Abstract
OipA is a phase-variable virulence factor of Helicobacter pylori. Mutations in oipA to turn the gene phase on in a cag pathogenicity island (PAI)-negative strain of H. pylori (J68) or phase off in a cag PAI-positive strain (26695) demonstrated that phase on oipA alleles in both strains had both increased oipA mRNA and human gastric adenocarcinoma (AGS) cell adherence compared to isogenic oipA phase off mutants. An oipA phase off mutant of H. pylori 26695 demonstrated decreased IL-8 secretion by AGS cells and failure to translocate the cag PAI effector CagA. Increased attachment by OipA expressing cag PAI-negative H. pylori J68 failed to alter secreted IL-8 levels. Thus, OipA is necessary but not sufficient for the induction of IL-8; however, it is necessary for translocation of the oncoprotein CagA. Perhaps the nearly invariant phase on status of oipA alleles among cag PAI-positive H. pylori isolates relates to the role of this outer membrane protein in effective translocation of CagA. oipA mRNA comparisons between AGS cell-adherent and non-adherent H. pylori 26695 revealed significantly greater levels in the adherent cells. This may allow H. pylori to adapt to conditions of host cell contact by altering expression of this virulence factor.
Keywords: phase variation, adhesin, CagA translocation, interleukin-8
This study demonstrates that expression of OipA of Helicobacter pylori is necessary for translocation of the oncoprotein CagA into gastric cells in vitro.
INTRODUCTION
Helicobacter pylori resides in the human stomach and is associated with chronic gastritis, gastric ulcers, duodenal ulcers and gastric cancer. The global prevalence of H. pylori infection is ∼50% of the population. Infection with strains of H. pylori containing the crucial virulence locus, the cag pathogenicity island (cag PAI), and thus the oncoprotein cytotoxin-associated antigen A (CagA) is strongly associated with more severe clinical outcomes (Hatakeyama 2004).
The cag PAI of H. pylori is a 40-kb chromosomal region, identifiable as a horizontally acquired DNA element by its lower GC content (Terry et al.2005). The cag PAI encodes a type IV secretion system (T4SS), and its presence in the H. pylori genome is positively associated with the presence of highly active alleles of the secreted vacuolating cytotoxin (VacA) (Tegtmeyer, Wessler and Backert 2011). VacA enhances colonization and pathogenesis in host cells by several experimentally documented activities (Foegeding et al.2016). There are >30 open reading frames in the cag PAI with cagA located at one end of the island. CagA is translocated into host epithelial cells via the T4SS upon attachment (Noto and Peek 2012). Such secretion systems involve conjugative structures to transport proteins through a complex channel structure directly through the membrane into the cytoplasm of the host cell (Backert and Tegtmeyer 2017). Once inside the cell, CagA is frequently tyrosine phosphorylated at EPIYA domains by a host kinase, and proceeds to interact with systems leading to host cell junction damage, cytoskeletal changes and cell proliferation although the non-phosphorylated form of CagA has specific cellular effects as well (Hatakeyama 2008; Backert, Tegtmeyer and Fischer 2015).
In a separate sequence of events, non-translocated proteins of the T4SS encoded by the cag PAI are involved in inducing proinflammatory cytokine production by host cells, including interleukin 8 (IL-8) (Fischer et al.2002). Secretion of IL-8 recruits an innate immune system response, resulting in inflammation of the gastric mucosa. The cag PAI is found disproportionately in H. pylori isolates from patients with chronic active gastritis, peptic ulcer disease and gastric cancer, indicating that it is an important virulence factor (Cover 2016). Thus, cag PAI-positive strains are innately more virulent, while cag PAI-negative strains lacking this PAI are much less virulent.
Outer membrane inflammatory protein A, or OipA, is an outer membrane protein unique to H. pylori. Approximately 4% of the H. pylori genome encodes a large set of outer membrane proteins, many with unique functions including adhesion and pH regulation (Oleastro and Ménard 2013). OipA, encoded by the gene oipA (previously hopH—HP0638), was originally named for its role in inducing inflammation in the host, as evidenced by high mucosal IL-8 levels; however, this finding is controversial as results from various studies indicate differing effects of OipA on inflammation (Dossumbekova et al.2006; Odenbreit et al.2009; Matsuo, Kido and Yamaoka 2017). Additionally, the particular means by which OipA might induce inflammation are unclear as the effects can often be attributed to cag PAI-mediated pathways.
In a more widely agreed upon manner, OipA has been shown to play a role in adherence to host cells and thus host colonization (Dossumbekova et al.2006). OipA is a member of the Hop (Helicobacter Outer Proteins) outer membrane family of proteins, several of which promote binding to the gastric epithelium (Oleastro and Ménard 2013). The gastric epithelial cell receptor for OipA is unknown, but has been hypothesized to be in the integrin family (Posselt, Backert and Wessler 2013).
OipA expression is regulated by slipped-strand mispairing (SSM) within a hypermutable CT dinucleotide repeat motif located in the 5΄ region of the gene (Miftahussurur and Yamaoka 2015). SSM is a mutagenic process that occurs during DNA replication of repetitive sequences (Torres-Cruz and van der Woude 2003). The poly CT repeat tract in oipA is just one of numerous occurrences of this repeat in H. pylori. Addition/deletion of CT repeats in oipA results in a frame shift that drives phase variation in protein expression. While there is great variation in the number of CTs in the dinucleotide repeat of oipA between H. pylori strains (Zhang et al.2014), SSM within oipA during in vitro growth appears to be less frequent than quantified in sabA, another outer membrane protein gene possessing a 5' poly CT tract than during similar in vitro growth (Yamaoka et al.2006; Goodwin et al.2008).
In a study of 410 H. pylori patient isolates, this CT sequence ranged from 3 to 12 repeats, with short repeats predominating in East Asia, where gastric cancer caused by H. pylori is much more prevalent compared to strains from Western countries (Yamaoka et al.2002). Studies show that a functional ‘phase on’ status of oipA is associated with increased risk for peptic ulcer disease and gastric cancer. The strong correlation of functional OipA and the virulence of the bacterium has made OipA a candidate for potential vaccines against H. pylori (Chen et al.2012). Additionally, functional OipA is associated with high H. pylori density in infected stomachs and severe neutrophil infiltration (Liu et al.2013). However, the positive association of oipA phase on status with the presence of the cag PAI and vacA s1m1 alleles makes the role of OipA with these disease states difficult to dissect.
OipA and the cag PAI are two important H. pylori virulence factors, and there is great interest in the relationship they have to one another. Ando et al. (2002) demonstrated that while the vast majority (>96%) of cag PAI-positive isolates contained oipA phase on alleles, none of the cag PAI-negative isolates in the study possessed phase on oipA, yet the gene was uniformly present and highly conserved. While both OipA and the cag PAI are thought to be involved in the secretion of cytokines such as IL-8 by host epithelial cells, most likely via a mechanism involving transcription factor NF-κΒ, how they work together (or if they work together) is largely unknown (Matsuo, Kido and Yamaoka 2017). This study aims in part to determine the relationship between oipA and the cag PAI, and to shed light on why oipA is so highly conserved in cag PAI-negative strains in which the protein is apparently seldom, if ever, expressed. We additionally sought to determine what pressure maintains oipA as a phase on allele in the presence of the cag PAI. Here, we show that OipA is necessary, but not sufficient for IL-8 secretion in vitro; however, it is necessary for translocation of CagA from H. pylori 26695 to human gastric adenocarcinoma (AGS) cells. We also demonstrate that oipA transcript levels are higher in cells in contact with gastric epithelial cells in vitro.
MATERIALS AND METHODS
Helicobacter pylori culture
Helicobacter pylori strains were cultured on tryptic soy agar II with 5% sheep's blood (BBL) for 24–48 h at 37°C in an ambient air/5% CO2 atmosphere. Liquid cultures of H. pylori were grown in Sulfite-Free Brucella Broth (SFBB) supplemented with 1X cholesterol (Gibco Life Technologies), 20 μg vancomycin/mL, shaking at 150 rpm.
Nucleotide sequence accession
The nucleotide sequence determined for oipA of H. pylori strain J68 has been deposited in GenBank and has the accession number BankIt2035716 Helicobacter MF576477.
Amplified fragment length polymorphism analysis
Amplified fragment length polymorphism (AFLP) was employed to determine natural variation in the oipA CT dinucleotide repeat tract of wild-type H. pylori strains grown in vitro as described in Goodwin et al. (2008) and Harvey et al. (2014). AFLP primers were designed to amplify ∼270 bp beginning upstream of oipA and including the CT dinucleotide repeat tract from both strains J68 and 26695 (Table S1, Supporting Information). Separate primers were designed to amplify a 294-bp control region of oipA from a non-repeat bearing region as a control. In each case, reverse primers contained fluorescent VIC tags on the 5΄ end (Applied Biosystems, Foster City, CA, USA)
rdxA mutant of cag PAI-negative Helicobacter pylori strain J68 for counterselection of oipA mutants
oipA mutant strains of H. pylori were generated using an antibiotic counterselection method designed at Vanderbilt University Medical Center (Loh et al.2011) to create markerless mutations in the oipA locus of H. pylori. We used a metronidazole-resistant (MtzR) strain of the cag PAI-positive strain of H. pylori 26695 (a gift of Drs Mark McClain and Timothy Cover) designated 26695 rdxA−. We created an analogous mutant of the cag PAI-negative strain J68 by deleting an internal portion of rdxA, thereby generating an H. pylori J68 MtzR mutant. This was accomplished by amplifying a 1556-bp amplicon containing the full-length rdxA gene (HP0954) using the HP0955 Fwd and HP0953 Rev primers (Table S1). The amplicon was cloned into pCR4 (Invitrogen) according to the manufacturer's protocol. The resultant plasmid prdxA (Table 1) was modified by inverse PCR to delete a 390-bp portion of rdxA using the inverse PCR rdxA Fwd and Rev primers (Table S1) with 5΄ phosphorylation to aid in ligation. The amplicon was purified using PCR Purification kit (IBI) and digested with DpnI restriction endonuclease to destroy the prdxA template, and then ligated using T4 DNA ligase (Quick Ligation Kit—New England Biolabs) and transformed into Escherichia coli DH5α. The confirmed plasmid construct was designated pΔrdxA (Table 1) and used in natural transformation of the cag PAI-negative, vacA s2/m2 strain, J68 (a gift from Dr Richard Peek Jr., Vanderbilt University Medical Center). MtzR colonies capable of growing on 5 μg metronidazole/mL SFBB 10% Newborn Calf Serum plates were selected, and the mutation was confirmed by PCR and DNA sequencing and the strain named J68 rdxA− (Table 2).
Table 1.
Plasmids used for creation of H. pylori mutants.
| Name | Description |
|---|---|
| prdxA | TOPO TA cloning vector pCR4 containing a1556 bp amplicon of the H. pylori strain 26695 rdxA gene |
| pΔrdxA | prdxA with a 390-bp deletion in the rdxA gene |
| pOipA (26695 and J68) | TOPO TA cloning vector pCR4 containing a 2300-bp amplicon including the entire oipA gene, as well as the untranslated regions both upstream and downstream |
| pOipA.BamHI (26695 and J68) | pOipA containing a BamHI site in the cloned allele of oipA |
| pMM672 | Helicobacter pylori 26695 plasmid in which the coding region of rdxA is deleted (Loh et al.2011) |
| pOipA::CAT-rdxA (26 695 and J68) | pOipA.BamHI with CAT-rdxA cassette cloned into the BamHI restriction site |
| p26695.oipAOFF | pOipA 26695 with CT repeat tract consisting of 5 CT repeats |
| pJ68.oipAON | pOipA J68 with CT repeat tract consisting of 11 CT repeats |
| pJ68oipAON.FLAG | pJ68.oipAON containing a FLAG tag |
| pJ68oipAOFF.FLAG | pOipA J68 containing a FLAG tag |
| pICB:CAT | Plasmid containing the cagPAI, including cagE with CAT inserted (Harvey et al.2014) |
Table 2.
Helicobacter pylori mutant strains.
| Name | Description |
|---|---|
| 26695 wild type | MtzS and CmS, contains oipA with a CT repeat tract consisting of 6 CT repeats (phase on) |
| J68 wild type | MtzS and CmS, contains oipA with a CT repeat tract consisting of 10 CT repeats (phase off) |
| 26695 rdxA− & J68 rdxA− | 26695 and J68 WT containing a 390-bp deletion in the rdxA gene. MtzR and CmS |
| 26695 oipA− and J68 oipA− | 26695 rdxA− and J68 rdxA− containing the oipA::CAT-rdxA cassette. MtzS and CmR |
| 26695 oipA phase Off | 26695 rdxA− containing mutant oipA with an altered CT repeat tract consisting of 5 CT repeats (phase off). MtzR and CmS |
| J68 oipA phase on | J68 rdxA− containing mutant oipA with an altered CT repeat tract consisting of 11 CT repeats (phase on). MtzR and CmS |
| J68 oipA phase on-FLAG | J68 rdxA−/oipA phase on::FLAG. MtzR and CmS |
| J68 oipA phase off-FLAG | J68 rdxA−/oipA phase off::FLAG. MtzR and CmS |
| 26695 cagE− | 26695 rdxA− containing cagE::CAT. MtzR and CmR |
| 26695 oipA phase off-cagE− | 26695 rdxA−/oipA phase off containing cagE::CAT. MtzR and CmR |
Cloning of rdxA complement/chloramphenicol-resistant plasmids
Subsequent to the creation of these MtzRH. pylori clones, a chloramphenicol (Cm) resistance gene (CAT or chloramphenicol acetyl transferase) together with an intact version of rdxA was inserted within the oipA gene of H. pylori strains 26695 rdxA− and J68 rdxA−. To accomplish this, the entire oipA gene from both strains 26695 and J68, including regions both upstream and downstream, was amplified using the oipA universal Fwd and Rev primers designed based on a consensus sequence built from several H. pylori sequenced genomes (Table S1). The resultant ∼2300 bp amplicons were cloned into pCR4 (Invitrogen), and the plasmids were designated pOipA 26695 and pOipA J68 (Table 1). GeneArt Site-Directed Mutagenesis System (Invitrogen) was utilized according to the manufacturer's protocol to insert a BamHI site into the cloned allele of oipA using the oipA BamHI Fwd and Rev mutagenic oligos (Table S1). Resulting plasmids, pOipA.BamHI 26695 and pOipA.BamHI J68 (Table 1), were digested with BamHI for cloning of the selectable markers CAT and rdxA. This two-gene cassette was purified as a BamHI fragment from pMM674 (Loh et al.2011), a gift of Dr Mark McClain and Dr Timothy Cover of Vanderbilt University Medical Center, and cloned into pOipA.BamHI, resulting in pOipA::CAT-rdxA 26695 and pOipA::CAT-rdxA J68 (Table 1). These plasmids were naturally transformed into H. pylori 26695 rdxA− or H. pylori J68 rdxA− with selection using 10 μg chloramphenicol/mL. Resultant strains were confirmed via PCR of the oipA locus and sequencing, and designated 26695 oipA− and J68 oipA−. These serve as both intermediates in the isolation of markerless mutants and oipA null mutants, and are each MtzS and CmR.
Cloning of oipA mutant plasmids
oipA mutagenic oligos were designed specific to each strain to alter the number of CT dinucleotide repeats in the 5΄ region of oipA in order to turn the gene phase off in the cag PAI-positive H. pylori strain 26695, and phase on in the cag PAI-negative H. pylori strain J68. This was accomplished by deleting one CT in strain 26695 to go from 6 CT repeats to 5, resulting in a frame shift in oipA from phase on to phase off and deleting one CT in the J68 oipA allele from 9 to 8 dinucleotide repeats (oipA phase off to phase on). Mutagenesis was performed using the 26695.oipAOFF and J68.oipAON mutagenic primers (Table S1), respectively, and clones were screened via sequencing. Mutant bearing plasmids were named p26695.oipAOFF and pJ68.oipAON (Table 1).
Confirmed plasmids were used in natural transformations and allelic exchange with the appropriate H. pylori oipA− strains to replace the oipA::CAT-rdxA locus on the recipient chromosome. Such allelic replacement mutants were selected for with SFBB plates containing 5 μg metronidazole/mL. These strains, 26695 oipA phase off and J68 oipA phase on (Table 2), are MtzR/CmS, and possess a mutated oipA gene with no antibiotic resistance genes left behind in the locus.
FLAG epitope-tagged oipA mutants
To determine OipA protein expression in the mutant H pylori strains via western blotting, we designed mutagenic primers containing the epitope tag FLAG (YKDDDKD) to insert this seven amino acid epitope encoding sequence into the gene oipA between codons 161 and 162 (Table S1). These primers were used in site-directed mutagenesis with pJ68.oipAON and pJ68.oipA (containing the wild-type phase off oipA allele). Successful plasmids were confirmed via sequencing and named with the appropriate strain and phase of oipA, e.g. pJ68oipAOFF.FLAG (Table 1). Plasmids were naturally transformed into the appropriate H. pylori strain oipA−, selected for MtzR CmS phenotype and confirmed by PCR and sequencing. These strains, J68 oipA phase on-FLAG and J68 oipA phase off-FLAG (Table 2), were used in western blotting using ANTI-FLAG antibodies (Sigma-Aldrich).
Western blotting
Mutant and control strains of H. pylori were grown 24–36 h on blood agar plates and 0.4 OD600 units of cells were denatured and reduced using Lamelli buffer (Bio-Rad) with 2-mercaptoethanol, and total proteins were separated by SDS-PAGE. Proteins were transferred from the gel to nitrocellulose blotting membranes (Bio-Rad). OipA expression was detected using monoclonal anti-FLAG M2 antibody (Sigma-Aldrich), followed by goat anti-mouse IgG-Peroxidase. Chemiluminescence was accomplished using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and visualized with X-ray film.
cagE null mutants
To render the cag PAI T4SS non-functional in the H. pylori strain 26695, we created cagE null mutants in both oipA phase on and phase off mutants. This was accomplished using pICB::CAT (Tummuru, Sharma and Blaser 1995), a plasmid possessing a chloramphenicol resistance gene in the gene cagE, previously named picB. This plasmid was naturally transformed into both H. pylori 26695 rdxA− with oipA unaltered and thus phase on, and H. pylori 26695 oipA phase off. Clones with the mutation were selected for using SFBB plates with 10 μg Cm/mL, and the CAT insertion within cagE was confirmed via PCR and DNA sequencing. Strains were designated H. pylori 26695 cagE− and H. pylori 26695 oipA off-cagE− (Table 2).
RNA extraction, cDNA synthesis and real-time quantitative PCR
Helicobacter pylori cells were inoculated in SFBB/10 μg vancomycin/mL containing 1X Cholesterol (Gibco/BRL) at ∼0.2 OD600 and incubated shaking at 150 rpm for 12–24 h in a 5% CO2/ambient air mixture. A total of ×108 cells were collected during exponential phase of growth (OD600 between 0.7 and 1.2) and suspended in 1 mL of RNAzol RT (Molecular Research Center, Inc.). Total RNA was extracted from each pellet according to the manufacturer's protocol. RNA samples were then used in cDNA synthesis. One microgram of purified RNA was combined with iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad), and cDNA was synthesized using the manufacturer's cDNA synthesis protocol. cDNA was used for RT-qPCR.
The relative quantity of oipA mRNA was determined as in Acio-Pizzarello et al. (2017) using a TaqMan Gene Expression assay (Life Technologies) performed on the Applied Biosystems StepOne apparatus. The assays were performed in technical triplicate for each gene and each strain according to the manufacturer's protocol using custom TaqMan Custom Gene Expression assays (Thermo Fisher), including the oipA.Taq, ftsZ.Taq and gyrB.Taq probes (Table S1). ftsZ was used as the normalizer. Relative expression of genes among the various mutants was calculated using the 2ΔΔCt method as described by Livak and Schmittgen (2001) and processed using the DataAssist software (Applied Biosystems).
AGS cell culture
AGS cells were a gift from Timothy Cover of Vanderbilt University Medical Center. Cells were grown in RPMI 1640 medium (Life Technologies), supplemented with 10 mM HEPES 10% NCS and penicillin/streptomycin (Gibco/BRL). Cultures were grown at 37°C in an ambient air/5% CO2 atmosphere in either six-well tissue culture treated plates (CytoOne) or T-75 tissue culture flasks (Thermo Fisher).
Adhesion assay and IL-8 ELISA
A total of 2.5 × 105 AGS cells were seeded in each well of a six-well plate for 24 h. Cells were washed three times with antibiotic-free RPMI 1640. Helicobacter pylori, suspended in this same medium, was introduced to AGS cells at a multiplicity of infection of 100:1 and co-cultured for 5 h. At the completion of adhesion assays, medium was collected for subsequent IL-8 ELISAs. Following three washes, AGS cells were lysed using 1 mL of PBS/0.1% saponin for 15 min with shaking at 50 rpm. Lysates were serially diluted and spotted onto blood agar plates, and titers were calculated at 5 days. Media from H. pylori-infected AGS cells were centrifuged at 3300 × g, and supernatant was assayed for IL-8 concentration. A 96-well plate (BioLegend) was coated with IL-8 capture antibody (BioLegend) overnight at 37°C. ELISA was performed according to the manufacturer's suggested protocol. Plates were read at 450 and 570 nm using a Bio-Rad iMark Microplate Reader.
CagA translocation assay
AGS cells were seeded in six-well plates as described above and incubated until ∼80% confluence. Helicobacter pylori strains to be assayed for CagA translocation into AGS cells were added at an MOI of 100 and infection allowed to proceed for 14 h. Non-adherent H. pylori cells were removed by aspiration and AGS monolayers were washed three times with PBS and then lysed using RIPA Buffer (25 mM Tris-HCl [7.5], 150 mM NaCl, 5mM EDTA, 1% Triton X-100, 0.1% SDS) with 2 mM Na3VO4 and Protease Inhibitor Cocktail (Thermo). Twenty micrograms of soluble protein was resolved on 7% TGX polyacrylamide gels (Bio-Rad) and western blotted to nitrocellulose. Translocated and tyrosine phosphorylated CagA was visualized using a rabbit monoclonal antibody to phospho-Tyrosine (Abcam), while total CagA was detected in identical blots, run simultaneously with identical protein loads, using a mouse monoclonal anti-CagA (Sigma-Aldrich). Detection utilized horseradish peroxidase-conjugated secondary anti-rabbit or anti-mouse IgG (Abnova) and SuperSignal West Pico Chemiluminescent Substrate (Thermo). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a protein loading control using a mouse monoclonal anti-GAPDH (Sigma Aldrich).
Statistical analysis
A Welch's unpaired t-test of unequal variance was used to determine statistically significant differences in gene transcription, adhesion and immunoassay studies. These statistics were calculated with α-level = 0.05.
RESULTS
Phase variation in the CT repeat tract of oipA is rare in vitro
Outer membrane inflammatory protein (OipA) is regulated via SSM in a CT dinucleotide repeat tract in the 5΄ end of the gene and results in phase on status in most cag PAI-positive strains of Helicobacter pylori, and phase off in cag PAI-negative strains, nearly without exception (Ando et al.2002). Typically, longer repeat sequences are more likely to vary in length during SSM that occurs during replication (Harvey et al.2014). In order to quantify the natural variation in this CT repeat length when strains of H. pylori are grown in vitro, we employed AFLP analysis.
Our results suggest that despite the ability of SSM in the CT tract to generate variability, there was little variation in the number of CT repeats present within in vitro grown wild-type H. pylori strains 26695 and J68 (Fig. 1). In the cag PAI-positive strain 26695, >97% of the amplicons generated from the poly CT region contained six CTs, consistent with a phase on oipA allele. In the cag PAI-negative H. pylori strain J68, while the majority of the amplicons generated from this same region possess nine CTs, indicating a phase off oipA allele, there were slightly more variants (∼10% amplicons with eight CTs, suggesting a phase on oipA allele) among the amplicons (Fig. 1). Overall, this apparent paucity of in vitro variation in the poly CT tract suggests that the relationship between the presence/absence of the cag PAI and the expression phase of oipA tends to be maintained, despite the repetitive sequence in the 5΄ region of oipA.
Figure 1.
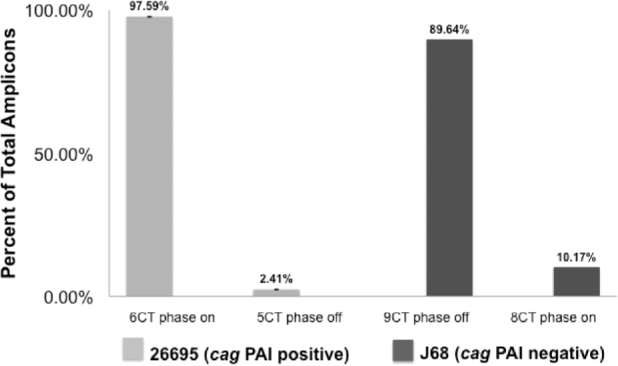
AFLP analyses demonstrates that phase variation in oipA in vitro is infrequent. Fluorescently labeled amplicons containing the poly CT repeat tract of oipA in both cag positive (26695) and cag PAI negative (J68) strains of H. pylori demonstrate relatively minor levels of variation in length, indicating that the expression phase status of oipA is quite stable when H. pylori is grown in vitro. Bars represent the mean ± standard deviation calculated using three biological replicates. Amplification of a similar sized amplicon from a non-repeat bearing region of oipA, as a PCR control, revealed no fragment polymorphisms (data not shown).
Experimental alterations to the CT tract result in changes in oipA expression
To quantify the effect of CT tract changes on oipA expression, RT-qPCR was used to quantify transcript levels, as phase status is known to affect mRNA levels (Acio-Pizzarello et al.2017). The deletion of an internal portion of rdxA in order to create a metronidazole-resistant control within wild type, phase on oipA in strain 26695 (Loh et al.2011) did not produce a significant difference in oipA transcript levels compared to the wild-type H. pylori strain, indicating that it is a reliable control (Fig. 2). Altering the number of CTs in oipA in strain 26695 such that the gene is phase off decreases the transcript levels of the gene by more than 80% compared to the isogenic phase on control. Additionally, the relative quantity of oipA transcript when the gene is phase off is not significantly different from the oipA− null mutant (Fig. 2).
Figure 2.
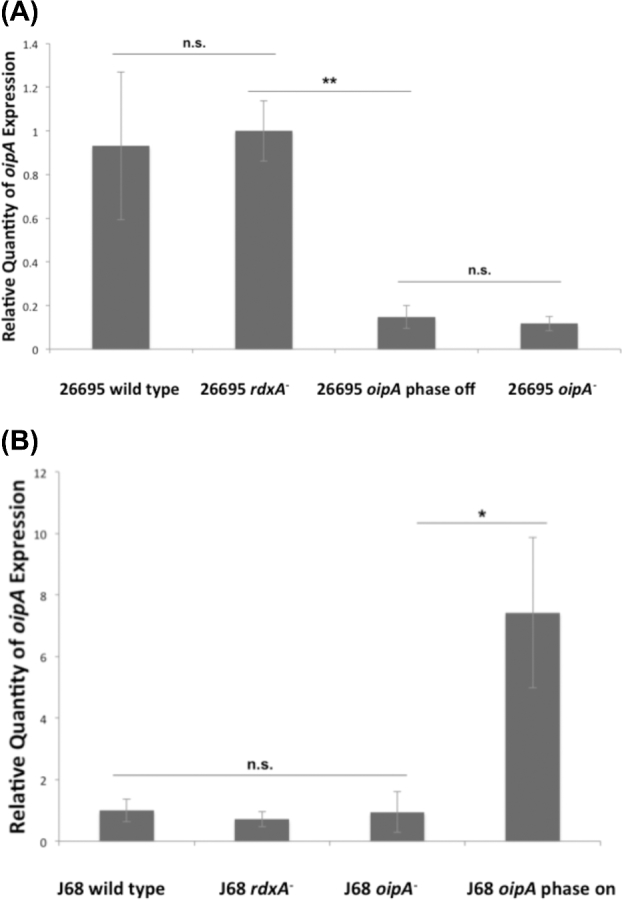
Phase off status of oipA in both cag PAI-positive and negative strains of H. pylori causes a significant decrease in the relative quantity of transcripts expressed compared to phase on status. RT-qPCR was used to determine the relative expression of oipA in the cag PAI-positive H. pylori 26695 wild-type and the isogenic rdxA− control strain, possessing oipA phase on, as well as to 26695 oipA phase off and oipA null mutants (A). Additionally, wild-type cag PAI-negative H. pylori strain J68 and isogenic rdxA− control, as well as to the J68 oipA phase on and J68 oipA null mutants (B). The data shown here are representative of the results obtained in three independent experiments, each conducted in technical triplicate. Error bars show standard deviation. Statistics were calculated using a Welch's unpaired t-test of unequal variance with 26695 rdxA− (oipA phase on) as the control (** = P ≤ 0.01, * = P ≤ 0.05, n.s. = P > 0.05).
Similarly, there was no significant difference in oipA mRNA levels between the H. pylori cag PAI-negative J68 rdxA− control and the wild-type strain J68 (oipA phase off), and the J68 oipA− null mutant (Fig. 2). However, when the J68 oipA CT dinucleotide repeat tract was mutated such that oipA was in frame and thus phase on, there was a significant increase in oipA transcript levels, further indicating that oipA mRNA levels are tied to its expression phase status.
Phase on oipA confers an increased host adherence phenotype
To demonstrate the increased oipA mRNA levels documented in the phase on H. pylori J68 mutant resulted in OipA protein expression, a 21-nucleotide sequence encoding the FLAG epitope was inserted into the oipA allele of H. pylori strain J68, and a monoclonal antibody to this epitope was used to detect OipA protein. In this cag PAI-negative and naturally oipA phase off H. pylori strain, when oipA was experimentally turned phase on, there was a novel ∼34 kDa protein detected (Fig. 3), indicating a successful switch from phase off to on results in the novel expression of OipA.
Figure 3.
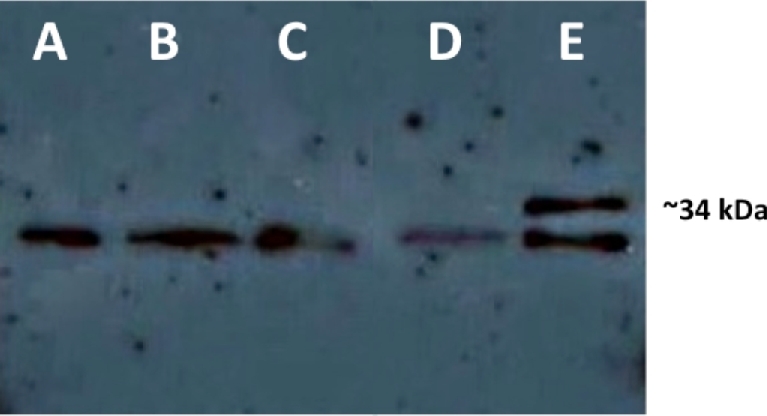
Phase on status of oipA in the cag PAI-negative H. pylori strain J68 results in expression of the protein OipA. An anti-FLAG western blot was performed using 0.4 OD600 units of (A) wild-type H. pylori J68 oipA phase off control, (B): H. pylori J68 oipA phase on mutant, (C): H. pylori J68 oipA phase off FLAG, (D): H. pylori J68 oipA phase on FLAG. This revealed the ∼34 kDa FLAG epitope tagged OipA only when oipA is phase on. An ∼30 kDa cross-reactive protein in H. pylori J68 appeared in all strains.
We next investigated whether the demonstrated decrease in oipA transcript level in the cag PAI-positive strain 26695 (Fig. 2) and OipA expression in the cag PAI-negative, oipA phase on mutant strain J68 would alter H. pylori adherence to gastric epithelial cells. When oipA was turned phase off in strain 26695, H. pylori attachment levels decreased by ∼80% (Fig. 4). oipA phase was also strongly correlated to H. pylori attachment capability in H. pylori strain J68, as attachment levels more than doubled when oipA was turned phase on. These results indicate that OipA has the ability to mediate attachment to host cells independent of the cag PAI and demonstrate that the cag PAI-negative strain J68 oipA allele is functional when phase on.
Figure 4.
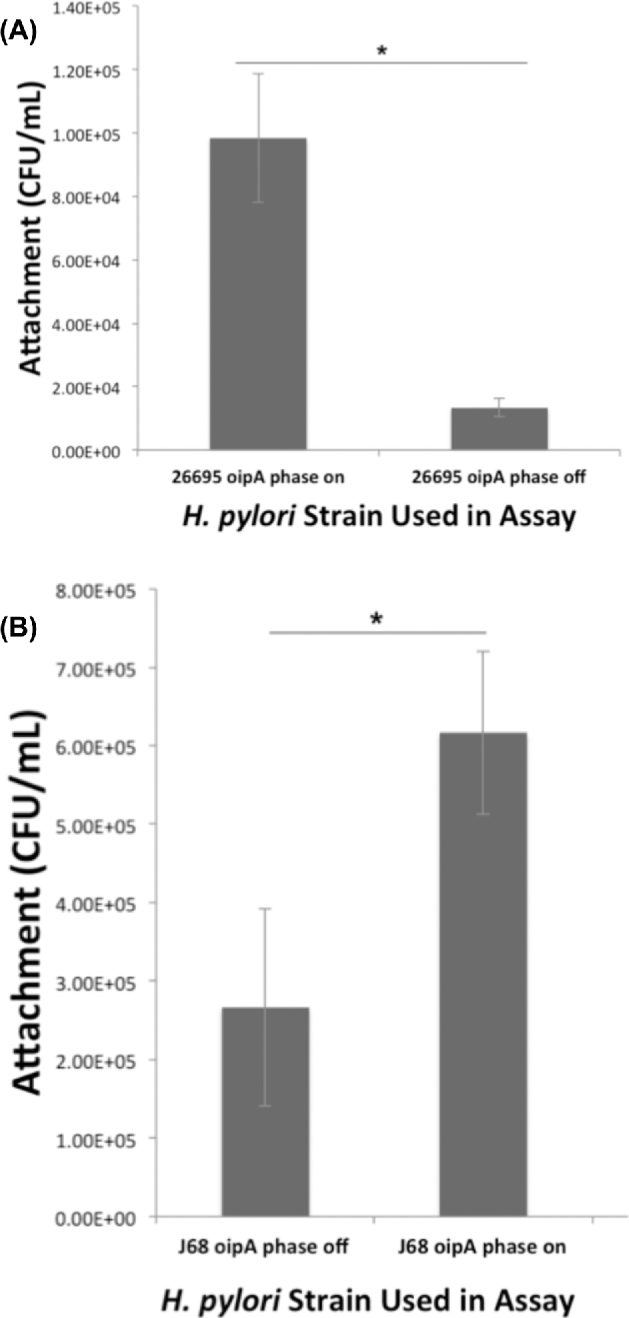
Helicobacter pylori exhibits increased adherence ability when oipA is phase on in both cag PAI-positive or negative strains. Attachment assays were performed to determine whether oipA phase was important in the ability of H. pylori to adhere to AGS cells, particularly in the cag PAI-negative strain, J68, in which OipA is not expressed. Panel A shows the cag PAI-positive strain 26695, with oipA in alternate expression phases and panel B shows H. pylori J68 with oipA in each expression phase. The data shown here are representative of results obtained in independent experiments, each conducted in technical triplicate. Error bars show standard deviation. Statistics were calculated using a Welch's unpaired t-test of unequal variance (* = P ≤ 0.05).
OipA effects on IL-8 production and CagA translocation
Due to the differences in attachment ability observed between H. pylori strains with oipA phase on and phase off, and the theorized involvement of OipA in the host inflammatory response, we hypothesized that the altered attachment phenotype might be associated with altered epithelial cell IL-8 production. To quantify alterations in IL-8 concentration, AGS cell attachment assay culture medium was used in ELISAs. AGS cells infected with the cag PAI-positive strain 26695 with oipA experimentally mutated to phase off produced ∼80% less IL-8 than those infected by isogenic H. pylori with oipA phase on (Fig. 5). This is nearly the same reduction in IL-8 secretion from AGS cells infected with a cagE null mutant that lacks a functional cag PAI T4SS. There was no significant difference in the amount of IL-8 produced by AGS cells infected with H. pylori strain 26695 possessing an oipA null mutation relative to the isogenic oipA phase off mutant, and both of these IL-8 concentrations were comparable to the amount of IL-8 produced by uninfected AGS cells (Fig. 5).
Figure 5.
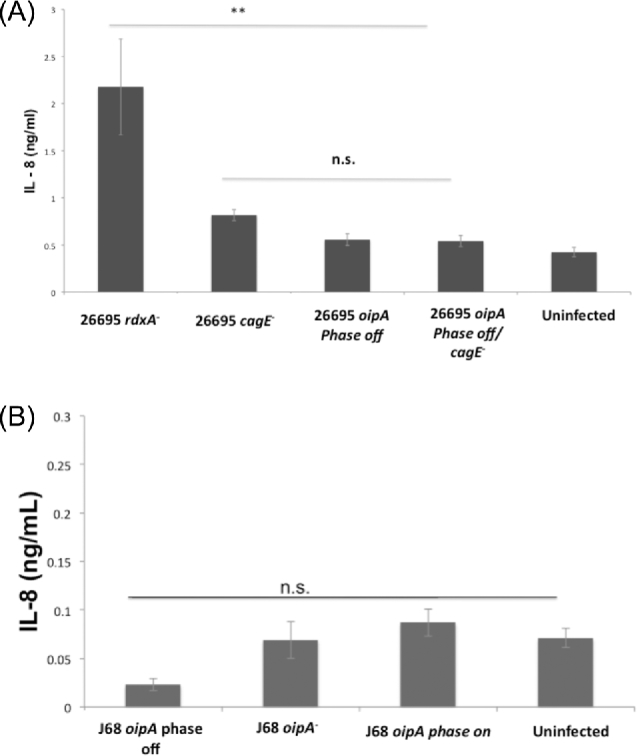
Phase on status of oipA in H. pylori mediates gastric epithelial cell IL-8 production only in the presence of the cag PAI. Cell medium from AGS cells infected with H. pylori 26695 oipA phase on (26695 rdxA−), oipA phase on/cagE null mutant (26695 cagE−), oipA phase off (26695 oipA phase off), oipA phase off/cagE null mutant (26695 oipA phase off/cagE−) as well as from uninfected AGS cells was collected after a 5-h infection and used in ELISA to quantify AGS cell IL-8 production. This is a representative of three independent experiments (A). The same assay was performed for the cag PAI-negative H. pylori strain J68 using oipA phase off (J68 rdxA−), oipA null mutant (J68 oipA−) and oipA phase on (J68 oipA phase on). Medium form uninfected cells after a 5-h incubation is also shown. The data shown here are representative of the results obtained in three independent experiments, each conducted in technical triplicate (B). Error bars show standard deviation. Statistics were calculated using a Welch's unpaired t-test of unequal variance (** = P ≤ 0.01, n.s. = P > 0.05).
In contrast, in H. pylori strain J68 there was no significant difference in IL-8 production by AGS cells, whether infected with oipA phase on, oipA phase off, oipA null mutant strains or left uninfected (Fig. 5). This stark contrast between cag PAI-positive and cag PAI-negative H. pylori strains indicates that while OipA has the ability to mediate IL-8 production in the presence of the T4SS encoded by the cag PAI, OipA cannot itself alone mediate the host inflammatory response in the absence of the cag PAI. Thus, OipA appears necessary but not sufficient for inducing IL-8 secretion by AGS cells.
Because the cag PAI-positive H. pylori strain 26695 was greatly altered in the ability to induce the secretion of IL-8 by AGS cells depending on the expression phase of OipA, we next asked how oipA phase variation affected the translocation of the cag PAI effector protein CagA into AGS cells. We visualized translocated, tyrosine phosphorylated CagA in western blots of 14-h H. pylori-infected AGS cells (Fig. 6). While oipA phase on H. pylori strain 26695 was capable of translocating CagA into the AGS cells, the isogenic oipA phase off strain could not. This indicates a role for OipA in the translocation of this oncoprotein from H. pylori to AGS cells in vitro.
Figure 6.
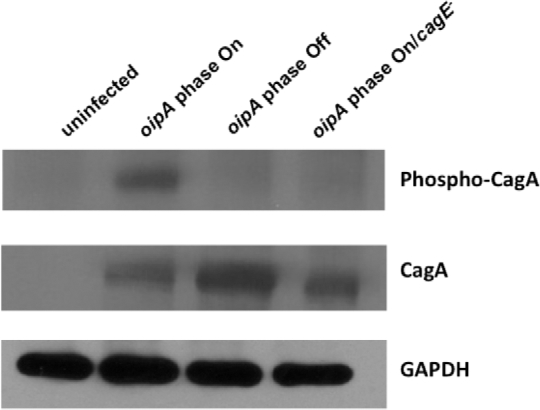
OipA is Involved in CagA Translocation from H. pylori 26695 into AGS cells. AGS cells were infected 14 h with H. pylori 26695 rdxA− (oipA phase on), H. pylori 26695/rdxA−/oipA phase off, H. pylori 26695/rdxA−/oipA phase on/cagE− or left uninfected. Adherent H. pylori and AGS cells were subjected to western blotting using either anti phosphotyrosine to visualize translocated CagA in its tyrosine phosphorylated form, anti-CagA, or anti glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control. CagA protein is translocated and tyrosine phosphorylated only in the presence of phase on oipA allele and a functional T4SS. Changing the expression phase of oipA to phase off or ablating the cagE gene of the T4SS prevented the translocation of this effector protein. These results are representative of the results obtained in two independent experiments.
oipA mRNA transcript levels are increased in AGS cell-adherent Helicobacter pylori compared to non-adherent
We next asked if there was a difference in the amount of oipA mRNA transcripts in H. pylori strain 26695 cells attached to AGS cells versus those that were also incubated with host cells, but remained unattached. In order to determine this, attachment assays were performed as before; however, after the 5-h incubation, infected AGS cell culture supernatants were collected and the non-adherent H. pylori population was harvested, as were adherent H. pylori after saponin-mediated lysis of infected AGS cells. RT-qPCR was used to quantify relative oipA transcript levels in both populations of H. pylori. Due to the variation in H. pylori cell numbers in these two populations and subsequently mRNA and cDNA amounts compared to traditional mRNA extraction preparations, a housekeeping gene was used in RT-qPCR experiments. gyrB was employed as a comparison to oipA.
Our results indicate that strain 26695 oipA phase on H. pylori cells that were attached to AGS cells after the 5-h infection had seven times the amount of oipA transcript compared to non-adherent cells, while housekeeping gene gyrB transcript levels did not vary (Fig. 7). Independent experiments using the H. pylori 26695 oipA phase off mutant also showed significantly increased oipA mRNA levels, relative to the non-adherent population. Genomic DNA preparations from adherent and non-adherent H. pylori cells from identical experiments were isolated and used in AFLP analyses described above. There was no detectable length polymorphism in the amplicons generated from these two populations (data not shown), indicating that neither was the non-adherent population of H. pylori was enriched for naturally occurring phase off oipA allele bearing cells nor were the adherent bacteria enriched for cells bearing phase on oipA alleles.
Figure 7.
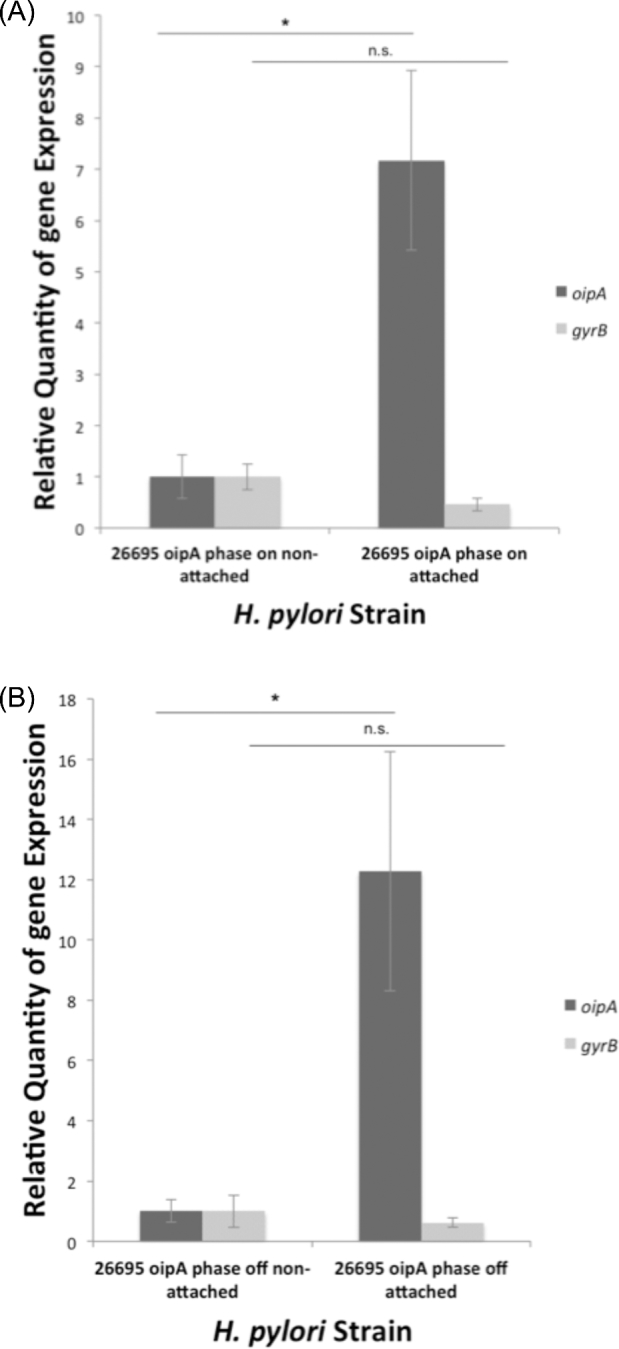
Helicobacter pylori attached to AGS cells possess higher levels of oipA transcript. Attachment assays were performed using 26695 oipA phase on (A) and 26695 oipA phase off (B). For each infection, H. pylori in the supernatant was collected separately from the bacteria attached to the AGS monolayer. RNA was extracted, and cDNA synthesized was used in RT-qPCR to determine relative quantities of oipA transcripts in attached cells compared to non-attached. The data shown here are representative of the results obtained in independent experiments, each conducted in technical triplicate. Error bars show standard deviation. Statistics were calculated using a Welch's unpaired t-test of unequal variance (* = P ≤ 0.05, n.s. = P > 0.05).
DISCUSSION
OipA and the cag PAI are two well-documented Helicobacter pylori virulence factors. The cag PAI encodes a T4SS that induces the production of proinflammatory cytokines by host gastric epithelial cells (Khatoon et al.2017). OipA acts as an adhesin; however, the role of OipA in eliciting a proinflammatory response, such as IL-8 secretion, is a subject of controversy. Some studies have claimed that while OipA is an adhesin, it has no effect on IL-8 secretion by host cells (Fischer et al.2002; Odenbreit et al.2002; Dossumbekova et al.2006). Studies have shown that the majority of cag PAI-positive strains of H. pylori possess an oipA allele that is phase on while cag PAI-negative strains contain a phase off, yet highly conserved, allele of oipA (Ando et al.2002; Matsuo, Kido and Yamaoka 2017). Despite this and the importance of both virulence factors in H. pylori virulence, the relationship between OipA and the cag PAI is somewhat enigmatic.
Despite the ability of oipA to undergo phase variation (Miftahussurur and Yamaoka 2015), our AFLP results suggest that there is little variation in the CT tract length in populations of H. pylori grown in vitro. This indicates that, while possible, phase variation at this locus may be rare. Rates of mutation in this poly CT tract length may vary between strains as Zhang et al. (2014) demonstrated that >75% of oipA alleles of western H. pylori isolates have relatively long poly CT tract lengths, most between 6 and 10 CT repeats. Helicobacter pylori isolates of Asian origin have much shorter CT repeat tract lengths, most having condensed CT repeat lengths of 3 (Ando et al.2002). These Asian H. pylori isolates are invariable oipA phase on. Our results showing that alterations in CT tract length are rare for oipA grown in vitro are in contrast to phase variation in other outer membrane proteins possessing poly CT tracts (Solnick et al.2004; Styer et al.2010), including sialic acid binding adhesin (SabA) (Harvey et al.2014). sabA alleles vary in lab-cultured populations such that there are three distinct subpopulations with varying CT tract lengths within a population of H. pylori strain 26695 with 60% possessing six CT repeats, 10% with seven repeats and 30% with eight (Goodwin et al.2008). This same study demonstrated similar phase variation in H. pylori strain J99 as well. This inconsistent phase variation between oipA and sabA may suggest that factors other than the repetitive nature of CT repeat sequences might impact SSM and thus phase variation. While Asian isolates of H. pylori have utilized a condensation of the CT repeat tract to minimize oipA phase variation and thus maintain oipA phase on status, western isolates, with longer poly CT tracts, yet reduced phase variation, may have utilized an alternate evolutionary strategy to limit phase variation. Perhaps epigenetic influences on this locus may impact phase variation of oipA.
The vast majority of bacterial genome sequences encode protein or structural RNA. Additionally, when bacterial species transition to permanent host interactions, they undergo genomic reduction (Moran 2002). This adds intrigue to the conundrum that phase off alleles of oipA are nearly always found in cag PAI-negative strains of H. pylori (Ando et al.2002), yet these strains have not lost the gene for this protein that is apparently unexpressed. We speculate that phase on and thus expressed alleles of oipA must serve an important function in these H. pylori strains of reduced virulence. In silico correction of oipA revealed that the protein produced when the CT tract is altered to generate a phase on allele in the cag PAI-negative strain used in this study, J68, it shares 97% amino acid identity with the protein produced by cag PAI-positive strain 26695 (data not shown). Based on this finding, we hypothesized that turning oipA phase on in cag PAI-negative strains of H. pylori would lead to the production of a full-length protein, and an increased ability of these cells to adhere to AGS cells and to elicit proinflammatory cytokine (IL-8) production. Our creation of a phase on allele of oipA in H. pylori strain J68 resulted in novel protein expression and increased attachment to AGS cells in vitro. However, this OipA expression in cag PAI-negative isolate J68 is not sufficient to elicit IL-8 secretion from AGS cells. Phase on oipA confers an increase in host adherence ability in both cag PAI-positive and negative strains. These findings show that OipA has the ability to mediate host attachment independent of the cag PAI. We hypothesize that the expression of OipA must be important under as yet undetermined circumstances during host infection by these reduced virulence strains, otherwise we would predict evidence of decay and perhaps deletion of the gene, rather than persistent conservation in the absence of apparent expression.
IL-8 induction experiments demonstrated that phase off status of oipA in the cag PAI-positive strain 26695 correlates not only with a decrease in host attachment, but also with decreased host production of this proinflammatory cytokine. It is striking that in the absence of OipA expression, there is a significant decrease in IL-8 production even in the presence of a functional cag PAI-encoded T4SS. This indicates that the cag PAI cannot mediate IL-8 induction independent of OipA. This finding is also supported by the fact that OipA expression does not elicit significant IL-8 secretion by AGS cells when infected by the cag PAI-negative strain J68 with oipA experimentally turned phase on. We hypothesize that OipA works in conjunction with the cag PAI T4SS to induce the host inflammatory response, and that both H. pylori surface structures must be present to stimulate host proinflammatory cytokine production.
Studies by Odenbreit et al. (2002) and Dossumbekova et al. (2006) concluded that IL-8 induction and CagA translocation are independent of OipA. In contrast, in the current study, we demonstrate that both IL-8 induction and CagA translocation are dependent on the expression of OipA. This discrepancy may be due to the use of different H. pylori strains or differing cell lines used in infections. Additionally, a counterselection method for introduction of markerless mutations initially described in a study by Loh et al. in 2011 (Ando et al.2002) was employed in our study. The different nature of mutations in oipA may help explain the differing results between these studies. While our study demonstrates the necessity of OipA for IL-8 induction, this outer membrane protein alone cannot stimulate this proinflammatory event. The novel experimental ability to express OipA in a cag PAI-negative strain in the absence of any cag PAI proteins or VacA s1/m1 may allow better investigation of the role of this outer membrane protein in altered host cell responses. OipA joins BabA and HopQ (Ishijima et al.2011; Javaheri et al.2016; Koniger et al.2016) as H. pylori outer membrane proteins encoded outside the cag PAI to facilitate the translocation of CagA into host cells. We believe that part of the explanation for the nearly uniform possession of phase on oipA alleles among cag PAI-positive H. pylori isolates lies in the association between OipA and the translocation of this oncoprotein. Selective pressure to maintain the expression of accessory proteins, such as OipA, in isolates of H. pylori bearing the cag PAI may explain why these more virulent isolates usually express OipA while the cag PAI-negative isolates would not experience this same pressure.
In vitro infection of AGS cells by H. pylori strain 26695 and subsequent RT-qPCR experiments revealed a significant increase in the quantity of oipA mRNA transcripts in the H. pylori cells adhered to AGS cells, compared to the non-adherent population in the same experiments. We initially proposed two alternative hypotheses regarding these results. First, we hypothesized that, despite the relative rarity of phase variation in oipA in vitro, there may be selection within the population of H. pylori used in the infection that allows cells expressing OipA to better adhere to host cells. However, our AFLP analyses on the poly CT region of oipA in both the adherent and non-adherent populations of H. pylori strain 26695 revealed neither enrichment for phase on oipA alleles in adherent H. pylori nor selection for phase off oipA alleles in the non-adherent population.
An alternative hypothesis is that contact with AGS cells may trigger increased transcription of oipA or increased stability of oipA mRNA as a response to the presence of the gastric epithelium. This hypothesis is supported by data showing the same increase in oipA transcript levels occurs in attached cells compared to non-attached, even when oipA is experimentally rendered phase off in 26695. The ability to regulate oipA expression may be adaptive in colonizing the stomach in the face of mucus shedding and epithelial cell turnover (Oleastro and Ménard 2013). Other investigators have demonstrated H. pylori gene expression alterations in response to gastric epithelial cell contact (Joyce et al.2001; van Amsterdam et al.2003; Silva et al.2017). Affected genes include genes of the cag PAI, the vacuolating cytotoxin and genes within the plasticity zone. The ability to sense and respond to host cell contact (Johnson, Gaddy and Cover 2012) is almost certainly an important part of the exquisite adaptation of H. pylori to the host microcosm that contributes to the decades long infection that is a hallmark of H. pylori pathogenesis.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSPD online.
Supplementary data are available at FEMSPD online.
Acknowledgements
The authors wish to thank Dr Richard Peek Jr. of Vanderbilt University Medical Center (VUMC) for the gift of the H. pylori strain J68. We also thank Dr Timothy Cover and Dr Mark McClain of VUMC for advice on the use of their counter selection markerless mutation system and Dr John Loh, also of VUMC, for advice on CagA translocation assays.
FUNDING
This work was supported by a grant to MHF from The National Institutes of Health, National Institute of Allergy and Infectious Diseases, R-15 AI053062. DNH was supported in part by an American Society for Microbiology Undergraduate Research Fellowship (ASM-URF) and a grant from the Howard Hughes Medical Institute Undergraduate Research Grant to the College of William & Mary Biology Department.
Conflict of Interest. None declared.
REFERENCES
- Acio-Pizzarello CR, Acio AA, Choi EJ et al. Determinants of the regulation of H. pylori adhesins include repeat sequences in both promoter and coding regions as well as the two component system, ArsRS. J Med Microbiol 2017;66:798–807. [DOI] [PubMed] [Google Scholar]
- Ando T, Peek RM, Pride D et al. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J Clin Microbiol 2002;40:239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Tegtmeyer N. Type IV secretion and signal transduction of Helicobacter pylori CagA through interactions with host cell receptors. Toxins (Basel) 2017;9, DOI: 10.3390/toxins9040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol 2015;10:955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lin M, Li N et al. Therapeutic vaccination with Salmonella-derived codon optimized outer inflammatory protein DNA vaccine enhances protection in Helicobacter pylori infected mice. Vaccine 2012;30:5310–5. [DOI] [PubMed] [Google Scholar]
- Cover TL. Helicobacter pylori diversity and gastric cancer risk. MBio 2016;7:e01869–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossumbekova A, Prinz C, Mages J et al. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: genetic and functional genomic analysis of hopH gene polymorphisms. J Infect Dis 2006;194:1346–55. [DOI] [PubMed] [Google Scholar]
- Fischer W, Püls J, Buhrdorf R et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol 2002;42:1337–48. [DOI] [PubMed] [Google Scholar]
- Foegeding JN, Caston RR, McClain SM et al. An overview of Helicobacter pylori VacA toxin biology. Toxins 2016;8, DOI: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AC, Weinberger DM, Ford CB et al. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology 2008;154:2231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey VC, Acio CR, Bredehoft AK et al. Repetitive sequence variations in the promoter region of the adhesin-encoding gene sabA of Helicobacter pylori affect transcription. J Bacteriol 2014;196:3421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer 2004;4:688–94. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. Linking epithelial polarity and carcinogenesis by multitasking Helicobacter pylori virulence factor CagA. Oncogene 2008;27:7047–54. [DOI] [PubMed] [Google Scholar]
- Ishijima N, Suzuki M, Ashida H et al. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem 2011;286:25256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaheri A, Kruse T, Moonens K et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat Microbiol 2016;2:16189. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Gaddy JA, Cover TL. Alterations in Helicobacter pylori triggered by contact with gastric epithelial cells. Front Cell Infect Microbiol 2012;2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EA, Gilbert J V, Eaton KA et al. Differential gene expression from two transcriptional units in the cag pathogenicity island of Helicobacter pylori. Infect Immun 2001;69:4202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatoon J, Prasad KN, Rai RP et al. Association of heterogenicity of Helicobacter pylori cag pathogenicity island with peptic ulcer diseases and gastric cancer. Br J Biomed Sci 2017;74:121–6. [DOI] [PubMed] [Google Scholar]
- Koniger V, Holsten L, Harrison U et al. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat Microbiol 2016;2:16188. [DOI] [PubMed] [Google Scholar]
- Liu J, He C, Chen M et al. Association of presence/absence and on/off patterns of Helicobacter pylori oipA gene with peptic ulcer disease and gastric cancer risks: a meta-analysis. BMC Infect Dis 2013;13:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- Loh JT, Shaffer CL, Piazuelo MB et al. Analysis of cagA in Helicobacter pylori strains from Colombian populations with contrasting gastric cancer risk reveals a biomarker for disease severity. Cancer Epidem Biomar 2011;20:2237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Kido Y, Yamaoka Y. Helicobacter pylori outer membrane protein-related pathogenesis. Toxins (Basel) 2017;9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur M, Yamaoka Y. Helicobacter pylori virulence genes and host genetic polymorphisms as risk factors for peptic ulcer disease. Expert Rev Gastroent 2015;9:1535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell 2002;108:583–6. [DOI] [PubMed] [Google Scholar]
- Noto JM, Peek RMJ. The Helicobacter pylori cag pathogenicity island. Methods Mol Biol 2012;921:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenbreit S, Kavermann H, Puls J et al. CagA tyrosine phosphorylation and interleukin-8 induction by Helicobacter pylori are independent from alpAB, HopZ and bab group outer membrane proteins. Int J Med Microbiol 2002;292:257–66. [DOI] [PubMed] [Google Scholar]
- Odenbreit S, Swoboda K, Barwig I et al. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect Immun 2009;77:3782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleastro M, Ménard A. The role of Helicobacter pylori outer membrane proteins in adherence and pathogenesis. Biology (Basel) 2013;2:1110–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posselt G, Backert S, Wessler S. The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun Signal 2013;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B, Nunes A, Vale FF et al. The expression of Helicobacter pylori tfs plasticity zone cluster is regulated by pH and adherence, and its composition is associated with differential gastric IL-8 secretion. Helicobacter 2017;22, DOI: 10.1111/hel.12390. [DOI] [PubMed] [Google Scholar]
- Solnick JV, Hansen LM, Salama NR et al. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. P Natl Acad Sci USA 2004;101:2106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer CM, Hansen LM, Cooke CL et al. Expression of the BabA adhesin during experimental infection with Helicobacter pylori. Infect Immun 2010;78:1593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J 2011;278:1190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry CE, McGinnis LM, Madigan KC et al. Genomic comparison of cag pathogenicity island (PAI)-positive and -negative Helicobacter pylori strains: identification of novel markers for cag PAI-positive strains. Infect Immun 2005;73:3794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cruz J, van der Woude MW. Slipped-strand mispairing can function as a phase variation mechanism in Escherichia coli. J Bacteriol 2003;185:6990–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummuru MK, Sharma SA, Blaser MJ. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol 1995;18:867–76. [DOI] [PubMed] [Google Scholar]
- van Amsterdam K, van Vliet AHM, Kusters JG et al. Induced Helicobacter pylori vacuolating cytotoxin VacA expression after initial colonisation of human gastric epithelial cells. FEMS Immunol Med Micr 2003;39:251–6. [DOI] [PubMed] [Google Scholar]
- Yamaoka Y, Kikuchi S, ElZimaity HMT et al. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology 2002;123:414–24. [DOI] [PubMed] [Google Scholar]
- Yamaoka Y, Ojo O, Fujimoto S et al. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut 2006;55:775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Qian J, Zhang X et al. Outer membrane inflammatory protein A, a new virulence factor involved in the pathogenesis of Helicobacter pylori. Mol Biol Rep 2014;41:7807–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSPD online.


