Abstract
In skeletal muscle, DNA methylation contributes to the suppression of gene expression in several biological processes and diseases. A protocol for the detection of methylated cytosine was thus established based on methylation-sensitive enzymes, immunoprecipitation, and bisulfite conversion. DNA methylation analysis, with bisulfite conversion and sequencing, enables the quantification of methylation at each single base position. Here, we describe a basic method of bisulfite sequencing that can be used to analyze local DNA methylation status to confirm genome-wide DNA methylation analysis or correlation of gene expression regulatory mechanisms.
Keywords: Epigenetics, DNA methylation, Methylcytosine, Bisulfite conversion, Bisulfite sequencing analysis
1. Introduction
To date, there are three established strategies to detect DNA methylation [1]. Methylation-sensitive enzymatic digestion was a simple way to define methylated DNA, but it is no longer widely used because of its sequence restriction and low working efficacy. Real-time PCR, following digestion with methylation-specific enzymes, enables the quantification of the methylation status of a few methylated-CpG regions; however, it cannot define the specific methylated CpG position. Several recent reports have combined real-time PCR with DNA microarray (or sequencing) technologies to perform genome-wide screening or profiling of DNA methylation; however, this enzymatic strategy is not compatible with high-throughput analysis [2]. On the other hand, combining high-throughput approaches with immunoprecipitation, which captures sheared DNA fragments around the methylated cytosine using anti-methyl-CpG binding protein (MeCP2) or anti-5-methyl cytosine antibodies, could achieve the desired result. However, each of these approaches remains limited by an inability to identify the exact quantity or position of methylated cytosine.
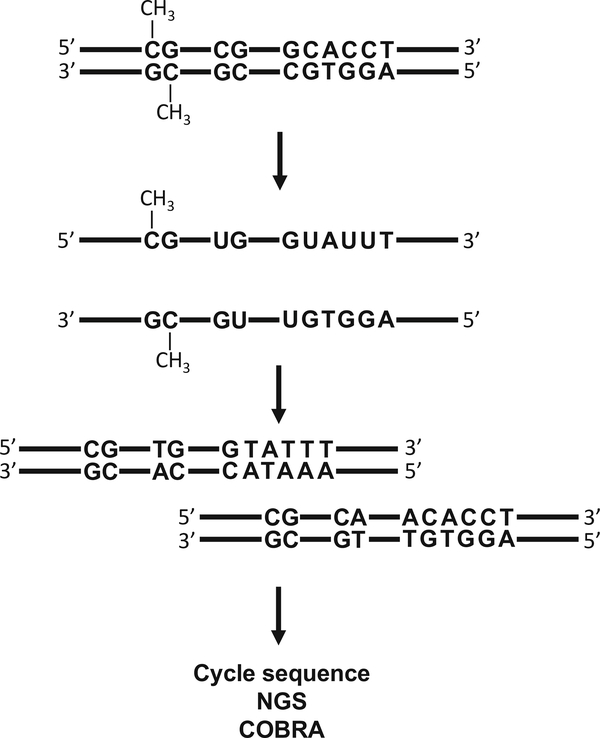
Bisulfite conversion is regarded as the gold standard in defining DNA methylation status. Treatment of DNA with sodium bisulfite converts unmethylated cytosine to uracil, which is subsequently converted to thymine during PCR amplification, while methylcytosine remains unchanged (Fig. 1) [3, 4]. In this regard, several commercial kits have proven useful and show high conversion rates. Leontiou et al. examined the performance of four such kits and accordingly ranked them by performance [5]. The MethylEdge Bisulfite Conversion System (Promega) emerged as the recommended kit to use due to the results obtained for bisulfite amplicon Sanger sequencing, but no significant difference in conversion rate was observed [5].
Fig. 1.
Methylation analysis based on Bisulfite conversion
Coupling bisulfite conversion with DNA sequence analysis enables the qualitative and quantitative detection of methylcytosine at a single base-pair resolution. Following bisulfite conversion, DNA is analyzed with next-generation transcriptomics to validate the genome-wide DNA methylation profile, and the regional methylation status established by Sanger sequencing. DNA methylation status can also be analyzed by treating bisulfite-converted DNA with restriction enzymes, more commonly referred to as Combined Bisulfite Restriction Analysis (COBRA). Digestion of PCR-amplified DNA with BstUI (which digests the sequence “CGCG”), followed by an analysis of DNA fragmentation with electrophoresis, is used to evaluate the approximate rate of cytosine methylation [6].
Using primary muscle progenitor cells, referred to as satellite cells (SCs), we demonstrate an example of the bisulfite sequencing method based on Sanger sequencing. Bisulfite-converted SC DNA was PCR amplified and subcloned into a cloning vector. Sanger sequence analysis was subsequently used to show the DNA methylation status at each single base position. The procedure contains DNA preparation (Subheading 3.1), bisulfite conversion (Subheading 3.2), PCR and subcloning (Subheading 3.3), and colony-direct sequencing (Subheading 3.4). Each bisulfite sequencing step does not contain any unique technique; thus, numerous alternative methods or protocols can be substituted. The isolation and culture of muscle SCs is described in “Note 1.”
2. Materials
2.1. DNA Preparation
AllPrep DNA/RNA Micro Kit (Qiagen).
AccuBlue High Sensitivity dsDNA Quantitation Kit (Biotium).
β-mercaptoethanol, ethanol, TE buffer.
Fluorometer.
2.2. Bisulfite Conversion
EpiTect Plus DNA Bisulfite Kit (Qiagen).
Thermal cycler.
Ethanol.
2.3. Primer Design and PCR
GO Taq master mix (Promega).
PCR primer.
Plasmid with analyzing sequence.
Agarose, TAE buffer.
Thermal cycler.
2.4. Subcloning and Sequencing
pGEM-T Easy Vector System (Promega).
LB agar plate, with Ampicillin.
Competent cells, SOC medium.
2% X-gal, dilute with dimethylformamide (DMF).
GO Taq master mix (Promega).
M13 forward (−20) primer, M13 reverse primer.
ExoSAP-IT For PCR Product Clean-Up (Affymetrix).
BigDye Terminator v3.1 Cycle sequencing Kit (Thermo Fisher Scientific).
T7 primer.
Water bath (42 °C), Incubator (37 °C).
ABI PRISM 3100 (Applied Biosystems).
3. Methods
3.1. DNA Preparation
To validate the gene expression profile of the harvested sample, extracting genomic DNA and total RNA from the same sample is highly recommended. For satellite cell culture conditions and isolation, see Note 1. All the centrifuge steps in this section were performed at room temperature.
3.1.1. Cell Collection and Lysis
Add 10 μL of β-mercaptoethanol to 1 mL of Buffer RLT Plus.
Discard cell culture medium and add 350 μL of Buffer RLT Plus directly to cells on culture dish.
Scrape the cells with chip, and harvest the entire cell suspension into a 1.5 mL tube.
Pass the lysate a minimum of five times through a blunt 20-gauge needle fitted to an RNase-free syringe.
Transfer the homogenized lysate to an AllPrep DNA spin column on a 2 mL collection tube and centrifuge for 30 s at 10,0 rpm (≥ 8,000 × g).
Use the flow-through for RNA purification and store the column at 4 °C on a new 2 mL collection tube for DNA purification later.
3.1.2. Extraction of DNA and RNA
Add 350 μL of 70% ethanol to the flow-through and mix well by pipetting.
Transfer the mixture into an RNeasy MinElute spin column.
Centrifuge for 15 s at 10,000 rpm (≥ 8,000 × g). Discard the flow-through.
Add 700 μL of Buffer RW1 to the column. Centrifuge for 15 s at 10,000 rpm (≥ 8,000 × g). Discard the flow-through.
Add 500 μL of Buffer RPE to the column. Centrifuge for 15 s at 10,000 rpm (≥ 8,000 × g). Discard the flow-through.
Add 500 μL of 80% ethanol to the column. Centrifuge for 15 s at 10,000 rpm (≥ 8,000 × g). Discard the flow-through.
Centrifuge for 5 min at 10,000 rpm (≥ 8,000 × g) to completely remove excess buffer or ethanol.
Place the column in a new 1.5 mL collection tube and add 14 μL RNase-free water directly to the membrane.
Centrifuge for 1 min at 10,000 rpm (≥ 8,000 × g) to elute the RNA.
Add 500 μL of Buffer AW1 to the AllPrep DNA spin column. Centrifuge for 15 s at 10,000 rpm (≥ 8,000 × g). Discard the flow-through.
Add 500 μL of Buffer AW2 to the column. Centrifuge for 15 s at 14,000 rpm. Discard the flow-through.
Centrifuge for 2 min at 14,000 rpm (20,000 × g) to remove buffer or ethanol completely.
Place the column in a new 1.5 mL collection tube and add 50 μL of EB buffer directly to the membrane.
Centrifuge for 1 min at 10,000 rpm (≥ 8,000 × g) to elute the DNA.
Add the eluted DNA into the same column again. Centrifuge for 1 min at 10,000 rpm (≥ 8,000 × g) to elute the DNA.
3.1.3. DNA Quantification
Measuring genomic DNA concentration was performed using a fluorescence-based method owing to its high sensitivity. In this section, we describe the method used for the AccuBlue High Sensitivity dsDNA Quantitation Kit (Biotium).
Warm the Quantification and 100× Enhancer solutions to room temperature and invert gently.
Dilute the sample 1:10 by adding 2 μL of sample to 18 μL of TE buffer.
Prepare the dsDNA concentration standards in TE buffer at 10, 8, 4, 2, 1, and 0.5 ng/μL.
To prepare the Working solution, add 2 μL of 100× Enhancer solution to 198 μL of Quantification solution in each well.
Add 10 μL of standards and samples into each well, and mix well by pipetting up and down.
Incubate at room temperature for 10 min in the dark.
Measure fluorescence using a microplate reader set to 485 nm excitation/530 nm emission maxima or other filter combination for detecting green fluorescence (e.g., FITC filter set).
3.2. Bisulfite Conversion
For bisulfite conversion, an EpiTect Plus DNA Bisulfite Kit was used. Methylated mouse genomic DNA as a control for bisulfite conversion can be purchased from several sources; however, unmethylated mouse genomic DNA is not available commercially. In place of an unmethylated DNA control, the plasmid DNA used to subclone the target DNA sequence was used as a positive control for bisulfite conversion. In the preparation for Subheading 3.2.2, 30 mL of 100% ethanol was added to Buffer BW and 27 mL of 100% ethanol was added to Buffer BD. Buffer BD and the MinElute DNA spin column are stored at 4 °C. Based on the cell culture size, low genomic DNA yields were acquired from in vitro cultured SCs in the 8-well chamber slide. A single bisulfite reaction typically made use of 300 ng of genomic DNA.
3.2.1. Bisulfite Reaction
Dissolve the Bisulfite Mix in 800 μL of RNase-free water, vortex, and warm to 60 °C until completely dissolved.
- Prepare the bisulfite reactions in 200 μL PCR tubes as below:
DNA sample (20 ng/μL) 15 μL RNase free water 25 μL Bisulfite mix 85 μL DNA protect buffer 15 μL Total volume 140 μL - Perform Bisulfite reaction by thermal cycler using the following protocol:
Denature 95 °C 5 min Incubation 60 °C 25 min Denaturation 95 °C 5 min Incubation 60 °C 85 min Denaturation 95 °C 5 min Incubation 60 °C 175 min Hold 4 °C
3.2.2. Purify Single Strand DNA (ssDNA)
Transfer the bisulfite reactions into 1.5 mL tubes.
Add 310 μL of Buffer BL to each sample. Vortex the solutions and spin down.
Add 250 μL of 100% ethanol to each sample. Vortex the solutions for 15 s and spin down.
Transfer the entire mixture into the MinElute DNA spin column.
Centrifuge the spin columns at 10,000 rpm (≥ 8,000 × g) for 1 min and discard the flow-through.
Add 500 μL of Buffer BW to each column and centrifuge at 10,000 rpm (≥ 8,000 × g) for 1 min. Discard the flow-through.
Add 500 μL of Buffer BD to each column. Close the lid completely and incubate for 15 min at room temperature.
Centrifuge at 10,000 rpm (≥ 8,000 × g) for 1 min. Discard the flow-through.
Add 500 μL of Buffer BW to each column and centrifuge at rpm (≥ 8,000 × g) for 1 min. Discard the flow-through.
Repeat step 9.
Add 250 μL of 100% ethanol to each column and centrifuge at 14.0 rpm (20,000 × g) for 1 min. Discard the flow-through.
Place the columns into new 2 mL collection tubes and centrifuge at 14,000 rpm (20,000 × g) for 1 min to remove any residual liquid.
Place the spin columns with open lids into clean 1.5-mL microcentrifuge tubes and incubate at 60 °C for 5 min in a heating block.
Place the spin columns into new 1.5 mL tubes.
Add 15 μL of Buffer EB directly onto the center of each spin-column membrane.
Incubate the columns at room temperature for 1 min.
Centrifuge for 1 min at 10,000 rpm (≥ 8,000 × g) to elute the ssDNA.
3.3. PCR and Subcloning
For the bisulfite sequence, primer design is most important to obtain reliable data. Since bisulfite-converted DNA was fragmented and the cytosine converted to thymine (except within CpG sites), the following tips are recommended: (1) amplicon should be short (around 100 bp); (2) CpG must be avoided; and (3) non-CpG cytosine should be contained. MethPrimer (http://www.urogene.org/methprimer/) is widely known as a useful and convenient web tool to design primers for DNA methylation analysis (see Notes 2–4) [7]. High-fidelity enzymes are also appropriate to amplify for methylation analysis, as PCR errors can distort the results. However, high-fidelity enzymes based on Pfu polymerase failed to amplify the bisulfite-converted DNA in many cases. To overcome this, the analyzed sequence was amplified by PCR and subcloned into a TA-vector (see Notes 5 and 6).
3.3.1. PCR
- Prepare the PCR mixture (total volume 20 μL) as below:
GO Taq master mix 10 μL 5 μM forward primer 1 μL 5 μM reverse primer 1 μL Water 6 μL Bisulfite-converted DNA 2 μL - Perform the PCR on a thermal cycler using the following protocol:
Denature 95 °C 2 min *Denature 95 °C 30 s *Annealing 55–60 °C 30 s *Extension 72 °C 30 s (repeat * for 30–35 cycles) Elongation 72 °C 5 min Hold 4 °C Check amplicon length by 2.5% agarose gel electrophoresis and estimate amplicon yield (see Note 6).
3.3.2. TA-Cloning
- For the TA-cloning, using the pGEM-T Easy vector, the reaction conditions are as follows;
Rapid ligation buffer 5 μL pGEM-T easy vector 1 μL (50 ng) PCR product 1–1.5 μL T7 DNA ligase 1 μL Water 1.5–2 μL Incubate at 4 °C overnight.
Add 2.5 μL of ligation product into 25 μL of DH5α competent cells and incubate on ice for 30 min.
Incubate at 42 °C for 45 s followed by incubation on ice for 2 min.
Add 250 μL of SOC medium to competent cells; incubate at 37 °C with shaking for 1 h.
Spread 50 μL of 2% X-gal on a dried LB agar plate containing ampicillin and dry plates again in the incubator for 30 min.
Plate 50 μL of cultured transformed cells on the LB-plates and incubate overnight.
3.4. Colony PCR and Direct Sequencing
The pGEM-T Easy vector cloning site is located between the T7 and SP6 promoter sites, with the M13 forward and reverse sequencing primer sites located outside of these. Colony PCR was therefore performed using the M13 forward and reverse primers, and the sequence reaction cycle was undertaken using T7 primers (see Note 7).
- Prepare the colony PCR mixture (total volume 10 μL) as shown below:
GO Taq master mix 5 μL 5 μM M13 forward primer 0.5 μL 5 μM M13 reverse primer 0.5 μL Water 4 μL - Perform the PCR on a thermal cycler using the following conditions:
Denature 95 °C 2 min *Denature 95 °C 30 s *Annealing 58 °C 30 s *Extension 72 °C 30 s (repeat * for 25 cycles) Elongation 72 °C 5 min Hold 4 °C Load 2 μL of the PCR product to confirm amplicon length by 2.5% agarose gel electrophoresis.
Add 1 μL of ExoSAP-IT into the remaining PCR product.
Incubate at 37 °C for 45 min, followed by an inactivation step at 85 °C for 15 min. Thereafter store at 4 °C.
- Prepare the cycle sequencing reaction mixture as follows:
ABI PRISM BigDye terminator 2 μL 5 × BigDye terminator buffer 2 μL T7 primer (1.6 pmol) 2 μL PCR product 1 μL Water 3 μL - Perform the cycle sequencing reaction using a thermal cycler set to the following protocol;
Denature 96 °C 3 min *Denature 96 °C 30 s *Annealing 50 °C 10 s *Extension 60 °C 4 min (repeat * for 25 cycles) Hold 4 °C Add 2.5 μL of 125 mM EDTA and 30 μL of 100% ethanol to each well and seal the plate.
Vortex and incubate at room temperature for 15 min.
Centrifuge the plate at 4,000 rpm(1,650 × g),4 °C for 30 min.
Remove the seal, invert the plate, and centrifuge at 300 rpm for 1 min.
Add 30 μL of 70% ethanol to each well.
Reseal the plate and centrifuge at 4,000 rpm (1,650 × g), 4 °C for 5 min.
Remove the seal, invert the plate, and centrifuge at 300 rpm for 1 min.
Add 10 μL of Hi-Di Formamide to each well.
Reseal the plate, spin down, and incubate at 95 °C for 2 min.
Place the reaction plate on ice in the dark until analysis.
Analyze with a Genetic analyzer (see Note 8).
Footnotes
In our previous study, muscle SCs were isolated according to a protocol based on extracting a single myofiber and culturing on collagen type I-coated dishes established by Dr. Naohiro Hashimoto [8 9]. We have also isolated the muscle SCs by a protocol based on cell sorting [10]. Muscle SCs were isolated as Ter119−/CD45−/CD31−/CD34+/α7-integrin+/Sca-1 cells. Isolated SCs were seeded on laminin-coated plates. For differentiation analysis, 26,000 cells/well were seeded on each well of a laminin-coated 8 well Nunc Lab-Teck Chamber slide, and cultured with Growth Medium (DMEM supplemented with 3% FCS + 7% FBS). Cells were subsequently switched to Differentiation medium (DMEM supplemented with 2% horse serum) once the cells were grown up to 80% confluence [11].
To generate the code of the bisulfite-converted sequence, Microsoft Word proved convenient. First, convert “CG” to “XX,” and then convert all “C” to “T.” This aids the design and visualization of the PCR primers as well as the position of the CpG sites.
A high-fidelity PCR enzyme is recommended for amplification in each step.
The PCR primer should contain as much of the thymine converted from non-CpG site cytosine as possible to avoid amplifying un-converted DNA.
If bisulfite PCR does not amplify sufficient PCR product, purification, and concentration (e.g., phenol extraction and ethanol precipitation) can be used to improve TA-cloning efficacy.
Purification of the PCR product may improve subcloning efficacy, but is not necessary.
Sequencing analysis with over 20 colonies is recommended for statistical analysis.
QUMA (http://quma.cdb.riken.jp) or BiQ Analyzer (http://biq-analyzer.bioinf.mpi-inf.mpg.de) was useful and convenient to quantify and visualize the bisulfite sequencing data.
References
- 1.Zuo T, Tycko B, Liu TM et al. (2009) Methods in DNA methylation profiling. Epigenomics 1:331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova M, Fan JB (2010) Genome-wide DNA methylation profiling. Wiley Interdiscip Rev Syst Biol Med 2:210–223 [DOI] [PubMed] [Google Scholar]
- 3.Frommer M, McDonald LE, Millar DS et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89:1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grunau C, Clark SJ, Rosenthal A (2001) Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 29:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leontiou CA, Hadjidaniel MD, Mina P et al. (2015) Bisulfite conversion of DNA: performance comparison of different kits and methylation quantitation of epigenetic biomarkers that have the potential to be used in non-invasive prenatal testing. PLoS One 10:e0135058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang AS, Estécio MR, Doshi K et al. (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431 [DOI] [PubMed] [Google Scholar]
- 8.Wada MR, Inagawa-Ogashiwa M, Shimizu S et al. (2002) Generation of different fates from multipotent muscle stem cells. Development 129:2987–2995 [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto N, Murase T, Kondo S et al. (2004) Muscle reconstitution by muscle satellite cell descendants with stem cell-like properties. Development 131:5481–5490 [DOI] [PubMed] [Google Scholar]
- 10.Mozzetta C, Consalvi S, Saccone V et al. (2013) Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol Med 5:626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albini S, Coutinho Toto P, Dall’Agnese A et al. (2015) Brahma is required for cell cycle arrest and late muscle gene expression during skeletal myogenesis. EMBO Rep 16:1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]