SUMMARY
The circadian clock operates as intrinsic time-keeping machinery to preserve homeostasis in response to the changing environment. While food is a known zeitgeber for clocks in peripheral tissues, it remains unclear how lack of food influences clock function. We demonstrate that the transcriptional response to fasting operates through molecular mechanisms that are distinct from time-restricted feeding regimens. First, fasting affects core clock genes and proteins, resulting in blunted rhythmicity of BMAL1 and REV-ERBα both in liver and skeletal muscle. Second, fasting induces a switch in temporal gene expression through dedicated fasting-sensitive transcription factors such as GR, CREB, FOXO, TFEB, and PPARs. Third, the rhythmic genomic response to fasting is sustainable by prolonged fasting and reversible by refeeding. Thus, fasting imposes specialized dynamics of transcriptional coordination between the clock and nutrient-sensitive pathways, thereby achieving a switch to fasting-specific temporal gene regulation.
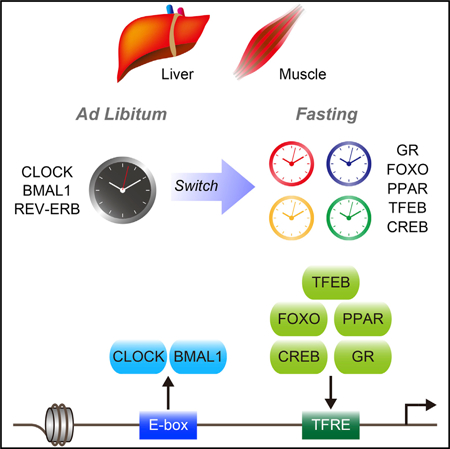
Graphical Abstract
In Brief
Kinouchi et al. reveal that fasting affects peripheral circadian clocks in the liver and skeletal muscle. Fasting operates by influencing the circadian clock and fasting-sensitive transcription factors, thereby cooperatively achieving fasting-specific temporal gene regulation.
INTRODUCTION
The mammalian circadian clock consists of a molecular machinery that operates in all cells and is fine-tuned by environmental cues to generate 24-hr rhythms in metabolism, physiology, and behavior (Bass and Takahashi, 2010; Schibler and Sassone-Corsi, 2002). The master pacemaker within the suprachiasmatic nucleus (SCN) is reset by light, while peripheral oscillators can be uncoupled from the SCN through food intake, highlighting the significance of temporal nutrient availability (Asher and Sassone-Corsi, 2015; Damiola et al., 2000; Stokkan et al., 2001).
Dietary regimens and time-restricted feeding have a profound impact on the clock in metabolic tissues (Eckel-Mahan et al., 2013; Hatori et al., 2012; Kohsaka et al., 2007; Mukherji et al., 2015; Tognini et al., 2017; Vollmers et al., 2009). Also, the clock regulates metabolic genes in a tissue-specific fashion, emphasizing the link between the circadian clock and metabolism (Dyar et al., 2013; Nakahata et al., 2009; Ramsey et al., 2009; Zhang et al., 2015). Although the circadian clock plays an important role in rhythmic gene expression, time-restricted feeding restores cyclic gene expression in arrhythmic Cry1—/—, Cry2—/— mutant mice, suggesting that nutrient-responsive transcriptional pathways contribute to the rhythmicity of circadian genes in a clock-independent manner (Vollmers et al., 2009). Finally, while evidence on how food intake is integrated into circadian transcriptional regulation is accumulating (Asher and Sassone-Corsi, 2015), how lack of food operates on the clock remains virtually unexplored.
Fasting is an adaptive state of metabolism when exogenous nutrient intake is limited (Longo and Mattson, 2014). In mammals, a drastic shift in metabolism takes place under low nutrient availability. Skeletal muscles undergo protein breakdown and provide amino acids for the liver to implement gluconeogenesis, producing glucose to maintain appropriate blood glucose levels. In parallel, the liver performs ketogenesis to supply ketone bodies to other vital organs, including the brain, by harnessing free fatty acids from adipose tissue (Longo and Mattson, 2014). Such metabolic shifts across different tissues are achieved by fasting-induced transcription factors (TFs) such as glucocorticoid receptor (GR), cyclic AMP responsive element binding protein (CREB), forkhead box TF class O (FOXO), TFEB, and peroxisome proliferator-activated receptors (PPARs) (Goldstein and Hager, 2015). Recent studies suggest that a fasting-mimicking diet and temporal feeding restriction have numerous health benefits, despite comparable calorie intake (Brandhorst et al., 2015; Hatori et al., 2012). Moreover, food restriction confers robustness to circadian rhythms, possibly mediating the protective effects of fasting against diverse diseases and aging (Hatori et al., 2012).
Although several studies have linked fasting to circadian rhythms (Kawamoto et al., 2006; Shavlakadze et al., 2013; Sun et al., 2015; Xie et al., 2016), it is still unclear how fasting impinges on the clock and fasting-induced TFs. We show that fasting affects daily rhythmic physiology by inducing various de novo oscillatory genes, which are distinct from those responsive to timed-feeding regimens. Fasting attenuates the rhythmicity of brain and muscle Arnt-like protein-1 (BMAL1) post-translational modification and REV-ERBα levels both in liver and skeletal muscle, leading to repression and derepression of their target genes, respectively. Also, specific classes of genes are temporally regulated by the clock and distinct fasting-sensitive TFs. Furthermore, a number of genes are induced by fasting in a BMAL1-dependent manner, so the clock modulates part of the fasting response. Lastly, the response to fasting is sustained during a 2-day period and reversed by refeeding. Thus, fasting uncovers a previously unappreciated coordination between the circadian clock and nutrient-sensing pathways leading to different classes of temporal gene expression.
RESULTS
Temporal Response to Fasting
We used 8-week-old male C57BL/6 mice fed normal chow ad libitum, subjected them to 24-hr fasting and then allowed refeeding for 24 hr. There was a reduction in oxygen consumption (VO2), respiratory exchange ratio (RER), and energy expenditure by fasting, which was completely abolished by refeeding (Figures S1A–S1F). Notably, RER during fasting was lower than the resting phase under ad libitum feeding, likely because mice are still feeding during the resting phase (Figures S1G and S1H). We then collected tissues every 4 hr from 24-hr fasted mice (FAST) or control mice fed ad libitum (FED) (Figure 1A). Body weight and epididymal white adipose tissue (eWAT) were reduced remarkably after fasting, while serum levels of corticosterone and β-hydroxybutyrate (β-OHB) were highly induced by fasting (Figures S1I–S1K).
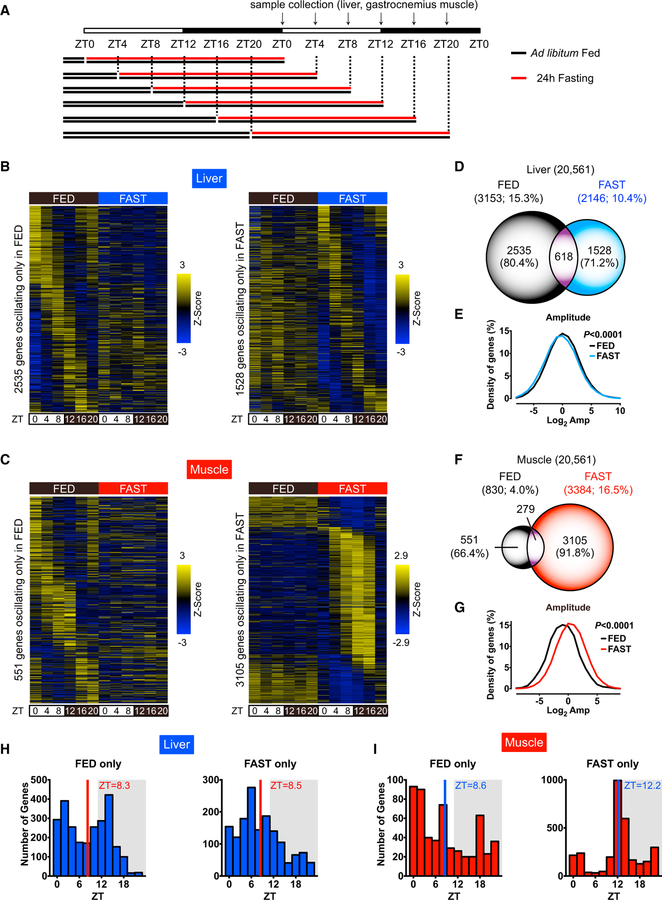
Figure 1. Temporal Response to Fasting in Liver and Muscle.
(A) Depiction of the experimental design for the 24-hr fasting and tissue collection at ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20.
(B and C) RNA-seq generated heatmaps of cycling transcripts only in FED (left) or in FAST (right) in (B) liver and (C) skeletal muscle (JTK_CYCLE p < 0.01).
(D) Venn diagram indicates the number of oscillating transcripts in the liver.
(E) Distribution of amplitude in oscillating transcripts in the liver.
(F) Venn diagram indicates the number of oscillating transcripts in skeletal muscle.
(G) Distribution of amplitude in oscillating transcripts in skeletal muscle.
(H and I) Peak phase distribution of hepatic and muscle transcripts oscillating only in FED (left) and FAST (right) in (H) liver and (I) skeletal muscle.
RNA sequencing (RNA-seq) analysis revealed that approximately 15% of hepatic transcripts and 4% of muscle transcripts were cyclic in FED mice (Hughes et al., 2010), which is consistent with a previous report (Figures 1B–1G; Tables S1, S2, S3, and S4) (Zhang et al., 2014). Of the rhythmic transcripts in FED mice, approximately 80% and 66% of hepatic and muscle genes, respectively, ceased oscillation after fasting (Figures 1D and 1F). A number of genes, particularly in skeletal muscle, gained oscillation after fasting, presumably reflecting a time-of-the-day-specific response to fasting (Figures 1B–1G). The overall amplitude of hepatic oscillating genes in FAST mice was dampened, whereas that of muscle genes was enhanced, as compared to FED mice (Figures 1E and1G). Phase analysis revealed that the peak phase in skeletal muscle from FAST mice was centered around zeitgeber time 12 (ZT12), while in liver it was more evenly distributed (Figures 1H and1I). Although some genes remained rhythmic both in FED and FAST mice (Figures S2A and S2B), peak phases were redistributed after fasting (Figures S2C and S2D). Thus, fasting elicits tissue-specific responses along the daily cycle.
Only a small portion of cyclic genes is common in liver and skeletal muscle in any condition, manifesting tissue specificity in rhythmic expression (Figures 2A, 2B, and S2E) (Masri and Sassone-Corsi, 2010). Gene Ontology analyses revealed unique biological processes enriched in a tissue-specific manner (Figures 2C and 2D). Notably, the electron transport chain was highly enriched in FAST liver, in keeping with the role of mitochondrial oxidative metabolism under fasting (Lin et al., 2005). Likewise, protein catabolic processes were enriched in FAST muscle, a well-documented metabolic adaptation upon fasting in the skeletal muscle (Milan et al., 2015). Circadian rhythm was enriched in a group of genes oscillating in both tissues and both conditions, underscoring the resilient nature of the core clock (Figure S2F).
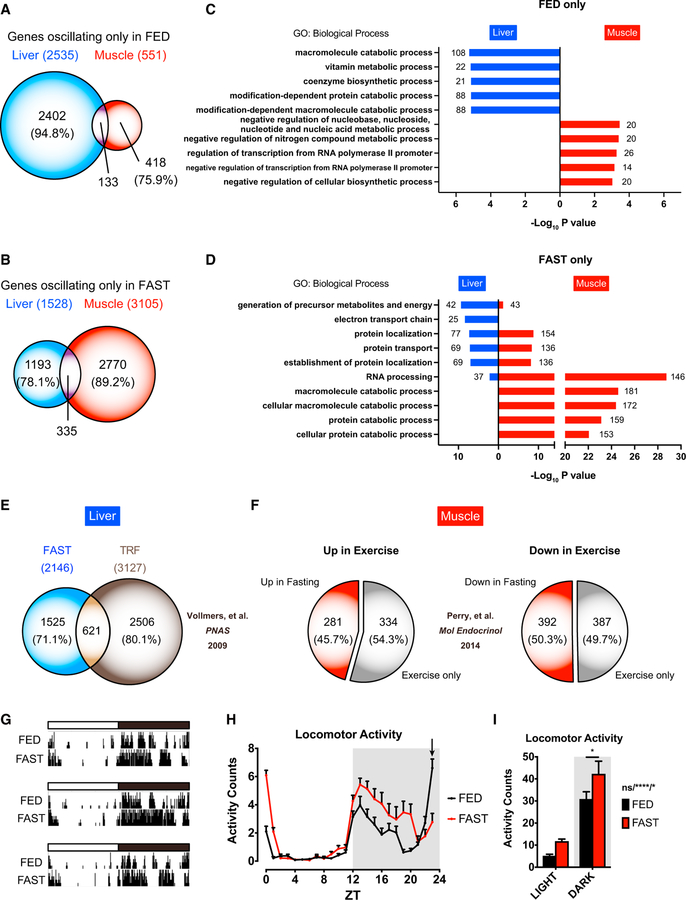
Figure 2. Temporal Response to Fasting in Distinct Group of Tissue-Specific Genes.
(A)Venn diagrams of hepatic and muscle cycling transcripts in FED.
(B) Venn diagrams of hepatic and muscle cycling transcripts in FAST.
(C) Gene Ontology analysis of top five biological processes enriched in rhythmic hepatic (blue) and muscle (red) genes only in FED, with the number of genes indicated on the graph.
(D) Gene Ontology analysis of top five biological processes enriched in rhythmic hepatic (blue) and muscle (red) genes only in FAST, with the number of genes indicated on the graph.
(E) Venn diagram of the number of hepatic cycling genes in fasting mice and in time-restricted fed mice (JTK_CYCLE p < 0.01) (Vollmers et al., 2009).
(F) Pie charts comparing genes induced in exercise (left) and repressed in exercise (right) (Perry et al., 2014) versus our fasting transcriptome at ZT12.
(G) Representative actograms from three individual mice under FED and FAST conditions.
(H) Locomotor activity under FED and FAST conditions. Mice under ad libitum normal chow for 24 hr were subsequently fasted for 24 hr. Arrow indicates time of food removal.
(I) Locomotor activity during light and dark phases under FED and FAST conditions. Data are shown as means + SEMs (n = 16 biological replicates). *p < 0.05 and ****p < 0.0001 by two-way ANOVA (interaction/phase/group) with Bonferroni post hoc tests. ns, non-significant.
We next compared hepatic genes cycling under time-restricted feeding (Vollmers et al., 2009) with genes oscillating in FAST mice. Only a few genes are common to both groups, indicating that the response to fasting is distinct from time-restricted feeding (Figure 2E). Furthermore, we investigated whether skeletal muscle from FAST mice shares transcriptional signatures with the skeletal muscle from treadmill-exercised mice (Perry et al., 2014). Approximately half of the exercise-induced genes and exercise-repressed genes in skeletal muscle were also induced and repressed, respectively, by fasting at ZT12 (Figure 2F). Supporting this notion, locomotor activity during fasting was enhanced particularly during the active phase (Figures 2G–2I).
Post-transcriptional regulation plays an important role in circadian gene expression in vivo. As Gene Ontology analysis showed that RNA processing is enriched in cycling genes upon fasting (Figure 2D), we implemented exon intron split analysis (EISA) on our RNA-seq dataset to compare intronic reads as a surrogate for precursor mRNA and exonic reads for mature mRNA (Gaidatzis et al., 2015). A large number of intronic and exonic transcripts displayed oscillatory expression in liver and muscle (Figures S3A–S3D). Approximately 77% of the cycling exonic transcripts in FED liver were oscillatory only in exonic reads, in agreement with a previous analysis (Figure S3E) (Koike et al., 2012). This proportion appeared to be conserved in FED skeletal muscle as well as in FAST liver and skeletal muscle, stressing the importance of post-transcriptional control (Figures S3F–S3H). We also focused on groups of genes oscillating both in intronic and exonic regions, and compared their peak phases to explore the delay in the phase of exonic transcripts in comparison to that of intronic transcripts. The lag of peak phase disappeared after fasting in the liver, while becoming present after fasting in skeletal muscle, which is suggestive of tissue-specific post-transcriptional control upon fasting (Figures S3I–S3L).
Fasting Targets Core Clock Components
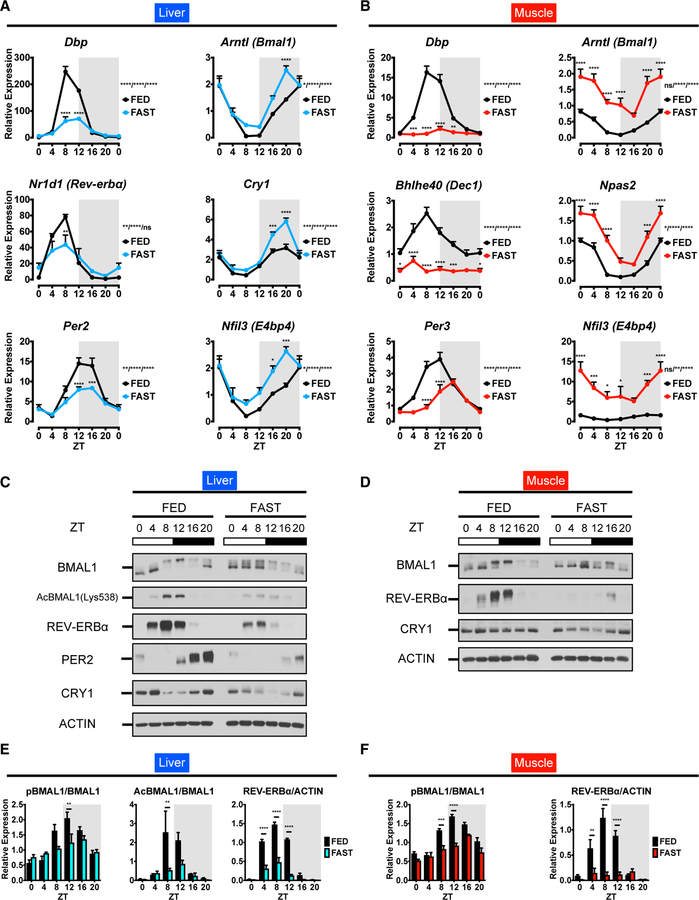
Differently from nutritional challenges (Eckel-Mahan et al., 2013; Tognini et al., 2017), we reasoned that lack of food may have a direct influence on core clock components. The expression of BMAL1-target genes was significantly attenuated (Dbp, Nr1d1, Bhlhe40, Per2, Per3), while REV-ERBα targets were derepressed both in liver and skeletal muscle (Arntl, Cry1, Npas2, Nfil3) (Figures 3A and 3B). The phosphorylation of BMAL1 displays robust rhythmicity in FED mice and is associated with the active transcription of target genes (Tamaru et al., 2009). Rhythmic BMAL1 protein phosphorylation is significantly dampened in FAST mice, both in liver and muscle, in keeping with the decreased expression of BMAL1-target genes (Figures 3C–3F). Furthermore, the rhythmic acetylation of hepatic BMAL1 was mitigated in FAST mice, presumably as a result of the increased deacetylation of BMAL1 by SIRT1 under fasting (Figure S4A) (Nakahata et al., 2008). In addition, REV-ERBα and cryptochrome circadian regulator 1 (CRY1) levels in liver and muscle, as well as hepatic PER2, were dampened in rhythmicity in FAST mice (Figures 3C–3F).
Figure 3. The Circadian Clock Fails to Sustain Robust Rhythmicity under Fasting Conditions.
(A and B) Expression profiles of core clock genes in (A) liver and (B) muscle. Transcripts were normalized to 18S rRNA and presented as means + SEMs (n = 5 replicates per time point per group). ZT0 is double plotted for visualization. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-way ANOVA (interaction/time/group) with Bonferroni post hoc tests.
(C and D) Core clock proteins in total lysates from liver (C) and muscle (D) of FED and FAST mice.
(E and F) Quantitation of the blot band density of the hepatic (E) and muscle (F) proteins presented as means + SEMs (n = 3 replicates per time point per group).
**p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-way ANOVA with Bonferroni post hoc tests.
Temporal Patterns of Fasting-Induced Differential Gene Expression
Fasting results in the activation of several nutrient-sensing factors, such as GR, CREB, FOXO, TFEB, and PPARs (Settembre and Ballabio, 2014). We examined the expression levels of these regulators along the diurnal cycle in FED or FAST mice, both in liver and muscle. By and large, fasting induces these TFs in both tissues with distinct diurnal patterns, although some TFs appear to be unaltered (Figures S4B–S4I). To decipher whether fasting-induced changes in the core clock impinge on temporal genome-wide expression in concert with fasting-sensitive TFs, we examined available datasets (see Experimental Model and Subject Details; Table S5). We analyzed gene targets for hepatic BMAL1 (Koike et al., 2012; Yang et al., 2016), REV-ERBα (Fang et al., 2014), and muscle BMAL1 (Dyar et al., 2013). In addition, we analyzed the targets for hepatic GR (Frijters et al., 2010), CREB (Ravnskjaer et al., 2013; Zhang et al., 2005), FOXO (Haeusler et al., 2014), TFEB (Settembre et al., 2013), PPARα (Montagner et al., 2016), and for muscle GR (Kuo et al., 2012), CREB (Pearen et al., 2009), FOXO (Milan et al., 2015), TFEB (Mansueto et al., 2017), and PPARδ (Gan et al., 2011). We also paralleled muscle histone deacetylase 3 (HDAC3)-target genes from muscle-specific Hdac3—/— mice, since muscle HDAC3 is primarily regulated by REV-ERBα, while HDAC3 exerts numerous REV-ERBα—independent actions (Hong et al., 2017). Most fasting-sensitive TF liver targets correspond to genes cycling in the FED condition, presumably because of the naturally occurring feeding-fasting cycle in ad libitum feeding (Figure 4A). Furthermore, hepatic GR-, CREB-, and FOXO-target genes oscillating in the FED condition were prone to peak at ZT8–ZT12, which is consistent with the fasting phase in ad libitum feeding (Figure 4B). Conversely, most fasting-sensitive TF targets in the muscle parallel a higher number of oscillatory genes in the FAST condition and peak sharply at ZT12, since the response to fasting in skeletal muscle appears highly rhythmic (Figures 1C, 4C, and 4D). Collectively, each fasting-responsive TF may contribute to rhythmic transcription under ad libitum feeding as well as the rhythmic response to fasting in a tissue-specific and phase-specific manner.
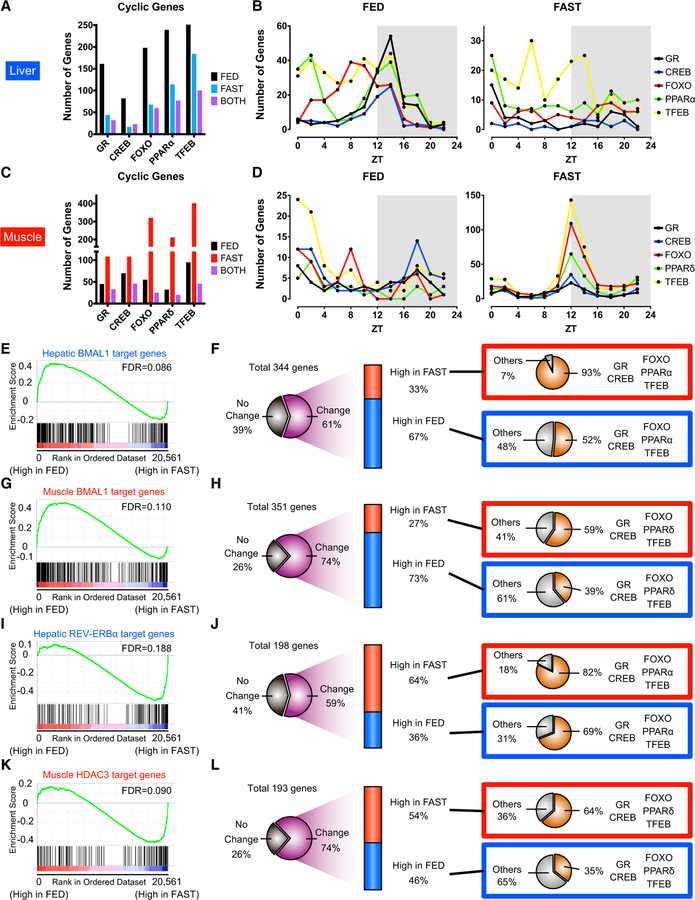
Figure 4. Temporal Pattern of Differential Gene Regulation by Clock and Fasting-Induced Transcription Factors.
(A) Number of oscillating genes among GR, CREB, FOXO, TFEB, and PPARα targets in liver under FED, FAST, or both FED and FAST conditions.
(B) Peak phase distribution of liver fasting-sensitive transcription factor (TF) target genes oscillating only in FED (left) and FAST (right).
(C) Number of oscillating genes among GR, CREB, FOXO, TFEB, and PPARδ targets in skeletal muscle under FED, FAST, or both FED and FAST conditions.
(D) Peak phase distribution of fasting-sensitive TF target genes in muscle oscillating only in FED (left) and FAST (right).
(E) Gene set enrichment analysis (GSEA) for hepatic BMAL1 target genes at ZT8 enriched in FED.
(F) Differentially expressed BMAL1 liver target genes at ZT8 illustrating fasting-sensitive TF coordinated regulation.
(G) GSEA for muscle BMAL1 target genes at ZT8 enriched in FED.
(H) Differentially expressed muscle BMAL1 target genes at ZT8 illustrating fasting-sensitive TF coordinated regulation.
(I) GSEA for hepatic REV-ERBα target genes at ZT12 enriched in FAST.
(J) Differentially expressed REV-ERBα liver target genes at ZT12 illustrating fasting-sensitive TF coordinated regulation.
(K) GSEA for muscle HDAC3 target genes at ZT12 enriched in FAST.
(L) Differentially expressed HDAC3 muscle target genes at ZT12 illustrating fasting-sensitive TF coordinated regulation. Gene sets with a false discovery rate (FDR) <0.25 are considered significantly enriched.
BMAL1 phosphorylation and acetylation are high in FED mice, indicating that BMAL1-target genes should be highly expressed. Similarly, REV-ERBα levels decrease under fasting, predicting that the expression of REV-ERBα-target genes may be higher (Figures 3C–3F). We focused at ZT8 and ZT12, times when BMAL1 and REV-ERBα activity is high, and carried out gene set enrichment analysis (GSEA) to test whether the expression of BMAL1 and REV-ERBα gene targets is actually higher in FED and FAST mice, respectively, at their peaks. Hepatic BMAL1 targets were significantly induced in FED mice at ZT8 (Figures 4E and S5A). Also, two-thirds of differentially expressed BMAL1-target liver genes displayed higher expression in FED mice at ZT8 and ZT12 (Figures 4F and S5B). Similarly, muscle BMAL1-target genes were significantly enriched in FED mice both at ZT8 and ZT12, and approximately 70% of differentially expressed muscle BMAL1 targets also exhibited higher gene expression in FED mice at both time points (Figures 4G, 4H, S5C, and S5D). Notably, approximately 30% of differentially expressed BMAL1-target genes in both liver and muscle showed higher expression under fasting. These groups of genes had a significantly higher proportion of GR, CREB, FOXO, TFEB, hepatic PPARα (or muscle PPARδ) targets than those demonstrating higher expression in FED mice, suggesting that a subgroup of BMAL1 targets is also jointly regulated by fasting-sensitive TFs (Figures 4F, 4H, S5B, and S5D). Of note, targets of fasting-sensitive TFs are present among BMAL1-target genes displaying higher expression in FED mice, which suggests that fasting-sensitive TFs are also involved in activation during the fed state.
Conversely, hepatic REV-ERBα and muscle HDAC3-target genes at ZT12 were significantly enriched in FAST mice, and approximately 60% of differentially expressed REV-ERBα/ HDAC3 targets displayed higher gene expression in FAST mice (Figures 4I–4L). These REV-ERBα-HDAC3-target genes whose expression is induced by fasting were proportionally more likely to be GR, CREB, FOXO, TFEB, hepatic PPARα (or muscle PPARδ) targets. Among these fasting-sensitive TFs, PPARα was particularly enriched in hepatic REV-ERBα—target genes, in keeping with the central role of REV-ERBα in lipid metabolism (Figure S5E) (Bugge et al., 2012; Cho et al., 2012). In fact, metabolic target genes of hepatic and muscle REV-ERBα-HDAC3 were derepressed under fasting (Figures S5F and S5G) (Hong et al., 2017; Zhang et al., 2015). REV-ERBα interacts with hepatocyte nuclear factor 6 (HNF6) in liver (Zhang et al., 2016), and several HNF6/REV-ERBα targets, including Apoa4 and Cd36, were upregulated under fasting in the liver (Figure S5H). Notably, hepatic Onecut1 (Hnf6) expression significantly declined upon fasting, which could also expedite derepression of HNF6/REV-ERBα targets (Figure S5I). Thus, the attenuation of BMAL1 and REV-ERBα by fasting mediates genome-wide changes in target gene expression.
Coordination between Clock and Fasting-Sensitive TFs Reveals Unique Classes of Responsive Genes
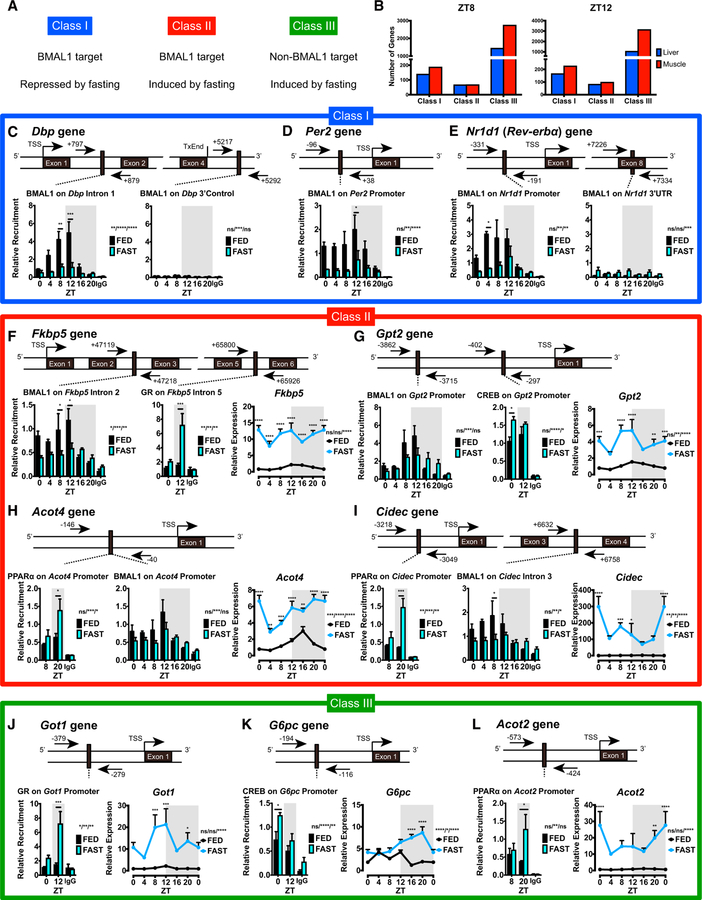
Fasting induces an enhancement in the expression of some BMAL1-target genes, while it leads to the repression of others both in liver and muscle. To discern how BMAL1 and fasting-sensitive TFs coordinate transcription upon fasting, we classified genes into three groups (Figures 5A and S6A–S6F):
Class I: BMAL1-target genes whose expression is repressed by fasting
Class II: BMAL1-target genes activated by fasting
Class III: non-BMAL1-target genes whose expression is activated by fasting
Figure 5. Altered Recruitment of BMAL1 and Fasting-Induced TFs after Fasting.
(A) Criteria for gene classification of BMAL1-target genes and differential gene expression after fasting.
(B) Number of genes identified for each class in liver and skeletal muscle at ZT8 (left) and ZT12 (right).
(C–E) Chromatin immunoprecipitation in liver for BMAL1 or immunoglobulin G (IgG) followed by RT-qPCR specific for class I genes (C) Dbp, (D) Per2, and (E) Nr1d1 (Rev-ERBα).
(F–I) Chromatin immunoprecipitation in liver for BMAL1, GR, CREB, PPARα, or IgG followed by RT-qPCR specific for class II genes (F) Fkbp5, (G) Gpt2, (H) Acot4, and (I) Cidec.
(J–L) Chromatin immunoprecipitation in liver for GR, CREB, PPARα, or IgG followed by RT-qPCR specific for class III genes (J) Got1, (K) G6pc, and (L) Acot2. Data are presented as means + SEMs (n = 3–4 replicates per time point per group). Diagram above shows regions of amplification by primers designed for analysis (arrows) and TSS (transcription start site). Gene expression profiles are shown next to ChIP-qPCR data for visualization. Transcripts were normalized to 18S rRNA and presented as means + SEMs (n = 5 replicates per time point per group). ZT0 is double plotted for visualization. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-way ANOVA (interaction/time/group) with Bonferroni post hoc tests.
The number of genes in each class was similar between ZT8 and ZT12 both in liver and muscle, although the number of class III genes is much higher than the other groups (Figure 5B). To examine whether this classification corresponds to transcriptional mechanisms specific for each class, we performed chromatin immunoprecipitation (ChIP) followed by qPCR. BMAL1 was recruited less to class I genes such as Dbp, Per2, and Nr1d1 (Rev-ERBα) under fasting conditions, consistent with hepatic Dbp, Per2, and Nr1d1 gene expression (Figures 3A and 5C–5E). BMAL1 recruitment to class II genes (e.g., Fkbp5, Gpt2, Acot4, Cidec) was reduced by fasting, despite the increased expression of those genes upon fasting (Figures 5F–5I). Similarly, BMAL1 recruitment to Per1 was decreased upon fasting, despite Per1 induction upon fasting (Figure S6G). Induction of class II genes or Per1 is achieved through GR, CREB, or PPARα, the recruitment of which was increased by fasting (Figures 5F–5I and S6G). Of note, fasting-induced genes are often regulated simultaneously by multiple fasting TFs, which likely accounts for Acot4 or Cidec induction by fasting, despite the similar recruitment of PPARα at ZT8 (Goldstein and Hager, 2015). Recruitment of GR, CREB, and PPARα to class III genes (e.g., Got1, G6pc, Acot2) was also increased by fasting (Figure 5J–5L). Although BMAL1 is known to occupy the G6pc genomic region, deletion of Bmal1 in the liver does not alter G6pc gene expression, indicating that BMAL1 binding to the G6pc site is unlikely to be functional and leading to the categorization of G6pc as a non-BMAL1-target gene (Koike et al., 2012; Yang et al., 2016).
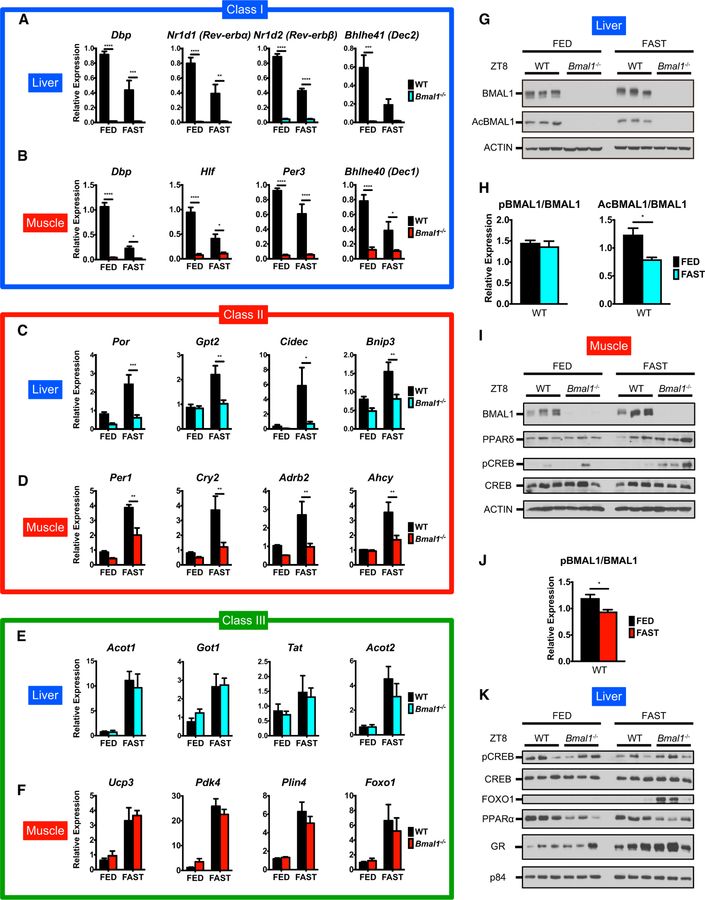
Next, we investigated the role of the clock in fasting-induced gene regulation in liver and muscle. We subjected Bmal1—/— mice and their wild-type (WT) littermates to 24-hr fasting (FAST) or ad libitum feeding (FED) and harvested liver and gastrocnemius muscle at ZT8, a time when BMAL1 recruitment is at its peak (Figure S7A). Body weight loss under fasting was comparable in both genotypes (Figures S7B and S7C). We explored the expression of genes belonging to the three identified classes in fasted Bmal1—/— mice. Expression of class I genes such as Dbp, Nr1d1 (Rev-ERBα), and Per3 was abrogated by Bmal1 ablation as expected (Figures 6A and 6B). Class II genes were activated by fasting in a BMAL1-dependent manner, despite the decreased recruitment of BMAL1 to their promoters under fasting (Figures 6C, 6D, and S7D). However, class III genes are activated by fasting in a BMAL1-independent fashion (Figures 6E, 6F, and S7E). To validate this notion, liver-specific Tfeb—/— (LiKO) mice were subjected to 24-hr fasting or fed ad libitum, and livers were collected at ZT8 (Figure S7F). While LiKO mice displayed similar expressions of clock genes, including class I in the liver, genes within classes II and III required TFEB for gene activation (Figures S7G–S7J). Thus, clock and fasting-sensitive TFs coordinate temporal gene transcription. We confirmed that BMAL1 protein levels were abolished both in Bmal1—/— liver and skeletal muscle (Figure 6G–6J). Under fasting, FOXO1 and GR expression in liver nuclear fraction and PPARδ and phosphorylated CREB (pCREB)/CREB expression in skeletal muscle appeared higher in Bmal1—/— mice (Figures 6I and 6K). Conversely, hepatic nuclear PPARα was lower in Bmal1—/— mice, presumably because it is a direct target of BMAL1 (Oishi et al., 2005) (Figure 6K). Thus, the clock modulates fasting-sensing pathways in a tissue-specific manner.
Figure 6. BMAL1 Modulates Fasting-Sensing Pathways.
(A and B) Representative gene expression profiles of (A) hepatic and (B) muscle BMAL1-target genes repressed by fasting (class I) in WT and Bmal1—/— mice at ZT8.
(C and D) Gene expression profiles of (C) hepatic and (D) muscle BMAL1-target genes induced by fasting (class II) in WT and Bmal1—/— mice at ZT8.
(E and F) Gene expression profiles of (E) hepatic and (F) muscle fasting-induced genes of non-BMAL1 targets (class III) in WT and Bmal1—/— mice at ZT8.
(G) BMAL1 in hepatic total lysates from WT and Bmal1—/— mice at ZT8 under ad libitum fed (FED) and 24-hr fasting (FAST) conditions.
(H) Quantitation of hepatic BMAL1 presented as means + SEMs (n = 3 replicates per group).
(I) Fasting-responsive proteins in muscle total lysates from WT and Bmal1—/— mice at ZT8 under FED and FAST conditions.
(J) Quantitation of muscle BMAL1 presented as means + SEMs (n = 3 biological replicates per group).
(K) Fasting-responsive proteins in hepatic nuclear extracts from WT and Bmal1—/— mice at ZT8 under FED and FAST conditions. Transcript levels were normalized to 18S rRNA and presented as means + SEMs (n = 4–5 replicates per genotype per group). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-way ANOVA with Bonferroni post hoc tests.
Response to Fasting Is Reversible by Refeeding
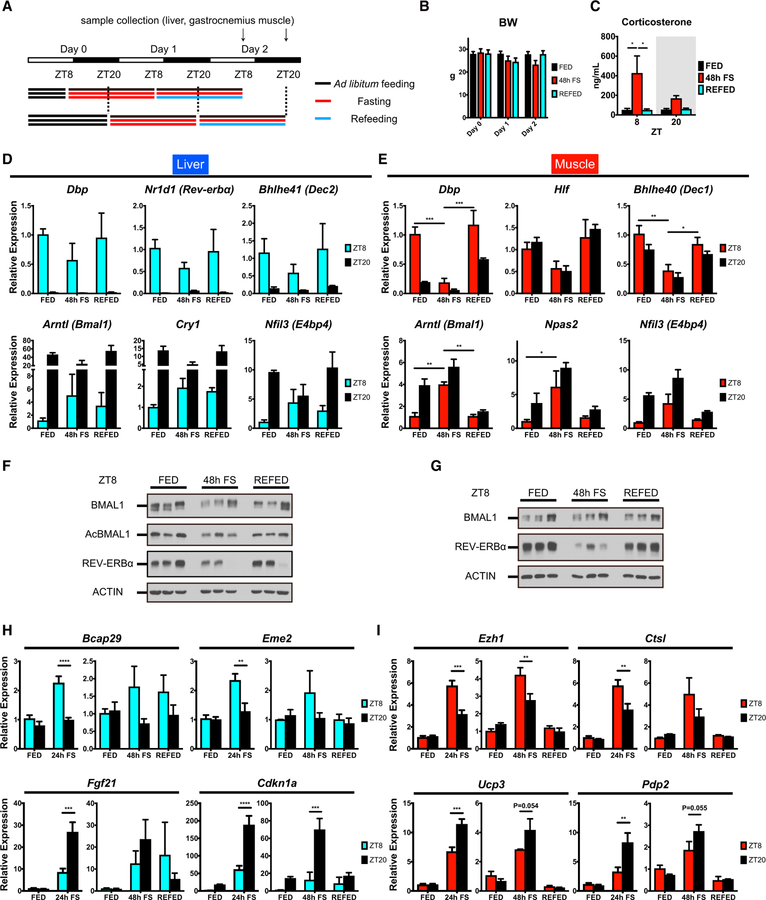
We next investigated whether the response to fasting is preserved for more than one daily cycle and whether it may be reversible by refeeding. Tissues were collected at ZT8 and ZT20 from 48-hr fasted mice (48h-FS), 24-hr fasted, and subsequently 24-hr refed (REFED) mice, or control mice fed ad libitum with normal chow (FED) (Figure 7A). Body weight was reduced by 48-hr fasting and regained by refeeding, while serum corticosterone, free fatty acids, and β-OHB levels were highly induced by 48-hr fasting and normalized by refeeding (Figures 7B, 7C, S1, and S7K). As for the 24-hr fasting, 48-hr fasting attenuated the expression of BMAL1 targets, whereas REV-ERBα-target genes were derepressed both in liver and skeletal muscle at ZT8, and both were reversed by refeeding (Figures 7D and 7E). Forty-eight-hour fasting affected BMAL1 phosphorylation and acetylation, as well as the levels of REV-ERBα protein, although the ratio of phosphorylation or acetylation to the total BMAL1 protein abundance was comparable due to the reduced BMAL1 expression by 48-hr fasting, especially in the liver. Refeeding basically restored the fed state profiles (Figures 7F, 7G, and S7L).
Figure 7. Response to Fasting Is Reversible by Refeeding.
(A) Experimental design of 48-hr fasting and 24-hr fasting with 24-hr refeeding. Tissues were collected at ZT8 and ZT20.
(B) Body weight before and after ad libitum fed (FED), 48-hr fasting (48h FS), or 24-hr fasting with 24-hr refeeding (REFED).
(C) Serum corticosterone levels under FED, 48h-FS, and REFED conditions. Data are shown as means + SEMs (n = 3 replicates per time point per group).
(D and E) Gene expression profiles of (D) hepatic and (E) muscle core clock genes normalized to 18S rRNA and presented as means + SEMs (n = 3 replicates per time point per group).
(F and G) Core clock proteins in total lysates from liver (F) and muscle (G) of FED, 48h-FS, and REFED mice.
(H and I) Gene expression profiles of (H) hepatic and (I) muscle genes displaying rhythmic response to 24-hr fasting (24h-FS) and 48h-FS.
Transcript levels were normalized to 18S rRNA and presented as means + SEMs (n = 3 replicates per time point per group). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-way ANOVA with Bonferroni post hoc tests.
Finally, we explored whether the rhythmic transcriptional response to 24-hr fasting is maintained for an extended cycle of 24-hr fasting (Figures 1C and 1F). By and large, the cyclic response was sustained under the 48-hr fasting condition, while it was virtually lost by refeeding both in liver and skeletal muscle (Figures 7H and 7I). Overall, the transcriptional response to fasting appeared to be sustainable by fasting per se and reversible by refeeding.
DISCUSSION
Food intake is considered as a major zeitgeber for peripheral clocks (Damiola et al., 2000; Stokkan et al., 2001; Vollmers et al., 2009). We sought to test whether ‘‘lack of food’’ merely reflects ‘‘free-running’’ conditions as in the SCN under constant darkness, or if it rather entrains a specific set of gene oscillations. We demonstrated that a significant number of genes display rhythmic response to fasting in a tissue-specific manner. Skeletal muscle gained almost twice as many newly oscillating genes upon fasting as the liver, while this ratio is reversed in FED mice. Of note, rhythmic transcriptional response to fasting appears sustained for a prolonged period. Supporting this notion, serum corticosterone, β-OHB, and hepatic Fgf21 expression displayed rhythmic response to fasting (corticosterone peaking at ZT8–ZT12, β-OHB and Fgf21 at ZT20; Figures 7C, 7H, S1K, and S7K), while GR and PPARα recruitment is also cyclic (GR peaking at ZT12, PPARα at ZT20; Figures 5F, 5H–5J, and 5L). Conversely, only 20% and 34% of hepatic and muscle transcripts in FED mice preserved their rhythmicity under fasting. Among them, a fraction of the genes were cyclic and in phase between FED and FAST (137 hepatic and 36 muscle genes, respectively). Presumably, these genes are driven by tissue-intrinsic clocks, SCN-derived signals, or a combination of both. Thus, fasting appears to be a strong metabolic cue to entrain rhythmic gene expression.
Our findings are consistent with previous studies (Kawamoto et al., 2006; Shavlakadze et al., 2013; Sun et al., 2015; Xie et al., 2016). Other reports present apparent differences that could be explained by various factors (Mukherji et al., 2015; Wuarin and Schibler, 1990). First, the clock in skeletal muscle appears to be more sensitive to fasting, resulting in larger changes in gene and protein expression compared to the liver. The liver response to fasting appears to display different degrees, although always consistent in our analyses. Second, younger mice are likely to be more susceptible to fasting; 8-week-old mice displayed increased attenuation of clock genes and proteins by fasting as compared to older mice (Figures 3, 6, 7, and S7). Lastly, different housing environments could influence mouse feeding behavior.
We staggered the starting times of fasting so that mice at any time point have undergone the same length of 24-hr fasting. This protocol is more appropriate than sequential time collection. First, sequential time collection results in different lengths of fasting. When starting fasting at ZT0 for all animals and harvesting tissues in a sequential manner, mice at ZT0 would receive 24-hr fasting, while mice at ZT20 would receive 44-hr fasting. Second, ad libitum-fed mice at ZT12 are not really ‘‘fasted.’’ Our analysis of metabolic cages shows that even during the light phase, mice engage in feeding activity, resulting in a significant increase in oxygen consumption, RER, and energy expenditure in comparison to fasted mice. Furthermore, if mice at ZT12 were fasted for 36 hr, then our transcriptome would have demonstrated the expression of fasting-induced genes peaking at ZT12, which was not the case (Figures 1B and 1C). Hepatic PPARα or REV-ERBα targets are expressed at lower levels at ZT12 (Figures S4D and S5F). Also, the feeding status of animals is likely to vary from peak feeding, which may contribute to rhythmicity. The experimental design used in the present study appears to be most appropriate to assess the time-of-the-day-specific response to fasting.
Fasting appears to be a suppressive signal for the circadian clock. This may be partly accomplished by increased nicotinamide adenine dinucleotide-positive (NAD+) levels and subsequent enhanced sirtuin activity by fasting (Cantó et al., 2010; Rodgers et al., 2005), which hinders BMAL1 recruitment to chromatin and gene activation (Masri et al., 2014; Nakahata et al., 2008). Moreover, fasting could control clock protein translation, post-translational modifications, and degradation. Per1 activation upon fasting is somewhat reminiscent of the effect elicited by light, although it is unclear whether Per1 induction by fasting contributes to the mitigation of BMAL1-dependent activation (Shigeyoshi et al., 1997). Feeding or insulin induces Per2, suggesting that a switch from fasting to feeding could act as an entrainment signal by activating Per1 and Per2 (Sato et al., 2014).
PPAR, CCAAT-enhancer-binding protein (C/EBP), GR, and CREB motifs are particularly enriched in hepatic fasting-induced enhancers (Goldstein et al., 2017). Moreover, CREB, FOX, and GR motifs are enriched in hepatic DNase I hypersensitive sites during the fasting phase of temporal feeding restriction even in Bmal1 null mice (Sobel et al., 2017). Therefore, fasting-sensitive TFs may work in concert or independently of the circadian machinery. We thereby investigated how GR, CREB, FOXO, TFEB, and PPARs may operate in concert with BMAL1 upon fasting. Although the protein levels of some of the fasting-induced TFs in FAST mice seemed comparable to FED mice, the recruitment of TFs such as hepatic CREB and PPARα was enhanced by fasting in a rhythmic manner. This led to a classification of fasting-dependent genes based on their differential gene expression. Our analysis revealed that there is a group of genes whose expression is dually controlled by fasting-sensitive TFs and BMAL1, both in liver and skeletal muscle. BMAL1 appears to play a dominant role in the expression of these genes in ad libitum fed condition, while fasting-responsive TFs ‘‘take over’’ BMAL1 upon nutritional deprivation to achieve highly efficient gene control. Notably, gene induction by fasting is dependent on BMAL1, presumably because it may contribute to a favorable chromatin state for the induction of class II genes by fasting-responsive TFs. Physiological fasting occurs at approximately ZT8–ZT12, so that BMAL1 and fasting-responsive TFs may cooperatively produce robust oscillation under dark phase restricted feeding. While we did not test whether BMAL1 and fasting-sensitive TFs physically interact, TFEB binds E-box sequences, suggesting competition or synergistic transcription (Settembre and Ballabio, 2014). Moreover, BMAL1 and feeding-responsive TFs such as sterol regulatory element binding proteins (SREBPs) and farnesoid X receptor (FXR) could cooperatively drive diurnal gene expression. The majority of genes activated by fasting are non-BMAL1 targets, thus oscillation is possible even without a functional clock (Vollmers et al., 2009).
Post-transcriptional control plays an important role in circadian gene expression (Fustin et al., 2013; Kojima et al., 2012; Terajima et al., 2017). A number of distinct genes were cycling only in exonic reads under fasting, indicating that nutrient-sensing signals could control the RNA processing machinery. RNA editing of Apob mRNA and the excretion of small-molecular-weight apolipoprotein B (ApoB) are suppressed after fasting, although fasting does not affect the relative levels of Apob mRNA, indicating that RNA editing could sense fasting signals (Leighton et al., 1990). Fasting also alters circadian alternative splicing and contributes to the temporal pattern of gene expression (McGlincy et al., 2012). Peroxisome proliferator-activated receptor g co-activator 1a (PGC-1a) interacts with components of the RNA processing machinery, suggesting that PGC-1a could mediate the fasting signal to post-transcriptional gene control (Monsalve et al., 2000). Since transcription and RNA processing are tightly coupled (Bentley, 2014), it is tempting to speculate that fasting-sensitive TFs such as PPARs and FOXO also modulate post-transcriptional gene regulation in the context of fasting by using co-activators and could use them to bridge transcription with post-transcriptional processes.
Our study shows that daily gene expression is highly responsive to fasting through temporal coordination of the circadian clock and fasting-sensitive TFs. This reorganization of gene regulation by fasting could prime the genome to a more permissive state to anticipate upcoming food intake and thereby drive a new rhythmic cycle of gene expression. Therefore, optimal fasting in a timed manner would be strategic to confer robust circadian oscillation that ultimately benefits health and protects against aging-associated diseases.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paolo Sassone-Corsi (psc@uci.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Age-matched male C57BL/6 mice (the Jackson Laboratory), male Bmal1—/— mice, and male liver-specific Tfeb—/— (Tfeb LiKO) mice were housed under a temperature-controlled, 12hr light/ 12hr dark schedule. Bmal1—/— mice and Tfeb LiKO mice were previously described (Kondratov et al., 2006; Settembre et al., 2013). All animal experiments were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at the University of California, Irvine.
METHOD DETAILS
Fasting Schedule
Male C57BL/6 mice, male Bmal1—/— mice, and male Tfeb LiKO mice were placed on a normal chow diet (2020X Teklad Global Extruded Rodent Diet). For the 24-hr fasting experiments, C57BL/6 mice at 8 weeks of age, Bmal1—/— mice at an average of 14.4 weeks of age, and Tfeb LiKO mice at an average of 18.5 weeks of age were randomly divided into an ad libitum fed group, or a fasting group. The fasted mice were placed in new cages without food 24-hr before sacrifice to prevent eating remnants of chow diet on the cage floor. For the 48-hr fasting and refeeding experiment, male wild-type mice to Bmal1—/— mice at an average of 14.9 weeks of age were randomly divided into ad libitum fed group, a fasting group, or a fasting with subsequent refeeding group. All mice were individually housed 1 week prior to harvesting tissue.
Metabolic Cage Analysis
Indirect calorimetry was carried out by negative-flow system cages Oxymax/CLAMS (Columbus Instruments). Feeding and lighting conditions in metabolic cages were the same as those in the normal cages. Feeder counts less than 0 g or more than 0.8 g per 10 min during ad libitum fed experiment were excluded from the study. Any feeder counts during the fasting experiment were regarded as noise and treated as null since mice were restricted from any access of food.
Locomotor Activity Analysis
Mice were individually housed under 12hr light/ 12hr dark schedule. Locomotor activity was measured using optical beam motion detection (Philips Respironics). Data were collected using the Minimitter VitalView 5.0 data acquisition software. Actograms and activity profiles were computed using Clocklab software (Actimetrics).
RNA Extraction and Reverse Transcription
Total RNA was extracted from liver and skeletal muscle with TRIzol (Invitrogen), followed by precipitation with isopropanol and ethanol. Complimentary DNA (cDNA) was obtained by reverse transcription of 1 mg RNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories).
Protein Extraction and Western Blot
Whole cell extracts of liver and skeletal muscle were obtained as follows: frozen liver and skeletal muscle tissues were homogenized in modified RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS, 5 mM MgCl2, and 1 mM PMSF) supplemented with Protease Inhibitor Cocktail (Roche), 1 mM DTT, 20 mM NaF, 1 mM Sodium Orthovanadate, 10 mM Nicotinamide, 330 nM Trichostatin A (T8552, Sigma). Samples were sonicated briefly, and centrifuged at 13200 rpm for 15 min at 4°C. Supernatants were stored as whole cell protein lysates. Nuclear extraction was carried out as follows: frozen liver tissues were homogenized in 1.5 mL of buffer A (10 mM HEPES [pH 7.8], 25 mM KCl, 0.5 mM spermidine, 1 mM EGTA, 1 mM EDTA, 320 mM sucrose, 0.3% Triton X-100, and 0.5 mM PMSF) supplemented with the same inhibitors for homogenization of whole cell extracts. Samples were centrifuged at 3300 rpm for 10 min at 4°C. Supernatants were stored as a cytoplasmic fraction. Pellets were resuspended in 1.5 mL of buffer A with the inhibitors. Samples were then spun down at 2000 rpm for 10 min at 4°C, and pellets were resuspended in 1 mL of low salt buffer (10 mM HEPES [pH 7.8], 25 mM KCl, 0.5 mM spermidine, 1 mM EGTA, 1 mM EDTA, 20% Glycerol, and 0.5 mM PMSF) with the same inhibitors for homogenization. Samples were centrifuged at 2000 rpm for 10 min at 4°C, and pellets were resuspended in 1 mL of low salt buffer with the inhibitors. Samples were further centrifuged at 2000 rpm for 5 min at 4°C, and pellets were resuspended in one volume (equal to the pellet size) of low salt buffer with the inhibitors. After measuring the total volume, two volumes of high salt buffer (10 mM HEPES [pH 7.8], 525 mM KCl, 0.5 mM spermidine, 1 mM EGTA, 1 mM EDTA, 20% Glycerol, and 0.5 mM PMSF) with the same inhibitors for homogenization were added to samples, followed by mixture and incubation for 1 h at 4°C. Samples were centrifuged at 12000 rpm for 20 min at 4°C, and supernatants were stored as nuclear fraction. 5–60 mg of protein lysates were separated on 6%–10% gels by SDS-PAGE, and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies and peroxidase-conjugated secondary antibodies at 4°C, and visualized by chemiluminescent HRP substrate (WBKLS0500, EMD Millipore). The blots were developed with autoradiography films (HyBlot CL, Denville Scientific). The films were scanned and densitometry was analyzed through ImageJ software.
Chromatin Immunoprecipitation
Approximately 250 mg of frozen liver tissues were homogenized in 4 mL PBS containing Protease Inhibitor Cocktail (Roche), 20 mM NaF, 0.5 mM PMSF, 10 mM Nicotinamide, and 330 nM Trichostatin A (T8552, Sigma). Liver homogenates were first cross-linked with 2 mM disuccinimidyl glutarate and 1 mM MgCl2 for 30 min, and further cross-linked with 1% formaldehyde for 5 min at room temperature. Cross-linking reaction was stopped by incubation with 125 mM glycine for 10 min. Samples were centrifuged at 1000 g for 5 min at 4°C, and pellets were resuspended in 4 mL PBS with the inhibitors. Samples were then centrifuged at 1000 g for 10 min at 4°C, and pellets were resuspended in 1 mL of cell lysis buffer (5 mM HEPES [pH 8.0], 85 mM KCl, 0.5% NP-40, and the same inhibitors for homogenization) and incubated on ice for 10 min. Samples were spun down at 3000 rpm for 5 min at 4°C, and pellets were resuspended in 1 mL of SDS lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS, and the same inhibitors for homogenization) and incubated on ice for 20 min. Samples were sonicated with Bioruptor (Diagenode), and centrifuged at 13200 rpm for 10 min at 4°C. Supernatants were diluted 1:4 in dilution buffer (50 mM Tris-HCl [pH 8.0], 167mM NaCl, 1.1% Triton X-100, 0.11% Sodium Deoxycholate, and the same inhibitors for homogenization). 1 mL of chromatin samples were first pre-cleared with 50 mL of 50% slurry, salmon sperm DNA-blocked Protein G Sepharose (P3296, Sigma-Aldrich) for 2 hr at 4°C. After removal of Protein G Sepharose by quick centrifugation, samples were immunoprecipitated with antibodies overnight at 4°C. 50 mL of 50% slurry Protein G Sepharose beads were then added to samples, followed by incubation for 3 hr at 4°C. The beads were washed with 1ml of low-salt RIPA (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% Sodium Deoxycholate, and 0.1% SDS), high-salt RIPA (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% Sodium Deoxycholate, and 0.1% SDS), LiCl wash (10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 1 mM EDTA [pH 8.0], 0.5% NP-40, and 0.5% Sodium Deoxycholate), and twice with TE wash (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA [pH 8.0]) buffers. Samples were reverse cross-linked with 200 mL of elution buffer (10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM EDTA [pH 8.0], and 0.5% SDS) overnight at 65°C, and subsequently treated with 2 mL of 2 mg/ml RNase A for 30 min at 37°C, and 1 mL of 10 mg/ml proteinase K for 1 hr at 55°C. After adding 2 mL of 20 mg/ml glycogen, samples were extracted twice with 210 mL of phenol/chloroform/isoamyl alcohol. Finally, DNA was precipitated with ethanol, and resuspended in TE wash buffer.
Quantitative real-time PCR analysis
cDNA and immunoprecipitated chromatin DNA were used for quantitative real-time PCR (RT-qPCR) using SsoAdvanced Universal SYBR Green Supermix and CFX96 Detection System (Bio-Rad Laboratories). All gene expression was normalized to 18S Ribosomal RNA. The sequences of primers used in RT-qPCR are listed in Table S6.
Antibodies
The antibodies used for western blots in this study are as follows. BMAL1 (ab93806, Abcam, 1:10000), Acetyl-BMAL1 (AB15396, EMD Millipore, 1:1000), REV-ERBα (#13418, Cell Signaling Technology, 1:1000), PER2 (PER21-A, Alpha Diagnostic International, 1:1000), CRY1 (A302–614A, Bethyl Laboratories, 1:2000), GR (#12041, Cell Signaling Technology, 1:2000), Phospho-CREB (#9198, Cell Signaling Technology, 1:1000), CREB (#9197, Cell Signaling Technology, 1:1000), FOXO1 (#2880, Cell Signaling Technology, 1:1000), PPARα (sc-9000, Santa Cruz Biotechnology, 1:1000), SIRT1 (07–131, EMD Millipore, 1:10000), PPARδ (PA1–823A, Thermo Fisher Scientific, 1:1000), TFEB (A303–673A, Bethyl Laboratories, 1:2000), ACTIN (ab3280, Abcam, 1:10000), a—TUBULIN (T5168, SIGMA-ALDRICH, 1:10000), p84 (GTX70220, GeneTex, 1:10000), secondary antibodies (12–348 and AP160P, EMD Millipore, 1:10000). The antibodies used for ChIP are as follows. BMAL1 (ab93806, Abcam, 2 µg), GR (#12041, Cell Signaling Technology, 5 µl), CREB (sc-186 X, Santa Cruz Biotechnology, 10 µg), PPARα (sc-9000 X, Santa Cruz Biotechnology, 8 µg), normal rabbit IgG (sc-2027, Santa Cruz Biotechnology).
Serum Corticosterone quantitation
Serum levels of corticosterone was quantified using ‘‘Corticosterone ELISA Kit’’ (ADI-900–097, Enzo Life Sciences) according to manufacturer’s instructions.
Serum free fatty acid quantitation
Serum levels of free fatty acid were determined using ‘‘Free Fatty Acid Quantitation Kit’’ (MAK044, SIGMA-ALDRICH) according to manufacturer’s instructions.
β-Hydroxybutyrate quantitation
Serum levels of β-Hydroxybutyrate were measured using ‘‘β-Hydroxybutyrate LiquiColor Assay’’ (2440–058, EKF DIAGNOSTICS-STANBIO) according to manufacturer’s instructions.
RNA sequencing
Total RNA extracted from liver and skeletal muscle as described previously was used for RNA sequencing (RNA-seq). Total RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip and Nanodrop absorbance ratios for 260/280 nm and 260/230 nm. Library construction was performed according to the Illumina TruSeq mRNA stranded protocol. The input quantity for total RNA was 0.75 ug and mRNA was enriched using oligo dT magnetic beads. The enriched mRNA was chemically fragmented for four minutes. First strand synthesis used random primers and reverse transcriptase to make cDNA. After second strand synthesis the ds cDNA was cleaned using AMPure XP beads and the cDNA was end repaired and then the 3’ ends were adenylated. Illumina barcoded adapters were ligated on the ends and the adaptor ligated fragments were enriched by nine cycles of PCR. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. The concentration for clustering on the flowcell was 12.75 pM. The 72 samples were multiplexed into three libraries of 24 samples each, and sequenced on an Illumina HiSeq 2500 instrument during two single-end 100 cycles sequencing runs by the Genomics High-Throughput Facility at the University of California, Irvine. The version of HiSeq control software was HCS 2.2.58 with real time analysis software, RTA 1.18.64. Each library of 24 samples was sequenced between four and five times on distinct flowcell lanes for a total of 13 flowcell lanes. The resulting sequencing data for each library were post-processed and demultiplexed to produce FastQ files using Illumina software CASAVA 1.8.2. Reads failing Illumina’s standard quality tests were not included in the FastQ files. The quality of the remaining sequences was further assessed using the PHRED quality scores produced in real time during the base-calling step of the sequencing runs.
Short reads alignment to reference genome and transcriptome
The sequencing reads from each replicate in the experiment were separately aligned to the reference genome mm10 and corresponding known splice junctions extracted from the RefSeq database using the short-read aligner ELAND v2e (Illumina). Reads with a non-unique best match or with more than two mismatches with the reference sequences were discarded from the analyses. The remaining uniquely aligned reads were used to estimate both mature RNA and precursor mRNA relative abundance in the next steps of the analyses.
Read counting methods
Gene expression results produced by standard RNA-seq analysis tools are obtained by normalizing the number of reads located inside exonic regions of known genes. To compare the rhythmicity of the pre-mRNAs and mRNAs in our datasets, we implemented the Exon Intron Split Analysis (EISA) method using a protocol similar to the one proposed by the authors of the EISA method (Gaidatzis et al., 2015). All the components of this analysis were performed using programs specifically developed in-house. This section describes the method used in these programs to decide if a read is exonic or intronic, based on the RefSeq gene annotations available at the time of the analysis as well as the alignment results.
Exonic reads are detected following a common protocol. In brief, all isoforms of a given gene are merged together to create a single meta-transcript for the gene, where each exonic position of the meta-transcript belongs to an exon of at least one gene isoform. Similarly, the union of all the splice junctions in all the gene isoforms is used to detect the exonic reads. The resulting meta-transcripts and splice junctions are further processed to discard any exonic position of a gene overlapped by any position of a different gene to remove the corresponding ambiguities. 23,989 gene meta-transcripts were extracted following this protocol. Sequencing reads located in the exons of a meta-transcript, or uniquely mapped to any of the splice junctions associated with the meta-transcript, were considered exonic and therefore contributed to the estimate of the mRNA relative abundance for the corresponding gene in each sample.
Intronic reads were detected following a protocol equivalent to the protocol reported in the EISA method (Gaidatzis et al., 2015). Similar to the protocol reported above to detect exonic reads, all isoforms of a given gene are merged together to create a single meta-transcript for the gene. A notable difference for the intronic positions of the curated meta-transcripts is that they belong to the intersection of the introns of all the gene isoforms, instead of the union which is used in the exonic case. This protocol ensures that there is no overlap between the exonic and intronic positions of the curated meta-transcripts. Intronic positions overlapped by any position of a different gene were discarded, as in the exonic case. Genes with no intron and genes with no intron longer than 100 bp in the corresponding meta-transcripts were discarded. This protocol resulted in 19,433 gene meta-transcripts suitable for the EISA analysis. Sequencing reads located in the introns of a meta-transcript were used to estimate the pre-mRNA relative abundance for the corresponding gene.
Normalization and gene expression results Reads
Per Kilobase of exon model per Million mapped reads (RPKM) values are commonly used in RNA-seq experiments to quantify the relative abundance of each mRNA in a sample and to perform differential gene expression analyses. The same metric can be used to quantify the relative abundance of each pre-mRNA in a sample simply by replacing the size of the exons in the meta-transcripts by the size of the introns in the meta-transcripts, and by replacing the exonic read counts by the intronic read counts in the calculation (Gaidatzis et al., 2015).
In order to study the mRNA relative abundance in each sample, we first normalized the exonic read counts by the size of the exons in the meta-transcripts and by the total number of exonic reads found in each sample for all 23,989 gene meta-transcripts extracted, as described in the previous section. This first set of gene expression results is equivalent to the gene expression results of a standard RNA-seq analysis. Since only 19,433 genes were found suitable for the EISA analysis, we then normalized both exonic and intronic read counts considering only these 19,433 genes in the normalization procedure in order to remove the bias that would be introduced by normalizing using a different set of genes. The first set of exonic expression results was used to further study the mRNA abundance in each sample, as described in the next sections. In addition, comparisons between mRNA and pre-mRNA abundance in each sample were performed using the second set of exonic expression results and the set of intronic expression results. The three sets of gene expression results for each sample are available for download from the GEO database.
QUANTIFICATION AND STATISTICAL ANALYSIS
Bioinformatic Analysis
Time series of expression levels were then used to determine circadian behaviors of transcripts using JTK-CYCLE (Hughes et al., 2010). Transcripts found with no expression in greater than six replicates were filtered before analysis of circadian behavior. A gene with p < 0.01 was considered significantly rhythmic over the circadian cycle. Heatmaps of the rhythmic transcripts were generated by R package ‘gplots’. Differential analysis of transcript expression levels between FED and FAST at a specific ZT was done using Cyber-T (Kayala and Baldi, 2012), a differential analysis program using a Bayesian-regularized t test. Fold change (FC) in the fasting study was calculated with RPKM values. Differential expression were determined with p < 0.05 in Bayes-regularized t test and FC ≥ 1.3. Gene Set Enrichment Analysis was performed to determine whether or not a specific subset of genes are enriched either in FED or FAST mice (Subramanian et al., 2005). Gene sets with false discovery rate of less than 25% were considered significantly enriched. The Database for Annotation, Visualization and Integrated Discovery (DAVID) pathway analysis tool was used to identify gene ontology terms related to biological process (https://david-d.ncifcrf.gov/). Analysis was done using pipelines implemented for the Circadiomics (Patel et al., 2012) database and web portal (circadiomics.ics.uci.edu) where all the transcriptomic data associated with this work is publicly available.
Analysis of Published Dataset
We obtained time-restricted fed, hepatic transcriptome by microarray (Vollmers et al., 2009) from GEO (GSE13060), hepatic BMAL1 ChIP-seq (Koike et al., 2012) from ‘‘Master peak lists for ChIP-seq experiments for BMAL1,’’ inducible Bmal1—/— liver transcriptome by RNA-seq (Yang et al., 2016) from GEO (GSE70497), Rev-ERBα—/— liver transcriptome by microarray and hepatic REV-ERBα GRO-seq (Fang et al., 2014) from ‘‘List of Differentially Expressed Genes Identified in Rev-ERBα—/— Livers,’’ prednisolone-induced liver transcriptome by microarray (Frijters et al., 2010) from GEO (GSE21048), cAMP (Zhang et al., 2005) and glucagon induced liver transcriptome by microarray (Ravnskjaer et al., 2013) from ‘‘The lists of cAMP responsive genes from hepatocytes’’ (http://natural.salk. edu/CREB/) and GEO (GSE47179), respectively, liver specific Foxo1,3,4—/— liver transcriptome by microarray (Haeusler et al., 2014) from GEO (GSE60527), liver specific PPARα—/— liver transcriptome by microarray (Montagner et al., 2016) from GEO (GSE73299), TFEB-overexpressed liver transcriptome by microarray (Settembre et al., 2013) from GEO (GSE35015), pre- and post-exercise gastrocnemius muscle transcriptome by microarray (Perry et al., 2014) from GEO (GSE61712), muscle specific Bmal1—/— tibialis anterior muscle transcriptome by microarray (Dyar et al., 2013) from GEO (GSE43071), muscle specific Hdac3 —/— muscle transcriptome by RNA-seq from ‘‘Differentially expressed genes from RNA-seq’’ (Hong et al., 2017), dexamethasone-treated C2C12 myotubes transcriptome by microarray (Kuo et al., 2012) from GEO (GSE28840), cAMP induced muscle transcriptome by microarray (Pearen et al., 2009) from GEO (GSE15793), muscle specific Foxo1,3,4—/— muscle transcriptome by microarray (Milan et al., 2015) from GEO (GSE52667), PPARδ overexpressing muscle transcriptome by microarray (Gan et al., 2011) from GEO (GSE29055), and TFEB overexpressed muscle transcriptome by microarray (Mansueto et al., 2017) from GEO (GSE62975). Calculation of FC in microarray was carried out by logarithmic normalization unless already logarithmic-transformed. Differential expression was determined by p < 0.05 in Bayes-regularized t test along with FC ≥ 1.3 in RNA-seq and LogFC ≥ 0.37 in microarray, except for Foxo1,3,4—/— muscle transcriptome where LogFC ≥ 1.5 was applied because of a single replicate. Specifically, among the genes in master peak lists of BMAL1 ChIP-seq (Koike et al., 2012), differentially expressed genes with FC (WT/iKO) ≥ 1.3 at CT8 and CT12 were selected as hepatic BMAL1 target genes at ZT8 and ZT12, respectively (Yang et al., 2016). ‘‘Derepressed genes in Rev-ERBα—/—’’ among ‘‘List of Differentially Expressed Genes Identified in Rev-ERBα—/— Livers’’ were used as hepatic REV-ERBα target genes (Fang et al., 2014). Differentially expressed genes with LogFC (shNS, Glucagon/shNS, Control) ≥ 0.37 were selected as glucagon induced genes in the liver (GSE47179). ‘‘The lists of cAMP responsive genes from hepatocytes’’ were selected as cAMP induced genes in the liver (Zhang et al., 2005). Genes included either in glucagon or cAMP induced genes in the liver were finally extracted as hepatic CREB target genes. Likewise, we selected differentially expressed genes with LogFC (Prednisolone-treated WT/Vehicle-treated WT) ≥ 0.37 as hepatic GR target genes (GSE21048), with LogFC (WT-fasted/L-FoxO1,3,4—/—-fasted) ≥ 0.37 as hepatic FOXO target genes (GSE60527), with LogFC (LWT_Fasted/PPARα-LKO_Fasted) ≥ 0.37 as hepatic PPARα target genes (GSE73299), with LogFC (Injected/CTL) ≥ 0.37 as hepatic TFEB target genes (GSE35015), with LogFC (WT post-exercise/WT pre-exercise) ≥ 0.37 as exercise-induced genes (GSE61712), with LogFC (WT pre-exercise/WT post-exercise) ≥ 0.37 as exercise-repressed genes (GSE 61712), with LogFC (Tibialis Anterior Muscle_Ctrl/Tibialis Anterior Muscle_mKO) ≥ 0.37 at ZT8 and ZT12 as muscle BMAL1 target genes at ZT8 and ZT12, respectively (GSE43071), with LogFC (myotube_Dex/myotube_ETOH) ≥ 0.37 either at 6 hr or at 24 hr after treatment as muscle GR target genes (GSE28840), with LogFC (Formoterol/Saline) ≥ 0.37 either at 1 hr or at 4 hr after treatment as muscle CREB target genes (GSE15793), with LogFC (wild-type, starved/Foxo 1,3,4 muscle specific knock-out, starved) ≥ 1.5 as muscle FOXO target genes (GSE52667), with LogFC (MCK-PPARb/Non-transgenic mouse) ≥ 0.37 as muscle PPARδ target genes (GSE29055), and with LogFC (Injected/Control) ≥ 0.37 as muscle TFEB target genes (GSE62975). Genes with FC (KO/WT) R1.3 among the ‘‘Differentially expressed genes from RNA-seq’’ was used as muscle REV-ERBα-HDAC3 target genes (Hong et al., 2017). Genes with JTK-CYCLE p < 0.01 were extracted as rhythmic genes under time-restricted feeding (GSE13060). Latest version of annotation files was downloaded from GEO and applied to microarray dataset. Updated version of gene symbol was obtained from BioGPS to overlay fasting transcriptome data (https://biogps.org).
Other Statistical Analysis
Experimental data were presented as means + SEM. For some points, where the error bars are shorter than the height of the symbol, the error bars were not drawn. The quantitation of enrichment in ChIP-qPCR was shown as relative recruitment, which was normalized % input relative to the first replicate of WT at ZT0. Comparisons with two variables were analyzed by two-way ANOVA with Bonferroni post hoc test (GraphPad Prism 7). Amplitude distribution of oscillating genes was analyzed by Mann-Whitney test. Evaluation of phase lag between intron and exon was carried out by Wilcoxon Signed Rank Test. In qPCR for Tfeb LiKO and ChIP-qPCR, any data more than 1.5 interquartile ranges above the third quartile or below the first quartile were defined as outliers and excluded from the analysis. Graphs were depicted using Microsoft Excel, GraphPad Prism 7, and Adobe Illustrator CS6.
DATA AND SOFTWARE AVAILABILITY
The GEO accession number for the RNA-seq data reported in this paper is GSE107787.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-BMAL1 | Abcam | Cat# ab93806; RRID: AB_10675117 |
| Rabbit polyclonal anti-acetyl BMAL1 (Lys538) | EMD Millipore | Cat# AB15396; RRID: AB_11212017 |
| Rabbit monoclonal anti-REV-ERBα | Cell Signaling Technology | Cat# 13418; RRID: AB_2630359 |
| Rabbit polyclonal anti-PER2 | Alpha Diagnostic International | Cat# PER21-A; RRID: AB_1620951 |
| Rabbit polyclonal anti-CRY1 | Bethyl Laboratories | Cat# A302–614A; RRID: AB_10555376 |
| Rabbit polyclonal anti-SIRT1 | EMD Millipore | Cat# 07–131; RRID: AB_10067921 |
| Rabbit monoclonal anti-Glucocorticoid Receptor | Cell Signaling Technology | Cat# 12041; RRID: AB_2631286 |
| Rabbit monoclonal anti-Phospho-CREB (Ser133) | Cell Signaling Technology | Cat# 9198; RRID: AB_2561044 |
| Rabbit monoclonal anti-CREB | Cell Signaling Technology | Cat# 9197; RRID: AB_331277 |
| Rabbit polyclonal anti-CREB-1 | Santa Cruz Biotechnology | Cat# sc-186 X; RRID: AB_2086021 |
| Rabbit monoclonal anti-FOXO1 | Cell Signaling Technology | Cat# 2880; RRID: AB_2106495 |
| Rabbit polyclonal anti-PPARα | Santa Cruz Biotechnology | Cat# sc-9000; RRID: AB_2165737 |
| Rabbit polyclonal anti-PPARα | Santa Cruz Biotechnology | Cat# sc-9000 X; RRID: AB_2165737 |
| Rabbit monoclonal anti-TFEB | Bethyl Laboratories | Cat# A303–673A; RRID: AB_11204751 |
| Rabbit polyclonal anti-PPARδ | Thermo Fisher Scientific | Cat# PA1–823A; RRID: AB_2165895 |
| Mouse monoclonal anti-ACTIN | Abcam | Cat# ab3280; RRID: AB_303668 |
| Mouse monoclonal anti-a-TUBULIN | SIGMA-ALDRICH | Cat# T5168; RRID: N/A |
| Mouse monoclonal anti-p84 | GeneTex | Cat# GTX70220; RRID: AB_372637 |
| Goat polyclonal anti-Rabbit IgG, HRP conjugate | EMD Millipore | Cat# 12–348; RRID: AB_390191 |
| Rabbit polyclonal anti-Mouse IgG, HRP conjugate | EMD Millipore | Cat# AP160P; RRID: AB_92531 |
| Normal Rabbit IgG | Santa Cruz Biotechnology | Cat# sc-2027; RRID: AB_737197 |
|
Chemicals, Peptides, and Recombinant Proteins | ||
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596018 |
| iScript Reverse Transcription Supermix | Bio-Rad Laboratories | Cat# 1708841 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad Laboratories | Cat# 1725271 |
| cOmplete, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 05056489001 |
| Trichostatin A | Sigma | Cat# T8552 |
| Protein Assay Dye Reagent Concentrate | Bio-Rad Laboratories | Cat# 5000006 |
| Nitrocellulose Membrane | Bio-Rad Laboratories | Cat# 1620115 |
| Immobilon Western Chemiluminescent HRP Substrate | EMD Millipore | Cat# WBKLS0500 |
| HyBlot CL Autoradiography Film | Denville Scientific | Cat# E3012 |
| DSG (disuccinimidyl glutarate) | Thermo Fisher Scientific | Cat# 20593 |
| Protein G Sepharose | Sigma-Aldrich | Cat# P3296 |
|
Critical Commercial Assays | ||
| Corticosterone ELISA Kit | Enzo Life Sciences | Cat# ADI-900–097 |
| Free Fatty Acid Quantitation Kit | SIGMA-ALDRICH | Cat# MAK044 |
| b-Hydroxybutyrate LiquiColor Assay | EKF DIAGNOSTICS-STANBIO | Cat# 2440–058 |
|
Deposited Data | ||
| Fasting circadian transcriptome | This paper | GEO: GSE107787 |
|
Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat# 000664; RRID: IMSR_JAX:000664 |
| Mouse: Bmal1—/— | (Kondratov et al., 2006) | N/A |
| Mouse: liver-specific Tfeb—/— | (Settembre et al., 2013) | N/A |
|
Oligonucleotides | ||
| See Table S6 for qPCR and ChIP-qPCR primer lists | N/A | N/A |
|
Software and Algorithms | ||
| VitalView 5.0 | Koninklijke Philips Electronics N.V. | N/A |
| Clocklab | Actimetrics | http://www.actimetrics.com/products/clocklab/ |
| Oxymax 4.52 | Columbus Instruments | http://www.colinst.com/ |
| JTK_CYCLE | (Hughes et al., 2010) | https://www.openwetware.org/wiki/HughesLab:JTK_Cycle |
| Bioconductor | N/A | https://www.bioconductor.org/ |
| Cyber-T | (Kayala and Baldi, 2012) | http://cybert.ics.uci.edu/ |
| Gene Set Enrichment Analysis | (Subramanian et al., 2005) | http://software.broadinstitute.org/gsea/index.jsp |
| The Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.7 | N/A | https://david-d.ncifcrf.gov/ |
| CircadiOmics | (Patel et al., 2012) | http://circadiomics.igb.uci.edu/ |
| BioGPS | N/A | https://biogps.org |
| Prism 7 | GraphPad Software | https://www.graphpad.com/ |
| Illustrator CS6 | Adobe Systems | N/A |
| Excel 2011 | Microsoft | N/A |
|
Other | ||
| Normal chow diet (Teklad global soy protein-free extruded) | Envigo | Cat# 2020X |
| CLAMS (Comprehensive Lab Animal Monitoring System) | Columbus Instruments | http://www.colinst.com/ |
| Mini Mitter | Philips Respironics | N/A |
| Q125 Sonicator | Qsonica | Cat# Q125–110 |
| CFX96 Touch Real-Time PCR Detection System | Bio-Rad Laboratories | N/A |
Highlights.
Transcriptional response to fasting is robustly rhythmic in liver and muscle
Lack of food fails to sustain ‘‘free-running’’ conditions of peripheral circadian clocks
Genes are temporally regulated by the clock and fasting-related transcription factors
Rhythmic response to fasting is reversible by refeeding
ACKNOWLEDGMENTS
We thank members of the Sassone-Corsi laboratory, as well as Melanie Oakes, Seung-Ah Chung, and Yuzo Kanomata at the Genomics High-Throughput Facility at the University of California, Irvine. K.K. was supported by a Japan Society for the Promotion of Science (JSPS) fellowship. C.M., N.C., Y.L., and P.B. were supported by grants from the Defense Advanced Research Projects Agency (DARPA; D17AP00002) and NIH (GM123558) to P.B. This study was supported by the NIH, a Novo Nordisk Foundation Challenge Grant, and INSERM (France).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and six tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.077.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Asher G, and Sassone-Corsi P (2015). Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92. [DOI] [PubMed] [Google Scholar]
- Bass J, and Takahashi JS (2010). Circadian integration of metabolism and energetics. Science 330, 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014). Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet 15, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, Dubeau L, Yap LP, Park R, Vinciguerra M, et al. (2015). A Periodic Diet that Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metab 22, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, and Lazar MA (2012). Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, and Auwerx J (2010). Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. (2012). Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, and Schibler U (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar KA, Ciciliot S, Wright LE, Biensø RS, Tagliazucchi GM, Patel VR, Forcato M, Paz MI, Gudiksen A, Solagna F, et al. (2013). Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab 3, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, and Sassone-Corsi P (2013). Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, and Lazar MA (2014). Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frijters R, Fleuren W, Toonen EJ, Tuckermann JP, Reichardt HM, van der Maaden H, van Elsas A, van Lierop MJ, Dokter W, de Vlieg J, and Alkema W (2010). Prednisolone-induced differential gene expression in mouse liver carrying wild type or a dimerization-defective glucocorticoid receptor. BMC Genomics 11, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, and Okamura H (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. [DOI] [PubMed] [Google Scholar]
- Gaidatzis D, Burger L, Florescu M, and Stadler MB (2015). Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol 33, 722–729. [DOI] [PubMed] [Google Scholar]
- Gan Z, Burkart-Hartman EM, Han DH, Finck B, Leone TC, Smith EY, Ayala JE, Holloszy J, and Kelly DP (2011). The nuclear receptor PPARβ/ δ programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev 25, 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, and Hager GL (2015). Transcriptional and Chromatin Regulation during Fasting - The Genomic Era. Trends Endocrinol. Metab 26, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Baek S, Presman DM, Paakinaho V, Swinstead EE, and Hager GL (2017). Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res 27, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Hartil K, Vaitheesvaran B, Arrieta-Cruz I, Knight CM, Cook JR, Kammoun HL, Febbraio MA, Gutierrez-Juarez R, Kurland IJ, and Accili D (2014). Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat. Commun 5, 5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. (2012). Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15, 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Zhou W, Fang B, Lu W, Loro E, Damle M, Ding G, Jager J, Zhang S, Zhang Y, et al. (2017). Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med 23, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, and Kornacker K (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Noshiro M, Furukawa M, Honda KK, Nakashima A, Ueshima T, Usui E, Katsura Y, Fujimoto K, Honma S, et al. (2006). Effects of fasting and re-feeding on the expression of Dec1, Per1, and other clock-related genes. J. Biochem 140, 401–408. [DOI] [PubMed] [Google Scholar]
- Kayala MA, and Baldi P (2012). Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res 40, W553–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, and Bass J (2007). High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6, 414–421. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Sher-Chen EL, and Green CB (2012). Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev 26, 2724–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, and Antoch MP (2006). Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 20, 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T, Lew MJ, Mayba O, Harris CA, Speed TP, and Wang JC (2012). Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc. Natl. Acad. Sci. USA 109, 11160–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton JK, Joyner J, Zamarripa J, Deines M, and Davis RA (1990). Fasting decreases apolipoprotein B mRNA editing and the secretion of small molecular weight apoB by rat hepatocytes: evidence that the total amount of apoB secreted is regulated post-transcriptionally. J. Lipid Res 31, 1663–1668. [PubMed] [Google Scholar]
- Lin J, Handschin C, and Spiegelman BM (2005). Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1, 361–370. [DOI] [PubMed] [Google Scholar]
- Longo VD, and Mattson MP (2014). Fasting: molecular mechanisms and clinical applications. Cell Metab 19, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D’Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, et al. (2017). Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab 25, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, and Sassone-Corsi P (2010). Plasticity and specificity of the circadian epigenome. Nat. Neurosci 13, 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Valomon A, Chesham JE, Maywood ES, Hastings MH, and Ule J (2012). Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol 13, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan G, Romanello V, Pescatore F, Armani A, Paik JH, Frasson L, Seydel A, Zhao J, Abraham R, Goldberg AL, et al. (2015). Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun 6, 6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, and Spiegelman BM (2000). Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6, 307–316. [DOI] [PubMed] [Google Scholar]
- Montagner A, Polizzi A, Fouche´ E, Ducheix S, Lippi Y, Lasserre F, Barquissau V, Re´ gnier M, Lukowicz C, Benhamed F, et al. (2016). Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 65, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, and Chambon P (2015). Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc. Natl. Acad. Sci. USA 112, E6683–E6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, and Sassone-Corsi P (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, and Sassone-Corsi P (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Shirai H, and Ishida N (2005). CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARαlpha) in mice. Biochem. J 386, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VR, Eckel-Mahan K, Sassone-Corsi P, and Baldi P (2012). CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat. Methods 9, 772–773. [DOI] [PubMed] [Google Scholar]
- Pearen MA, Ryall JG, Lynch GS, and Muscat GE (2009). Expression profiling of skeletal muscle following acute and chronic beta2-adrenergic stimulation: implications for hypertrophy, metabolism and circadian rhythm. BMC Genomics 10, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MC, Dufour CR, Tam IS, B’chir W, and Giguère V (2014). Estrogen-related receptor-a coordinates transcriptional programs essential for exercise tolerance and muscle fitness. Mol. Endocrinol 28, 2060–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskjaer K, Hogan MF, Lackey D, Tora L, Dent SY, Olefsky J, and Montminy M (2013). Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. J. Clin. Invest 123, 4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, and Puigserver P (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118. [DOI] [PubMed] [Google Scholar]
- Sato M, Murakami M, Node K, Matsumura R, and Akashi M (2014). The role of the endocrine system in feeding-induced tissue-specific circadian entrainment. Cell Rep 8, 393–401. [DOI] [PubMed] [Google Scholar]
- Schibler U, and Sassone-Corsi P (2002). A web of circadian pacemakers. Cell 111, 919–922. [DOI] [PubMed] [Google Scholar]
- Settembre C, and Ballabio A (2014). Lysosome: regulator of lipid degradation pathways. Trends Cell Biol 24, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. (2013). TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol 15, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavlakadze T, Anwari T, Soffe Z, Cozens G, Mark PJ, Gondro C, and Grounds MD (2013). Impact of fasting on the rhythmic expression of myogenic and metabolic factors in skeletal muscle of adult mice. Am. J. Physiol. Cell Physiol 305, C26–C35. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, and Okamura H (1997). Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Sobel JA, Krier I, Andersin T, Raghav S, Canella D, Gilardi F, Kalantzi AS, Rey G, Weger B, Gachon F, et al. ; CycliX Consortium (2017). Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol 15, e2001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, and Menaker M (2001). Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Dang F, Zhang D, Yuan Y, Zhang C, Wu Y, Wang Y, and Liu Y (2015). Glucagon-CREB/CRTC2 signaling cascade regulates hepatic BMAL1 protein. J. Biol. Chem 290, 2189–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K, and Sassone-Corsi P (2009). CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol 16, 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima H, Yoshitane H, Ozaki H, Suzuki Y, Shimba S, Kuroda S, Iwasaki W, and Fukada Y (2017). ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet 49, 146–151. [DOI] [PubMed] [Google Scholar]
- Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, Baldi P, and Sassone-Corsi P (2017). Distinct Circadian Signatures in Liver and Gut Clocks Revealed by Ketogenic Diet. Cell Metab 26, 523–538.e5. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, and Panda S (2009). Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA 106, 21453–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, and Schibler U (1990). Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell 63, 1257–1266. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, et al. (2016). Metabolic Regulation of Gene Expression by Histone Lysine b-Hydroxybutyrylation. Mol. Cell 62, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, and FitzGerald GA (2016). Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med 8, 324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. (2005). Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102, 4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, and Hogenesch JB (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA 111, 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. (2015). GENE REGULATION. Discrete functions of nuclear receptor Rev-ERBα couple metabolism to the clock. Science 348, 1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Damle M, Guan D, Li Z, Kim YH, Gannon M, and Lazar MA (2016). HNF6 and Rev-ERBα integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev 30, 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for the RNA-seq data reported in this paper is GSE107787.