Abstract
Background & aims
Hepatitis B virus (HBV) RNA can undergo alternative splicing but the relevance of this post-transcriptional regulation remains elusive. Here, the mechanism of HBV alternative splicing regulation and its impact on liver pathogenesis were investigated.
Methods
HBV RNA-interacting proteins were identified by RNA pull-down combined to mass spectrometry analysis. HBV splicing regulation was investigated in chemically and surgically induced liver damage in whole HBV genome transgenic mice and in hepatoma cells. Viral and endogenous gene expression were quantified by RT-qPCR, western blot and ELISA. Resident liver immune cells were studied by FACS.
Results
HBV pregenomic RNA-interacting proteins were identified and 15% were directly related to the splicing machinery. Expression of these splicing factors was modulated in HBV transgenic mice under liver injuries and contributed to an increase of the HBV spliced RNA encoding for HBV splicing-generated protein (HBSP). HBSP transgenic mice exhibited an attenuated hepatic damage under chemically induced liver fibrosis. The protective effect of HBSP resulted from a decrease of inflammatory monocyte/macrophage recruitment through a downregulation of CCL2 expression produced by hepatocytes. In human hepatoma cells, ability of HBSP to control CCL2 expression was confirmed and maintained in a whole HBV context. Finally, viral spliced RNA detection related to a decrease of CCL2 expression in liver of HBV chronic carriers underscored this mechanism.
Conclusion
Microenvironment modified by liver injury increased HBSP RNA expression through splicing factors regulation, which in turns controlled hepatocyte chemokine synthesis. This feedback mechanism suggests a novel insight in liver immunopathogenesis during HBV infection.
Lay summary
Hepatitis B virus persists during decades in liver of chronic infected patients. One of the main mechanisms developed by this virus to persist is to escape immune response. Our study highlights how the crosstalk between virus and liver infected cells may contribute to this immune escape.
Keywords: HBV, liver fibrosis, HBSP, alternate splicing, macrophages, CCL2
Introduction
Constitutive splicing is an essential step for eukaryotic gene expression, generating mature mRNAs by removing introns and allowing exons to be joined together accurately. In contrast, alternate splicing (including exon skipping, intron retention, alternate 3’ or 5’ splice sites) contributes to the diversity of eukaryotic proteome[1] and involves in the control of expression of regulatory proteins in the viral context[2]. The splicing process is coordinated by the spliceosome, a large ribonucleoprotein complex, on specific RNA domains in association with ubiquitous transregulatory splicing factors including serine–arginine-rich (SR) and heterogeneous nuclear ribonucleoprotein (hnRNPs) families[3].
The ability of Hepatitis B Virus (HBV) transcripts to undergo to alternate splicing (AS) has been well described in vitro and in the liver of patients with chronic infection (CHB)[4, 5]. The 3.5 kb pregenomic RNA (pgRNA) of HBV, which encodes for capsid and polymerase proteins, and constitutes the template for viral genome replication, can be alternatively spliced[6]. The major pgRNA spliced variant, SP1RNA, has one third of the viral genome deleted (“intron 2447/489”) and may account for up to 30% of total HBV pgRNA. SP1RNA can be packaged in core particles, reverse-transcribed and secreted. While pgRNA packaging leads to wild-type Dane particle secretion (wtHBV), the shorter SP1RNA constitutes a matrix for defective HBV circulating particles (dHBV), varying in proportion from 0% to more than 50% of HBV forms[7–10]. The regulation of SP1RNA during the course of liver disease remains poorly understood. However, recent reports in HBV infected patients have shown that the proportion of dHBV relates to viral replication, to interferon therapy failure[11] and increases with liver disease progression towards hepatocellular carcinoma (HCC)[8, 9].
SP1RNA allows the expression of HBV splicing-generated protein (HBSP). HBSP protein shares the N-terminal 46 amino-acids sequence of the viral polymerase fused to an original viral sequence constituting its C-terminal part (65 amino-acids). HBSP has been identified in liver tissues from patients with CHB[12], in whom it can induce an immune response[13]. The function of HBSP protein in liver pathogenesis remained uncertain, although in vitro studies had suggested an impact on cell viability, proliferation and migration [12, 14] and more recently on hacking the TNF-α signaling pathway [15].
The previous observation that dHBV particles increase with liver disease progression prompted us to investigate whether and how regulation of HBV AS and liver pathogenesis are mechanistically linked. Because, the lack of robust animal models of liver pathogenesis induced by HBV infection, we decided to investigate the viral post-transcriptional regulation in HBV transgenic mice[16]. Here we report that alteration of spliceosome machinery in HBV expressing cells is switched on by liver injury and enables a striking reduction in liver monocyte/macrophage recruitment through HBSP expression. Our findings reveal a novel paradigm whereby AS can generate a viral product able to inhibit immune-mediated inflammation and thereby down-modulate organ damage.
Materials and Methods
Mice and liver pathogenesis models
Inbred C57BL/6J HBV-transgenic mouse lineage 1.3.32 (TgHBV) has been previously described[17]. TgHBV mice express and replicate HBV in the liver under the control of HBc promoter/enhancer-II. The two independent transgenic HBSP1 and HBSP2 mice strains carry HBSP gene (333 bp) encoding for the HBSP protein under the control of the HBx promoter/enhancer I (nt 832/1371)[15]. A rabbit β-globin intron was inserted between the HBx promoter/enhancer-I sequence and HBSP gene (genotype A). HBSP1 and HBSP2 transgenic mice were generated and expanded by back-crossing against the C57BL/6J strain (>12 back-crossings). C57BL/6J invalidated for CCR2 gene expression (CCR2KO) mice were also used [18]. All experiments were performed on 2-months old, heterozygous TgHBV, TgHBSP1, TgHBSP2 or homozygous CCR2 KO and corresponding littermate (WT) male mice.
WT and TgHBV mice were treated for 7 weeks with carbon tetrachloride intraperitoneal injections (CCl4, 1ml/kg in oil, Sigma) twice a week to induce chronic liver fibrosis. Fulminant liver injury was generated by surgical bile duct ligation (BDL) using double sutures of bile duct near liver junction; WT and TgHBV mice were monitored for 2 days before sacrifice. Lipopolysaccharide intraperitoneal injections (LPS, 2.5mg/kg, Sigma) three times a week for 2 weeks were performed on WT and TgHBV mice to induce liver inflammation. TgHBSP, CCR2 KO and corresponding WT mice were treated twice a week with CCl4 (1ml/kg in oil, Sigma) for 2 and/or 10 weeks. Acute liver injury was also performed in TgHBSP1 mice by a single intraperitoneal injection of CCl4 (1ml/kg in oil). Thioacetamide intraperitoneal injections (TAA, 250mg/kg in PBS, Sigma) three times a week were performed on WT and TgHBSP1 mice for 2 weeks to induce liver fibrosis. TgHBV, TgHBSP and littermate WT were sacrificed 48 to 72 hours after the last injection. Mice livers were perfused with PBS-2% FCS and removed for analysis. Animals were maintained in pathogen–free conditions on 12-hour dark/light cycles and treated in accordance with EU regulations on animal care (Directive 86/609/EEC). All procedures were approved by the local animal care and use committee (Agreement A75-14-08; n° 02890 and 02891). Alanine aminotransferase (ALT) in serum was measured on a Cobas® 6000 analyzer (Roche), according to the manufacturer’s instructions.
Human HBV infected liver samples
Patients were recruited from Mustapha Hospital (Alger, Algeria) and The Royal London Hospital (London, UK). Full ethical approval was obtained for the study; agreements by the National Health Ministry (according to legal article about clinical trials n°387 et 388 of 31/07/2006) and the Local Ethical Review Board at the Royal London Hospital. All patients recruited gave written informed consent. Liver samples, from biopsies surplus to diagnostic requirements, were obtained from 12 HBV-monoinfected patients. Patients were anti-HCV, anti-HDV and anti-HIV antibody negative and treatment naive. All patients had evidence of moderate liver damaged; scored by a pathologist according to the Metavir stage. ALT, HBeAg and HBsAg titres were determined using commercial assays (Roche, Biorad and Diasorin, respectively).
Additional methods are provided as supporting information.
Results
Increase in HBV alternate splicing upon liver injury
To assess whether liver injury impacts on HBV splicing regulation, TgHBV were treated with either CCl4 (7 weeks/IP) or LPS (2 weeks/IP) to induce liver fibrosis or inflammation, respectively (FigS1 and Fig.S2). The amount of HBV pgRNA was reduced by a 5.3 fold and a 3.9 fold in LPS and CCl4 treated TgHBV mice, respectively (Fig.1A) as well as HBV viral load (FigS3). In contrast, HBV AS was significantly increased (Fig.1A). Indeed, quantification of HBV pgRNA and SP1RNA-derived forms showed a significant increase of the SP1RNA proportion in both CCl4 and LPS-treated TgHBV mice compared to control mice (oil-TgHBV vs CCl4-TgHBV: 32.5±9.6% vs 49.7±17.2%, p=0.04; PBS-TgHBV vs LPS-TgHBV: 27.6±8.7% vs 42.7±10.1%, p=0.01). Additionally, HBV AS upregulation was also observed in TgHBV mice with a BDL-induced fulminant hepatitis (control-TgHBV vs BDL-TgHBV: 30.6±4.9% vs 69.0±3.0%, p=0.001) (Fig.S4). These results may support an influence of liver damage on HBV post-transcriptional regulation.
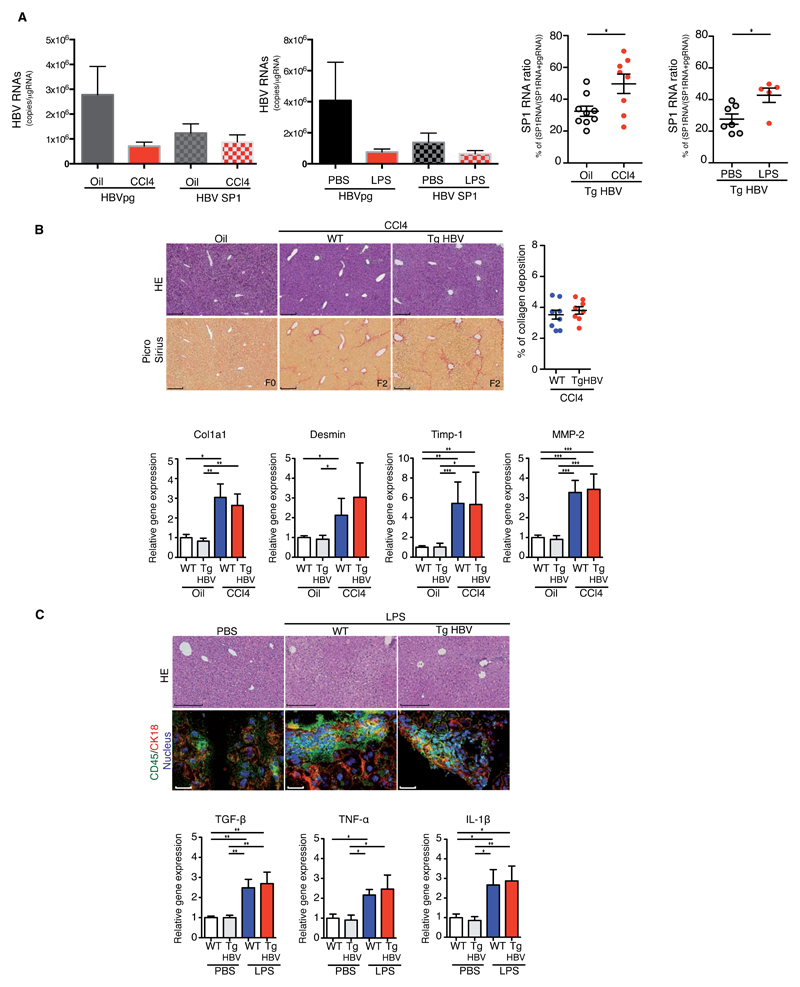
Fig.1. Liver injury increases HBV pgRNA alternate splicing in HBV transgenic mice.
(A) HBV pgRNA and SP1RNA expression (left) and SP1RNA proportion (right) quantified in the liver of TgHBV mice treated with oil/CCl4 for 7 weeks or with PBS/LPS for 2 weeks. (B) Representative H&E and Picro-Sirius red stained histological liver section (left, bar 250μm) and quantification of collagen deposition (right) from 7 weeks oil, CCl4-WT and CCl4-TgHBV treated mice (upper panel). Fibrosis-related mRNA expression in the liver, each dot represents a mouse, experiments were performed on oil-WT (n=7), oil-TgHBV (n=7), CCl4-WT (n=9) and CCl4-TgHBV (n=8) mice (lower panel). C) Representative H&E histological staining (upper, bar: 250μm); CD45 and Cytokeratin 18 (CK18) immunofluorescence staining on liver section (lower, bar: 50μm) from 2 weeks PBS, LPS-WT and LPS-TgHBV treated mice (upper panel). Cytokines mRNA expression in the liver of 2 weeks PBS or LPS-treated WT and TgHBV mice. Each dot represents a mouse; experiments were performed on PBS-WT (n=6), PBS-TgHBV (n=7), LPS-WT (n=5), and LPS-TgHBV (n=5) treated-mice (lower panel). Mann-Withney U-tests: *p<0.05, **p<0.01 and ***p<0.001.
Previous clinical studies suggested a correlation of HBV alternative splicing and the course of liver disease[8, 9] but the relevance of HBV pgRNA splicing to liver pathogenesis remains unknown. We therefore compared liver fibrosis or inflammation following CCl4 or LPS treatment of TgHBV and their littermate wild-type (WT) control mice. Despite the well-known activities of HBV proteins on signaling pathways involved in inflammatory and fibrotic processes[19], CCl4-treated TgHBV and WT mice displayed a comparable low stage of liver fibrosis (F2) on histological section (Fig.1B). This was confirmed by quantification of collagen deposition and the expression of genes involved in liver fibrosis. Similarly, liver histology, staining for CD45+ cells and quantification of inflammation-inducible genes confirmed an equivalent degree of liver injury in LPS-treated WT and TgHBV mice (Fig.1C and Fig.S5).
Regarding that several viral proteins may impact on liver disease, it was difficult to define the role of HBV AS and related HBSP expression in this experimental model. However, the lack of increase in CCl4-induced liver injury in TgHBV mice, expressing the complete HBV genome, contrasted with the exacerbations previously observed in transgenic mice expressing a subgenomic HBV sequence that is unable to express SP1RNA[20]. Considering the high level of SP1RNA in the liver of TgHBV mice, we therefore hypothesized that HBV AS, triggered by liver injury could limit the ensuing liver inflammation and fibrosis. To understand the role of SP1RNA during liver pathogenesis, we first investigated its regulation.
Identification of HBV alternate splicing factors
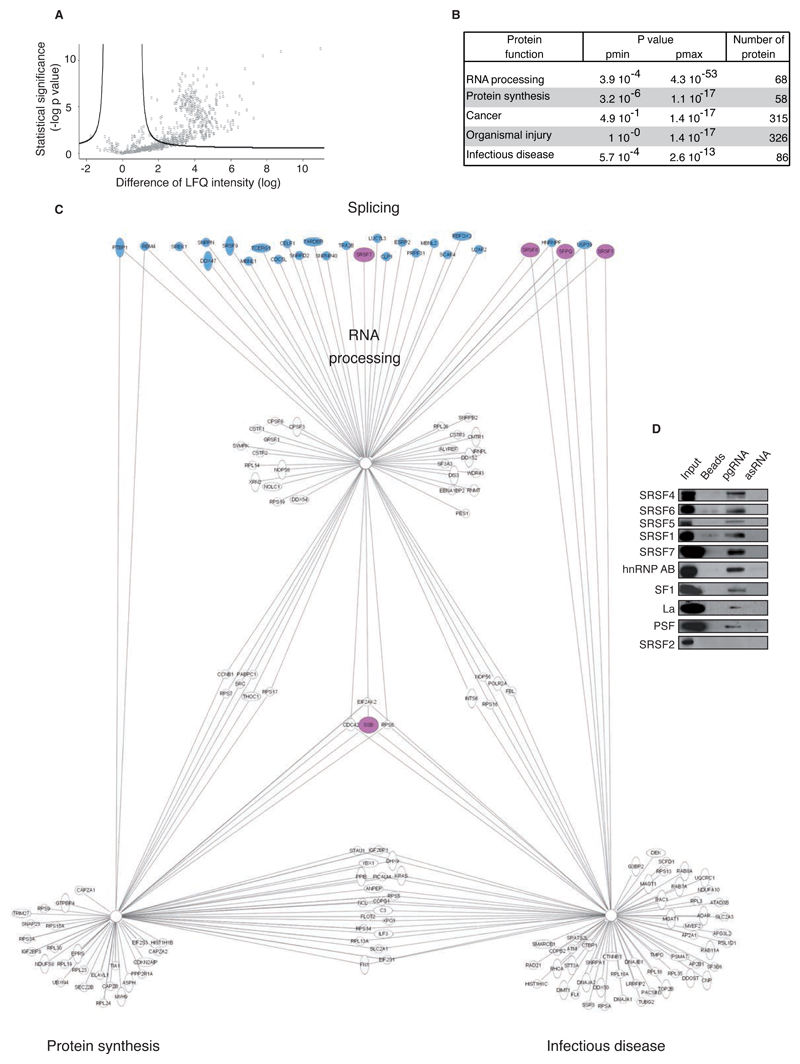
Characterization of pgRNA transregulatory splicing proteins remained a crucial prerequisite to investigate HBV AS regulation. Thus, HepG2 nuclear proteins interacting with HBV pgRNA were explored by RNA pull-down assay. Identification and relative quantification by Orbitrap mass-spectrometry showed in six independent experiments that 389 proteins were significantly associated with viral pgRNA (Fig.2A and Table S1). Classification by gene ontology according to the biological functions allowed to distinct 5 groups representing 88% of associated-proteins (12% proteins remaining classified in group “other”) (Fig.2B). The most significant group was the “RNA processing” family, which was mainly composed of factors directly involved in RNA splicing (28/68 proteins; stained in Fig.2C). Moreover, 30 additional proteins involved in RNA splicing and comprising hnRNPs proteins, were classified in further groups (Table S1). As illustrated, the proteins belonging to the “RNA processing” group have additional biological functions, which may contribute to the liver disease progression during infectious disease. Specific interaction of 9 transregulatory splicing factors, hnRNPAB, SF1, La, PSF and SRSF1,4,5,6,7 were confirmed in the pgRNA pull-down extract and not in controls (using antisense HBV pgRNA and empty beads) by western blot (Fig.2D). The splicing factors SRSF2 and hnRNPH, absent or not significantly linked to pgRNA, were used as controls.
Fig. 2. Identification of cellular splicing factors interacting with HBV pgRNA.
(A) Volcano plot representation of HBV pgRNA-interacting proteins (700/1442) detected in 6 independent RNA pull-down assays (beads used as control) and identified by mass spectrometry (nanoRSLC-LTQ Orbitrap Velos). Proteins showing a significant interaction with HBV pgRNA (n=389/700) are depicted uppers the black line according to significance analysis. X-axis shows the difference of the average of the logarithm of Label Free Quantification (LQF) intensities between pgRNA and control; y-axis shows the negative logarithmic of t-test Welsh p-value. (B) Biological function groups of HBV pgRNA binding proteins identified. (C) Schematic network presenting the main protein families binding to HBV pgRNA according to Ingenuity software. Transregulatory splicing factors are colored in blue and the ones selected for further characterization are colored in pink. (D) HBV pgRNA pull-down extract analysis by western blot confirming a specific interaction of 9 selected trans-regulatory splicing factors with pgRNA (antisense pgRNA (asRNA), beads and SRSF2 protein were used as negative control).
Splicing proteins are implicated in the regulation of HBV alternate splicing
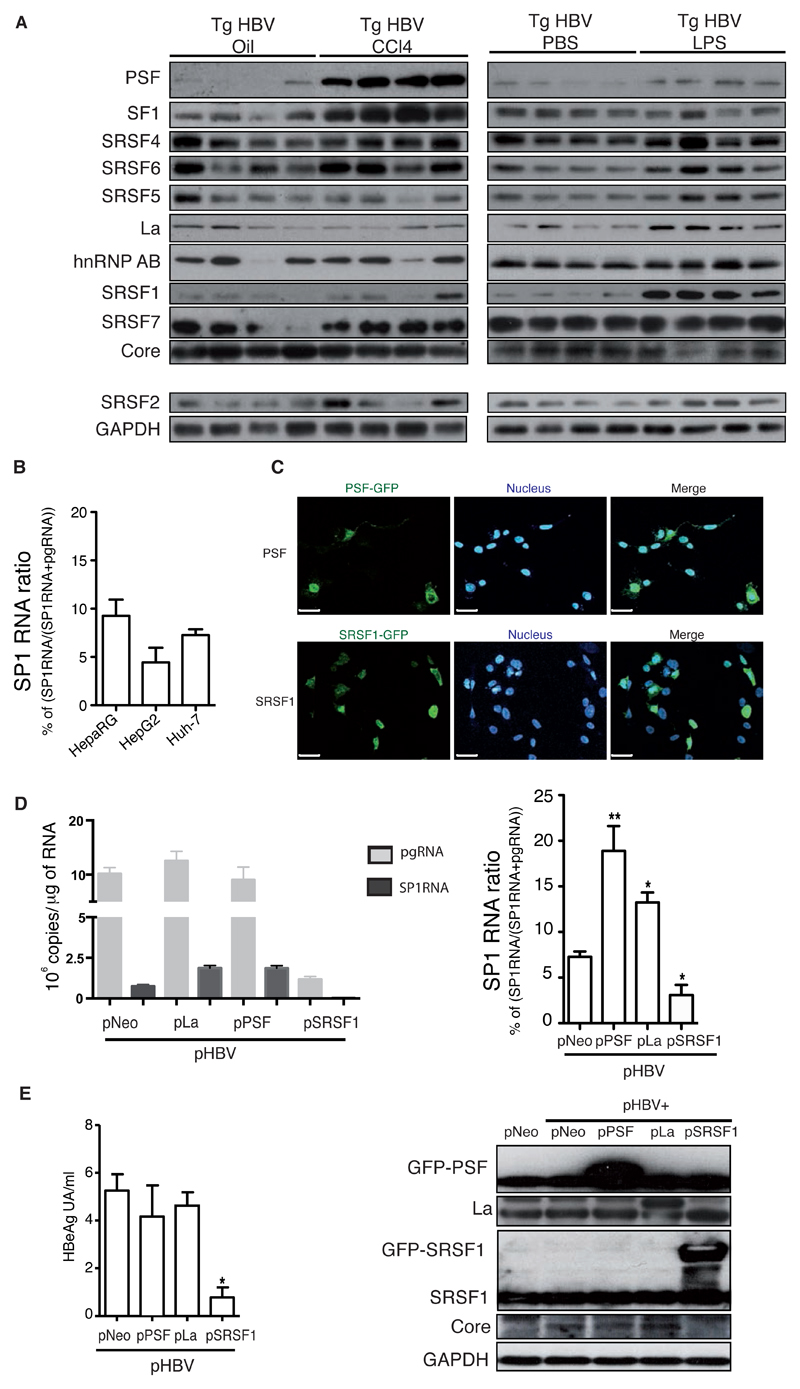
The modulation of expression of HBV pgRNA interacting transregulatory splicing proteins (conserved inter-species) was investigated in liver extracts of TgHBV mice. Chemical induction of fibrosis led to an overexpression of PSF and SF1 proteins, whilst LPS-induced liver inflammation increased the expression of SRSF1 and La proteins (Fig.3A). Despite the differential signature of splicing factors expression in both liver disease models, a comparable increase of SP1RNA proportion was observed. It was considered that AS regulation in eukaryotic cells has largely been driven by the alteration of cis-regulatory elements. These data provide additional evidence suggesting AS regulation and trans-regulatory proteins expression according to liver damage [21].
Fig. 3. SP1RNA regulation is dependent on splicing-proteins expression.
(A) Splicing regulatory proteins expression in the liver of TgHBV mice treated with oil/CCl4 for 7 weeks or with PBS/LPS for 2 weeks (SRSF2 and hnRNPH1 used as controls). (B) SP1RNA proportion (SP1RNA/(SP1RNA+pgRNA)x100) in Huh7, HepG2 and HepaRG cells expressing HBV after transient transfection of pHBV vector. (C) Huh7 co-tranfected cells with pHBV and PSF-GFP or SRSF1-GFP; PSF-GFP and SRSF1-GFP visualization and nucleus were counter stained with Hoescht (middle, bar 50μm); (D) pgRNA and SP1RNA quantification in HBV expressing Huh7 cells co-transfected with Neo (control), PSF, La or SRSF1 plasmids (left panel), and (right panel) HBV SP1RNA proportion in these cells (n≥3 experiments). (E) HBeAg cell supernatant quantification (left panel) and PSF, SRSF1, La and Core expression by western blot (right panel). Mann-Withney U-tests: *p<0.05, **p<0.01 and ***p<0.001.
Conservation of splicing factors in mammals led us to investigate their involvement on HBV AS in vitro. Once different human hepatoma cells were tested, impact of PSF, SRSF1 and La expression in viral AS regulation was investigated (Fig.3 B-D). In Huh7 expressing HBV cells, La and PSF proteins increased SP1RNA proportion, whereas SRSF1 led to a decrease of viral splicing efficiency (Fig.3E). This paradoxical decrease may be attributable to the fact that SRSF1 expression also considerably reduced HBV transcription, core and HBeAg expression (Fig.3D), suggesting an impact on HBV RNA synthesis or metabolism. Taken together, these data emphasized for a possible influence of the microenvironment altered by liver injury on the expression of the HBV-regulatory splicing factors we had defined, driving regulation of the SP1RNA and consequently HBSP expression.
HBSP protein partially prevents CCl4-induced liver fibrosis
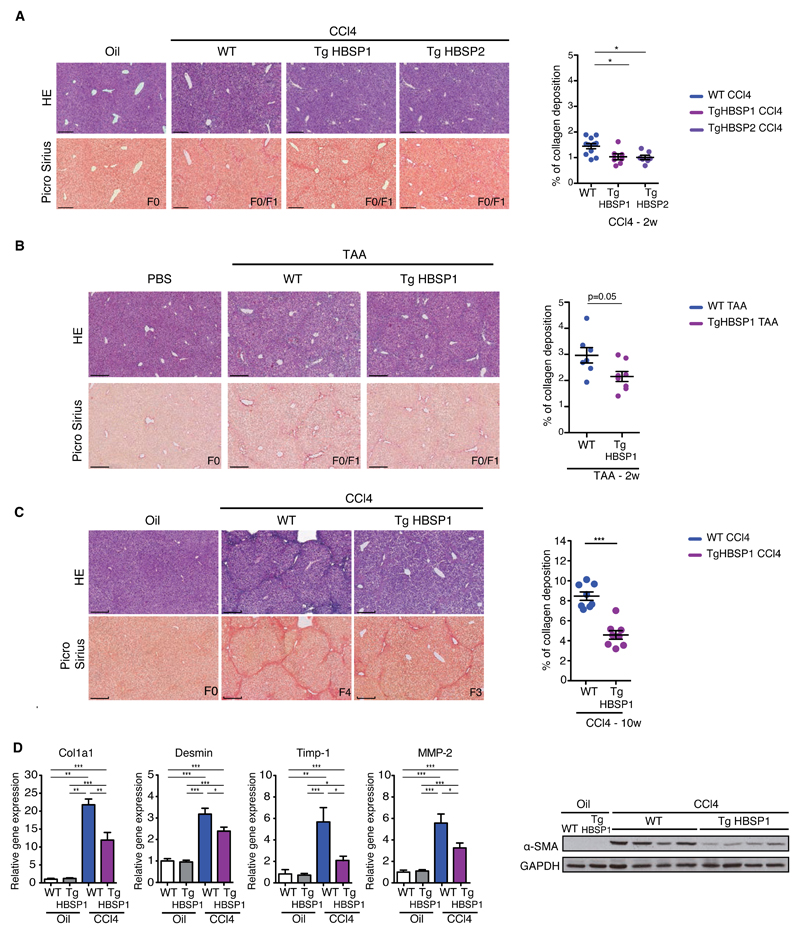
The impact of the HBV alternative splicing product HBSP on liver pathology was investigated in two transgenic mouse strains TgHBSP1&2 expressing the HBSP gene under the control of the weak hepato-specific HBx promoter (Fig.S6). The amount of HBSP RNA in TgHBSP mice was up to 10 fold lesser than SP1RNA expression in TgHBV mice (1.2x105 ±3.4x104 vs 1.2x106±9.9x105 copies/μg of liver RNAs, respectively) and remained stable with aging (FigS5)[15]. Since TgHBSP did not spontaneously develop liver pathogenesis, WT and TgHBSP mice were challenged for 2 weeks with CCl4 or TAA to induce liver fibrosis (Fig.4A, 4B, S6). Surprisingly, and despite a low fibrosis score (F0/F1) for all mice, TgHBSP mice treated with CCl4 or TAA revealed a reduction in collagen deposition compared to their WT littermates.
Fig.4. HBSP expression protects against chemical-induced liver fibrosis.
Representative H&E and Picro-Sirius red stained histological liver section (bar 250μm) (left) and quantification of collagen deposition (right) from: (A) WT, TgHBSP1 and TgHBSP2 mice treated for 2 weeks with Oil (n=4), CCl4-WT (n=11), CCl4-TgHBSP1 (n=7) and CCl4-TgHBSP2 (n=7) mice. (B) WT and TgHBSP1 mice treated for 2 weeks with PBS (n=4 for both) or TAA-WT (n=7) and TAA-TgHBSP1 (n=8) mice. (C) WT and TgHBSP1 mice treated for 10 weeks with oil (n=6 for both WT and TgHBSP1) or CCl4 (n=8 for both WT and TgHBSP1). (D) Liver fibrosis-related gene mRNA and α-SMA protein expression in the liver of WT and TgHBSP1 mice treated for 10 weeks with oil or CCl4. Each dot represents a mouse. Mann-Withney U-tests: *p<0.05, **p<0.01 and ***p<0.001.
To evaluate whether HBSP limited the extent of chemically-induced fibrogenesis and to investigate its biological impact on advanced stages of liver fibrosis, WT and TgHBSP1 mice were challenged with CCl4 for 10 weeks (Fig.S6). As expected, all WT treated mice displayed severe fibrosis with periportal septa and developed cirrhosis (F4) (n=8/8) (Fig.4C). In contrast, TgHBSP1 treated-mice exhibited less marked fibrosis, with 75% animals having a F2 (n=2/8) and F3 (n=4/8) fibrosis score and only 25% reaching the F4 cirrhotic state (n=2/8). This observation was corroborated by a significant two-fold decrease in collagen deposition in TgHBSP1 liver tissue compared to WT mice upon CCl4 treatment (Fig.4C; 4.5±1.2% vs 8.4±1.2%, p=0.0002, respectively). The expression of pivotal fibrosis-related genes Col1a1, Desmin, Timp-1 and MMP-2 confirmed the lower sensitivity of HBSP mice to 10 weeks CCl4-induced fibrosis (Fig.4D). In accordance with these results, α-SMA protein expressed by activated hepatic stellate cells (HSCs) was detected in a higher amount in the liver of WT compared to TgHBSP1 mice.
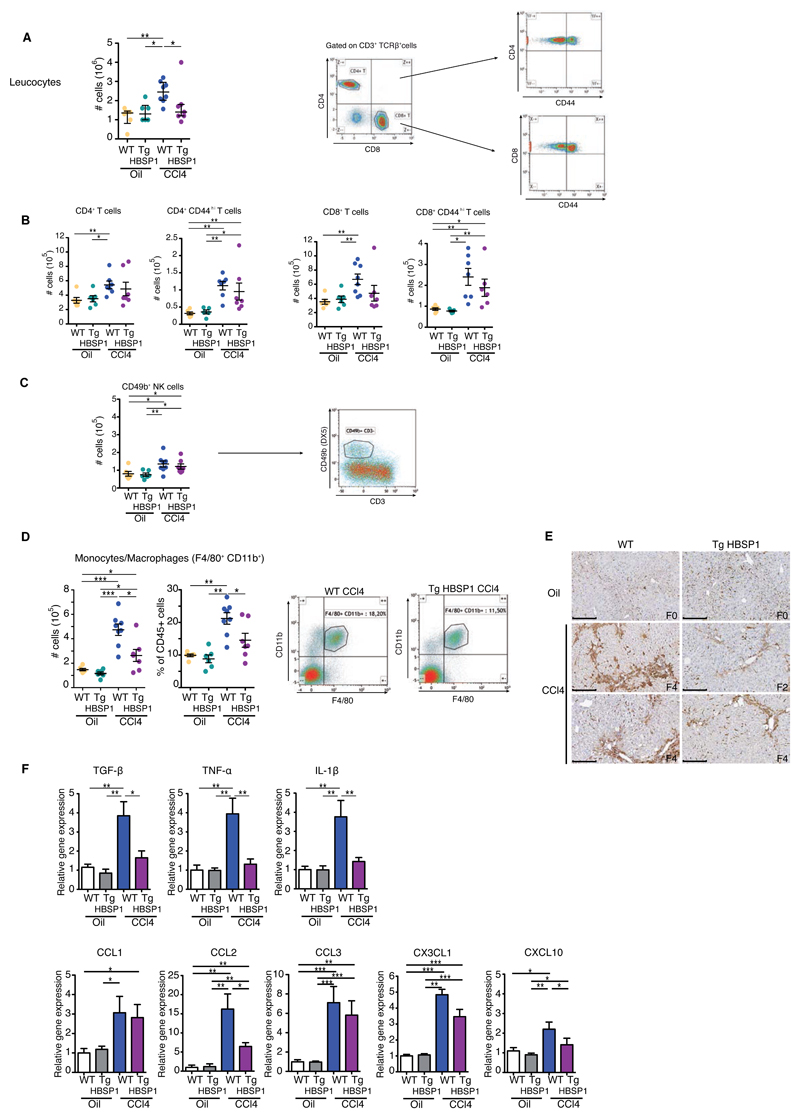
Recruitment of immune cells to the liver plays a central role in hepatic fibrogenesis[22]. To investigate whether this was altered by the expression of HBSP, intrahepatic leucocytes from 10 weeks CCl4-treated mice were isolated and characterized by flow cytometry. The CCl4-induced liver leucocyte infiltrate was significantly decreased in TgHBSP1 compared to WT mice (Fig.5A). No significant difference in the intrahepatic upregulation of CD49b+ NK, CD4+ and CD8+ T (±CD44hi) cells was observed between WT and TgHBSP1 mice treated with CCl4 (Fig.5B, 5C). In contrast, the number and proportion of liver monocytes/macrophages, defined as CD45+ F4/80+ CD11b+ cells, were significantly reduced in TgHBSP1 compared to WT mice treated with CCl4 (Fig.5D; 2.6±0.5x105 vs 4.7±0.5x105 cells, p=0.02 and 14.5±2.1% vs 21.2±1.8% of CD45+, p=0.02, respectively) (Fig.5D). In addition, CCl4-challenged WT mice exhibited a higher periportal infiltrate of F4/80+ monocytes/macrophages than TgHBSP1 treated mice on stained liver sections (Fig.5E).
Fig.5. HBSP expression reduces intrahepatic monocytes/macrophages infiltrate.
WT and TgHBSP1 mice were treated for 10 weeks with oil or CCl4. Intrahepatic leucocytes were isolated and analysed by flow cytometry. (A) Intrahepatic leucocyte cells number (left) and representative gating of CD4+ and CD8+ T cells (right). (B) CD4+ T (7-AAD-, CD3+,TCRβ+,CD4+) and CD44hi effector/memory CD4+ T cells number. CD8+ T (7-AAD-, CD3+,TCRβ+,CD8+) and CD44hi effector/memory CD8+ T cells number (left). (C) NK (7-AAD-,CD49b+,CD3-) cells number and representative gating. (D) Intrahepatic monocytes/macrophage cells number, proportion and representative gating of (7-AAD, CD45+, F4/80+, CD11b+) from WT and TgHBSP1 CCl4-treated mice (right). (E) Representative monocytes/macrophages F4/80 staining on histological liver section (bar 250μm). (F) Cytokines (upper panel) and chemokines (lower panel) mRNA expression in the liver. Each dot represents a mouse; experiments performed on 10 weeks oil-WT (n=6), oil-TgHBSP1 (n=6), CCl4-WT (n=8) and CCl4-TgHBSP1 (n=8) treated mice. Mann-Withney U-tests: *p<0.05, **p<0.01 and ***p<0.001.
Monocytes/macrophages contribute to liver fibrosis through the secretion of inflammatory cytokines, which directly activate HSCs[23]. In line with this, we confirmed that upon CCl4-induced chronic fibrosis, TgHBSP1 displayed a less pro-fibrotic inflammatory environment than WT mice, with a significant decrease in TGF-β, TNF-α and IL-1β genes expression (Fig.5F) without differential distribution of monocyte/macrophage ontogeny markers CD163 and Marco (Fig.S8) in WT and TgHBSP mice [24]. We next investigated the postulate that the decrease in monocyte/macrophage infiltrate in the HBSP-expressing liver could result from the impaired production of a chemokine involved in monocyte/macrophage survival and/or recruitment within the liver[25]. CCL1, CCL2, CCL3, CXCL10 and CX3CL1 transcripts were quantified and only CXCL10 and CCL2 were significantly reduced in the liver of TgHBSP1 compared to WT treated mice (Fig.5F). Together, these results show that HBSP expression restricts partially chronic induced liver fibrosis.
HBSP impairs the monocyte/macrophage recruitment during liver injury
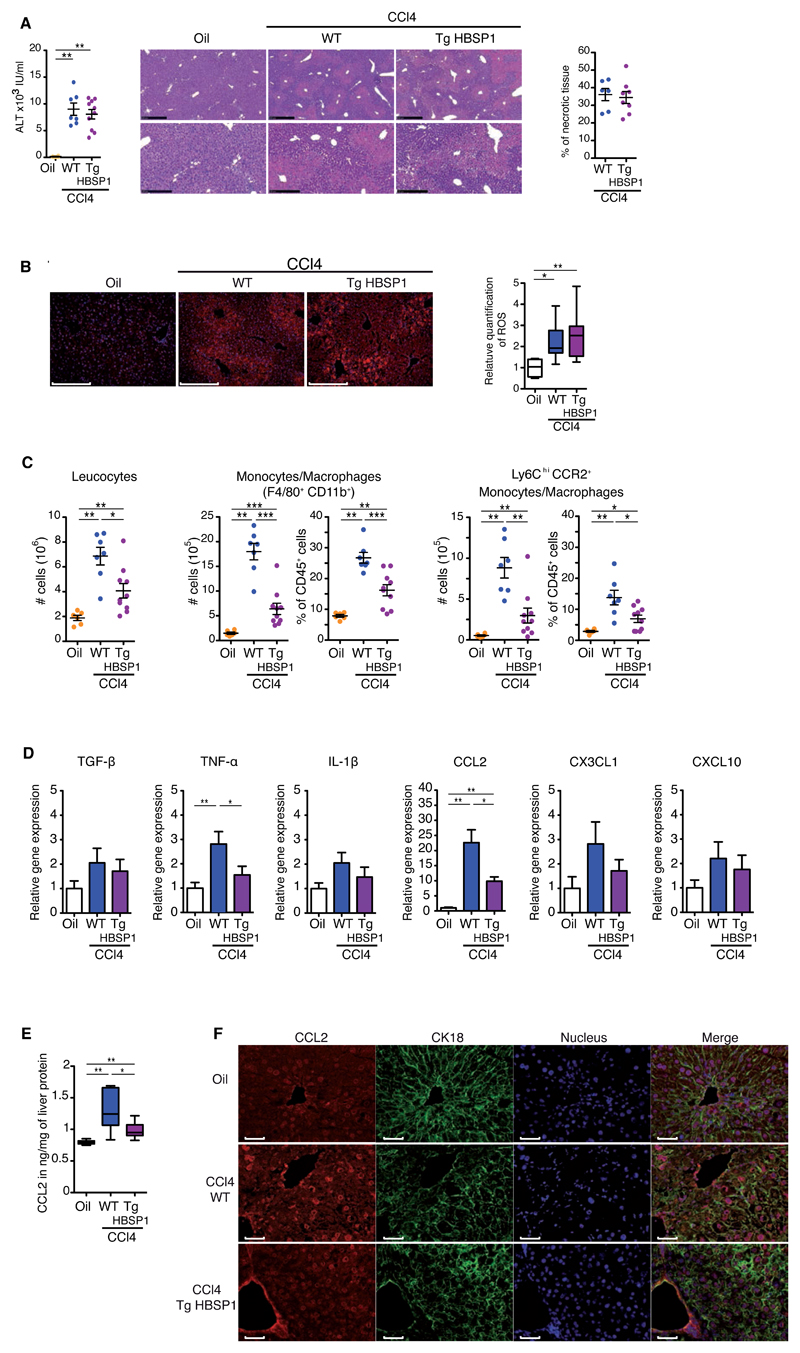
One explanation for the limited liver pathogenesis observed in TgHBSP1 mice could be a resistance of HBSP-expressing hepatocytes to CCl4-induced cytotoxicity. To investigate this possibility, WT and TgHBSP1 mice were treated with a single CCl4-injection to induce acute liver injury. In the context of acute hepatitis, TgHBSP1 and WT mice displayed comparable degrees of liver injury, with a similar increase in circulating ALT and equivalent central and periportal necrosis (Fig.6A). The metabolism of CCl4 in hepatocytes leads to reactive oxygen species (ROS) production and hepatocyte necrosis through lipid peroxidation accumulation. Consistent with this, ROS content assessment on liver extracts revealed a similar enhancement in TgHBSP1 and WT treated mice (Fig.6B). In mouse strains, 48h after a single CCl4 injection, liver apoptosis was rarely observed compared to necrosis and neither Desmin transcripts nor α-SMA protein expression were up-regulated, suggesting that HSCs were not yet activated (Fig.S9). Taken together, these results did not support a direct impact of HBSP expression on CCl4 hepatocyte toxicity as an explanation for the reduced susceptibility of TgHBSP1 mice to liver damage in the chronic exposure model.
Fig.6. HBSP has no impact on CCl4-induced hepatocyte acute damage but decreases intrahepatic monocyte/macrophage recruitment and CCL2 expression.
WT and TgHBSP1 mice treated with a single injection of oil or CCl4. (A) Sera ALT and liver necrotic tissue quantification on H&E stained histological section, representative H&E staining are presented (upper: bar 500μm; lower: bar 250μm). (B) ROS staining on liver frozen section (left, bar 100μm) and ROS content quantification in liver extract with H2-DCFDA probe (right). (C) Intrahepatic cells number (left) and proportion of monocytes/macrophages (middle; 7-AAD, CD45+, F4/80+, CD11b+) and pro-inflammatory Ly6Chi CCR2+ subpopulation (right). (D) Liver cytokines and chemokines mRNA expression. (E) CCL2 quantification in liver protein extract. (F) CCL2 and CK18 immunofluorescence staining on liver sections, nucleus were labelled with Hoescht (bar 50μm). Each dot represents a mouse; experiments were performed on oil (n=6), CCl4-WT (n=7) and CCl4-TgHBSP1 (n=10) single injected mice. Mann-WithneyU-tests: *p<0.05, **p<0.01 and ***p<0.001.
Instead we hypothesised that HBSP expression directly down-regulated hepatocyte chemokines content and thus monocyte/macrophage recruitment, accounting for the relative resistance to liver fibrosis in TgHBSP1 treated mice. Therefore, monocyte/macrophage recruitment was investigated by flow-cytometry in the acute CCl4 hepatitis model. As previously observed during chronic exposure, the influx of intrahepatic leucocytes and monocytes/macrophages was significantly reduced in HBSP-expressing compared to WT-mice with acute liver injury (Fig.6C). The monocytes/macrophages decreased in TgHBSP1 mice included the inflammatory Ly6Chi CCR2+ subpopulation (Fig.6C right panel: CCl4-WT vs CCl4-TgHBSP1: 8.8±0.1x105 vs 2.9±0.9x105 cells, p=0.003 and 13.8±2.3% vs 6.9±1.1% of CD45+ cells, p=0.01, respectively), recruited through a CCL2-dependent mechanism under CCl4 treatment[26]. The diminished recruitment of monocytes/macrophages in TgHBSP1 acute liver damage was only coupled to a decrease in CCL2 transcripts (Fig.6D) and to a down-regulation of CCL2 protein in the liver (Fig.6E). Reduced infiltration of monocytes/macrophages, known to be a major source of TNF-α[23], might also account for the decrease of TNF-α expression observed. The hepatic cells types producing CCL2 in the acute hepatitis model were further explored by immunofluorescence. CCL2 staining revealed a high signal around blood vessels and unexpectedly hepatocytes (counterstained by cytokeratin 18) were also clearly positive for CCL2 (Fig.6F).
These data suggested that HBSP expression impacted on liver monocyte/macrophage recruitment through a down-regulation of hepatocyte CCL2 expression upon acute liver injury.
HBSP modulates CCL2 expression in hepatocytes
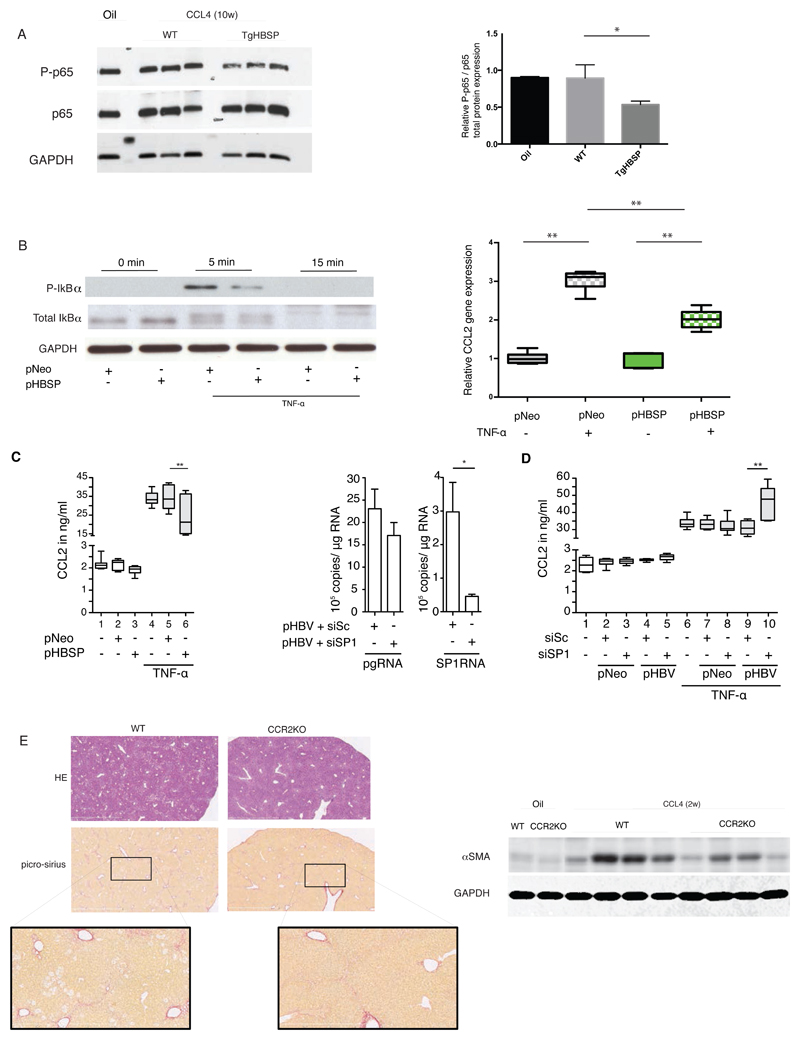
Our results suggested that CCL2 production by hepatocytes contributes to the recruitment of monocytes/macrophages and consequently to liver damage. Next, we investigated the ability of HBSP to control the expression of CCL2. In line with our previous results[15] we showed that p65 (RelA) phosphorylation was impaired in the liver of TgHBSP1 compared to WT mice challenged for 10 weeks with CCl4 (Fig.7A). This result suggested an impact of NF-κB activation on CCL2 expression. Then, we observed in HeLa expressing HBSP cells a reduced level of IκBα phosphorylation at 5 minutes after TNF-α, treatment (Fig.7B, left panel), together with a significant reduction of CCL2 transcription level compared to control cells 30 minutes post-treatment with TNF-α (Fig.7B, right panel). A significant decrease of CCL2 secretion was also displayed in the supernatant of HepaRG expressing HBSP cells treated by TNF-α (Fig.7C left panel; lanes 6 vs 5, respectively: 24.9±3.7 vs 33.7±2.3 ng/ml; p=0.008). In order to further explore CCL2 modulation by HBSP in a whole HBV genome context, a siRNA targeting the SP1RNA encoding HBSP was designed. In HBV-transfected HepaRG, siSP1RNA efficiently knocked down SP1RNA (6-fold decrease compared to scramble siRNA) without cross-reactivity on pgRNA expression (Fig.7C middle panel). Upon TNF-α stimulation, CCL2 secretion was significantly higher in HepaRG cells in which SP1RNA had been knocked down than in those transfected with a scramble siRNA (Fig.7D, lanes 10 vs 9: 46.3±4.0 vs 31.1±1.6 ng/ml, p=0.008).
Fig.7. Association between SP1RNA, HBSP and CCL2 expression in vitro.
(A, left panel) Western blot detection of p65 (RelA) total and its phosphorylated form (P-p65) on Ser536 in liver of TgHBSP and control mice treated by vehicle or CCl4 during 10 weeks. (A, right panel) Quantification of P-p65 related expression in liver of TgHBSP and control mice treated by vehicle (n=4), by CCl4 during 10 weeks in control (n=3) and TgHBSP (n=3) mice. (B, left panel) Total IκBα and phosphorylation level in HeLa cells transfected with pNeo or pHBSP plasmids and treated with TNF-α (25ng/ml) for 0, 5, 15 minutes. (B, right panel) Relative CCL2 mRNA quantification (normalized with GAPDH) by RTqPCR measured in HeLa expressing HBSP or control (pNeo) cells and treated or not with TNF-α (25ng/ml) during 30 minutes (n=5 experiments). (C, left panel) Secreted CCL2 protein quantification by HepaRG cells (stimulated or not with 50 ng/ml TNF-α) expressing pNeo (control) or pHBSP plasmid (n=7 experiments). (C, middle panel) HBV pgRNA and SP1RNA quantification from HepaRG cells co-transfected with pHBV plasmid and scramble (siSc) or SP1-siRNA (siSP1) (n=7 experiments). (D) Secreted CCL2 protein quantification by HepaRG cells (stimulated or not with TNF-α) expressing pNeo or pHBV plasmid with siSc or siSP1-siRNAs (n=6 experiments). (E, upper left panel) Representative H&E and Picro-Sirius red stained histological liver section (bar 2mm) and (E lower left panel) focus of Picro-Sirius red stained (bar 800µm) of WT and CCR2Ko mice treated for 2 weeks with CCl4-WT (n=) and CCl4. (E right panel) Fibrosis-related α-SMA protein expression in the liver of WT and CCR2KO mice treated for 2 weeks with oil (n=2) or CCl4 (WT with n=4 out of 5 and CCR2KO with n= 4 out of 5). Mann-Withney U-tests: *p<0.05 and **p<0.01.
To assess whether the reduction of CCL2 expression, through NF-κB, in HBSP expressing cells may account for the weaker liver fibrogenesis observed in TgHBSP, CCR2 invalidated (CCR2KO) (n=5) and control (n=5) mice were treated with CCl4. After 2 weeks of treatment, picro-sirius staining and western blot analysis of α-SMA expression revealed less marked liver fibrosis in CCR2KO mice, highlighting the main influence of this chemokine on fibrogenesis (Fig.7E).
CCL2 expression relies on SP1RNA in liver of HBV chronic infected patients
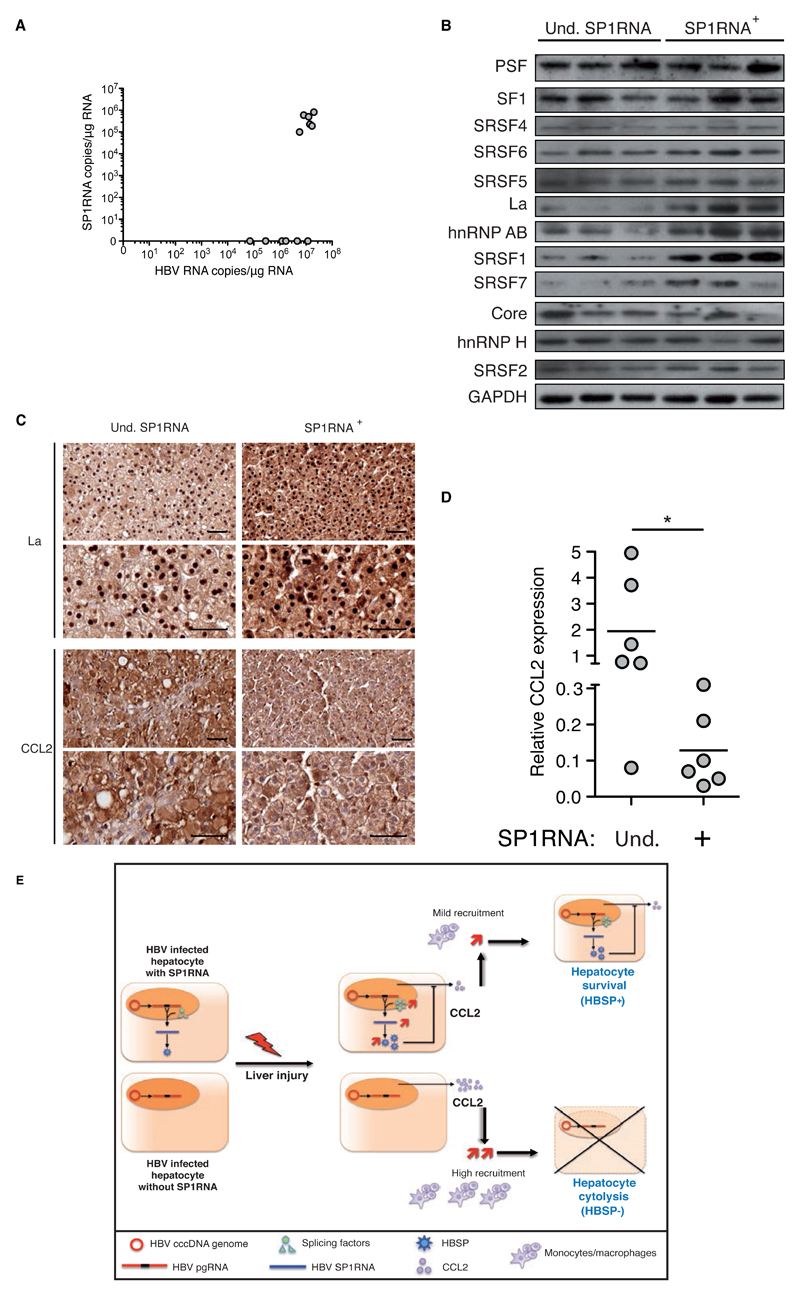
Several lines of evidence indicate that subjects with CHB have alterations in chemokines that may drive pathogenesis[27, 28]. However the contribution of CCL2 recruitment of monocytes/macrophages has not been addressed in this context. Only few studies showed no correlation of circulating CCL2 levels with HBV disease stage but without information on CCL2 hepatic expression[28, 29]. Furthermore, although HBV splicing regulation was previously suggested during liver disease[8–10, 30], the contribution of splicing factors expression was never considered. Therefore we investigated splicing factors and CCL2 expression in liver tissue surplus to diagnostic requirements available from 12 HBV monoinfected treatment-naive patients who were either undetectable or positive for HBV SP1RNA (Table S2). All had low-mild liver fibrosis (F0:F1:F2 fibrosis score in SP1RNA+ n=1:4:1, in SP1RNA- n=0:6:0) with a high level of HBV RNA expression (SP1RNA+ vs SP1RNA- groups: 1.3x107±5.3x106 vs 3.3x106±4.6x106 copies of HBV RNA/µg of liver RNA) (Fig.8A). For 3/6 patients from each group, liver sample size allowed the additional study of the expression of transregulatory splicing proteins. Despite exhibiting a similar stage of liver disease, SP1RNA positive compared to negative liver samples displayed a higher expression of hnRNPAB together with La and SRSF1 splicing factors (Fig.8B), which were also up-regulated in LPS treated HBV transgenic mice. In SP1RNA negative liver samples, La protein expression was mainly observed by immunohistochemistry in nucleus while it was increased and gathered in both nuclear and cytoplasmic compartments in SP1RNA positive liver samples (Fig.8C), highlighting the shuttling of La protein [31]. Regarding CCL2 expression on patient liver sections, a differential staining was noticed with an increase in SP1RNA negative hepatocytes (Fig.7D). The level of intrahepatic CCL2 transcripts was further quantified and SP1RNA positive liver samples displayed a 14 fold lower CCL2 expression compared to SP1RNA negative tissues (Fig.8D; 0.13±0.04 vs 1.87±0.80, p=0.01, respectively).
Fig.8. Association between SP1RNA, CCL2 and transregulatory splicing factors in HBV chronic carriers.
(A) HBV RNA and SP1RNA quantification in the liver of HBV chronic carriers (n=6 for undetectable (Und.) and n=6 for detectable SP1RNA groups). (B) Splicing factors expression in the liver of HBV chronic carriers with undetectable or detectable SP1RNA. (C) La and CCL2 immunochemistry on serial liver section counterstained with hematoxylin (upper and lower bar 50μm) from chronic HBV carrier with undetectable or detectable SP1RNA. (D) CCL2 mRNA quantification in the liver of HBV chronic carriers with undetectable (n=6) or detectable SP1RNA (n=6). (E) Schematic model for SP1RNA synthesis and HBSP activity during liver damage. Under liver damage SP1RNA/HBSP expressing hepatocyte through decreasing liver inflammatory microenvironment have a selective advantage leading to the increase of SP1RNA proportion during the course of HBV infection. Mann-Withney U-tests: *p<0.05 and **p<0.01.
In conclusion, enhancement of transregulatory splicing proteins expression and HBV pgRNA splicing in the liver of patients with CHB was associated with a reduction in CCL2 expression, as previously observed in transgenic mice and in vitro. Our findings support the critical role of infected hepatocyte through chemokine production in liver disease and suggest a novel feedback loop regulation displayed by HBV to counteract liver pathogenesis.
Discussion
Alternate splicing regulation of HBV transcripts was first described more than 20 years ago but its role in HBV disease has remained poorly defined[4, 5]. The proportion of dHBV has been shown to fluctuate considerably in chronic carriers and to be increased with the severity of liver pathogenesis[8, 9]. However the regulation of HBV AS and its impact on liver immunopathogenesis had not been investigated.
To decipher the mechanism of post-transcriptional regulation of HBV pgRNA, hepatic disease was induced in a HBV transgenic mouse model. In contrast to HBV infection models, transgenic mice present the benefice of a stable viral genome variability eliminating any potential interference of HBV cis-regulation on pgRNA/protein interactions. This model is sufficient to decipher the influence of liver damage on the viral post-transcriptional regulation even if the relevance of data obtained on liver pathogenesis is limited due to the transgene inducing HBV tolerance and to the constant amount of viral transcripts (including SP1RNA) and proteins. Interestingly, we showed that chemical liver damage led to a significant increase of HBV AS, suggesting an influence of the microenvironment on the splicing machinery. Indeed, the expression of transregulatory splicing proteins interacting with HBV pgRNA was affected by liver injuries in mice but also in liver of chronic HBV infected patients (Fig.3/8). Transcriptomic analysis from liver diseases samples including hepatocarcinoma have previously suggested the role of such dysregulations of AS factors[32], here we reported an impact on HBV AS in vitro, in TgHBV mice and in CHB liver patients. Whilst the modulation of transregulatory splicing protein expression differed in liver inflammation and fibrosis in TgHBV treated mice, both liver damages triggered the same activation of viral AS. Characterization of molecular pathways[33] involved in the control of trans-regulatory splicing proteins and consequences on genome expression of hepatocyte during liver injury will be important to investigate in further clinical studies.
The main impact of AS regulation is an enhancement of SP1RNA, which encodes HBSP. Unexpectedly, HBSP expression in transgenic mice reduced the liver fibrosis induced by chemical treatment. Our results showed that HBSP expression in hepatocytes of treated mice downregulated their CCL2 expression and consequently reduced the recruitment of monocytes/macrophages. However, an effect on T cells could not be excluded considering trends observed in cytometry and cytokines expression data. These tendencies may result either from a domino effect on monocyte/macrophage recruitment or from an independent direct activity of HBSP on the adaptative immune system. Nevertheless, the direct impact of HBSP on hepatocyte CCL2 expression was confirmed in cells expressing HBSP, as well as in a whole HBV genome context after SP1RNA knockdown. As previously reported[15], once activated by inflammation, the NF-kB signaling pathway is partially hijacked by HBSP and as observed in vitro, may account for the downregulation of CCL2 expression. However, TgHBV displayed a similar fibrotic disease to WT mice, despite a non-significant 1.4 fold decrease of CCL2 expression (data not shown) and enhancement of viral AS. In parallel, CCL2-downregulation was found in SP1RNA positive liver patients in HBV chronic carriers with comparable liver injury. These data suggest the capacity of HBSP protein to antagonize liver fibrogenesis in a dynamic regulation of immune-mediated liver damage during the course of HBV infection.
Chemokines and their receptors orchestrate the chronology of immune responses during liver injury. Among these chemokines, CCL2 plays a central role in the hepatic recruitment of activated inflammatory monocytes/macrophages expressing Ly6Chi CCR2+ markers[23, 26, 34]. Damaged hepatocytes have previously been suggested to be able to produce CCL2[35, 36] but were considered a minor source compared to activated HSCs and immune cells[35, 37]. Our data highlight the significant contribution of CCL2 produced by hepatocytes in liver damage, since CCL2 downregulation in TgHBSP mice had a direct impact on liver pathogenesis. Therefore, reduced monocyte/macrophage infiltrates may account for the differential expression of cytokines and also for the recruitment of other immune cells in 10 weeks CCl4-treated mice, which seemed also partially affected, compared to control mice. Nevertheless, the main role of CCL2 downregulation was supported by the data obtained from the single CCl4-injection in TgHBSP mice as well as the similar impairment of liver fibrosis to those observed in CCR2KO treated mice, although altered immune composition at baseline may limit conclusion for this last model. Influence of monocytes/macrophages during HBV pathogenesis has been poorly explored even if the modulation of noncytolytic antiviral activity via their cytokine secretion was reported [38–40]. Additionally, monocytes/macrophages could participate in HBV specific adaptive immunity through the formation of intrahepatic myeloid-cell aggregates enabling local CD8+ T cells expansion (iMATEs)[41, 42]. Thus, by countering monocyte/macrophage recruitment, HBSP might support the maintenance of a tolerogenic environment and favour immune escape[43]. Furthermore, the relationship between CCL2 and human HCC survival emphasized the clinical observation of HBV splicing enhancement prior to development of HCC and speculate about the role of HBSP in liver disease progression [44].
Strengthened by previous clinical data reporting an increase of HBV defective circulating particles related to SP1RNA with liver disease severity[8–10], our present study suggests a new feedback loop involving HBV AS to counteract liver injury. As proposed in Figure 8E, the hepatic microenvironment altered by liver damage may contribute to enhance splicing factors involved in HBV SP1RNA production, and thus HBSP expression. Hepatocytes expressing HBSP exhibit reduced CCL2 secretion and consequently attenuated monocyte/macrophage recruitment, thereby constraining inflammation. Although speculative at this stage, it is possible that hepatocytes expressing HBV SP1RNA are preferentially maintained by inhibition of local antiviral immunity, constituting a novel mechanism for viral persistence during chronic liver injury. To date, whether efficient viral suppression by antiviral therapy is accessible for the management of HBV infection, eradication of intracellular virus remains a challenge. CCL2 targeted immunotherapy in combination with nucleoside analog should contribute to facilitate an efficient HBV cure through a disruption of immune tolerance established during viral infection.
Supplementary Material
Acknowledgments
We thank N. Stadler, C. Cordier, B. Kane and F. Marliot for technical assistance; and Drs D. Auboeuf, C. Bourgeois, T. Heise, O. Ann for providing SRSF1 and PSF plasmids. We thank Drs ML. Michel, S. Briechler, H. Strick-Marchand, F. Pagès, D. Scott-Algara, A. Coulomb, J. Pol and T. Capiod for helpful scientific discussion. We thank animal facilities platform from Pitié and Broussais Universities. We thank FACS, proteomic and histological platforms of Broussais institute.
Financial support: This work was supported by grants from INSERM, UPMC, ANRS (n° N14015DR) and PHC-Tassili (11MDU826). MD was supported by ANRS (grant ASA14013DRA). YM was supported by French Ministry for Higher Education and Research and by the Ligue contre le Cancer (grant n° GB/MA/VSP-10504).
Abbreviations
- SR
serine-arginine rich protein
- hnRNP
heterogeneous nuclear ribonucleoprotein protein
- HBV
Hepatitis B Virus
- AS
Alternative splicing
- CHB
Chronic hepatitis B
- pgRNA
HBV pregenomic RNA
- SP1RNA
HBV single spliced pregenomic RNA
- wtHBV
Wild type HBV circulating particles
- dHBV
defective HBV circulating particles
- HCC
Hepatocarcinoma
- HBSP
Hepatitis B splicing generated protein
- TgHBV
HBV transgenic mouse
- TgHBSP
HBSP transgenic mouse
- WT
wild-type
- KO
knockout
- IP
Intraperitoneal
- CCl4
Carbon tetrachloride
- LPS
Lipopolysaccharide
- TAA
Thioacetamide
- BDL
Bile Duct ligation
- PBS
phosphate-buffered saline
- ALT
Alanine aminotransferase
- RTqPCR
reverse transcription polymerase chain reaction
- HBeAg
HBe antigen
- HBsAg
HBs antigen
- Fig
figure
- ROS
reactive oxygen species
Footnotes
Author contributions: Conception and design of these experiments MD, YM, DK and PS. Conduction the experiments: MD, YM, BL, HW, AS, FR, VF, JA, IQ, CC and PS. Analysis of the data: MD, YM, BL, CG, AB, CC, DS, ML, MM and PS. Provide biological samples: SG, UG, PK, AB, MM and ND. Wrote the paper: MD, MM and PS.
All authors have no conflict of interest.
References
- [1].Pohl M, Bortfeldt RH, Grutzmann K, et al. Alternative splicing of mutually exclusive exons--a review. Biosystems. 2013 Oct;114(1):31–38. doi: 10.1016/j.biosystems.2013.07.003. [DOI] [PubMed] [Google Scholar]
- [2].Hernandez-Lopez HR, Graham SV. Alternative splicing in human tumour viruses: a therapeutic target? Biochem J. 2012 Jul 15;445(2):145–156. doi: 10.1042/BJ20120413. [DOI] [PubMed] [Google Scholar]
- [3].Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010 Sep 15;430(3):379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- [4].Susuki T, Masui N, Kajino K, et al. Detection and mapping of spliced RNA from a human hepatoma cell line transfected with the hepatitis B virus genome. Proc Nat Acad Sci. 1989;86:8422–8426. doi: 10.1073/pnas.86.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu H-L, Chen P-J, Tu S-J, et al. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected Hep G2 cells. JVirol. 1991;65:1680–1686. doi: 10.1128/jvi.65.4.1680-1686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015 May;479–480C:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Terre S, Petit MA, Brechot C. Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J Virol. 1991 Oct;65(10):5539–5543. doi: 10.1128/jvi.65.10.5539-5543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bayliss J, Lim L, Thompson AJ, et al. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol. 2013 Nov;59(5):1022–1028. doi: 10.1016/j.jhep.2013.06.018. [DOI] [PubMed] [Google Scholar]
- [9].Redelsperger F, Lekbaby B, Mandouri Y, et al. Production of hepatitis B defective particles is dependent on liver status. Virology. 2012 Sep 15–30;431(1–2):21–28. doi: 10.1016/j.virol.2012.05.008. [DOI] [PubMed] [Google Scholar]
- [10].Soussan P, Pol J, Garreau F, et al. Expression of defective hepatitis B virus particles derived from singly spliced RNA is related to liver disease. J Infect Dis. 2008 Jul 15;198(2):218–225. doi: 10.1086/589623. [DOI] [PubMed] [Google Scholar]
- [11].Chen J, Wu M, Wang F, et al. Hepatitis B virus spliced variants are associated with an impaired response to interferon therapy. Sci Rep. 2015;5 doi: 10.1038/srep16459. 16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Soussan P, Garreau F, Zylberberg H, et al. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J Clin Invest. 2000 Jan;105(1):55–60. doi: 10.1172/JCI8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bayard F, Godon O, Nalpas B, et al. T-cell responses to hepatitis B splice-generated protein of hepatitis B virus and inflammatory cytokines/chemokines in chronic hepatitis B patients. ANRS study: HB EP 02 HBSP-FIBRO. J Viral Hepat. 2012 Dec;19(12):872–880. doi: 10.1111/j.1365-2893.2012.01611.x. [DOI] [PubMed] [Google Scholar]
- [14].Chen JY, Chen WN, Jiao BY, et al. Hepatitis B spliced protein (HBSP) promotes the carcinogenic effects of benzo [alpha] pyrene by interacting with microsomal epoxide hydrolase and enhancing its hydrolysis activity. BMC Cancer. 2014;14:282. doi: 10.1186/1471-2407-14-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pol JG, Lekbaby B, Redelsperger F, et al. Alternative splicing-regulated protein of hepatitis B virus hacks the TNF-alpha-stimulated signaling pathways and limits the extent of liver inflammation. FASEB J. 2015 Jan 28;5:1879–1889. doi: 10.1096/fj.14-258715. [DOI] [PubMed] [Google Scholar]
- [16].Cheng L, Li F, Bility MT, et al. Modeling hepatitis B virus infection, immunopathology and therapy in mice. Antiviral Res. 2015 Sep;121:1–8. doi: 10.1016/j.antiviral.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guidotti LG, Matzke B, Schaller H, et al. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995 Oct;69(10):6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rodero MP, Poupel L, Loyher PL, et al. Immune surveillance of the lung by migrating tissue monocytes. Elife. 2015 Jul 13;4:e07847. doi: 10.7554/eLife.07847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao J, Zhang Z, Luan Y, et al. Pathological functions of interleukin-22 in chronic liver inflammation and fibrosis with hepatitis B virus infection by promoting T helper 17 cell recruitment. Hepatology. 2014 Apr;59(4):1331–1342. doi: 10.1002/hep.26916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen Y, Wei H, Sun R, et al. Increased susceptibility to liver injury in hepatitis B virus transgenic mice involves NKG2D-ligand interaction and natural killer cells. Hepatology. 2007 Sep;46(3):706–715. doi: 10.1002/hep.21872. [DOI] [PubMed] [Google Scholar]
- [21].Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014 Oct;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pellicoro A, Ramachandran P, Iredale JP, et al. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014 Mar;14(3):181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- [23].Marra F, Tacke F. Roles for chemokines in liver disease. Gastroenterology. 2014 Sep;147(3):577–594 e571. doi: 10.1053/j.gastro.2014.06.043. [DOI] [PubMed] [Google Scholar]
- [24].Beattie L, Sawtell A, Mann J, et al. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J Hepatol. 2016 Oct;65(4):758–768. doi: 10.1016/j.jhep.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Karlmark KR, Zimmermann HW, Roderburg C, et al. The fractalkine receptor CX(3)CR1 protects against liver fibrosis by controlling differentiation and survival of infiltrating hepatic monocytes. Hepatology. 2010 Nov;52(5):1769–1782. doi: 10.1002/hep.23894. [DOI] [PubMed] [Google Scholar]
- [26].Baeck C, Wei X, Bartneck M, et al. Pharmacological inhibition of the chemokine C-C motif chemokine ligand 2 (monocyte chemoattractant protein 1) accelerates liver fibrosis regression by suppressing Ly-6C(+) macrophage infiltration in mice. Hepatology. 2014 Mar;59(3):1060–1072. doi: 10.1002/hep.26783. [DOI] [PubMed] [Google Scholar]
- [27].Ren YY, Liu YZ, Ding YP, et al. Immune characteristics of different immune phases in natural course of chronic HBV infection. Hepatogastroenterology. 2013 Jun;60(124):789–795. doi: 10.5754/hge12907. [DOI] [PubMed] [Google Scholar]
- [28].Tan AT, Koh S, Goh W, et al. A longitudinal analysis of innate and adaptive immune profile during hepatic flares in chronic hepatitis B. J Hepatol. 2010 Mar;52(3):330–339. doi: 10.1016/j.jhep.2009.12.015. [DOI] [PubMed] [Google Scholar]
- [29].Lian JQ, Yang XF, Zhao RR, et al. Expression profiles of circulating cytokines, chemokines and immune cells in patients with hepatitis B virus infection. Hepat Mon. 2014 Jun;14(6):e18892. doi: 10.5812/hepatmon.18892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Soussan P, Tuveri R, Nalpas B, et al. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J Hepatol. 2003 Mar;38(3):343–348. doi: 10.1016/s0168-8278(02)00422-1. [DOI] [PubMed] [Google Scholar]
- [31].Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010 May-Jun;1799(5–6):365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chettouh H, Fartoux L, Aoudjehane L, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013 Jul 1;73(13):3974–3986. doi: 10.1158/0008-5472.CAN-12-3824. [DOI] [PubMed] [Google Scholar]
- [33].Mallinjoud P, Villemin JP, Mortada H, et al. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res. 2014 Mar;24(3):511–521. doi: 10.1101/gr.162933.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dambach DM, Watson LM, Gray KR, et al. Role of CCR2 in macrophage migration into the liver during acetaminophen-induced hepatotoxicity in the mouse. Hepatology. 2002 May;35(5):1093–1103. doi: 10.1053/jhep.2002.33162. [DOI] [PubMed] [Google Scholar]
- [35].Saiman Y, Friedman SL. The role of chemokines in acute liver injury. Front Physiol. 2012;3:213. doi: 10.3389/fphys.2012.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ziraldo C, Vodovotz Y, Namas RA, et al. Central role for MCP-1/CCL2 in injury-induced inflammation revealed by in vitro, in silico, and clinical studies. PLoS One. 2013;8(12):e79804. doi: 10.1371/journal.pone.0079804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ait-Goughoulte M, Lucifora J, Zoulim F, et al. Innate antiviral immune responses to hepatitis B virus. Viruses. 2010 Jul;2(7):1394–1410. doi: 10.3390/v2071394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Boltjes A, Movita D, Boonstra A, et al. The role of Kupffer cells in hepatitis B and hepatitis C virus infections. J Hepatol. 2014 Sep;61(3):660–671. doi: 10.1016/j.jhep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- [39].Boltjes A, van Montfoort N, Biesta PJ, et al. Kupffer cells interact with hepatitis B surface antigen in vivo and in vitro, leading to proinflammatory cytokine production and natural killer cell function. J Infect Dis. 2014 Apr 15;211(8):1268–1278. doi: 10.1093/infdis/jiu599. [DOI] [PubMed] [Google Scholar]
- [40].Hosel M, Quasdorff M, Wiegmann K, et al. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009 Dec;50(6):1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- [41].Huang LR, Wohlleber D, Reisinger F, et al. Intrahepatic myeloid-cell aggregates enable local proliferation of CD8(+) T cells and successful immunotherapy against chronic viral liver infection. Nat Immunol. 2013 Jun;14(6):574–583. doi: 10.1038/ni.2573. [DOI] [PubMed] [Google Scholar]
- [42].Nakamoto N, Kanai T. Role of toll-like receptors in immune activation and tolerance in the liver. Front Immunol. 2014;5:221. doi: 10.3389/fimmu.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Heymann F, Peusquens J, Ludwig-Portugall I, et al. Liver inflammation abrogates immunological tolerance induced by Kupffer cells. Hepatology. 2015 Mar 21; doi: 10.1002/hep.27793. [DOI] [PubMed] [Google Scholar]
- [44].Li X, Yao W, Yuan Y, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2015 Oct 9; doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- Author names in bold designate shared co-first authorship
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.