ABBREVIATIONS
- BET
brain extraction tool
- CMS
comparative marker selection
- FDR
false discovery rate
- FLAIR
fluid-attenuating inversion recovery
- GBM
Glioblastoma Multiforme
- GLCM
gray level co-occurrence matrix
- IPA
Ingenuity Pathway Analysis
- MRI
magnetic resonance imaging
- TCGA
the cancer genome atlas
- TCIA
the cancer imaging archive
- T1WI
T1-weighted images
- VOI
volume of interest
Imaging is a routine tool for the diagnosis of most types of disease processes; in oncology, imaging is key to identify primary and metastatic neoplasms, provide differential diagnoses, evaluate tumors at baseline, and assess treatment response. Despite increasing refinements of modalities such as magnetic resonance imaging (MRI) and the enormous amount of imaging data generated everyday, imaging is not typically used for tumor characterization beyond location, size, and gross appearance. Unlocking the full spectrum of data contained in routine imaging studies would greatly enhance tumor characterizations, accelerate the transition towards imaging-based personalized diagnostics and enable the identification of more accurate, noninvasive, clinically relevant biomarkers.1,2 Toward this end, radiomics refers to microscale (on a pixel or voxel level) imaging features extracted from standard medical images.
Glioblastoma Multiforme (GBM) is characterized by a few hallmark mutations: TP53, PTEN, EGFR, IDH1, NF1, RB1, PIK3CA, and PIK3R1.3 An ever changing and rapidly evolving mutational landscape makes it almost impossible to capture the entire genomic temporal and spatial tumor heterogeneity in a single biopsy. Further, in analogy to Heraclitus's concept of “Panta Rhei,” that we can never step twice into the same river due to its constant flow,4 we also can never look at the same tumor twice due to the highly proliferative nature and rapid progression of niche biology across the heterogeneous tumor landscape; thus, it becomes apparent that a single snapshot microarray genomic test at time of surgery or biopsy is not enough to capture a very dynamic tumor environment over time. For the most part, numerous biopsies and repeated genomic testing would carry a high morbidity and likely be limited by cost of care at most medical centers. Thus, we recognize that a more efficient and high-throughput, clinically applicable way to identify key molecular tumor markers across the entire tumor landscape and over multiple time points is needed. This MRI-based noninvasive radiogenomic testing can be termed “radiopsy,” and might serve as an adjunct to invasive genomic testing. Further, it can be of importance for in-depth monitoring of molecular and phenotypic tumor behavior over time, as MRI is a clinically accepted, FDA approved, and cost effective way to screen patients at multiple time points throughout the cancer treatment paradigm.
Recently, quantitative parameters, such as volumetric and radiomic imaging features, have been linked with genomic data.5-7 The linkage of imaging information with genomic data has been termed radiogenomics (or imaging-genomics).8
In this study, we sought to apply the power of radiomic analysis to GBM, the most common and most aggressive primary brain cancer in adults, and correlate imaging characteristics with underlying key genomic aberrations. Our objectives were to determine whether there is a relationship between the 3 most frequently mutated genes (TP53, PTEN, EGFR) promoting GBM oncogenicity and imaging characteristics as assessed by radiomic textural analysis and demonstrate that extracted radiomic features carry a similar complexity as microarray-based genomic information. This can be anticipated to further clinically applicable radiomic test methods to noninvasively identify hallmark mutations in glioblastoma.
METHODS
The collection of the original material and data provided by The Cancer Genome Atlas (TCGA) project was conducted in compliance with all applicable laws, regulations, and policies for the protection of human subjects, and any necessary approvals, authorizations, human subject assurances, informed consent documents, and IRB approvals were obtained.
Patients
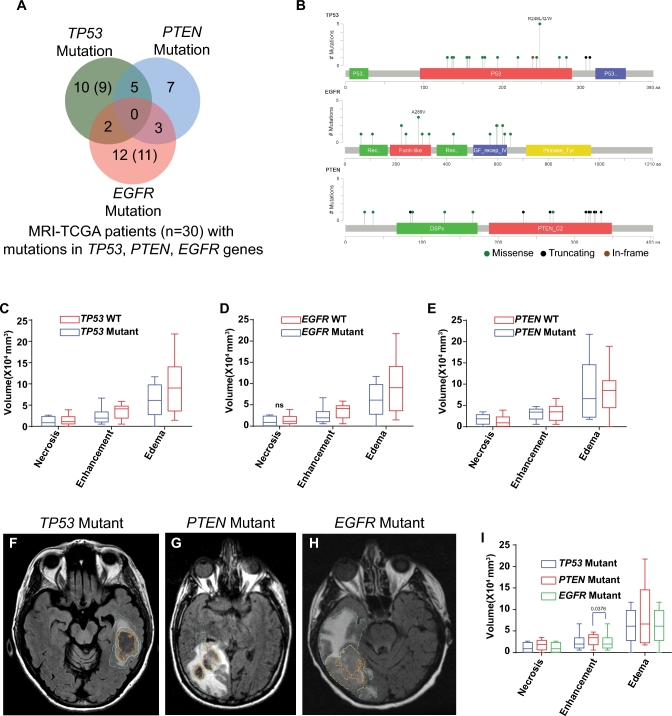
A total of 29 TCGA patients underwent radiomic analysis. The preoperative MRI studies of the TCGA patients were available for public download from The Cancer Imaging Archive (TCIA). Of 29 patients, as shown in Figure 1A, 10 patients had TP53 mutations and no mutations in PTEN and EGFR genes; 7 patients had PTEN mutations and no mutations in TP53 and EGFR genes, and 12 patients had EGFR mutation and no mutations in TP53 and PTEN genes. Five patients had TP53 and PTEN mutations, 2 patients had mutations in EGFR and TP53, and 3 patients had EGFR and PTEN mutation. Detailed information of specific mutations in each gene are presented in Tables, Supplemental Digital Content 1.
FIGURE 1.
TCGA/TCIA GBM patients with known gene mutation and their cognate MRI volumetric information. A, Venn diagram depicting patients’ association with specific gene mutation. Numbers show patients with imaging data while numbers within the brackets are patients with whole genome expression profiles. B, Schematic representation of mutation type and frequency across specific domains of genes. Gene names are shown at the left top corner of graphs and color codes for mutation type are shown at the bottom right corner. C-E, MRI volumes for patients with specific gene mutation. Volumes in mm3 of specific MRI phenotype is plotted on y-axis. Pair-wise comparisons (mutant vs Wild Type (WT)) are shown on x-axis. P-values are shown above the bar graphs. ns: P-value not-significant. F-H, Representative MRI scans with volumes for TP53 mutant F, PTEN mutant G, and EGFR mutant H, patients. Necrosis, contrast enhancing, and FLAIR areas are demarcated in orange, yellow, and blue lines, respectively. I, MRI volumetric comparisons across mutations. Statistically significant MRI volume is identified with number above the pair of box plots.
Image Registration and Segmentation
Segmentation was performed on the conventional MR images, namely precontrast T1-weighted images (T1WI), postcontrast T1WI, and fluid-attenuating inversion recovery (FLAIR) images. Tumor delineation was performed in a semi-automated fashion, on a slice-by-slice basis, using 3D Slicer (https://www.slicer.org), an open-source image analytics platform. The segmented images were reviewed in consensus by 2 board-certified neuroradiologists (RRC; 9 yr of experience, AJK, 35 yr of experience). Prior to image segmentation, all 3 images were coregistered into the same geometric space using affine registration (12 degrees of freedom). Registration was implemented using the General Registrations (BRAINS) Toolbox in 3D Slicer. After image registration, we used postcontrast T1WI and FLAIR images for tumor delineation. Postcontrast T1WI were used to determine the borders of the enhancement (ie, active tumor) and nonenhancing central component (ie, necrosis). The edema/invasion component was segmented using the FLAIR image. The precontrast T1WI was used to exclude hemorrhage from the delineated tumor. Additionally, a region in the contralateral normal-appearing white matter was also segmented for within-sequence normalization of the data. The outlines of the 3 segmented phenotypes (active tumor, necrosis, edema/invasion) and contralateral normal-appearing white matter were saved in a label volume and further used in radiomic analysis.
Radiomic Analysis
The volume of interest (VOI) corresponding to each phenotype, as well as the combined phenotype representing the whole GBM, were used for radiomic analysis. Multiple invariant and volume-independent radiomic features were extracted from each VOI. Our in-house radiomic analysis pipeline for MR images consists of 3 steps: (i) skull-stripping, (ii) image intensity normalization, (iii) radiomic feature extraction. In the first step, the Functional Magnetic Resonance Imaging of the Brain’s Brain Extraction Tool (BET; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET) was used to remove nonbrain tissue from the anatomical MR images. To account for scanner and within-sequence protocol differences, we applied an intensity normalization algorithm to standardize the intensity scales across MR images of the same contrast. Finally, radiomic features were extracted based on histogram analysis and the Gray Level Co-occurrence Matrix (GLCM). Histogram analysis provides information about the intensity distribution within a VOI; 10 histogram-based features were extracted, namely minimum, maximum, mean, standard deviation, skewness, kurtosis, and percentiles (1%, 5%, 95%, and 99%). GLCM is a tabulation of how often different combinations of gray levels co-occur in a region of interest and in a specific direction and provides information about the distribution of pairs of voxels separated by a given distance at a specific direction. Three parameters were defined for the calculation of the GLCM: direction of spatial relationship, distance between the reference and neighboring pixel, and number of gray levels. In our implementation, we considered 4 directions of spatial relationship (ie, in-plane rotations; horizontal, vertical, diagonal up, diagonal down) and a distance of 1 pixel (ie, co-occurrences between neighboring pixels); thus, 4 GLCMs were calculated per VOI. Given that there is no literature support of the appropriate number of gray levels, multiple gray levels were examined by re-quantization of the initial image (l = 8, 16, 32, 64, 256 gray levels). For each GLCM, the following 20 features were extracted: autocorrelation, contrast, correlation, cluster shade, cluster prominence, dissimilarity, energy, entropy, homogeneity, maximum probability, variance, sum average, sum variance, sum entropy, difference variance, difference entropy, information measure of correlation 1 and 2, inverse difference moment, and normalized inverse difference moment. To obtain rotation invariant measures of the GLCM-based features, the average, range, and angular variance of the features calculated for different θ were obtained, thereby resulting in 60 invariant GLCM-based features for each VOI for a specific gray level. Accounting for multiple gray levels, phenotypes, and sequences, a total of 2480 were obtained for each patient. An additional 620 texture features from the contralateral normal-appearing white matter were extracted from human MRIs and used for within-sequence normalization purposes.
Analysis of Texture Features and Genes Associated with Patients with Specific Gene Mutation
Significant texture features associated with 3 groups of patients with a mutually exclusive mutation in either TP53, PTEN, or EGFR (n = 29), were selected using the comparative marker selection (CMS) module of GenePattern (Broad Institute, Boston, Massachusetts). Features with false discovery rate (FDR) less than 0.05 were included. Heatmap using these significant texture features values across patients were generated using the GENE-E software from Broad Institute. GENE-E converts values to heatmap colors using the mean and maximum values for each row or the standard deviations from the row mean for each row. Genomic data was unavailable for one of the EGFR and TP53 mutant patients. Gene expression analyses were performed with whole genome expression profiles of patients with mutation either in TP53 (n = 9), EGFR (n = 11), or PTEN (n = 7) gene. Briefly, level 3 whole genome expression profiles were obtained from the public TCGA data portal (https://gdc-portal.nci.nih.gov/). Gene expression profiles were grouped into TP53 mutant vs TP53 WT, PTEN mutant vs PTEN WT, and EGFR mutant vs EGFR WT groups and analyzed by using the CMS module as above. Heatmap using these significant genes expression levels across patients were generated using the GENE-E (Broad Institute); and patients were grouped by unsupervised hierarchical clustering. Genes with significant P-values (P < .05) were selected for network/pathway analyses by Ingenuity Pathway Analysis (IPA). Consensus clustering of patients based on significant-CMS filtered texture features and genes (TP53: n = 342; PTEN: n = 18; EGFR: n = 97) were performed using GENE-E, where columns (with patient ID and mutant/WT annotations) were clustered based on 1 minus spearman rank correlation for distance metric with average linkage. Hierarchical clustering algorithm was used with 100 resampling iteration, with row-wise (values of selected texture features and genes across patients) normalization.
RESULTS
We identified patients in the TCGA database that also had corresponding MR imaging present in the TCIA and had nonoverlapping mutations in either TP53 (n = 10), PTEN (n = 7), or EGFR (n = 12; Figure 1A). These were either missense, truncating, or in-frame mutations (Figure 1B). We then sought to determine if there was a difference in MRI appearance of those tumors carrying mutually exclusive mutations in TP53, PTEN, or EGFR genes. Conventional volumetric MRI analysis for areas of necrosis, contrast enhancement, and peritumoral edema did not reveal any significant differences between WT and respective mutation except that EGFR WT as compared to mutant (P = .029) and PTEN mutant as compared to EGFR mutant (P = .0376) tumors showed slightly larger areas of contrast enhancement (Figures 1C-1E). Representative images are shown for TP53, PTEN, and EGFR mutated tumors (Figures 1F-1I).
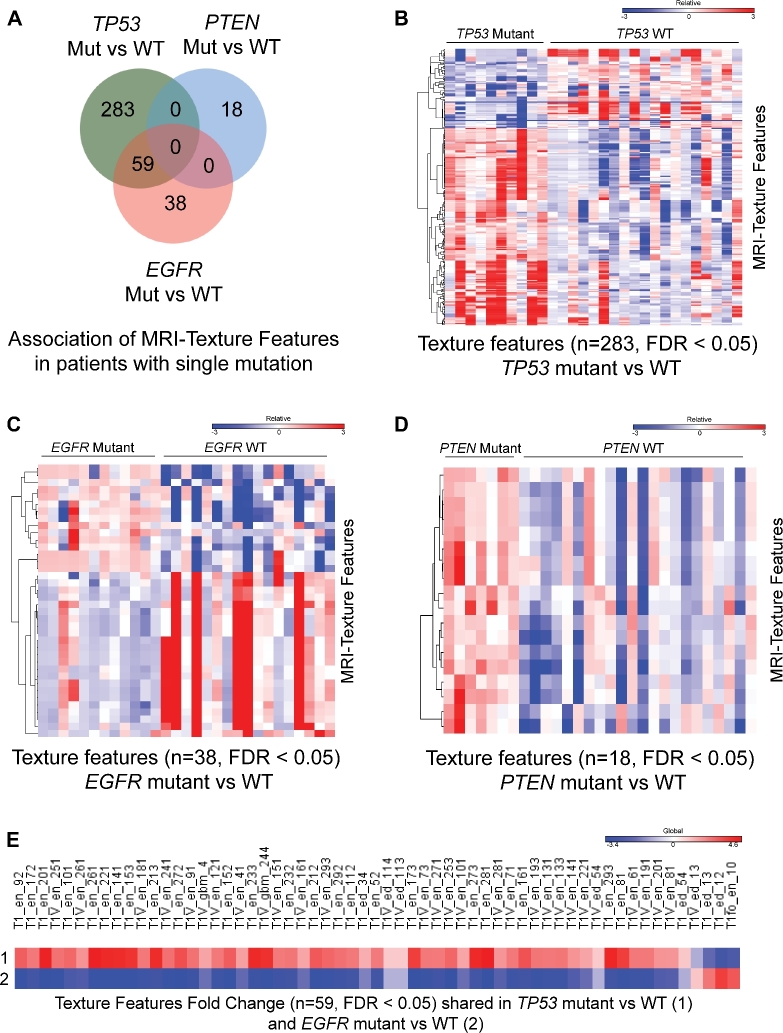
We then identified the significant radiomic features for the 3 genotypes using the CMS tool,9 this in analogy as previously used for gene expression analysis by our group.8,10 We identified 283 unique radiomic features for TP53 mutated tumors, 18 for PTEN mutated tumors, and 38 features for EGFR mutated tumors that were nonoverlapping (Figure 2A). Heatmap representation of the radiomic texture features of mutated vs WT tumors for the respective 3 mutations demonstrates the complexity and depth of radiomic data (Figures 2B-2D). Similar to gene expression data, radiomic data also shows overlapping features that are highly correlated with multiple traits of interest, in this case TP53 and EGFR mutated tumors (n = 59 features; Figure 2E).
FIGURE 2.
MRI texture feature signatures are uniquely associated with patients with TP53, PTEN, and EGFR gene mutations. A, Venn diagram depicting numbers of MRI texture features, their unique and overlapping association with GBM patients with specific gene mutation. B-D, Heatmap generated using uniquely associated and significant texture features (FDR < 0.05) in TP53 mutant vs WT (n = 283) B, EGFR mutant vs WT (n = 38) C, and PTEN mutant vs WT (n = 18) D. Color scale bar above heatmap show range of texture feature values across patients. E, Heatmap generated using fold change values of shared and significant texture features (n = 59; FDR < 0.05) in TP53 mutant vs WT and EGFR mutant vs WT. Color scale above heatmap show range of fold change values for specific texture feature.
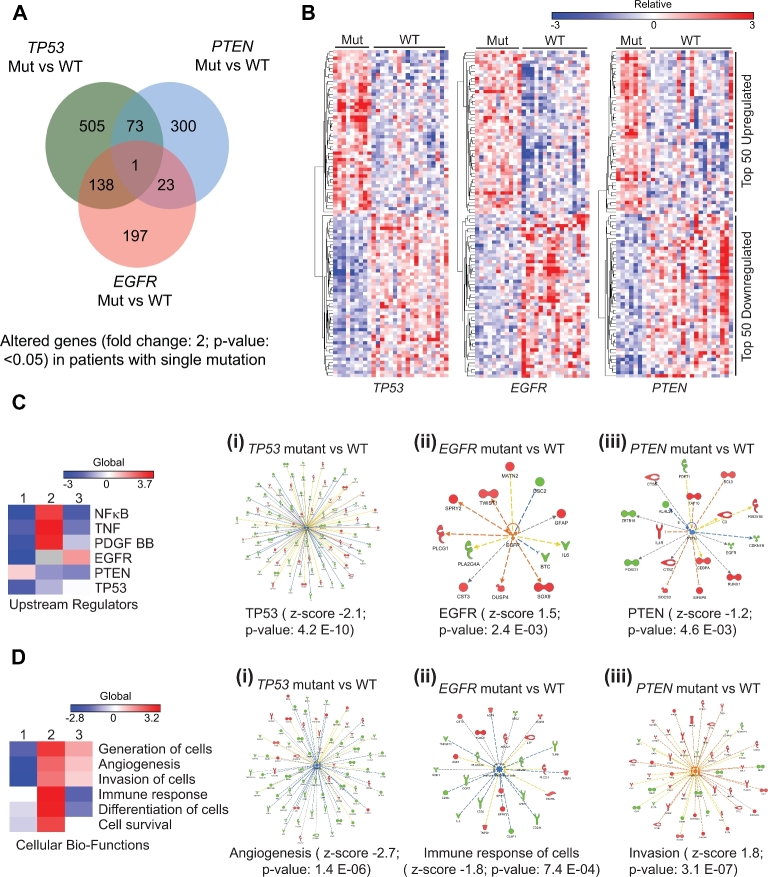
We then sought to determine TP53, PTEN, and EGFR mutation defining gene expression profiles to mirror the analysis as done for the radiomic features. We identified 505 unique and nonoverlapping genes significantly upregulated in TP53 mutated tumors, 300 for PTEN mutated tumors, and 197 genes for EGFR mutated tumors (Figure 3A). The latter gene expression profiles and heatmaps for mutational versus WT defining gene expression profiles (P < .05) demonstrate a slightly more complex, but similar pattern as for genotype defining radiomic feature sets (Figure 3B). Combined analysis using shared and significant genes revealed significantly differentially expressed upstream regulating molecules: NFKB, TNF, PDGFBB, EGFR, PTEN, and TP53, the latter 3 serving as correct positive control for the initial selection criteria (Figure 3C). We then determined the major associated TP53 (P-value 4.2E-10), PTEN (P-value 4.6E-03), or EGFR (P-value 2.4E-03) defining gene expression clusters (Figure 3C, i, ii, iii, Tables, Supplemental Digital Content 2, 3, and 4, respectively). A similar analysis was performed for the main cellular biofunctions defined by the most highly differentially regulated TP53, PTEN, and EGFR associated gene networks, which revealed functions such as: generation of cells, angiogenesis, invasion of cells, immune response, differentiation of cells, and cell survival, all hallmarks of cancer and GBM in particular (Figure 3D). The major biofunction and defining gene clusters associated with each mutant genotype were as follows: TP53-angiogenesis (P-value 1.4E-06), PTEN-invasion (P-value 3.1E-7), and EGFR-immune response (P-value 7.4E-04; Figure 3D, i, ii, iii, Tables, Supplemental Digital Content 5, 6, and 7, respectively).
FIGURE 3.
Specific gene signatures are uniquely associated with patient tumors carrying mutually exclusive TP53, EGFR, and PTEN gene mutations. A, Venn diagram with numbers of significantly altered genes in patients with specific gene mutation vs WT comparisons. B, Heatmap generated using uniquely associated and significant genes (top 50 upregulated and top 50 downregulated genes with fold change ≥2.0 and P < .05) in TP53 mutant vs WT (left), EGFR mutant vs WT (middle) and PTEN mutant vs WT (right). Color scale bar above heatmap show range of gene expression levels. C, Heatmap generated using activation z-scores of upstream regulators in mutant vs WT analyses. 1: TP53 mut vs WT; 2: PTEN mut vs WT; 3: EGFR mut vs WT. Color scale bar shown above heatmap show range of texture feature values. C: i, Network of TP53 regulated genes in TP53 mut vs WT analysis. Z-score and P-value are shown under the network. C: ii, Network of EGFR regulated genes in EGFR mut vs WT analysis. Z-score and P-value are shown under the network. C: iii, Network of PTEN regulated genes in PTEN mut vs WT analysis. Z-score and P-value are shown under the network. D, Heatmap generated using activation z-scores of cellular bio-functions in mutant vs WT analyses. 1: TP53 mut vs WT; 2: PTEN mut vs WT; 3: EGFR mut vs WT. Color scale bar shown above heatmap show range of texture feature values. D: i, Status of angiogenesis and associated gene network in TP53 mut vs WT analysis. Z-score and P-value are shown under the network. D: ii, Status of immune response of cells and associated gene network in EGFR mut vs WT analysis. Z-score and P-value are shown under the network. D: iii, Status of invasion and associated gene network in PTEN mut vs WT analysis. Z-score and P-value are shown under the network.
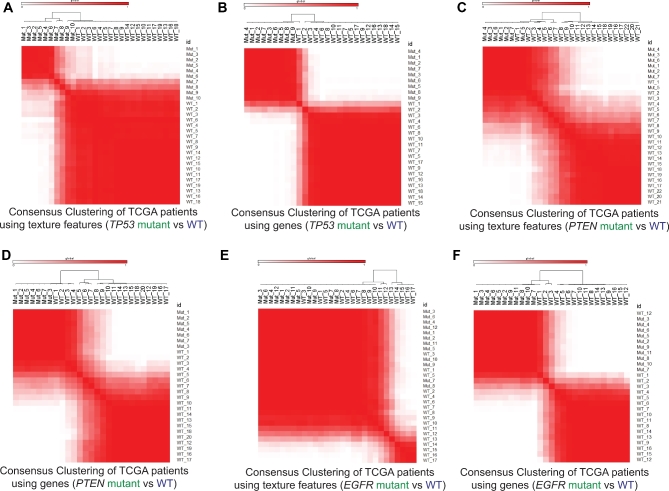
Side by side consensus cluster analysis demonstrated similar correlation matrices for TP53 mutant vs WT radiomic texture features (Figure 4A) as for the corresponding gene expression results (Figure 4B). This was similarly seen for PTEN mutant vs WT (Figures 4C and 4D) and to a lesser degree for EGFR mutant vs WT (Figures 4E and 4F).
FIGURE 4.
Consensus clustering of GBM patients on the basis of texture features and genes. A, Consensus clustering of patients based on significant altered texture features in TP53 mutant vs WT analysis. B, Consensus clustering of patients based on significantly altered genes in TP53 mutant vs WT analysis. C, Consensus clustering of patients based on significant altered texture features in PTEN mutant vs WT analysis. D, Consensus clustering of patients based on significantly altered genes in PTEN mutant vs WT analysis. E, Consensus clustering of patients based on significant altered texture features in EGFR mutant vs WT analysis. F, Consensus clustering of patients based on significantly altered genes in EGFR mutant vs WT analysis. Range of patient correlations are shown color scale bar above each plot.
DISCUSSION
In this study, we demonstrate that there are distinct MRI radiomic texture feature signatures associated with the TP53-PTEN-EGFR mutational landscape. We show that, unlike conventional quantitative volumetric image analysis, the extracted radiomic feature sets containing 4800 data points per tumor carry extra depth of information. Our results demonstrated that radiomic features are now approaching the complexity of whole genome microarray expression data. These findings have important clinical implication, since radiomic analysis is noninvasive and performed using routine imaging obtained in the everyday clinical setting. If prospectively validated, it can serve as cost effective adjunct radiogenomic diagnostic test method and advance personalized patient care. The field of medical radiomics, particularly cancer radiomics, is rapidly evolving with numerous studies now showing that radiomic data are highly correlated with genomic data across various types of cancers.11-15
However, for the field of radiogenomics to evolve, causality and biologic genetic validation is required with gain- and loss-of-function studies, this is currently underway in our laboratory. Nevertheless, the herein presented data are unique in showing that the 3 hallmark mutations in GBM3 have very distinctly corresponding and partially nonoverlapping radiomic feature sets (Figure 2). The radiomic data structure visibly approaches the complexity of genomic microarray data, and there is a quantifiable relationship between genotypic-specific radiomic features and gene expression clusters (Figures 3). Side by side analysis shows that consensus clustering of both radiomic and genomic data yields similar appearing graphical representation and separation of mutant vs WT TP53, PTEN, and EGFR glioblastoma (Figure 4). Altogether, these data suggest that radiomic analysis does unlock inherent information present in routine imaging that previously was inaccessible. Furthermore, these data carry quantifiable information that can be utilized towards the prediction of complex genomic events. In analogy to a surgically obtained biopsy, this type of analysis can yield voxel size based micro- to macroscopic radiomic tumor sampling of any size and location within a tumor and can be termed “radiopsy” as we suggest. It follows that radiopsy is not limited by physical constraints such as biopsy needle size and length, trajectory chosen and morbidity such as iatrogenic bleeding, infection, or parenchymal damage caused.
In the field of pathology, mainstream implementation of the microscope in the early nineteenth century completely disrupted the field from gross pathologic specimens (organs) to accessing microscopic depth with resolutions down to the cellular level.16 Surprisingly, in radiology, a similar paradigm shift is just happening now. Radiomics allows several-folds increase in the depth of analysis and yields a “microscopic” image appearance with thousands of data points instead of conventional radiologic descriptors related to lesion size, location, and appearance, reminiscent of conventional gross organ pathology. Interestingly, since, the field of pathology has evolved much further and by means of whole genome sequencing, we now have capabilities to zoom into single cells down to the very molecular level. The herein mentioned novel radiologic radiomic methods are advancing the field, but are just approaching that very depth down to the “cellular and molecular” level until the radiome has been fully sequenced, studied, and made sense of. This will require unified methods, large prospective trials with spatially matched voxel-biopsy radiogenomic analysis and basic laboratory functional genomic and radiogenomic validation using RNA interference and genetically engineered animal models.
Certainly, the above-mentioned obstacles must be overcome, and until then radiogenomics remains at the correlative stage only. Another pitfall is the lack of analysis methods for complex radiomic data; as of today, for the most part, gene expression microarray analyses methods have been adopted and tailored to fit the radiomic data structure, but no unified analysis method is used for data quality control, noise reduction, normalization, batch effect control across MRI scanner or field strength comparison just to name a few. However, the herein presented radiomic texture extraction method attempts to address those latter pitfalls and presents a robust texture analysis method that validated across a very diverse set of source images.
In summary, our results show that the high resolution radiomic brain cancer landscape is approaching its genomic counterpart in complexity and heterogeneity. It also demonstrates the evolution of the field of radiology and image analysis is similar to the advent of the microscope in pathology, now unlocking previously inaccessible microscale data in routine imaging. This study will further the development of the MRI-based noninvasive biopsy or radiopsy and advance personalized molecular therapy as adjunct to invasive genomic testing.
Disclosures
This work was supported by John S. Dunn Sr. Distinguished Chair in Diagnostic Imaging Fund, MD Anderson Cancer Center startup funding. Radiological Society of North America Scholar Grant (RSCH11506). Cancer Prevention and Research Institute of Texas (CPRIT) grant (RP160150) (RRC). This work was also supported by NIH R25 Baylor College of Medicine, Department of Neurosurgery Research Grant and NREF (Neurosurgery Research and Education Foundation) (338703) (POZ). The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this work.
Supplementary Material
Supplemental digital content is available for this article at www.neurosurgery-online.com.
REFERENCES
- 1. Hassan I, Kotrotsou A, Bakhtiari AS et al. Radiomic texture analysis mapping predicts areas of true functional MRI activity. Sci. Rep. 2016;6:25295 doi: 10.1038/srep25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nicolasjilwan M, Hu Y, Yan C et al. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42(4):212-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brennan CW, Verhaak RG, McKenna A et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beris AN, Giacomin AJ. “παντα ρεĩ: everything flows. Appl Rheol. 2014;24(5), 52918, 1-, 13. [Google Scholar]
- 5. Zinn PO, Sathyan P, Mahajan B et al. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PLoS One. 2012;7(8):e41522 10.1371/journal.pone.0041522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aerts HJ, Velazquez ER, Leijenaar RT et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006 doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2014;275(1):215-227. [DOI] [PubMed] [Google Scholar]
- 8. Zinn PO, Majadan B, Sathyan P et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PloS One. 2011;6(10):e25451 https://doi.org/10.1371/journal.pone.0025451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22(15):1924-1925. [DOI] [PubMed] [Google Scholar]
- 10. Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2015;275(1):215-227. [DOI] [PubMed] [Google Scholar]
- 11. Itakura H, Achrol AS, Mitchell LA et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015;7(303):303ra138-303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Y, Li H, Guo W et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci. Rep. 2015;5:17787 doi: 10.1038/srep17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo W, Li H, Zhu Y et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging (Bellingham). 2015;2(4):041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parmar C, Leijenaar RTH, Grossmann P et al. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci. Rep. 2015;5:11044 doi: 10.1038/srep11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van den Tweel JG, Taylor CR. A brief history of pathology: preface to a forthcoming series that highlights milestones in the evolution of pathology as a discipline. Virchows Arch. 2010;457(1):3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.