Abstract
The innate immune system is crucial for eventual control of infections, but may also contribute to pathology. Listeria monocytogenes is an intracellular gram-positive bacteria and a major cause of food-borne disease. However, important knowledge on the interactions between L. monocytogenes and the immune system is still missing. Here we report that Listeria DNA is sorted into extracellular vesicles (EV)s in infected cells and delivered to bystander cells to stimulate the cGAS-STING pathway. This was also observed during infections with Francisella tularensis and Legionella pneumophila. We identify the multivesicular body protein MVB12b as a target for TBK1 phosphorylation, which is essential for sorting of DNA into EVs and stimulation of bystander cells. EVs from Listeria-infected cells inhibited T cell proliferation, and primed T cells for apoptosis. Collectively, we describe a pathway for EV-mediated delivery of foreign DNA to bystander cells, and suggest that intracellular bacteria exploit this pathway to impair anti-bacterial defense.
Keywords: Innate immunology, DNA sensing, cGAS-STING pathway, extracellular vesicles, inter-cellular communication
Introduction
Listeria monocytogenes is an intracellular gram-positive bacteria and the causative agent of listeriosis, which often occurs during pregnancy, immunosuppression or extremes of age 1. L. monocytogenes is taken up into cellular vacuoles, from which the bacteria escape through the action of the cytolysin listeriolysin O, thus allowing replication in the cytoplasm 2. The immune system is essential for control of infection, with both innate and adaptive components being important. For instance, mice lacking the cytokine tumor necrosis factor (TNF)α or MyD88, a central adaptor in induction of TNF expression, are highly susceptible to L. monocytogenes infection 3,4. Likewise, T cells are essential for sterilizing immunity, and for long-term protection 5.
In addition to the protective actions of the immune system, it also contributes to pathology. The cytokine interferon (IFN)β, components in the IFNβ-induction pathway, and the IFNα/β receptor are known to increase susceptibility to Listeria disease 6–9. Therefore, full understanding of the mechanisms that govern the IFN pathway during Listeria infection may provide knowledge that can be used therapeutically. Nucleic acids are potent stimulators of production of type I IFNs 10. Nucleic acids can be sensed in endosomes by Toll-like receptors (TLR), with TLR3 and 7/8 detecting RNA, and TLR9 detecting DNA 11. In the cytoplasm, RNA is detected by the DEAD-box helicases RIG-I and MDA5, and signal via the adaptor protein MAVS 12,13, while DNA is detected by cGAS and signals via STING 14,15. Downstream of the adaptor protein, the pathways merge at the kinase TBK1, which phosphorylates the transcription factor IFN regulatory factor 3 (IRF3) to activate transcription of type I IFN genes. In T cells, the cGAS-STING pathway induces little or no type I IFN expression 16–18 but inhibits proliferation and induces cell death 17–19.
We previously reported that L. monocytogenes induces IFNβ expression in human macrophages through the cGAS-STING pathway 20, and other reports have suggested that bacterial cyclic-di-nucleotides and bacterial RNA can also stimulate IFNβ expression 21,22. Thus, cells infected with L. monocytogenes, can be activated through a range of innate immune pathways. However, there is limited knowledge on whether L. monocytogenes infection stimulates innate immune responses in bystander cells, what mechanisms may be involved, and what the functional impact is.
Results
Supernatants from cells infected with intracellular bacteria contain IFN-inducing potential
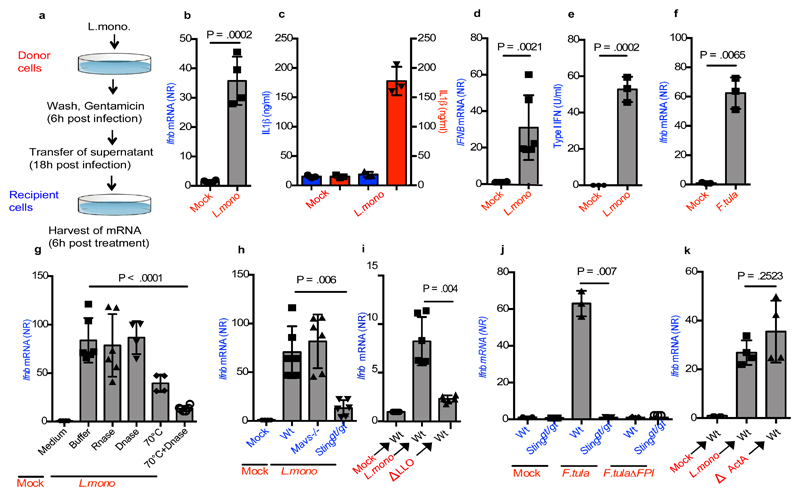
We were interested in exploring whether infected cells were able to send signals to non-infected cells, thus propagating immune responses. To this end, we used a setup where one set of cells (called donor cells, red) were infected with L. monocytogenes, and supernatants were subsequently harvested and transferred to a second set of cells (called recipient cells, blue), which were analysed for immune activation. (Figure 1a). The supernatants from the infected cells induced strong Ifnb expression in wild type (Wt) recipient MEFs, despite the lack of live bacteria in the supernatants and irrespective of whether donor cells were treated with chloramphenicol or gentamicin (Figure 1b and Supplementary Figure 1a, 1b). We observed minimal cell death in the donor cells under these experimental conditions (Supplementary Figure 1c), and treatment of donor cells with the pan-caspase inhibitor z-VAD-fmk during infection did not affect the stimulation of recipient cells (Supplementary Figure 1d). Initiation of gentamicin treatment as early as 1 h post infection of donor cells did not affect the ability of supernatants to stimulate recipient cells (Supplementary Figure 1e). In contrast to the induced Ifnb expression, interleukin (IL) 1β production was not induced in cells receiving supernatants from Listeria-infected cultures (Figure 1c). The observed induction of Ifnb mRNA, and Ifna4 mRNA, in recipient cells was dependent on the presence of cells in the donor cell tissue dishes (Supplementary Figure 1f), and was not explained by transfer of bacteria or bacterial products targeting TLRs (Supplementary Figure 1g).
Figure 1. Supernatants from cells infected with intracellular bacteria contain IFN-inducing potential.
(a) Schematic representation of the experimental set-up. (b) Relative Ifnb mRNA levels in MEFs treated with supernatants from cells infected with L. monocytogenes (MOI 200) or receiving mock treatment (n=4). (c) IL1β levels in cultures from BMDCs treated with supernatants from mock- or L. monocytogenes-infected cells (MOI 200) 3 or infected directly with L. monocytogenes 53 (n=3). (d) IFNB mRNA levels in PBMCs stimulated with supernatants from THP1 cells infected with L. monocytogenes (n=6). (e) Type I IFN bioactivity levels in PBMC recipient cells stimulated with supernatants from L. monocytogenes-infected donor PBMCs (n=3). (f) Relative Ifnb mRNA levels in MEFs stimulated with supernatants from cells infected with F.tularensis (MOI 400) or receiving mock treatment (n=3). (g) Ifnb mRNA was measured in Recipient cells stimulated with supernatants subjected to treatment with RNase, DNase, heat, or heat and DNase prior to transfer to recipient cells (n=6,6,4,4,6). (h) Induction of Ifnb mRNA in Wt, Mavs-/- and Stinggt/gt MEFs receiving supernatants from mock- and L. monocytogenes-infected donor MEFs (n=6). (i, j) Induction of Ifnb mRNA in Wt and Stinggt/gt cells, receiving supernatants from donor MEFs given mock treatment or infected with wt L. monocytogenes or F.tularensis or the respective mutants unable to escape into the cytoplasm: ΔLLO L. monocytogenes and ΔFPI F.tularensis. (n=4,3) (k) Induction of Ifnb mRNA in Wt cell receiving supernatants from donor MEFs infected with wt or ΔActA L. monocytogenes (n=4). The presented data are representative of at least 3 independent experiments. The Ifnb/IFNB mRNA levels were normalized to bactin/BACTIN mRNA levels and shown as relative levels compared to mock. Data are shown as mean ± SD. NR, normalized ratio. P values were calculated using 2-tailed unpaired students t-test.
The ability of bacteria-infected cells to transfer IFN-inducing potential to bystander cells was dose dependent, since we observed correlation between the infection dose and (i) the degree of bacterial infection in the donor cells, (ii) the induction of Ifnb expression in donor cells, and (iii) the induction of Ifnb expression in recipient cells (Supplementary Figure 1h-j). Importantly, supernatants from human THP1 cells and PBMCs infected with L. monocytogenes stimulated IFNB expression and type I IFN bioactivity, respectively, in recipient cells (Figure 1d-e). Finally, we found that the phenomenon was not restricted to L. monocytogenes infection, since supernatants from MEFs infected with Francisella tularensis or Legionella pneumophila also induced Ifnb expression in recipient cells (Figure 1f and Supplementary Figure 1k).
To investigate the nature of the transferable IFN-inducing entity, supernatants from infected MEFs were treated with DNase or RNase (Supplementary Figure 1l). This treatment did not alter induction of Ifnb expression in recipient cells (Figure 1g). Although nuclease treatment alone did not alter induction of Ifnb mRNA in recipient cells, the response was modestly reduced upon heat treatment of the supernatants prior to addition onto recipient cells, and further reduced when heat was combined with DNase treatment (Figure 1g).
In order to confirm the role for DNA in IFNβ induction in bystander cells, supernatants from L. monocytogenes-infected donor cells were used to stimulate Wt, Mavs-/- and Stinggt/gt recipient cells. Ifnb expression in the recipient cells was found to be dependent on STING but not MAVS (Figure 1h). The phenotypic differences between the cellular genotypes tested were unlikely to be due to clonal abnormalities (Supplementary Figure 1m-1r). Moreover, the ability of supernatants from L. monocytogenes- or F. tularensis-infected cells to induce Ifnb expression in recipient cells was dependent on escape from vacuoles into the cytosol in donor cells (Figure 1i, 1j). Finally, we found that infection of donor cells with a L. monocytogenes ActA-deletion mutant unable to mediate cell-cell spread via efferocytosis23 did not alter the Ifnb induction in recipient cells (Figure 1k).
Foreign intracellular DNA stimulates IFNβ expression in bystander cells via EVs
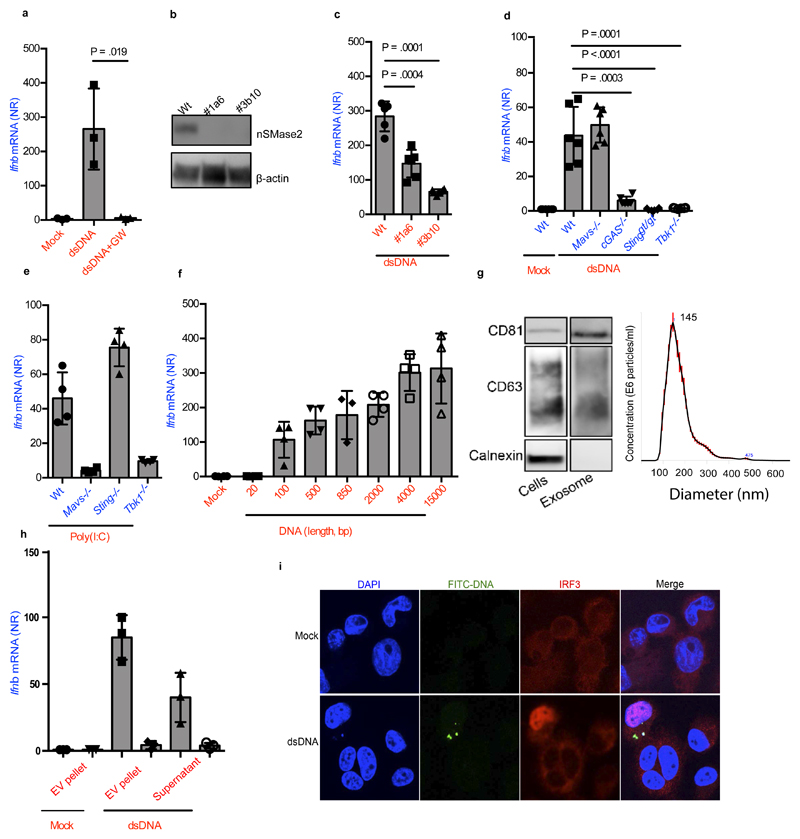
Based on these results, we decided to examine the role for EVs, such as exosomes, as the system responsible for delivery of IFN-stimulating DNA to recipient cells. Exosomes are vesicles derived from the multivesicular bodies (MVB) that contain cytoplasmic molecules, sorted in a controlled manner 24. Wt bone-marrow derived macrophages (BMM)s were transfected with double-stranded DNA, and cultures were incubated overnight in the presence or absence of GW4869, an inhibitor of neutral sphingomyelinase (nSmase2) required for exosome biogenesis 25,26 (Supplementary Figure 2a). Interestingly, Ifnb expression was induced in Wt BMM recipient cells stimulated with supernatant from DNA-transfected or electroporated donor cells, and this response was abrogated by treatment of donor cells with GW4869 (Figure 2a, Supplementary Figure 2b). GW4869 alone did not inhibit DNA-transfection-stimulated Ifnb expression per se (Supplementary Figure 2c). In agreement with the data based on the nSmase2 inhibitor, supernatants from DNA-transfected MEFs stably knocked-out for nSmase2 had significantly reduced capacity to induce Ifnb expression in Wt MEF recipient cells and exosomes from these cells had significantly less levels of CD63 and CD81 compared to Wt (Figure 2b, 2c, Supplementary Figure 2d, 2a). Supernatants from DNA-transfected Wt cells retained the capacity to induce Ifnb expression in IFNα/β receptor deficient recipient cells, and accordingly recombinant IFNβ only marginally affected Ifnb expression in MEFs (Supplementary Figure 2e-f). Together, these data argue for a role for EVs, and exclude a role for type I IFN, in the stimulation of IFNβ in recipient cells. Similar to bystander-activation by Listeria-infected cells, recipient cells lacking STING were unable to induce Ifnb expression upon treatment with supernatants from DNA-transfected cells (Figure 2d). Additionally, we tested cells deficient for cGAS and TBK1, which were incapable of inducing a response to the supernatant, whereas MAVS-deficient cells retained a normal response (Figure 2d). We noted that supernatants from donor cells transfected with RNA were also able to stimulate IFNβ expression in recipient cells, and this was dependent on MAVS and TBK1 (Figure 2e). Although TBK1 is also activated by cytokines and TLRs 27, stimulation of these pathways in DNA-transfected Sting-/- donor cells enabled Ifnb induction in Wt recipient cells (Supplementary Figure 2g), suggesting the observed phenomenon to be a specific feature of cytoplasmic nucleic acid sensors. Finally, stimulation of Ifnb expression in recipient cells correlated with the length of the DNA transfected into donor cells (Figure 2f).
Figure 2. Foreign intracellular DNA stimulates IFNβ expression in bystander cells through EVs.
(a) Ifnb induction in BMMs stimulated for 6 h with supernatants isolated from BMMs 18 h after Lipofectamine transfection with dsDNA (1μg/ml) in the presence or absence of GW4869 (10μM) (n=3). (b) Immunoblot analysis of two nSmase2-/- MEF clones targeted with two different gRNAs (1a6, 3b10). (c) Ifnb mRNA levels in recipient Wt MEFs treated with supernatants from Wt or nSmase2-/- MEFs transfected with DNA (n=6). (d) Induction of Ifnb mRNA in Wt, Mavs-/-, cGas-/-, Stinggt/gt and Tbk1-/- cells upon stimulation with supernatants from Wt MEFs transfected with DNA (n=6). (e) Induction of Ifnb in Wt, Mavs-/-, Stinggt/gt and Tbk1-/- MEFs upon stimulation with supernatant from cells transfected with poly(I:C) (1 μg/ml) (n=4). (f) Ifnb mRNA induction in MEFs stimulated with supernatants from donor cells transfected with dsDNA of the shown sizes (n=4). (g) Cellular lysates and isolated EVs were analyzed by Immunoblotting for exosomal markers CD81 and CD63, and the ER marker calnexin. The EVs were also subjected to Nanoparticle Tracking Analysis for evaluation of size distribution. The Red error bars indicate one standard error of the mean (+/-), while the black curve represents the mean of three independent measurements. (h) Induction of Ifnb mRNA in Wt MEFs stimulated with EVs from dsDNA- or mock transfected MEFs (+/- GW4869), and with the remaining supernatant from the EV isolation procedure (n=4). (i) PMA-differentiated THP1 cells treated for 4 hours with supernatants from Wt MEFs stimulated with FITC-labelled DNA were subjected to confocal microscopy for visualization of FITC and IRF3. Nuclei were stained with DAPI. Merge includes the bright field image. The presented data are representative of at least 3 independent experiments. The Ifnb mRNA levels were normalized to bactin mRNA levels and shown as relative levels compared to mock. Data are shown as mean ± SD. P values were calculated using 2-tailed unpaired students t-test.
To further examine whether EVs from DNA-transfected cells could deliver pathogen-associated molecular pattern (PAMP)s to adjacent cells, EVs were isolated from supernatants of transfected donor cells (Figure 2g). The EV-containing pellet and the remaining supernatants were used to stimulate Wt MEF recipient cells. Importantly, recipient cells treated with the resuspended EV pellet exhibited high Ifnb expression, and this was abrogated if EVs were isolated from DNA-transfected donor cells treated with GW4869 (Figure 2h), or if the recipient cells lacked cGAS (Supplementary Figure 2h). The residual Ifnb induction observed in response to supernatant is likely explained by the fact that not all EVs are pelleted upon ultracentrifugation (Supplementary Figure 2i) 28. In further support of foreign DNA being sorted into EVs in donor cells, we were able to detect FITC in CD63-positive vesicles collected from FITC-DNA transfected CD63-RFP MEFs (Supplementary Figure 2j). Finally, we observed that recipient cells receiving supernatants from FITC-DNA-transfected donor cells were positive for FITC staining, and that it was the FITC+ cells that contained nuclear IRF3 (Figure 2i). Collectively, these data, together with the requirement for cGAS in recipient cells, demonstrate that EVs are responsible for delivering DNA as the IFN-stimulating PAMPs from DNA-transfected donor cells to recipient cells.
Listeria infection activates EV-dependent stimulation of bystander cells
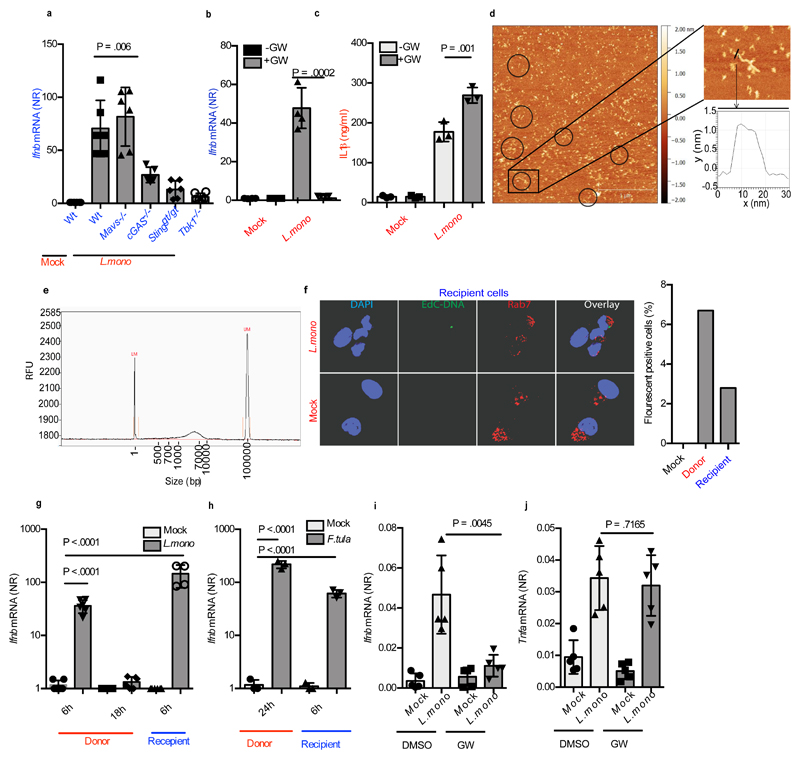
The results above demonstrate that DNA is the PAMP in EVs from donor cells stimulated with synthetic DNA. However, L. monocytogenes produces cyclic-di-AMP, which can stimulate STING directly 21,29. Therefore, we examined the requirement for cGAS in recipient cells receiving supernatants from infected cells, and also whether bacterial DNA was delivered to recipient cells. When supernatants from L.monocytogenes-infected Wt cells were used to stimulate recipient cells deficient in either STING, cGAS, MAVS or TBK1, a high induction of Ifnb was observed in Wt and MAVS deficient cells, but not in cGAS, STING or TBK1 deficient cells (Figure. 3a). Furthermore, when L.monocytogenes infection occurred in the presence of GW4869, supernatants from the donor cells failed to stimulate Ifnb expression in recipient cells (Figure 3b). GW4869 treatment did not affect L.monocytogenes replication in donor cells (Supplementary Figure 3a). Intriguingly, inhibition of exosome biogenesis augmented IL1β secretion by macrophages infected with L.monocytogenes (Figure 3c), suggesting a crosstalk between the cGAS and inflammasome pathways.
Figure 3. Listeria infection activates EV dependent stimulation of type I IFN expression in bystander cells.
(a) Ifnb mRNA levels in Wt, Mavs-/-, cGas-/-, Stinggt/gt and Tbk1-/- cells stimulated for 6h with supernatants from MEFs infected with L. monocytogenes (MOI 200) for 18 h (n=5). (b) Induction of Ifnb in MEFs (n=4) and (c) IL1β in the supernatants of BMMs (n=3) stimulated with supernatants from L. monocytogenes-infected MEFs in the presence or absence of GW4869 (10μM). (d) DNA extracted from EVs from supernatants of MEFs infected with L. monocytogenes (MOI 200) were analyzed by AFM. Circles are shown around extended structures with a width and height similar to DNA. Scale bar 500nm. The boxed part of the image is magnified in the image to the right for measurement of the height of exosomal DNA (~1.5 nm). (e) Fragment analyzer electrophoresis of DNA extracted from EVs from supernatants of MEFs infected with L.monocytogenes. Markers, 1 bp (left) and 100,000 bp (right). (f) Supernatants from Wt MEFs infected with EdC-labelled L. monocytogenes for 18 h, were transferred to recipient cells for 2h. The cells were stained for the early endosome marker Rab7 and EdC-labelled bacterial DNA was visualized using Click-it chemistry. The cells were analyzed by confocal microscopy. Nuclei were stained with DAPI. The graph to the right represents quantification of cells with positive fluorescent signal. For each treatment, more than 200 cells were examined (blinded). (g) Induction of Ifnb mRNA in MEFs infected with L. monocytogenes for 6h or 18h or treated for 6h with Gentamicin-treated supernatant from L. monocytogenes-infected MEFs (n=4). (h) Induction of Ifnb mRNA in MEFs infected with F.tularensis for 24h or treated for 6h with Gentamicin-treated supernatant from F.tularensis -infected MEFs (n=3). (i, j) Ifnb and Tnfa mRNA levels in spleens of mice left untreated or infected with L. monocytogenes (1x106cfu) for 24 h in the presence of GW4869 (GW, 0.125 μg per gram bodyweight) (n=5 mice). The presented data are representative of at least 3 independent experiments. The Ifnb and Tnfa mRNA levels were normalized to bactin mRNA levels and shown as relative levels compared to mock. Data are shown as mean ± SD. P values were calculated using 2-tailed unpaired students t-test.
To examine whether bacterial DNA was sorted into EVs, we first isolated DNA from EVs in the supernatants from L.monocytogenes-infected cells, and visualized the DNA by atomic force microscopy (AFM). The linear topography of DNA with a measured height of ˜1.5 nm was observed for the extended linear structures (Figure 3d, Supplementary Figure 3b), similar to that observed for plasmid DNA (Supplementary Figure 3c). When analyzing DNA from EVs of bacteria-infected cells using the automated fragment analyzer gel electrophoresis, we observed the DNA to be 700-10000 bp and 400-1500bp depending on the markers used (Figure 3e and Supplementary Figure 3d). This analysis also showed that degradation of the DNA in the EVs was dependent on combined detergent and DNAse treatment, thus confirming DNA to be located inside the EVs (Supplementary Figure 3d). Finally, L.monocytogenes with EdC-labeled genomic DNA was used for infection of donor cells (Supplementary Figure 3e). Using CLICK chemistry in recipient cells, we were able to detect transfer of bacterial DNA to the cytoplasm of recipient cells, with no detectable colocalization with the early endosome marker Rab7 (Figure 3f).
To assess the relative contribution of infected versus bystander cells to the total Ifnb expression, we infected MEFs with L.monocytogenes. Cells were harvested 6h and 18h post infection. From another set of donor cells, supernatants were harvested 18h post infection and used to stimulate Wt recipient MEFs. Measurement of Ifnb mRNA in the different cell populations showed that the response evoked in the recipient cells was comparable to the response in the donor cells that were infected with the bacteria (Figure 3g). Similar results were obtained for F.tularensis- infection and DNA transfection (Figure 3h, Supplementary Figure 3f). Finally, to evaluate whether EVs also contributed to the Ifnb induced by L.monocytogenes in vivo, mice were treated with GW4869 before and during infection, and Ifnb expression was measured in the spleen. Importantly, inhibition of exosome biogenesis significantly reduced the levels of Ifnb mRNA and IFN-stimulated gene Mx1 in the spleen of infected animals (Figure 3i, Supplementary Figure 3g), but not Tnfa mRNA (Figure 3j). The effect of GW4869 on Ifnb mRNA induction was not due to a general suppressive effect on Ifnb transcription, since Ifnb mRNA levels in the spleen after TLR9 agonist treatment was not affected (Supplementary Figure 3h). Collectively, these results demonstrate that EV-mediated DNA transfer to bystander cells occurs during infection with intracellular bacteria.
EVs mediate STING-dependent apoptosis in T lymphocytes
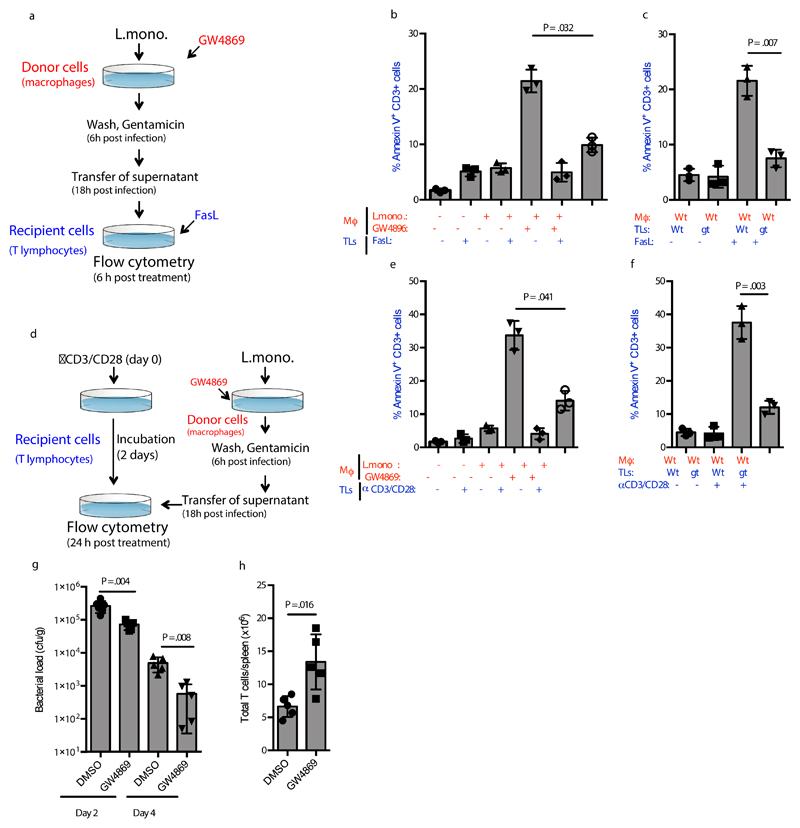
The pro-bacterial activity of the type I IFN system has been ascribed partly to a pro-apoptotic activity in T lymphocytes 6–9. In addition, STING-dependent signaling in T lymphocytes has been recently reported to impair proliferation and to induce cell death in this cell type 18. To investigate whether DNA-containing EVs from L.monocytogenes-infected cells were able to modulate T lymphocyte phenotypes through STING-dependent paracrine signaling, we treated T lymphocytes with Fas ligand (FasL) in the presence or absence of EVs from infected macrophages (Figure 4a). Interestingly, supernatants from L.monocytogenes-infected macrophages augmented FasL-stimulated apoptosis in splenic T lymphocytes, in a manner inhibited by GW4869 treatment of the macrophages and dependent on cGAS and STING expression in the T lymphocytes (Figure 4b, 4c, Supplementary Figure 4a, 4b). The T cells did express cGAS and STING (Supplementary Figure 4c). In a separate experimental setup, T lymphocytes were activated with anti-CD3/CD28 and subsequently treated with supernatants from L.monocytogenes-infected macrophages (Figure 4d). These experiments showed that EVs from infected cells also promoted apoptosis in activated T lymphocytes, and this was also dependent on STING expression in the lymphocytes (Figure 4e, f). Thus, activation of STING signaling in T lymphocytes by EVs from L.monocytogenes-infected cells promotes apoptosis.
Figure 4. EVs from Listeria-infected cells augments apoptosis in T lymphocytes.
(a) Illustration of the experimental set-up for the data shown in panel b and c. (b, c) Apoptosis in Wt and Stinggt/gt Splenic T lymphocytes treated for 6 h with FasL (5 ng/ml) and supernatants from BMMs infected with L. monocytogenes (MOI 200) in the presence or absence of GW4869 (10μM) (n=3). (d) Illustration of the experimental set-up for the data shown in panel e and f. (e, f) Apoptosis in Wt and Stinggt/gt Splenic T lymphocytes activated by CD3/CD28 for 48 followed by treatment for 24 h with supernatants from BMMs infected with L. monocytogenes (MOI 200) in the presence or absence of GW4869 (10μM) (n=3). The cells were evaluated for Annexin V staining and data are shown as % positive cells. (g) Bacterial load in spleens of mice infected with L. monocytogenes (1x106cfu) for 48 or 96 h in the presence of GW4869 (0.3125 µg per gram bodyweight). n = 6,5,5,5, mice. (h) Total number of splenic T cells in mice infected for 96 h with L. monocytogenes (1x106cfu) for 96 h in the presence of GW4869 (0.3125 µg per gram bodyweight) (n= 5 mice). The presented data are representative of 3 independent experiments. Data are shown as mean ± SD. P values were calculated using 2-tailed unpaired students t-test. Mϕ, macrophage. TLs, T lymphocytes. Gt, Stinggt/gt.
To explore whether treatment with the nSMase2 inhibitor in vivo affected bacterial growth, we used a low dose of GW4869 which was tolerated by mice over the course of an eight-day period, and still led to reduced Ifnb mRNA expression in the liver after L.monocytogenes infection (Supplementary Figure 4d, e). Importantly, GW4869 treatment also enabled the mice to better control the bacteria and was associated with significantly more expansion of T cells (Figure 4g, h). Thus, targeting of EVs through inhibition of nSMase2 reduces lymphocyte apoptosis and promotes anti-bacterial activity.
Foreign intracellular DNA is sorted into EVs in a STING-TBK1-dependent manner
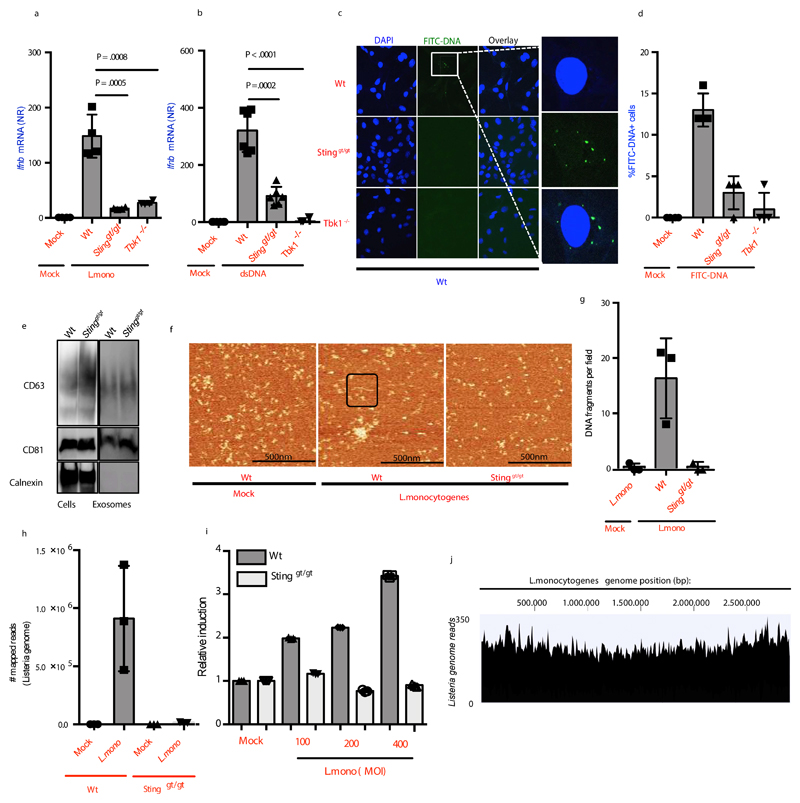
Next, we started to explore which cellular proteins were involved in sorting of cytoplasmic DNA into EVs in donor cells. We observed that supernatants from DNA-transfected or L.monocytogenes-infected STING or TBK1 deficient MEFs failed to stimulate Ifnb and Ifna4 mRNA expression or type I IFN bioactivity in Wt recipient cells (Figure 5a, 5b, Supplementary Figure 5a-5b). This phenotype was not rescued by pretreatment of the donor cells with IFNβ Supplementary Figure 5c. The requirement for STING in donor cells was also observed after infection with L.monocytogenes in human THP1 cells and after infections with F.tularensis in MEFs (Supplementary Figure 5d, 5e). Furthermore, FITC-labeled DNA was observed in Wt recipient cells that received supernatant from Wt donor cells transfected with FITC-labeled DNA, but not in Wt recipient cells stimulated with supernatant from STING and TBK1 deficient donor cells (Figure 5c, 5d), although the donor cells were transfected to similar efficiencies (Supplementary Figure 5f). The data suggest that the STING-TBK1 axis is required for sorting of cytoplasmic DNA into EVs.
Figure 5. Sorting of Foreign DNA sorting into EVs requires STING and TBK1.
(a, b) Ifnb mRNA levels in Wt MEFs treated with supernatants from Wt, Stinggt/gt or Tbk1-/- MEFs (a) infected with L.monocytogenes (n=4), or (b) transfected with DNA (2μg/ml). (c) Wt MEFs treated for 2h with supernatants from Wt, Stinggt/gt or Tbk1-/- MEFs stimulated with FITC-labelled DNA (1μg/ml, 18h) were subjected to confocal microscopy for visualization of FITC. Nuclei were stained with DAPI (n=6). (d) Quantification of the data shown in panel c. 100 cells were evaluated per group. (e) Immunoblot analysis of cell lysates and EVs isolated from Wt and Stinggt/gt cells. (f) Representatives images of AFM of DNA extracted from EVs from Wt and STING-deficient cells left untreated or infected with L. monocytogenes. Exosomal DNA in black box. (g) Quantification of the data shown in panel f (n=3). (h) Bacterial DNA in EVs isolated from Wt and Stinggt/gt MEFs infected with L.monocytogenes for 18 h. The data are represented as mapped number of bacterial genome reads from deep sequencing analysis (n=3). (i) Quantification of DNA in EVs isolated from Wt and Stinggt/gt MEFs infected with L.monocytogenes for 18 h using Picogreen (n=3). (j) Merged tracks of mapped bacterial DNA reads in EV DNA from L.monocytogenes-infected MEFs. The results are shown as tracked reads versus position in the L.monocytogenes genome. The presented data are representative of at least 2 independent experiments. The Ifnb mRNA levels were normalized to bactin mRNA and shown as relative levels compared to mock. Data are shown as mean ± SD. P values were calculated using 2-tailed unpaired students t-test.
To further investigate the role of STING in EV-mediated DNA transfer, we sought to determine if STING was required for the actual formation of EVs. To this end, we isolated and characterized EVs from both Wt and STING deficient cells, but observed comparable levels of exosomal markers CD63 and CD81 (Figure 5e). Given that exosomes originate from multivesicular endosomes and because STING can dock on perinuclear endosomes 14, we investigated if STING was required for the packaging of DNA into intraluminal vesicles in the endosome. DNA was extracted from EVs of Wt and STING deficient MEF cells infected with L.monocytogenes. We found that EVs derived from Wt cells packaged bacterial DNA to a significantly greater extent than EVs derived from infected STING-deficient cells, as evaluated by AFM (Figure 5f, 5g), deep sequencing (Figure 5h, Supplementary Figure 5g, Supplementary Table 1), and Picogreen DNA quantification (Figure 5i). Deep sequencing also revealed that the entire bacterial genome was represented in the EVs isolated from infected Wt cells (Figure 5j). Collectively, these data demonstrate that sorting of foreign DNA into EVs is dependent on STING.
MVB12b is an MVB phosphorylation target of TBK1 required for transfer of DNA by EVs
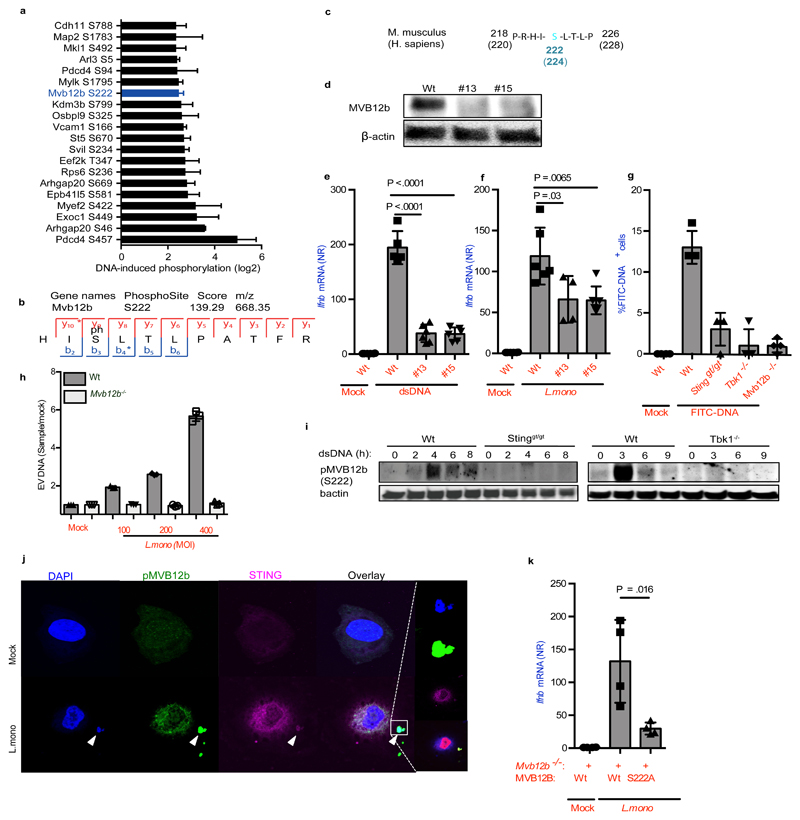
Given the requirement for the STING-TBK1 axis in donor cells for sorting of bacterial DNA into EVs and for stimulation of bystander cells, we wanted to identify proteins in MVBs phosphorylated by TBK1. Therefore, we performed phosphoproteomics on MEFs transfected with dsDNA in comparison to untreated cells using stable isotope labeling with amino acids in cell culture (SILAC) 30. Figure 6a shows the top 20 phosphorylated sites upon dsDNA stimulation. The full dataset is available in Supplementary Table 2. Interestingly, among the most strongly phosphorylated proteins we identified MVB12b/FAM125b, which is part of the endosomal sorting complexes required for transport (ESCRT) I 31. The ESCRT-I complex, which consists of Vps23, Vps28, Vps37, and MVB12, is involved in the generation of MVBs, and recognition of cargo 31. MVB12b was phosphorylated at Serine 222 (Figure 6b), which is conserved between human and mouse (Figure 6c). There is 96% amino acid homology between human and mouse MVB12b, suggesting findings in the mouse system also to reflect human biology.
Figure 6. Sorting of foreign DNA into EVs require TBK1 mediated phosphorylation of MVB12b.
(a) Top 20 induced phosphorylated peptides upon dsDNA transfection in MEFs plotted as median with range from four experiments. (b) MS/MS spectrum of the identified phosphorylation of serine 222 on Mvb12b. (c) Amino acid sequence of protein murine MVB12b from amino acid 218-226 flanking TBK1 phospho-target Serine 222. For comparison, human MVB12b is also shown. (d) Immunoblot for MVB12b and β actin on cell lysates from Wt and two Mvb12b-/- clones (made with independent gRNAs). (e, f) Induction of Ifnb mRNA in Wt MEFs stimulated with supernatants from Wt and Mvb12b-/- MEFs transfected with dsDNA (2μg/ml) (n=6) or infected with L.monocytogenes (MOI 200) (n=4). (g) Quantification of % FITC-positive recipient Wt MEFs after treatment with supernatants from the indicated MEF donor cells, transfected with FITC-DNA (1ug/ml) for 6h (n=4). (h) Quantification of DNA in EVs isolated from Wt and Mvb12b-/- MEFs infected with L.monocytogenes for 18 h using Picogreen (n=3). (i) Phosphorylation of MVB12b in Wt MEFs upon dsDNA stimulation compared to Stinggt/gt and Tbk1-/- MEFs. β actin was used as loading control. (j) Co-localization of STING and phospho-MVB12b 6 h after infection with L.monocytogenes. Nuclei were stained with DAPI. (k) Induction of Ifnb mRNA in Wt recipient MEFs stimulated with supernatants from L.monocytogenes-infected Mvb12b-/- donor MEFs reconstituted with Wt or S222A mutants (n=4). The presented data are representative of at least 2 independent experiments. The Ifnb mRNA levels were normalized to bactin mRNA and shown as relative levels compared to mock. Data are shown as mean ± SD. P values were calculated using 2-tailed unpaired students t-test.
To examine the role of MVB12b in mediating bystander activation of the cGAS-STING pathway, we generated MVB12b-deficient MEFs (Figure 6d). Deletion of MVB12b did not impair the ability of cells to secrete EVs or to activate STING-dependent signaling (Supplementary Figure 6a, 6b). Importantly, supernatants from Mvb12b-/- MEFs infected with L.monocytogenes or transfected or electroporated with DNA induced significantly less activation of Ifnb expression in recipient cells (Figure 6e, 6f, Supplementary Figure 6c, 6d). In agreement with this, supernatants from Mvb12b-/- donor MEFs transfected with FITC-DNA failed to deliver DNA into recipient Wt MEFs, and EVs from infected Mvb12b-/- donor MEFs packaged reduced levels of DNA (Figure 6g, 6h, Supplementary Figure 6e). To examine whether the phosphorylation of MVB12b was dependent on STING and TBK1 we compared phospho-S222 MVB12b levels in lysates from WT, Stinggt/gt, and Tbk1-/- cells. MVB12b was indeed phosphorylated after delivery of DNA into the cytoplasm, and this was dependent on STING and TBK1 (Figure 6i, Supplementary Figure 6f). Moreover, phospho-MVB12b colocalized with STING and DNA in the cytoplasm of L.monocytogenes-infected and DNA-transfected cells (Figure 6j and Supplementary Figure 6g). To ascertain the role of S222 in MVB12b in activating the cGAS-STING pathway, MVB12b-deficient MEFs were reconstituted with Wt or S222A MVB12b, and infected with L.monocytogenes or transfected with DNA. Importantly, supernatants from cells reconstituted with Wt MVB12b induced more Ifnb expression in recipient cells than supernatants from cells reconstituted with S222A MVB12b (Figure 6k and Supplementary Figure 6h). Thus, TBK1 phosphorylates Mvb12b to facilitate sorting of cytoplasmic DNA into EVs.
Discussion
In this work we show that DNA from intracellular bacteria is sorted into EVs through a STING-TBK1-MVB12b pathway and delivered to bystander cells to stimulate the cGAS-STING pathway. This enables danger signaling to spread across tissues prior to the infecting pathogen. In case of L. monocytogenes, it is known that type I IFNs contribute to the pathology of infection 6–8, and we found that EVs from L. monocytogenes-infected macrophages primed T cells for FasL-mediated apoptosis and also induced apoptosis in activated T cells. Thus, intracellular bacteria may exploit the STING-TBK1-MVB12b pathway to impair the antibacterial immune response, promoting establishment of infection.
Full orchestration of immune responses requires extensive interaction between cells. In addition to cytokines, there is an emerging appreciation of intercellular signals carrying information on the stress and infection status of donor cells. For instance, cGAMP can spread to adjacent bystander cells through gap junctions 32, or be packaged into virus particles33,34. In addition, EVs are capable of delivering cellular 35,36, as well as viral 37 components across tissues, thereby designing the ensuring immune response. However, the exosome system can be exploited by pathogens. For instance, herpesviruses and hepatitis B virus deliver miRNAs to surrounding cells to shape the immune response and viral replication microenvironment 38,39. Our finding that EVs from L.monocytogenes-infected cells promote lymphocyte death and impair anti-bacterial activity in vivo suggests that intracellular bacteria take advantage of the EV machinery to dampen the immunological activity of T cells. The in vivo relevance of stimulation of the cGAS-STING pathway in recipient cells may go beyond T cell death, since the type I IFN system has been reported to counter-act production of IL17A, which promotes neutrophil expansion during F.tularensis infection 40.
In addition to stimulation of type I IFN production through the cGAS-STING pathway, cytosolic DNA can activate the AIM2 and NLRP3 inflammasomes 41,42. Inflammasome activation leads to maturation of IL1β and activation of pyroptosis. The AIM2 and NLRP3 inflammasomes have been reported to be important for control of infections with L.monocytogenes and F.tularensis infection 43,44. Since both the AIM2 and cGAS pathways are stimulated by DNA, but activate downstream responses with opposing effects towards many intracellular bacterial infections, one important question relates to what determines whether a given species or population of DNA activates cGAS or AIM2? A comparative characterization of the requirements for activation of cGAS versus AIM2 could reveal regulatory differences between the two pathways that the bacteria may exploit to skew the DNA-activated responses towards a cGAS response. In support of this hypothesis, we found that levels of IL1β in cultures from L.monocytogenes-infected murine macrophages and in spleens from infected mice were elevated if treated with GW4869, and that the DNA-containing EVs did not induce IL1β production.
We found the mechanism of sorting of nucleic acids into EVs to be dependent on STING and TBK1. Moreover, we identified phosphorylation of S222 of MVB12b in donor cells to be essential for stimulation of STING signaling in recipient cells. These data strongly suggest that activated TBK1 targets ESCRT I to promote sorting of DNA into EVs. The mechanistic details on how cytoplasmic DNA is sorted into EVs through a specific mechanism dependent on TBK1-mediated phosphorylation of MVB12b at S222 remains to be uncovered. It was reported that sorting of hepatitis C virus RNA into exosomes was dependent on Annexin A2, which has RNA-binding properties, and is involved in membrane vesicle trafficking 37.
In conclusion, we report that bacterial DNA in the cytoplasm can be sorted into EVs and delivered to bystander cells, leading to stimulation of the cGAS-STING pathway. The sorting of foreign DNA into EVs proceeds through a pathway dependent on STING, TBK1, and MVB12b. This paracrine signalling driven by bacterial DNA promotes death of T lymphocytes and impairs anti-bacterial defence. Further understanding of the interplay between the AIM2-inflammasome and STING-IFN pathways may uncover how intracellular bacteria modulate DNA-stimulated immune responses for their own benefit.
Experimental Procedures
Mice
For in vivo treatment of mice with the exosome maturation inhibitor GW4869 (Sigma), 8-12 weeks old C57BL/6N mice were injected intraperitoneally with GW4869 (0.3125 or 1.25µg per gram bodyweight) or carrier control once per day for five consecutive days. On day six, mice were intraperitoneally infected with 1x106 L. monocytogenes (strain LO28). Bacterial inoculum was prepared as previously described 45. At 24 hours p.i., mice were sacrificed and spleens were harvested. For cfu assays, livers and spleens were weighed, homogenized in PBS, and 1:10 serial dilutions were plated on Oxford agar plates (Merck). Colonies were counted after approximately 30 hours at 37°C. Some mice were treated with ODN1826 (InvivoGen) at a dose of 10 μg per mouse. Animals were housed under specific pathogen-free conditions according to FELASA guidelines and animal experiments have been approved by the Vienna University of Veterinary Medicine institutional ethics committee and performed according to protocols approved by the Austrian law (BMWF 68.205/0032-WF/II/3b/2014).
Cell culture, transfections and transfer experiments
Wt or gene-modified MEFs were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% Fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. Murine bone marrow derived macrophages were generated as described previously 46. Briefly, bone marrow from mouse femurs and tibia were isolated and differentiated in the presence of GM-CSF (40ng/ml) in 10cm dishes. The media was replenished with RPMI with 10%FBS and GM-CSF on day 3 and day 7 post isolation. PBMCs were isolated and cultured as previously described 47. L929 cells were cultured in DMEM with 5% FBS and 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. THP1 cells were cultured in RPMI media supplemented with 10% Fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. Cells were differentiated using PMA (200nM) for 24 h. The PMA was washed off and cells were used for experiments 24 h later. Transfection of cells was performed using Lipofectamine-2000 or Amaxa 4D-nucleofector according to manufacturer’s instructions. The double-stranded DNA used was a 60mer HSV dsDNA. DNA fragments of varying sizes (NoLimits DNA fragments) were obtained from ThermoFisher. 2x105 cells/ml were seeded onto 24-well cell culture plates in 500µl media. For transfer experiments, producer cells were washed 3 times 6 h after DNA transfection and fresh media was added. The supernatants were collected 18h post transfection, centrifuged at 300g for 5min and transferred to recipient cells. Mock indicates uninfected or lipofectamine transfected cells. When indicated, cells were treated with 10μM GW4869 (D1692, Sigma). Only experiments where positive and negative controls gave the expected results were further analyzed and included in the study.
Culture and infection with L. monocytogenes, F. tularensis, and L. pneumophila
To culture Listeria monocytogenes (L028, 10403S, 2473, ΔLLO, and ΔActA 48, a single clone from a blood agar plate was used to inoculate 5ml of Brain-heart infusion (BHI) media. For Click-it chemistry, bacteria were grown in BHI media containing EdC. Optical density was measured at 600nm. An OD of 1 was considered equivalent to 2x109 CFU/ml. Cells infected with L.monocytogenes (MOI 200) for 6 hours were treated with 50µg/ml of Gentamicin (Sandoz, Cat.no. 010256) for an hour to kill extracellular bacteria, after which the cells were washed 3 times and the media was replaced with media containing 10µg/ml Gentamicin and incubated overnight. In some experiments we used chloramphenicol (C0378, Sigma Aldrich) instead of Gentamicin. To inhibit caspase activation, cells were treated with z-VAD-fmk (10μM, Invivogen) 30 min prior to infection. The compound was found to inhibit lipopolysaccharide + nigericin induced IL1β by more than 90% (not shown). F. tularensis subspecies novicida WT (U112) or ΔFPI mutant 49 strains were cultured in Tryptic Soy Broth (TSB) supplemented with cysteine (0.1%w/v). MEFs were infected with a MOI of 400:1 by spinoculation (15 minutes, 2 000g at room temperature). At 4 h post-invasion, cells were washed twice with PBS and the medium was replaced with medium containing 5 µg/ml Gentamicin and incubated for an additional 20 h. L. pneumophila Corby wild type was grown on buffered charcoal-yeast extract agar plates at 37 °C for three days. Bacterial material was then inoculated in an appropriate volume of PBS and optical density at 600 nm (OD600) was determined by an Ultrospec 10 Cell Density Meter (GE Healthcare Europe, Freiburg, Germany). An OD600 of one corresponded to 2 x 109 bacteria/mL. The desired multiplicity of infection was obtained by serial dilutions in PBS and lastly, in cell specific medium.
Genetic modification of MEFs by CRISPR/Cas9 and stable ectopic expression
Guide RNAs of the following sequences nSmase2a (1a6: TGCCCTCCACGCCGTGTCCT; 3b10: GCGACGAGGCTGCCAACGGC); MVB12b (TCCCGACACTACCGGCTACCTTCCG) were cloned into p lentiCRISPR-v2. 3rd generation lentiviral particles were used to transduce MEFs and clones were selected using puromycin at a final concentration of 2μg/ml.
For MEFs stable expressing CD63-RFP, cells were transduced with 3rd generation lentiviral particles expressing plasmid pCT-CD63-RFP (CYTO120-PA1-SBI, Biocat) and sorted for RFP+ cells using a FACS ARIA III cell sorter. The cells were cultured under puromycin selection (2μg/ml) and clones were picked.
Evaluation of cell death
MEFs were infected with L.monocytogenes (MOI 200) for 6h after which the cells were washed and treated with gentamicin (50µg/ml) for 1h, the cells were then incubated overnight in media containing (10µg/ml) gentamicin. The next day, cells were trypsinized and stained with AnnexinV and PI (Dead Cell Apoptosis Kit with Annexin V Alexa Fluor™ 488 & Propidium Iodide (PI), ThermoFisher) according to manufacturer’s instructions. Fluorescence was measured using a Novocyte flow cytometer and analysis was performed using FlowJo v.10 (Tree Star).
Induction of apoptosis in T cells and analysis for Annexin V
CD3+ cells were isolated from mouse spleens using EasySep kit from STEMCELL Technology. To evaluate the effects of EVs from L.monocytogenes-infected cells on apoptosis in bystander cells, resting, apoptosis-induced (low concentration of FasL, 5 ng/ml, Sigma-Aldrich, catalog no. F0427), or activated (Human T-Activator CD3/CD28 Dynabeads™, ThermoFisher Scientific catalog no 11161D) T lymphocytes were treated with supernatants from macrophages infected for 18h with L.monocytogenes (+/- GW4869). Apoptosis was evaluated by flow cytometry analysis Annexin V staining (ThermoFisher).
Murine splenic cells were stained with CD3-PE and T cells were sorted using FACS ARIA III sorter. Isolated murine T cells were activated with Mouse T-activator CD3/CD28 (11456D, ThermoFisher) for 48h and lysed using RIPA buffer (89901, ThermoFisher). The lysates were subjected to Western blotting.
Flow cytometry
Spleen cells were stained using fluorescently conjugated Abs against CD19 (D3) and CD3 (17A2) (eBioscience). Fluorescence was measured using LSRFortessa flow cytometer (BD Biosciences), and analysis was performed using FlowJo v.10 (Tree Star).
ELISA
Supernatants from cells, were analyzed for cytokine levels by ELISA for murine cytokine IL-1β using matched Ab pairs obtained from R&D Systems as described elsewhere 50.
ImageStream
Fluorescence of EVs was detected and acquired using the ImageStream MK II Imaging Flow Cytometer (Amnis, Co., Seattle, WA, USA). Isolated EVs were re-suspended in 300μl PBS and run at a low flow rate setting. Particles for each measurement were excited at 200mW using 561 nm laser for CD63-RFPmRuby and the 488 nm laser for FITC. A 60X magnification was used for all samples. Brightfield images were acquired on channel 1, FITC on channel 2, CD63-RFP on channel 4 and side scatter images on channel 6. Data was analyzed using IDEAS software v6.2 (Amnis Corporation).
Enzyme treatment of supernatants and EVs
Supernatants of infected cells were centrifuged and treated with 10μg RNase (EN0531, ThermoFisher) or 10 units of recombinant DNase I (Roche 04716728001) in incubation buffer at 37°C for 30min before transferring to recipient cells. Supernatants that were heated at 70°C were cooled to room temperature before treating with DNase as mentioned above. Mock was treated with DNase incubation buffer.
For analysis by fragment analyzer, EVs were treated with DNase I (18068-015, ThermoFisher) according to manufacturer’s instructions. DNase I was inactivated with EDTA at 65°C for 10min. The sample was divided in 4 parts and treated with 1% NP-40 for 15min on ice or 600μg/ml Proteinase K for 20min at 40°C and inactivated at 70°C for 20min. The samples were then subjected to DNase treatment as mentioned above before DNA isolation using the Qiagen DNA micro kit (56304).
Confocal microscopy
Experiments were carried out on cells seeded on cover slips. Cells were fixed with ice-cold methanol for 5 min at -20°C, washed with PBS and stained with DAPI, following which they were mounted onto glass slides using Prolong Gold. For nuclear translocation and co-localization assays, cells were fixed and permeabilized with 4% paraformaldehyde and 0.1% Triton-X. The cells were then blocked with 2.5% BSA and stained with the following antibodies: Rab7 (9367, Cell Signaling.), IRF3 (Cell Signaling, 4302), STING (AF6516), phospho S222 MVB12b (GenScript, Antibody production service) and corresponding secondary antibodies. Images were obtained on Zeiss LSM 710 confocal microscope using a 63× 1.4 oil-immersion objective and data were analyzed using ImageJ software. For click-it chemistry, EdC-labeled (T511307,Sigma) DNA of bacteria were detected in fixed cells according to manufacturer’s instructions (ThermoFisher, C10337) and AlexaFluor488 was detected under the microscope.
Phospho proteomics
MEFs were SILAC labeled and stimulated with dsDNA. Cells were lysed in 4% SDS, 10 mM Hepes, pH 8.0 for 15 min at room temperature with sonication. Proteins were reduced with 10 mm DTT for 30 min and then subjected to alkylation for 45 min with 55 mm iodoacetamide in the dark. To remove detergent, acetone (−20 °C) was added to a final concentration of 80% v/v, and proteins were precipitated for at least 2 h at −20 °C. The protein pellets were dissolved in 8 M urea, 10 mM Hepes, pH 8.0. Digestion with LysC was carried out for 3 h at room temperature. Samples were diluted with 4 volumes of 50 mM ammonium bicarbonate and further digested with trypsin overnight at room temperature. Peptides of DNA stimulated and untreated cells with distinct isotopic labels were mixed 1:1. Samples were desalted with C18, and incubated with TiO2 beads (MZ-Analysentechnik) pre-incubated with dihydrobenzoic acid (Sigma). After incubation, beads were washed with 30% acetonitrile and 0.5% (v/v) trifluoroacetic acid (TFA) in water followed by a second wash with 80% acetonitrile with 0.1% TFA. Phosphopeptides were eluted from beads with 15% NH3 and desalted on C18 StageTips.
We separated peptides on a Thermo Scientific EASY-nLC 1000 HPLC system (Thermo Fisher Scientific, Odense, Denmark). Columns (75-μm inner diameter, 50-cm length) were in-house packed with 1.9-μm C18 particles (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Peptides were loaded in buffer A (0.5% formic acid) and separated with a gradient from 5% to 30% buffer B (80% acetonitrile, 0.5% formic acid) for 120 min at 250 nl/min. The column temperature was set to 50 °C. A quadrupole Orbitrap mass spectrometer (34) (Q Exactive, Thermo Fisher Scientific) was directly coupled to the liquid chromatograph via a nano-electrospray source. The Q Exactive was operated in a data-dependent mode. The survey scan range was set to 300 to 1,650 m/z, with a resolution of 70,000 at m/z 200. Up to the 10 most abundant isotope patterns with a charge of ≥2 were subjected to higher-energy collisional dissociation with a normalized collision energy of 25, an isolation window of 3 Th, and a resolution of 17,500 at m/z 200. To limit repeated sequencing, dynamic exclusion of sequenced peptides was set to 20 s. Thresholds for ion injection time and ion target value were set to 20 ms and 3 × 10E6 for the survey scans and to 120 ms and 1E5 for the MS/MS scans. Data were acquired using Xcalibur software (Thermo Scientific).
To process MS raw files, we employed MaxQuant software (v. 1.5.3.34) 51. We used the Andromeda search engine (10.1021/pr101065j) integrated in MaxQuant, to search MS/MS spectra against the UniProtKB FASTA database (version from May 2014). Enzyme specificity was set to trypsin allowing cleavage N-terminal to proline and up to two miscleavages. Peptides required a minimum length of seven amino acids for identification. Carbamidomethylation was set as a fixed modification, and acetylation (N terminus), methionine oxidation as well as serine, threonine and tyrosine phosphorylation as variable modifications. A false discovery rate (FDR) cutoff of 1% was applied at the peptide and protein levels. Initial precursor mass deviation of up to 4.5 ppm and fragment mass deviation up to 20 ppm were allowed.
Furthermore, identifications were filtered for common contaminants (247 proteins) and identifications solely based on a modified site. Median SILAC ratios (of DNA treated versus untreated cells) for phosphopeptides were calculated from four individual experiments (two with label swap). Phosphopeptides, which were quantified in less than two experiments or those with a standard deviation >2 were excluded from the analysis.
Real time qPCR
mRNA was isolated using High Pure mRNA isolation kit (Roche) according to manufacturer’s instructions. qPCR was performed on extracted mRNA using Taqman primers Ifnb1 (Mm00439552), Ifna4 (Mm00833969), Tnfa (Mn00443258_m1), FlaA (FlaA Fwd GTTCAATCTTGCAACGTATGCGTC, FlaA Rev CCACTACCTAAAGTGATTGTTCCAGCA), beta actin (Mm00607939). Relative fold induction of a sample was calculated relative to mock treated sample using the ΔΔCt method.
Bioassays for human and murine type I IFN
Bioactive type I IFN was measured on cell supernatants by use of HEK-Blue™ IFN-α/β cells as reporter cells according to the manufacturer instructions (Invivogen). Type I IFN was measured using a cell-based assay. Briefly, L929 cells were cultured in DMEM 5%FCS and seeded into 96 well plates. Supernatant from stimulated BMDC’s were added as serial dilutions to the cells for 24h at 37°C alongside a positive control of muIFNα (100IU/ml). The next day, cells were infected with VSV/V10 and the cells were incubated until plaques were visible under the microscope. A 50% protection of virus-induced cell death in a well was used to define 1 U/ml of Type I IFN.
Western blotting
Cells were lysed using RIPA buffer in the presence of protease and phosphatase inhibitors. The centrifuged supernatants were subjected to SDS-PAGE (Criterion™ TGX™) and immunoblotting. Transfer to PVDF membranes was carried out using Trans-Blot Turbo™ Transfer System®. The blots were blocked with 2.5% BSA in PBS containing 0.1% Tween20 before incubating with the following primary antibodies: CD81 (sc-9158, Santa Cruz), CD63 (sc-5275, Santa Cruz), calnexin (ab-22595, Abcam), nSmase2a (sc-166637, Santa Cruz), cGAS (SAB2100310, Sigma-Aldrich), STING (AF6516, R&D), vinculin (V9131, Sigma), HRP-conjugated actin (ab-49906, Abcam). Corresponding peroxidase-conjugated secondary antibodies were used (Jackson ImmunoResearch). Membranes were developed using Super Signal West Dura extended duration substrate (Thermo Scientific, 34076). Original blots are shown in Supplementary Figure 7.
Nanoparticle Tracking Analysis
Samples were measured using the NanoSight LM10 system (NanoSight, Malvern Instruments,Malvern UK). Videos and data were analysed and recorded using the NTA software (version 3). Samples were diluted in particle-free PBS to a final volume of 0.4ml. Measurements were recorded with camera level 12 and detection threshold 5, three times, from which the final histogram was averaged.
EV isolation
EVs were isolated from the cell supernatant 52. Briefly, the supernatant was subjected to sequential centrifugation at 300g for 10 min at 4°C. The supernatant was then subjected to centrifugations at 2000g for 30 min followed by centrifugation at 20,000g at 45 min, both at 4°C. The supernatant was ultracentrifuged at 100,000g for 90min and the pellet was washed again in PBS and EVs pelleted at 100,000g for 90min.
Quantification of DNA using Picogreen
Isolated DNA was quantified using Quanti-it PicoGreen dsDNA reagent and kit (Invitrogen) according to manufacturer’s instructions. Briefly, total DNA was diluted in the TE buffer supplied. The DNA concentrations were determined using λ-DNA diluted to generate a high-range standard curve. Following incubation of samples with Picogreen for 5minutes, the florescence was measured at 485nm. Data are represented as fold induction relative to mock-infected cells.
MVB12b reconstitution
Plasmids encoding for Wt or S222A in the coding region for MVB12b were synthesized by Integrated DNA technologies. The coding sequence was cloned into the pCCLPGKpuro vector and was either transfected directly into MEFs or transduced following lentiviral production using 2nd generation lentiviral constructs. Transduced cells were passaged twice before seeding for the experiment.
Atomic Force Microscopy
The topology of exosomal DNA was visualized by AFM (Agilent AFM series 5500). The topography of DNA is a linear structure, whereas other cellular components such as proteins are non-linear. The topology was recorded by tapping mode AFM. Silicon nitride cantilevers (OMCL-TR400PSA) were purchased from Olympus. DNA samples were mixed with immobilization buffer (1XTAE-Mg2+-Ni2+: 1XTAE containing 12.5 mM MgCl2 and 5 mM NiCl2) prior to depositing on a mica surface. The sample (2µl) was immobilized on a freshly cleaved mica surface for 1 minute. After that, 400 ul of imaging buffer (1XTAE-Mg2+-Ni2+) was added into the liquid cell. All AFM images were analyzed by Gwyddion software.
Fragment Analyzer
DNA samples were run on a fragment analyzer using the High sensitivity large fragment analysis kit (DNF-493-0500 or DNF-464-0500, Advanced Analyticals) according to manufacturer’s instructions.
Deep sequencing
Total DNA was isolated from EVs using Qiagen DNA micro kit (56304), and sequenced on Illumina® sequencing systems using Miseq-NexteraTM XT (Department of Molecular Medicine, Aarhus University Hospital). The reads from each sample were mapped against the mouse genome (mm38) using CLCBIO workbench. The distribution and average coverage of the mapped reads in each chromosome in the mouse genome were normalized against the total number of reads in each sample, which were found to be comparable across all samples (Supplementary Table 1). The remaining unmapped reads from the previous step were mapped against the Listeria genome (NC_017544.1) for all samples.
Statistical analysis and reproducibility of results
The data are shown as means of biological replicates ± SD. Statistically significant differences between groups were determined using two-tailed Student's t-test when the data exhibited normal distribution and Wilcoxon rank-sum test when the data set did not pass the normal distribution test. The data shown are from single experiments. All experiments were performed at least 3 times with similar results.
Supplementary Material
Acknowledgements
The technical assistance of Kirsten Stadel Petersen, the NGS facility at the Department of Molecular Medicine, Aarhus University Hospital and FACS Core facility, Aarhus University is greatly appreciated. This work was funded by The Danish Medical Research Council (12-124330) (S.R.P.), The Novo Nordisk Foundation (NNF18OC0030274) (S.R.P.), The Lundbeck Foundation (R198-2015-171) (S.R.P.), The European Research Council (786602) (S.R.P.), EU FP7 Mobilex programme (DFF – 5053-00011) (R.N.), The BMBF (JPI-AMR - FKZ 01Kl1702) and DFG (SFB/TR-84 TP C01) (B.S.) and the Austrian Science Fund (FWF) through grant P 25186-B22 (T.D.).
Footnotes
Author Contributions
R.N. and S.R.P. conceived the idea and designed the experiments. R.N., R.T., F.M., T.P., A.K., B.Z, S.A., A.M., W.B., A.A., T.K., and M.K.T. performed the experiments. FM designed, performed and analyzed the phosphoproteomics experiment. E.F analyzed the NGS data. C.K.H., B.S., K.A.H., V.H., T.H., K.V.G., T.D. and S.R.P. supervised experiments. R.N. and S.R.P. wrote the manuscript.
Data Availability
The full NGS dataset is available at ENA (European Nucleotide Archive) with the identifier ‘ena-STUDY-AARHUS UNIVERSITY 12-12-2018-17:12:31:528-124’, under accession number ‘PRJEB30324’. The full mass spectrometry dataset is available at http://www.ebi.ac.uk/pride/archive/projects/PXD012135.
Original immuno blots are shown in Supplementary Figure 7.
Competing Interests
The authors declare no competing interests.
References
- 1.Barbuddhe SB, Chakraborty T. Listeria as an enteroinvasive gastrointestinal pathogen. Curr Top Microbiol Immunol. 2009;337:173–195. doi: 10.1007/978-3-642-01846-6_6. [DOI] [PubMed] [Google Scholar]
- 2.Cossart P, et al. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989;57:3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothe J, et al. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature. 1993;364:798–802. doi: 10.1038/364798a0. [DOI] [PubMed] [Google Scholar]
- 4.Edelson BT, Unanue ER. MyD88-dependent but Toll-like receptor 2-independent innate immunity to Listeria: no role for either in macrophage listericidal activity. J Immunol. 2002;169:3869–3875. doi: 10.4049/jimmunol.169.7.3869. [DOI] [PubMed] [Google Scholar]
- 5.Ladel CH, Flesch IE, Arnoldi J, Kaufmann SH. Studies with MHC-deficient knock-out mice reveal impact of both MHC I- and MHC II-dependent T cell responses on Listeria monocytogenes infection. J Immunol. 1994;153:3116–3122. [PubMed] [Google Scholar]
- 6.Stockinger S, et al. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- 7.Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. The Journal of experimental medicine. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. The Journal of experimental medicine. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell RM, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. The Journal of experimental medicine. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luecke S, Paludan SR. Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine. 2016 doi: 10.1016/j.cyto.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg RK, et al. T cells detect intracellular DNA but fail to induce type I IFN responses: implications for restriction of HIV replication. PLoS One. 2014;9:e84513. doi: 10.1371/journal.pone.0084513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin B, et al. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J Immunol. 2017;199:397–402. doi: 10.4049/jimmunol.1601999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerboni S, et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. The Journal of experimental medicine. 2017;214:1769–1785. doi: 10.1084/jem.20161674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulen MF, et al. Signalling strength determines proapoptotic functions of STING. Nature communications. 2017;8 doi: 10.1038/s41467-017-00573-w. 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen K, et al. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdullah Z, et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czuczman MA, et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature. 2014;509:230–234. doi: 10.1038/nature13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schorey JS, Cheng Y, Singh PP, Smith VL. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015;16:24–43. doi: 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 26.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. The Journal of biological chemistry. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 28.Jeppesen DK, et al. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J Extracell Vesicles. 2014;3:25011. doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem Sci. 2017;42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bridgeman A, et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349:1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentili M, et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 35.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 37.Dreux M, et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell host & microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, et al. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol. 2017;91 doi: 10.1128/JVI.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry T, et al. Type I IFN signaling constrains IL-17A/F secretion by gammadelta T cells during bacterial infections. J Immunol. 2010;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaidt MM, et al. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell. 2017 doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature immunology. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. European journal of immunology. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stockinger S, et al. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS pathogens. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm CK, et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nature communications. 2016;7 doi: 10.1038/ncomms10680. 10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogunjimi B, et al. Inborn errors in RNA polymerase III underlie severe varicella zoster virus infections. The Journal of clinical investigation. 2017;127:3543–3556. doi: 10.1172/JCI92280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. The Journal of cell biology. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss DS, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horan KA, et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 52.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3 22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 53.Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.