Abstract
Chlamydia are gram-negative obligate intracellular bacteria that replicate within a discrete cellular vacuole, called an inclusion. Although it is known that Chlamydia require essential nutrients from host cells to support their intracellular growth, the molecular mechanisms for acquiring these macromolecules remain uncharacterized. In the present study, it was found that the expression of mammalian cell glucose transporter proteins 1 (GLUT1) and glucose transporter proteins 3 (GLUT3) were up-regulated during chlamydial infection. Up-regulation was dependent on bacterial protein synthesis and Chlamydia-induced MAPK kinase activation. GLUT1, but not GLUT3, was observed in close proximity to the inclusion membrane throughout the chlamydial developmental cycle. The proximity of GLUT1 to the inclusion was dependent on a brefeldin A-sensitive pathway. Knockdown of GLUT1 and GLUT3 with specific siRNA significantly impaired chlamydial development and infectivity. It was discovered that the GLUT1 protein was stabilized during infection by inhibition of host-dependent ubiquitination of GLUT1, and this effect was associated with the chlamydial deubiquitinase effector protein CT868. This report demonstrates that Chlamydia exploits host-derived transporter proteins altering their expression, turnover and localization. Consequently, host cell transporter proteins are manipulated during infection as a transport system to fulfill the carbon source requirements for Chlamydia.
Keywords: glucose transporter protein, pathogenesis, intracellular parasite
Manipulation of GLUT1 and GLUT3 by Chlamydia.
INSTRUCTION
Chlamydia are obligate intracellular pathogens that infect a number of different eukaryotic cells and cause a wide range of diseases in humans, including sexually transmitted diseases, infectious blindness and respiratory tract infection (Kuo et al.1995; Haggerty et al.2010; Hu et al.2010; Torrone et al.2014). Chlamydiae infect host cells as metabolically inactive elementary bodies (EB) and then differentiate into a metabolically active, but non-infectious reticulate bodies (RB) that replicate within cellular vacuoles called inclusions (Moulder 1991). Thus, the entire infectious developmental cycle is physically isolated within an inclusion vacuole in the host cell. Following their replication at a late developmental stage, RB re-differentiate to infectious EB. Mature EB infect neighboring cells after exit from their host cells by lysis or are translocated by extrusion of intact inclusion vacuoles from vital host cells (Hybiske and Stephens 2008).
One of the major features for intracellular parasites is that they require various nutrients from host cells to support their growth (Naderer, Heng and McConville 2010; MacRae et al.2012; Steele et al.2013). As one of the most successful intracellular pathogens, Chlamydia contains incomplete gene sets for various metabolic pathways and has the capacity to acquire a variety of metabolic precursors and intermediates from their host cells (Stephens et al.1998). Chlamydia acquire host-derived lipids, including cholesterol, triglycerol phospholipids and sphingomyelin for intracellular growth and development (Hackstadt, Scidmore and Rockey 1995; van Ooij et al.2000) by intersecting host cellular pathways (Su et al.2004; Cocchiaro et al.2008; Elwell et al.2011; Boncompain et al.2014). Supported by the genomic analysis, Chlamydia require a number of essential amino acids from host cells to support their growth; thus, depletion of amino acids from cell culture medium results in aberrant chlamydial developmental forms and inhibits chlamydial growth in vitro (Coles et al.1993).
All cells need a carbon source for energetic metabolism and that source is often glucose. Early studies showed that infection of L cells with Chlamydia caused an increase in glucose consumption in host cells (Moulder 1970). Consistent with this study, chlamydial infection results in elevated ATP levels and glucose consumption, along with increased glucose transporter-1 (GLUT1) expression in Hela cells (Ojcius et al.1998). Furthermore, it has been shown that Chlamydia has the capability to utilize both cellular glucose and glutamate to support their intracellular growth (Weigent and Jenkin 1978; Iliffe-Lee and McClarty 1999); nevertheless, glucose is a singular preferred carbon source for optimal growth (Iliffe-Lee and McClarty 1999). The conclusion is supported by the observation that, unlike many other bacteria, chlamydiae do not respond with shifts in gene expression toward utilization of alternative carbon sources when glucose is limited (Nicholson, Chiu and Stephens 2004). These studies lead to the proposition that chlamydiae enhance host cell energy metabolic pathways during infection to promote chlamydial survival and growth.
Although there is an understanding of which nutrients Chlamydia require from host cells, the chlamydial inclusion membrane is not passively permeable to even low-molecular-weight molecules (Heinzen and Hackstadt 1997; Kleba and Stephens 2008). Thus, one of the challenging questions is how these required nutrients are transported across the inclusion vacuole membrane from the host cytosol to become available to support chlamydial viability. Although Chlamydia encode several general membrane transporters that are predicted to be involved in transport of nutrients from the periplasm to the chlamydial cytosol, there is no evidence to show the localization of these bacterial transporters in the inclusion membrane (Stephens et al.1998; Braun et al.2008; Fisher, Fernandez and Maurelli 2013). These data suggest that acquisition of nutrients from host cells is likely mediated by other uncharacterized mechanisms or pathways. We hypothesized that chlamydiae specifically intersect the host cell glucose pathway to insure acquisition of an essential carbon source. In the present study, we found that the expression of GLUT1 and glucose transporter-3 (GLUT3) from host cells was highly induced during Chlamydia trachomatis infection, and that GLUT1 protein was present in close proximity to the chlamydial inclusion. Knockdown of GLUT1 and GLUT3 with small interfering RNA (siRNA) significantly impaired chlamydial development and infectivity. GLUT1 protein was stabilized by deubiquitination during chlamydial infection and CT868, a chlamydial protein with deubiquitinase activity (Misaghi et al.2006; Le Negrate et al.2008; Claessen et al.2013; Fischer et al.2017), was shown to deubiquitinate GLUT1. Our findings provided evidence that Chlamydia intersects host transporter proteins that support microbial intracellular growth.
MATERIALS AND METHODS
Reagents
Restriction enzymes, Taq DNA polymerase and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA, USA). Mouse anti-GLUT1 (ab40084) and rabbit anti-GLUT3 (ab53095) were purchased from Abcam (Cambridge, MA, USA). Mouse anti-β Tubulin (G-8) was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Mouse anti-ubiquitin (clone P4D1) were purchased from Biolegend (San Diego, CA, USA). 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (2-NBDG), mouse anti-GFP (clone 3E6), goat anti-mouse Alexa 594 and goat anti-rabbit 594 were purchased from Invitrogen (Grand Island, NY, USA). Antibodies against phospho-p38 MAPK, p38 MAPK, phospho-ERK1/2, ERK1/2, phospho-JNK, JNK, phospho-AKT, and AKT were purchased from Cell Signaling Technology (Beverly, MA, USA). Mouse anti-Flag and anti-Flag M2 Magnetic beads were purchased from Sigma (St. Louis, MO, USA). IRDye 800CW goat anti-mouse IgG (H+L) and IRDye 680RD goat anti-rabbit IgG (H+L) were purchased from LI-COR Biosciences (Lincoln, NE, USA). Merifluor Chlamydia was purchased from Meridian Diagnostics, Inc. (Cincinnati, OH, USA). Cycloheximide, chloramphenicol, brefeldin A (BFA), SB202190, PD98059, SP600125, LY-294002 hydrochloride, phloretin, 4΄,6-diamidino-2-phenylindole (DAPI) and MG132 were purchased from Sigma (St. Louis, MO, USA). Power SYBR Green cells to CT kit was obtained from Ambion, USA. Dynabeads® Protein G, Lipofectmine 2000 and lipofectmine RNAiMAX were obtained from Invitrogen (Grand Island, NY, USA).
Cell lines and strains
Hela and McCoy cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Hyclone). L929 cells were maintained in RPMI1640 medium supplemented with 10% FBS. All of the cell lines were grown at 37°C in an atmosphere containing 5% CO2.
XL10-Gold Escherichia coli cells were obtained from Agilent Technologies. Dam−/Dcm− competent E. coli K12 cells were obtained from New England Biolabs (USA). Chlamydia trachomatis lymphogranuloma venereum serovar L2 (L2/434/Bu) EB were purified from L929 cells as described previously (Abromaitis and Stephens 2009).
Construction of plasmids
To construct the transfection plasmids EGFP-GLUT1, EGFP-GLUT3 and 3xFlag-GLUT1, total RNA from Hela cells was isolated by using RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and then used to synthesize the cDNA with ThermoScript RT-PCR system (Invitrogen, USA) according to the manufacturer's instruction. The full-length cDNAs of GLUT1 and GLUT3 were amplified and cloned into mammalian expression vector EGFP-N1 or p3XFlag-Myc-CMV-26. To construct the transfection plasmids 3xFlag-CT867 and 3xFlag-CT868, CT867 and CT868 genes were amplified from C. trachomatis L2 DNA and cloned into mammalian expression vector p3XFlag-Myc-CMV-26. All inserts were verified by sequencing. Expression was confirmed by immunoblotting or immunofluorescence assay. The primers used in this study were described in Table S1, Supporting Information.
Real-time RT-PCR
GLUT gene mRNA expression levels were detected by real-time RT-PCR. Briefly, Hela cells were infected with C. trachomatis L2 strain for 24 h and 48 h, cDNA was prepared by using Power SYBR Green Cells to CT kit according to manufacturer's instruction. Real-time RT-PCR was carried out with Power SYBR Green PCR master mix (Ambion) by using the specific primers (Table S1, Supporting Information) in a 7500 real-time PCR system (Applied Biosystems). Relative transcription levels were calculated by using the ΔΔCt method.
Transfection
Transfection of Hela cells was performed by lipofectamine 2000 according to the manufacturer's instruction. Briefly, monolayers of Hela cells with 50% confluence were plated on 4-well chamber slides or 6-well plates and incubated for overnight. Cells were then transfected with the indicated plasmids by lipofectamine 2000 and then infected with C. trachomatis at an Multiplicity of Infection (MOI) of 5 at 24 h post-transfection.
For siRNA knockdown, lipofectamine RNAiMAX reagent was mixed with indicated specific siRNAs or negative control siRNA in Opt-MEM medium, and the mixture was added into 4-well chamber slides or 24-well plates. Monolayers of Hela cells with 50% confluence were then plated on each well containing transfection mixture. Cells were infected with C. trachomatis at an MOI of 5 at 48 h post-transfection. The siRNA knockdown efficiency was evaluated by immunoblotting or real-time RT-PCR.
Immunofluorescence and microscopy
Indirect immunofluorescence was performed as described previously (Cocchiaro et al.2008).
Glucose trafficking assay
For detection of glucose uptake during the infection, infected cells were incubated with fluorescent glucose analog 2-NBDG (100 μmol) in phosphate-buffered saline (PBS) for 20 min at 37°C. After incubation, cells were washed two times with PBS, and the fluorescence was immediately detected by fluorescence microscopy under 40× oil objective.
To analysis the glucose uptake in chlamydial inclusions or extrusions in the presence of GLUT inhibitor phloretin, infected cells were treated with phloretin (50 μM) or Dimethyl sulfoxide (DMSO) for 2 h at 36 or 60 h post-infection (hpi), and then incubated with PBS buffer containing 2-NBDG (100 μM) and phloretin (50 μM) for another 20 min. Cells were washed two times with PBS, and immediately visualized by fluorescence microscopy. To quantitation of glucose accumulation in the inclusion, mean fluorescence intensity of the inclusions was measured by defining regions of interest (ROI) within the inclusions, and the data were corrected for background by subtracting the fluorescence intensity of cell-free region with the same size of ROIs. To compare the glucose uptake for different treatment samples, the mean fluorescence intensity of same size of ROIs within inclusions was measured in each field of samples. At least 4 fields were examined, and values were obtained from a total of 70 inclusions.
Immunoblotting and immunoprecipitation
Hela cells grown in 6-well plates were infected with L2 strain and collected at 24 and 48 hpi, respectively. The preparation of samples and subsequent immunoblotting were performed as previously described (Wang and Hardwidge 2012). For immunoprecipitation experiments, cells were lysed with a modified Radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP40, 0.25% deoxycholic acid and 1 mM Ethylenediaminetetraacetic acid (EDTA)) supplemented with protease inhibitor cocktail (Thermo Scientific) and incubated on ice for 30 min. The cell lysates were collected by centrifugation at 4°C and then incubated with anti-Flag beads, or Dynabeads Protein G with anti-GFP antibody, and the immunoprecipitation assay was performed according to the manufacturer's instructions.
Quantitation of infectious progeny and inclusion size
Hela cells were transfected with GLUT1- and GLUT3-specific siRNA or control siRNA using lipofectmine RNAiMAX reagent according to manufacturer's instructions. After 48 h post-transfection, the cells were infected with C. trachomatis and incubated for additional 48 h. To quantify production of infectious progeny in different siRNA transfected cells, infected cells were scraped into media at 48 hpi, lysed with 27 1/2 gauge needles and 10-fold serial dilutions were used to infect fresh McCoy monolayers that had been plated on 24-well plate for 24 h. The production of infectious progeny was detected by immunofluorescent staining of chlamydial inclusions with Merifluor Chlamydia according to manufacturer's instructions. The inclusion sizes at 30 hpi were analyzed by measuring the surface area of inclusions with image analysis software Volocity.
Statistical analysis
Statistical analyses were performed using Student's t-tests. Values are expressed as mean ± standard deviation (SD) of at least three independent experiments. A P value of ≤0.05 was considered statistically significant.
RESULTS
Transcription and translation of GLUT1 and GLUT3 are up-regulated during C. trachomatis infection
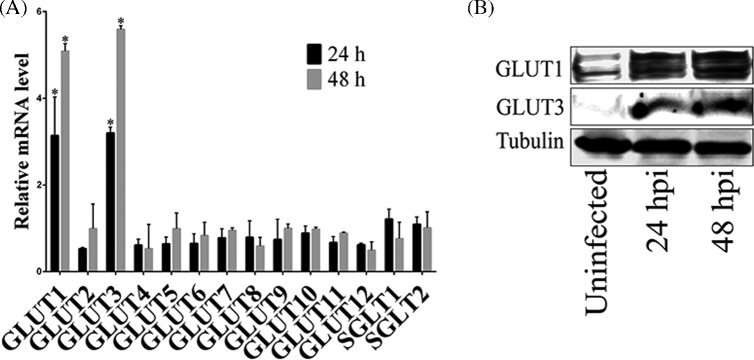
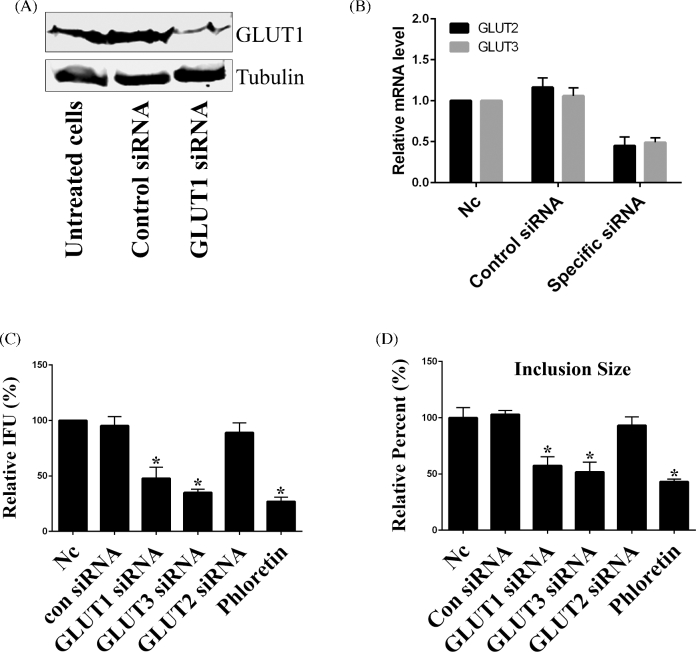
GLUTs are a family of membrane proteins that facilitate the transport of glucose and other substrates across plasma membranes, and have essential functions in carbohydrate homeostasis (Mueckler and Thorens 2013). It was previously shown that GLUT1 expression is increased during chlamydial infection (Ojcius et al.1998). To investigate whether Chlamydia affects the transcription of other GLUT-family genes, Chlamydia trachomatis-infected cells were evaluated for mRNA expression of GLUT genes at different times post-infection. It was found that infection of host cells with Chlamydia significantly increased the mRNA level of GLUT1 and GLUT3 at early and late times post-infection compared to uninfected cells, whereas there was no effect on mRNA transcription of 10 other detected GLUT genes (Fig. 1A). Consistent with the mRNA data, it was additionally shown that protein expression of GLUT1 and GLUT3 was also markedly increased after chlamydial infection (Fig. 1B). These data suggested that protein expression of GLUT1 and GLUT3 in host cells was specifically induced by Chlamydia during infection. Given the important roles of GLUT1 and GLUT3 in glucose metabolism in mammalian cells (Pascual et al.2004; Simpson et al.2008), induction of expression of GLUT1 and GLUT3 proteins may be important to meet the metabolic demands of infected cells during chlamydial growth and development.
Figure 1.
The expression of GLUT1 and GLUT3 is up-regulated during C. trachomatis infection. (A) Hela cell monolayers were grown in 24-well plates and infected with C. trachomatis for 24 or 48 h prior to testing. Gene expression of different GLUT genes was measured by real-time RT-PCR and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. The data are presented as relative mRNA levels compared to uninfected cells and shown as the mean ± SD. Each experiment was performed in triplicate and repeated at least three times. Asterisks indicate significantly different values compared with uninfected cells (P < 0.05, Student's t-test). (B) Hela cells grown in 100 mm culture dish were infected with C. trachomatis for 24 and 48 h, respectively. The protein levels of GLUT1 and GLUT3 were evaluated by immunoblotting. The experiment was repeated at least three times with similar results.
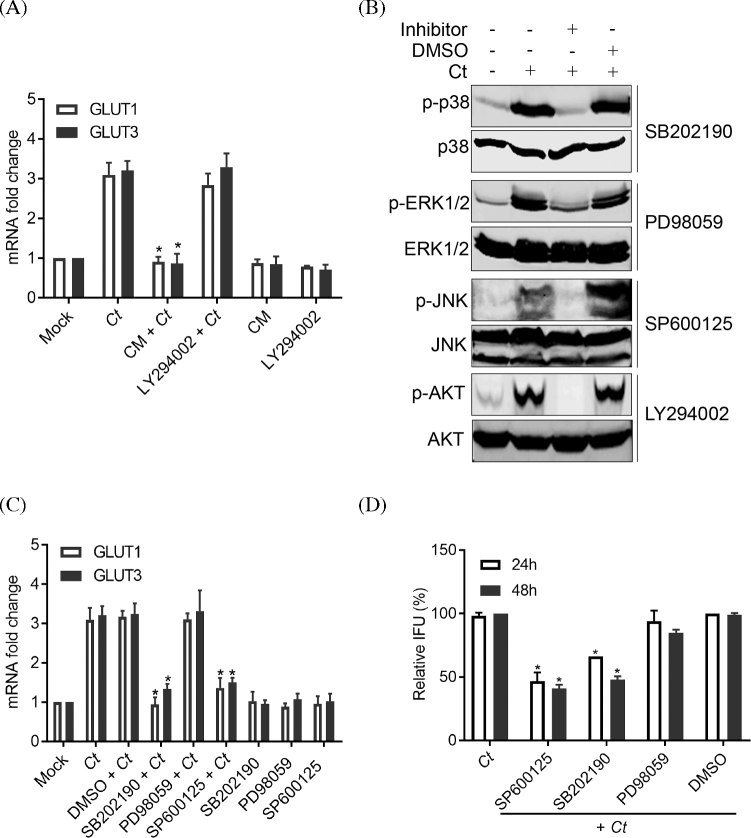
To investigate the mechanisms of up-regulation of GLUT1 and GLU3 expression during Chlamydia infection, infected cells were treated with chloramphenicol for 6 h at 24 hpi, and the mRNA levels of GLUT1 and GLUT3 were evaluated. Treatment of infected cells with chloramphenicol, a bacterial protein synthesis inhibitor, significantly blocked the increased expression of GLUT1 and GLUT3 compared with untreated cells (Fig. 2A). This finding shows that the induction of expression of GLUT1 and GLUT3 coincident with chlamydial infection was dependent on chlamydial protein synthesis. Host MAPK signaling pathways and PI3K/AKT pathway are involved in the regulation of gene expression (Whitmarsh 2007; Hemmings and Restuccia 2012), and it has been shown that both PI3K/AKT and MAPK signaling pathways can regulate the expression of GLUT1 and GLUT3 in mammalian cells (Hajduch et al.1998). We, herein, and others (Verbeke et al.2006; Subbarayal et al.2015) have shown that both PI3K/AKT and MAPK signaling pathways are activated during chlamydial infection, and activation of these pathways are triggered by Chlamydia-derived pathogen-associated molecular patterns (PAMPs) proteins (Bastidas et al.2013). We therefore investigated whether modulation of gene expression of GLUT1 and GLUT3 was mediated by Chlamydia-induced activation of these signaling pathways. Consistent with previous reports (Verbeke et al.2006; Buchholz and Stephens 2007; Zhou et al.2013), infection of Hela cells with Chlamydia significantly induced the phosphorylation of AKT, p38, ERK1/2 and JNK MAP kinases (Fig. 2B). Inhibition of PI3K/AKT signaling activation with a specific inhibitor, LY294002, did not affect the induction of GLUT1 or GLUT3 mRNA levels during chlamydial infection suggesting that the PI3K/AKT pathway was not involved (Fig. 2A). Likewise the ERK1/2 arm of the MAPK pathway was not implicated (Fig. 2C). In contrast, inhibition of p38 and JNK MAP kinase activation significantly affected the up-regulated gene expression for GLUT1 and GLUT3 (Fig. 2C). These data show that following infection, chlamydial components (e.g. PAMPs) modulate the MAPK pathway induction of GLUT1 and GLUT3 gene expression.
Figure 2.
The up-regulation of GLUT1 and GLUT3 is mediated by the activation of p38 and JNK MAPK pathways during C. trachomatis infection. (A) Hela cells infected with C. trachomatis were treated with either LY294002 (20 μM) or chloramphenicol (100 μg/ml) for 4 h at 24 hpi. Gene expression levels of GLUT1 and GLUT3 were measured by real-time RT-PCR and normalized to GAPDH expression. (B) Activation of host MAPK and PI3k/AKT signaling pathways during the chlamydial infection. Hela cells infected with C. trachomatis were treated with, or without, p38 inhibitor SB202190 (20 μM), ERK1/2 inhibitor PD98059 (20 μM), JNK inhibitor SP600125 (20 μM) or PI3K inhibitor LY294002 (20 μM) for 4 h at 24 hpi. The phosphorylated form of p38, ERK1/2, JNK or AKT was detected by immunoblotting. (C) Hela cells infected with C. trachomatis were treated with SB202190 (20 μM), PD98059 (20 μM), SP600125 (20 μM) or DMSO for 4 h at 24 hpi. Gene expression levels of GLUT1 and GLUT3 were measured by real-time RT-PCR and normalized to GAPDH expression. The data shown in panel B and C are presented as relative mRNA levels compared to untreated cells and shown as mean ± SD. (D) Hela cells were infected with C. trachomatis and treated with MAPK inhibitors SB202190 (2.5 μM), PD98059 (2.5 μM), SP600125 (2.5 μM) or vehicle for either 24 h at 24 hpi or 48 h at 2 hpi. The progeny formation of infectious EB was analyzed as described in Materials and Methods. Data are presented as percentage of chlamydial infection. IFU, inclusion forming units. Each experiment was performed in triplicate and repeated at least three times. Asterisks indicate significantly different compared with the data from infection only (P < 0.05, Student's t-test).
If the MAPK p38 and JNK pathway activation is required for up-regulation of GLUT1 and GLUT3, and if they are necessary for chlamydial growth, then inhibition of these arms of the MAPK pathway may be expected to inhibit chlamydial growth. To test whether the activation of MAPK pathway played a role in chlamydial infection, infected cells were treated with several MAPK inhibitors for 24 h at 24 hpi, or treated for 48 h after infection and the production of infectious EB was measured. It was found that the treatment of host cells with either a specific p38 inhibitor (SB202190) or with a JNK1/2 inhibitor (SP600125) significantly reduced the production of infectious chlamydial progeny compared to untreated cells or DMSO vehicle treatment, whereas treatment of infected cells with ERK1/2 inhibitor PD98059 had no effect on bacterial growth (Fig. 2D). These data show that the activation of p38 and JNK pathways are important for GLUT 1 and GLUT3 up-regulation and for chlamydial growth. These data are supported by a recent report that has shown inhibition of JNK pathway by specific inhibitor SP600125 affected the growth of C. trachomatis (Olive et al.2014).
GLUT1, but not GLUT3, is in close proximity to the chlamydial inclusion membrane
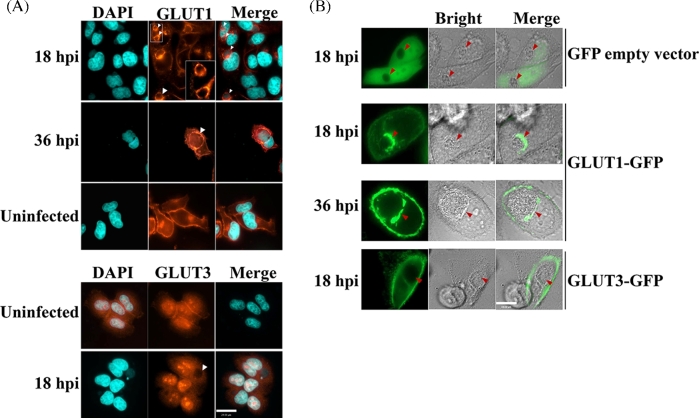
In uninfected cells, both GLUT1 and GLUT3 are localized to the plasma membrane where they function as transporters for the movement of extracellular glucose into mammalian cells. It was reasoned that if GLUT up-regulation was biologically meaningful, it may be a strategy for meeting the increased metabolic demands of chlamydial metabolism. As such, we tested whether chlamydia infection up-regulated GLUT1 and 3 were localized to the plasma membrane. The localization of these two proteins during the chlamydial developmental cycle was detected by immunofluorescence microscopy. Both GLUT1 and GLUT3 proteins were localized to the plasma membrane and host cytoplasm in uninfected cells (Fig. 3A), which was consistent with previous reports (Mueckler and Thorens 2013). Unexpectedly, in infected cells, up-regulated GLUT1, but not GLUT3, was additionally present in large amounts in close proximity with the chlamydial inclusion membrane by 18–36 h post-infection (Fig. 3A), whereas GLUT3 remained highly expressed at the host cell plasma membrane and cytoplasm (Fig. 3A).
Figure 3.
Recruitment of GLUT1 to inclusion membrane during the C. trachomatis infection. (A) Hela cell monolayers grown in 8-well chamber slides were infected with C. trachomatis for indicated time course, and then fixed and stained with GLUT1- (Red) or GLUT3-specific antibodies as indicated. Host cell nuclei and C. trachomatis (DNA, blue) were labeled with DAPI. Scale Bar, 24 μm. (B) Hela cell monolayers expressing GFP, GLUT1-GFP or GLUT3-GFP were infected with C. trachomatis, and the localization of GFP, GFP-GLUT1 or GFP-GLUT3 was detected by live imaging, respectively. Scale Bar, 19 μm. The arrowheads in all panels indicated the chlamydial inclusions. All of the experiments were repeated at least three times with similar results.
The immunofluorescence data were validated by C. trachomatis infection of Hela cells expressing GLUT1 or GLUT3 fused to a GFP reporter. Following infection, GFP-GLUT1 and GFP-GLUT3 were localized to the host cell plasma membrane (Fig. 3B); however, GFP-GLUT1 alone was concomitantly and predominantly observed in close proximity to the chlamydial inclusion (Fig. 3B). Thus, both immunofluorescence detection and GFP-tagging of GLUT1 demonstrated that GLUT1 uniquely co-localizes to the plasma membrane and adjacent to the inclusion during chlamydial infection.
Recruitment of GLUT1 in close proximity of chlamydial inclusion membrane is independent of de novo chlamydial protein synthesis but is dependent on BFA-sensitive pathway
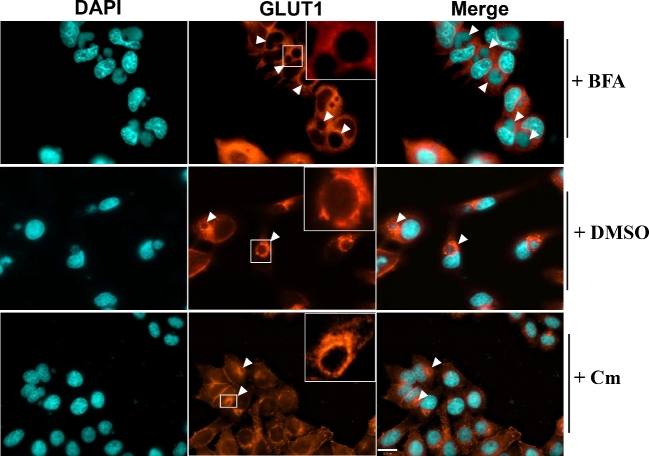
To test whether the unique re-distribution of GLUT1 adjacent to the inclusion was mediated by chlamydial proteins, Chlamydia-infected cells were treated with chloramphenicol for 8 h at 16 hpi, and the localization of GLUT1 was detected by immunofluorescence assay. It was shown that inhibition of chlamydial protein synthesis did not affect the localization of GLUT1 to the proximity of the chlamydial inclusion membrane (Fig. 4). These data suggest that while up-regulation of GLUT1 is dependent upon chlamydial protein synthesis, the re-distribution of GLUT1 was not mediated by chlamydial virulence factors produced after 16–24 hpi. Thus, the redistribution of GLUT1 is linked to the interaction of the native host secretion pathway with the chlamydial inclusion, whereas GLUT3 localization was not disrupted or redirected.
Figure 4.
The localization of GLUT1 in the presence of inhibitors during the chlamydial infection. Hela cell monolayers grown in 8-well chamber slides were infected with C. trachomatis for 6 h, and then incubated with fresh medium containing 1 μg/ml of brefeldin A (BFA), DMSO or 100 μg/ml of chloramphenicol (Cm) for another 12 h. All of the experiments were repeated at least three times with similar results. The arrowheads indicate the chlamydial inclusions. Scale bar, 22.00 μm.
The trans-Golgi network (TGN) serves as a sorting station to direct newly synthesized proteins either to the plasma membrane or to endosomal compartments (Traub and Kornfeld 1997). To investigate whether GLUT1 was also redirected to close proximity of chlamydial inclusion through the same pathway, BFA, a specific inhibitor of trans-Golgi-mediated secretion in mammalian cells, was employed to treat Chlamydia-infected cells at 16 hpi. Treatment of infected cells with 1 μg/ml of BFA significantly blocked the localization of GLUT1 to the plasma membrane and the recruitment of GLUT1 near the chlamydial inclusion membrane (Fig. 4), indicating that the close proximity of GLUT1 with inclusion membranes was dependent on a BFA-sensitive pathway. Because MAPK kinases p38 and JNK regulated the expression of GLUT1 during chlamydial infection, we also investigated the localization of GLUT1 in the presence of MAPK inhibitors during infection. The result of immunofluorescence showed that treatment of infected cells with MAPK inhibitors did not affect the localization or GLUT1 expression (Fig. S1, Supporting Information). We conclude that GLUT1 is trafficked by the canonical trans-Golgi secretion pathway but nevertheless GLUT1 is singularly redirected to predominantly localize adjacent to the inclusion.
GLUT1 and GLUT3 are important for chlamydial growth and infectivity
To determine whether induction of expression of GLUT1 and GLUT3 and subsequent localization of GLUT1 adjacent to the inclusion membrane are important for chlamydial infection and not a confounding artifact of host cell disruption, the expression of endogenous GLUT1 or GLUT3 in Hela cells was targeted by specific siRNA. The reduced expression of GLUT1 protein was confirmed by immunoblotting (Fig. 5A), and the reduced transcription of GLUT2 and GLUT3 by their specific siRNA was confirmed by real-time RT-PCR, but not immunoblotting due to their low expression at protein level (Fig. 5B). It was shown that knockdown of endogenous GLUT1 or GLUT3 expression significantly reduced the infectious chlamydial progeny as an over 50% reduction in infectivity was observed compared to untreated cells or control siRNA-treated cells (Fig. 5C). In addition, it was shown that knockdown of GLUT1 or GLUT3 by siRNA resulted in a 42% and 48% reduction in chlamydial inclusion size, respectively, (Fig. 5D) suggesting an inhibition of chlamydial growth. Phloretin is a specific GLUT inhibitor that binds competitively to the mammalian glucose-binding site (Salas-Burgos et al.2004). Consistent with the siRNA knockdown data, it was shown that treatment of infected cells with 50 μM of phloretin significantly reduced the infectious chlamydial progeny and inclusion size (Fig. 5C and D). As a control, knockdown of GLUT2 with specific siRNA had no effect on chlamydial growth and infectivity (Fig. 5C and D). Our data indicate that GLUT1 and GLUT3 are essential for full support of chlamydial growth and development during the infection by delivering glucose to the infected cell.
Figure 5.
GLUT1 and GLUT3 are important for chlamydial infectivity and development. (A) GLUT1 protein was transiently silenced by transfection of specific siRNA (100 nM), and the knockdown efficiency at 60 h post-transfection was evaluated using immunoblotting. (B) Hela cells were transfected with negative control siRNA (100 nM), GLUT2 siRNA (100 nM) or GLUT3 siRNA (100 nM), and the knockdown efficiency of GLUT2 and GLUT3 was evaluated by real-time RT-PCR. The data were presented as relative mRNA levels compared to nontransfected cells and shown as the mean ± SD. Each experiment was performed in triplicate and repeated at least three times. Asterisks indicate significantly different values compared with nontransfected cells (P < 0.05, Student's t-test). (C) Hela cells were depleted of GLUT1, GLUT2 or GLUT3 for 48 h, infected with C. trachomatis for 48 h or infected cells were treated with phloretin for 24 h at 24 hpi. The progeny infectious EB formation was analyzed as described in Materials and Methods. Data (mean ± SD, independent experiments) were presented as percentage of nontransfected cells (Nc). IFU, inclusion forming units. Asterisks indicate significantly different compared with un-transfected cells (P < 0.05, Student's t-test). (D) Hela cells were depleted of GLUT1, GLUT2 or GLUT3 for 48 h, infected with Chlamydia for 30 h or infected cells were treated with phloretin for 12 h at 18 hpi, and the inclusion size was as described in Materials and Methods. Data (mean ± SD, independent experiments) were shown as percentage of un-transfected samples (Nc). Asterisks indicate values that are significantly different from those of the control (P < 0.05, Student's t-test).
GLUT1 protein is deubiquitinated during chlamydial infection
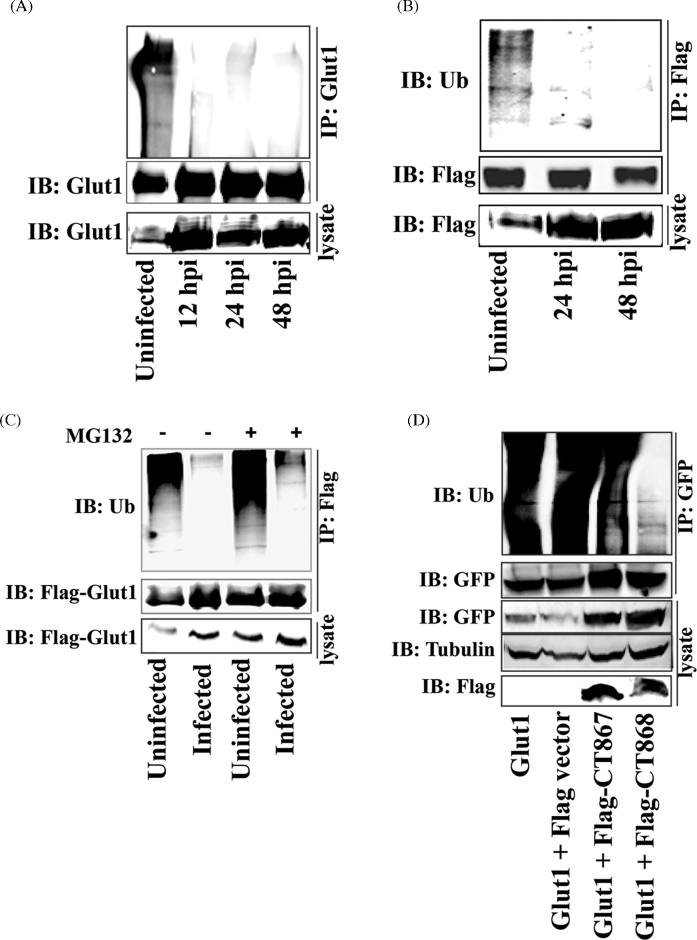
The increased expression of GLUT1 and GLUT3 during infection suggested that Chlamydia need to maintain a high level of these transporter proteins to transport adequate glucose from the extracellular environment to the host cells; however, cycloheximide, an inhibitor of host cell protein synthesis, does not inhibit chlamydial growth. To meet an increased glucose transport requirement, an alternative mechanism could be the stabilization of nascent GLUT1 and GLUT3 during chlamydial infection. Since GLUT1 is degraded by ubiquitin–proteasome pathway (Fernandes et al.2004), we therefore tested whether the ubiquitination of GLUT1 was altered during the chlamydial infection. Hela cells were infected with C. trachomatis and GLUT1 protein was immunoprecipitated from infected cell lysates at 12, 24 and 48 h post-infection with GLUT1-specific antibody. It was shown that infection of host cells with C. trachomatis significantly reduced the ubiquitination level of GLUT1 compared to the uninfected cells (Fig. 6A). This result was confirmed by examining the ubiquitination of overexpressed Flag-GLUT1 in host cells during chlamydial infection (Fig. 6B). Unlike GLUT1, we did not observe the ubiquitin-mediated posttranslational modification of GLUT3 in either uninfected or infected host cells (data not shown). Furthermore, it was shown that treatment of infected cells with the proteasome inhibitor MG132 (10 μM) did not recover the ubiquitination level of GLUT1 in infected cells (Fig. 6C). Blocking host protein synthesis with cytoheximide (CHX) significantly inhibited the protein synthesis of GLUT1 (Fig. S2A, Supporting Information). If chlamydial infection can stabilize the GLUT1, treatment of infected cells with CHX will not dramatically reduce the GLUT1 protein level. As expected, treatment of infected Hela cells with CHX (5 μg/ml) did not significantly reduce the GLUT1 level in either the inclusion membrane or the host cell plasma membrane (Fig. S2B, Supporting Information). These data suggest that GLUT1 protein was stabilized during chlamyidlal infection by altering the ubiquitination processing of GLUT1.
Figure 6.
Stabilization of GLUT1 by CT868-mediated deubiquitination during the chlamydial infection. (A) Hela cell monolayers were infected with C. trachomatis for 12, 24 or 48 h. Endogenous GLUT1 from infected cell lysates was immunoprecipitated using rabbit anti-GLUT1 antibody and protein A/G Magnetic beads, and then immunoblotted and probed with rabbit anti-ubiquitin and rabbit anti-GLUT1 antibodies. (B) Hela cells overexpressing Flag-GLUT1 were infected with C. trachomatis for 24 or 48 h. Flag-GLUT1 from cell lysates was immunoprecipitated using mouse anti-Flag M2 antibody-conjugated beads, and then immunoblotted and detected with anti-ubiquitin and anti-Flag antibodies. The expression of Flag-GLUT1 from cell lysates was immnoblotted and detected by anti-Flag antibody. (C) Hela cell monolayers expressing Flag-GLUT1 were infected with C. trachomatis for 20 h, and then treated in the presence or absence of MG132 (10 μM) for 4 h. GLUT1 from cell lysates was immunoprecipitated using mouse anti-Flag M2 antibody-conjugated beads, and then immunoblotted and stained using anti-ubiquitin and anti-Flag antibodies. (D) Hela cell monolayers stably expressing GFP-GLUT1 were transfected with p3xFlag-CMV vector, Flag-CT867 or Flag-CT868 expressing constructs. At 30 h post-transfection, GLUT1 from cell lysates was immunoprecipitated using anti-GFP antibody and then immunoblotted and stained with anti-ubiquitin and anti-GFP antibodies. The expression of CT867 and CT868 from cell lysates was detected by immunoblotting using anti-Flag antibody. All of the experiments were repeated at three times and a representative data was shown in each experiment.
Chlamydia encode two proteins (CT867 and CT868) with deubiquitinating and deneddylating activities (Misaghi et al.2006; Fischer et al.2017). The putative role of CT867 and CT868 in stabilization of GLUT1 was evaluated. Hela cells were co-transfected with GFP-GLUT1 and 3XFlag-CT868 or 3XFlag-CT867. GFP-GLUT1 was immunoprecipitated with GFP antibody-conjugated protein G beads after 30 h transfection, and the ubiquitination of GLUT1 was detected using anti-ubiquitin antibody. Our data showed that CT868, but not CT867, significantly suppressed the ubiquitination level of GLUT1 (Fig. 6D), suggesting that chlamydial protein CT868 can specifically target GLUT1 ubiquitination. A recent study has been shown that CT868 is localized to the inclusion membrane and its deubiquitinating enzyme domain is accsessible to host cytosol (Fischer et al.2017), thus it would be expected to be available to interact with host cell substrates at that interface.
DISCUSSION
Glucose transporters are a family of membrane proteins that facilitate the transport of glucose and other substrates across the plasma membrane in mammalian cells. Thirteen GLUT-family members have been identified (Zhao and Keating 2007). GLUT1 has been extensively studied and is responsible for maintaining the basal level of glucose uptake in most cell types and tissues (Bell et al.1990; Pascual et al.2004), whereas GLUT3 is highly expressed by neurons, although specific roles for glucose transport in other cell types have also been reported (Simpson et al.2008). The present study discovered that both GLUT1 and GLUT3 have fundamental roles in the glucose-related energy metabolism that supports chlamydial growth during the infection.
Our data demonstrated that GLUT1 and GLUT3 are specifically up-regulated during chlamydial infection. GLUT3 shows a higher affinity for glucose than other important glucose transporters like GLUT1, 2 or 4 (Simpson et al.2008). Given its singular plasma membrane localization, the role for GLUT3 is likely to maintain a high level of exogenous glucose transport for chlamydial infected cells. Since knockdown of GLUT3 with specific siRNA impaired chlamydial infectivity and development, it is speculated that knockdown of GLUT3 impaired the cellular energy metabolism and subsequent ATP production in host cells due to the reduced ability of glucose transport from the environment, which may indirectly affect chlamydial growth. Alternatively, GLUT3 may be required to transport sufficient glucose for the direct carbon needs of growing chlamydiae.
Unlike GLUT3, GLUT1 was observed in close proximity to the inclusion during infection. It has been shown that Chlamydia manipulate multiple host cell organelles and proteins to obtain host lipids. For example, Chlamydia infection induces Golgi fragmentation to form the Golgi ministacks surrounding the chlamydial inclusion (Heuer et al.2009), whereas recruitment of ER tubules in close proximity of inclusion membrane suggest a direct interaction between the ER and the chlamydial inclusion (Derre, Swiss and Agaisse 2011; Dumoux et al.2012). Although our observation could not unequivocally differentiate the organelle localization, the recruitment of only GLUT1 in close proximity of inclusion membrane suggested a unique role of GLUT1 for transportation of glucose or other substrates to Chlamydia. Although there is no evidence to show that Chlamydia can directly utilize glucose as a carbon source due to the absence of hexokinase, we found that glucose can be transported into the chlamydial inclusion. 2-NBDG is a fluorescent glucose analog that has been successfully used to detect the trafficking of glucose (Zou, Wang and Shen 2005). We discovered that 2-NBDG can efficiently translocate into inclusion lumen at early and late infection stages after incubating the infected cells with 100 μM of 2-NBDG for 20 min (Fig. S3A, Supporting Information). Inhibition of GLUT1 and GLUT3 activity by treatment of infected cells with phloretin significantly blocked the trafficking of 2-NBDG into inclusions compared to untreated cells (Fig. S3B, Supporting Information). As a control, DMSO treatment had no effect on 2-NBDG trafficking (Fig. S3B, Supporting Information). This is an unexpected finding, but which implies that the translocated glucose may play some uncharacterized roles during chlamydial infection. Interestingly, a recent study demonstrated that host transporter SLC35D2 was recruited to chlamydial inclusion membrane to import UDP-Glc into the inclusion lumen (Gehre et al.2016), which sheds light on a novel strategies utilized by Chlamydia to support their growth while sequestered within their inclusion vacuole.
Bacterial pathogens manipulate their host cellular functions by pathogen-specific virulence factors. However, our data demonstrated that recruitment of GLUT1 in proximity of inclusion membranes is not mediated by concurrent synthesis of chlamydial effector proteins. An effort to perform pull down assays also failed to identify bacterial binding partners of GLUT1 from Chlamydia-infected cells (data not shown). Alternatively, the vesicle-trafficking inhibitor BFA blocked the accumulation of GLUT1 to the proximity of the inclusion membrane, suggesting that Chlamydia has evolved strategies to redirect the trafficking of host GLUT1 by exploiting the host's native secretion pathway. It has previously been shown that chlamydial infection causes the Golgi fragmentation to form Golgi ministacks surrounding the inclusion membrane (Heuer et al.2009) and treatment of infected cells with BFA also abrogated the localization of Golgi ministacks to inclusion, it is possible that the enrichment and localization of GLUT1 was associated with the induced Golgi stacks during infection.
Protein turnover by the ubiquitination pathway is essential to maintain normal cellular homeostasis. Remarkably, chlamydial infection interfered with the ubiquitination of GLUT1. The targeting of GLUT1 turnover by Chlamydia may be mediated by a chlamydial effector deubiquitinase protein. CT868 has been shown to target host cellular functions with its deubiquitinase activity, and although the initial report suggested it was a cytosolic protein (Le Negrate et al.2008), recently it has been shown to be localized to the chlamydial inclusion membrane (Fischer et al.2017). Ubiquitin-mediated proteolytic system is one of the important host defense mechanisms against intracellular pathogens, in which the unwanted components and intracellular pathogens are tagged by ubiquitination system and further trigger the selective autophagy (Fujita and Yoshimori 2011; Choi et al.2014). Bacterial pathogens have evolved different strategies to evade this host defense response (Ashida, Kim and Sasakawa 2014), although it is unknown whether cytosolic exposed inclusion membrane proteins (Lutter, Martens and Hackstadt 2012) can be targeted by a ubiquitin-mediated host defense mechanism. Given the inclusion membrane localization of CT868 in host cells during chlamydial infection (Fischer et al.2017), it is possible that this deubiquinase protein may protect the chlamydial vacuolar compartment against ubiquitin-mediated host defense system as well as modify the turnover and perhaps localization of host cell proteins such as GLUT1.
This report demonstrated that Chlamydia manipulates the amount and localization of host glucose transporters GLUT1 and GLUT3 to support the metabolic demands for intracellular chlamydial growth. It is known that chlamydiae obtain most of their nutrients from their host cell as they lack complete biosynthetic pathways (Stephens et al.1998); however, how chlamydiae exploit the rich host cell cytosol is unknown as they are sequestered from the cytosol by the inclusion vacuole membrane. Chlamydiae ensure stable amounts of GLUT1, despite disruption of its synthesis or its secretory pathway, by deubiquination of GLUT1. These novel findings advance our understanding about how Chlamydia manipulates host cellular functions to manage its competitive and parasitic requirements for essential nutrients from host cells during the course of infection. Given that Chlamydia obtains many of its essential nutrients from host cells and is faced with the challenge of transporting these factors across the inclusion membrane, our findings for chlamydial modulation of GLUT1 reveal a novel strategy orchestrated by Chlamydia to support their intracellular growth.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSPD online.
Supplementary Material
Supplementary data are available at FEMSPD online.
Acknowledgements
We thank Dr. Jean Lee (Brigham and Women's Hospital and Harvard Medical School) for her immense support in providing space to Dr. XW for finishing some experiments in her laboratory. We thank Dr. Michael Starnbach (Department of Microbiology and Immunology, Harvard Medical School) for providing the C. trachomatis L2 serovar.
FUNDING
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI091851 to R.S.S.
Conflict of Interest. None declared
REFERENCES
- Abromaitis S, Stephens RS. Attachment and entry of Chlamydia have distinct requirements for host protein disulfide isomerase. Plos Pathog 2009;5:e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Kim M, Sasakawa C. Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol 2014;12:399–413. [DOI] [PubMed] [Google Scholar]
- Bastidas RJ, Elwell CA, Engel JN et al. Chlamydial intracellular survival strategies. Cold Perspect Med 2013;3:a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI, Kayano T, Buse JB et al. Molecular biology of mammalian glucose transporters. Diabetes Care 1990;13:198–208. [DOI] [PubMed] [Google Scholar]
- Boncompain G, Muller C, Meas-Yedid V et al. The intracellular bacteria Chlamydia hijack peroxisomes and utilize their enzymatic capacity to produce bacteria-specific phospholipids. Plos One 2014;9:e86196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun PR, Al-Younes H, Gussmann J et al. Competitive inhibition of amino acid uptake suppresses chlamydial growth: involvement of the chlamydial amino acid transporter BrnQ. J Bacteriol 2008;190:1822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz KR, Stephens RS. The extracellular signal-regulated kinase/mitogen-activated protein kinase pathway induces the inflammatory factor interleukin-8 following Chlamydia trachomatis infection. Infect Immun 2007;75:5924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park S, Biering SB et al. The parasitophorous vacuole membrane of Toxoplasma gondii Is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 2014;40:924–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Witte MD, Yoder NC et al. Catch-and-release probes applied to semi-intact cells reveal ubiquitin-specific protease expression in Chlamydia trachomatis infection. Chembiochem 2013;14:343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER et al. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. P Natl Acad Sci USA 2008;105:9379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles AM, Reynolds DJ, Harper A et al. Low-nutrient induction of abnormal chlamydial development - a novel component of chlamydial pathogenesis. FEMS Microbiol Lett 1993;106:193–200. [DOI] [PubMed] [Google Scholar]
- Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog 2011;7:e1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoux M, Clare DK, Saibil HR et al. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic 2012;13:1612–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Jiang SB, Kim JH et al. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. Plos Pathog 2011;7:e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes R, Carvalho A, Kumagai A et al. Downregulation of retinal GLUT1 in diabetes by ubiquitinylation. Molecular Vision 2004;10:618–28. [PubMed] [Google Scholar]
- Fischer A, Harrison KS, Ramirez Y et al. Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. Elife 2017;6:e21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DJ, Fernandez RE, Maurelli AT. Chlamydia trachomatis transports NAD via the Npt1 ATP/ADP translocase. J Bacteriol 2013;195:3381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshimori T. Ubiquitination-mediated autophagy against invading bacteria. Curr Opin Cell Biol 2011;23:492–7. [DOI] [PubMed] [Google Scholar]
- Gehre L, Gorgette O, Perrinet S et al. Sequestration of host metabolism by an intracellular pathogen. Elife 2016;5:e12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid-metabolism In chlamydia trachomatis-infected cells - directed trafficking of Golgi-derived sphingolipids to the Chlamydial inclusion. P Natl Acad Sci USA 1995;92:4877–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty CL, Gottlieb SL, Taylor BD et al. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis 2010;201:S134–55. [DOI] [PubMed] [Google Scholar]
- Hajduch E, Alessi DR, Hemmings BA et al. Constitutive activation of protein kinase B alpha by membrane targeting promotes glucose and system A amino acid transport, protein synthesis, and inactivation of glycogen synthase kinase 3 in L6 muscle cells. Diabetes 1998;47:1006–13. [DOI] [PubMed] [Google Scholar]
- Heinzen RA, Hackstadt T. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect Immun 1997;65:1088–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Perspect Biol 2012;4:a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 2009;457:731–5. [DOI] [PubMed] [Google Scholar]
- Hu VH, Harding-Esch EM, Burton MJ et al. Epidemiology and control of trachoma: systematic review. Trop Med Int Health 2010;15:673–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Exit strategies of intracellular pathogens. Nat Rev Microbiol 2008;6:99–110. [DOI] [PubMed] [Google Scholar]
- Iliffe-Lee ER, McClarty G. Glucose metabolism in Chlamydia trachomatis: the ‘energy parasite’ hypothesis revisited. Mol Microbiol 1999;33:177–87. [DOI] [PubMed] [Google Scholar]
- Kleba B, Stephens RS. Chlamydial effector proteins localized to the host cell cytoplasmic compartment. Infect Immun 2008;76:4842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CC, Jackson LA, Campbell LA et al. Chlamydia pneumoniae (TWAR). Clin Microbiol Rev 1995;8:451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Negrate G, Krieg A, Faustin B et al. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol 2008;10:1879–92. [DOI] [PubMed] [Google Scholar]
- Lutter EI, Martens C, Hackstadt T. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp Funct Genom 2012;2012:362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae JI, Sheiner L, Nahid A et al. Mitochondrial metabolism of glucose and glutamine Is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe 2012;12:682–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S, Balsara ZR, Catic A et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol 2006;61:142–50. [DOI] [PubMed] [Google Scholar]
- Moulder JW. Glucose metabolism of L-cells before and after infection with Chlamydia psittaci. J Bacteriol 1970;104:1189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 1991;55:143–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 2013;34:121–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderer T, Heng J, McConville MJ. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. Plos Pathog 2010;6:e1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson TL, Chiu K, Stephens RS. Chlamydia trachomatis lacks an adaptive response to changes in carbon source availability. Infect Immun 2004;72:4286–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojcius DM, Degani H, Mispelter J et al. Enhancement of ATP levels and glucose metabolism during an infection by Chlamydia - NMR studies of living cells. J Biol Chem 1998;273:7052–8. [DOI] [PubMed] [Google Scholar]
- Olive AJ, Haff MG, Emanuele MJ et al. Chlamydia trachomatis-induced alterations in the host cell proteome are required for intracellular growth. Cell Host Microbe 2014;15:113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual JM, Wang D, Lecumberri B et al. GLUT1 deficiency and other glucose transporter diseases. Eur J Endocrinol 2004;150:627–33. [DOI] [PubMed] [Google Scholar]
- Salas-Burgos A, Iserovich P, Zuniga F et al. Predicting the three-dimensional structure of the human facilitative glucose transporter Glut1 by a novel evolutionary homology strategy: Insights on the molecular mechanism of substrate migration, and binding sites for glucose and inhibitory molecules. Biophys J 2004;87:2990–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson IA, Dwyer D, Malide D et al. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol-Endoc M 2008;295:E242–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele S, Brunton J, Ziehr B et al. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. Plos Pathog 2013;9:e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 1998;282:754–9. [DOI] [PubMed] [Google Scholar]
- Su H, McClarty G, Dong F et al. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J Biol Chem 2004;279:9409–16. [DOI] [PubMed] [Google Scholar]
- Subbarayal P, Karunakaran K, Winkler AC et al. EphrinA2 receptor (EphA2) is an invasion and intracellular signaling receptor for Chlamydia trachomatis. PLoS Pathog 2015;11:e1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrone E, Papp J, Weinstock H et al. Prevalence of Chlamydia trachomatis genital infection among persons aged 14-39 years-United States, 2007-2012. MMWR-Morbid Mortal W 2014;63:834–8. [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Kornfeld S. The trans-Golgi network: a late secretory sorting station. Curr Opin Cell Biol 1997;9:527–33. [DOI] [PubMed] [Google Scholar]
- van Ooij C, Kalman L, van Ijzendoorn S et al. Host cell-derived sphingolipids are required for the intracellular growth of Chlamydia trachomatis. Cell Microbiol 2000;2:627–37. [DOI] [PubMed] [Google Scholar]
- Verbeke P, Welter-Stahl L, Ying S et al. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog 2006;2:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hardwidge PR. Enterotoxigenic Escherichia coli prevents host NF-kappaB activation by targeting IkappaBalpha polyubiquitination. Infect Immun 2012;80:4417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigent DA, Jenkin HM. Contrast of glycogenesis and protein-synthesis in monkey kidney cells and Hela-cells infected with Chlamydia trachomatis lymphogranuloma venereum. Infect Immun 1978;20:632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim Biophys Acta 2007;1773:1285–98. [DOI] [PubMed] [Google Scholar]
- Zhao FQ, Keating AF. Functional properties and genomics of glucose transporters. Curr Genomics 2007;8:113–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Huang Q, Li Z et al. PORF5 plasmid protein of Chlamydia trachomatis induces MAPK-mediated pro-inflammatory cytokines via TLR2 activation in THP-1 cells. Sci China Life Sci 2013;56:460–6. [DOI] [PubMed] [Google Scholar]
- Zou CH, Wang YJ, Shen ZF. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J Biochem Bioph Meth 2005;64:207–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSPD online.