015;5:960–71. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med. 2014;6:224ra26. doi: 10.1126/scitranslmed.3007947. [DOI] [PubMed] [Google Scholar]
- 35.Larkin J, Ascierto PA, Dréno B, Atkinson V, Liszkay G, Maio M, et al. Combined Vemurafenib and Cobimetinib in BRAF -Mutated Melanoma. N Engl J Med. 2014;371:1867–76. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 36.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 37.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 38.Atreya CE, Van Cutsem E, Bendell JC, Andre T, Schellens JHM, Gordon MS, et al. Updated efficacy of the MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in patients (pts) with BRAF V600E mutated (BRAFm) metastatic colorectal cancer (mCRC). ASCO Annual Meeting Proceedings; 2015. p. 103. [Google Scholar]
- 39.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–55. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 40.Yoshino Y, Ishioka C. Inhibition of glycogen synthase kinase-3 beta induces apoptosis and mitotic catastrophe by disrupting centrosome regulation in cancer cells. Sci Rep. 2015;5 doi: 10.1038/srep13249. 13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCubrey JA, Steelman LS, Bertrand FE, Davis NM, Sokolosky M, Abrams SL, et al. GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014;5:2881–911. doi: 10.18632/oncotarget.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemieux E, Cagnol S, Beaudry K, Carrier J, Rivard N. Oncogenic KRAS signalling promotes the Wnt/β-catenin pathway through LRP6 in colorectal cancer. Oncogene. 2015;34:4914–27. doi: 10.1038/onc.2014.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tape CJ. The Heterocellular Emergence of Colorectal Cancer. Trends Cancer Res. 2017;3:79–88. doi: 10.1016/j.trecan.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tauriello DVF, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97–119. doi: 10.1002/1878-0261.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yaffe MB. Sci Signal. Vol. 6. AAAS; 2013. The Scientific Drunk and the Lamppost: Massive Sequencing Efforts in Cancer Discovery and Treatment. scisignal.2003684v1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.