Abstract
CYP2A5 is a major enzyme responsible for nicotine and cotinine metabolism in mice. Nicotine and cotinine enhance alcoholic fatty liver in wild type (WT) mice but not in CYP2A5 knockout (KO) mice, and reactive oxygen species (ROS) generated during the CYP2A5-mediated metabolism contributes to the enhancing effect. In combination with ethanol, nicotine and cotinine increased lipid peroxidation end product 4-hydroxynonenal (HNE) in WT mice but not in KO mice. In ethanol-fed KO mice, only 5 and 10 genes were regulated by nicotine and cotinine, respectively. However, in ethanol-fed WT mice, 59 and 104 genes were regulated by nicotine and cotinine, respectively, and 7 genes were up-regulated by both nicotine and cotinine. Plin 2 and Cdkn1a are among the 7 genes. Plin2 encodes adipose differentiation-related protein (ADRP), a lipid droplet-associated protein, which was confirmed to be increased by nicotine and cotinine in WT mice but not in KO mice. Cdkn1a encodes P21 and elevated P21 in nuclei was also confirmed. HNE can increase P21 and P21 inhibit cell proliferation. Consistently, hepatocyte proliferation markers proliferating cell nuclear antigen (PCNA) and Ki67 were decreased in WT mice but not in KO mice by nicotine/ethanol and cotinine/ethanol, respectively. These results suggest that inhibition of liver proliferation via a ROS-HNE-P21 pathway is involved in nicotine- and cotinine-enhanced alcoholic fatty liver.
Keywords: ROS, HNE, P21, Plin2, Ki67, PCNA, CYP2A5
INTRODUCTION
Alcohol-associated liver disease (AALD) is a spectrum of liver disorders caused by heavy alcohol drinking. After alcohol drinking, fatty liver (hepatic steatosis) is induced by inhibiting peroxisome proliferator-activated receptor α activity, by decreasing the activities of AMP-activated protein kinase and sirtuin-1, or by inducing sterol regulatory element-binding protein-1 activity (1). Initially, steatosis was considered benign, later it was recognized that steatosis may make liver vulnerable to other hepatotoxins or further develop inflammation (steatohepatitis) followed by fibrosis and cirrhosis (2). The development and progression of AALD might be caused by inhibiting natural killer cells or activating Kupffer cells and complements (3).
Being induced by chronic ethanol consumption, microsomal ethanol oxidizing system is mainly composed of cytochrome P450s especially CYP2E1 (4). CYP2E1-mediated oxidative stress contributes to AALD (5,6). Alcoholic fatty liver was developed in wild type mice but not in the CYP2E1 knockout mice (7); in the CYP2E1 knockout mice reconstituted with human CYP2E1, alcoholic fatty liver was recovered and alcoholic steatohepatitis was developed (8), suggesting an essential role of CYP2E1 in the development of AALD. It is generally accepted that CYP2E1 is a risk factor for the development of AALD (5, 6).
Human CYP2A6 and its mouse ortholog CYP2A5 can also be induced by alcohol consumption (9,10). CYP2A6 and CYP2A5 are major enzymes that metabolize nicotine (11–15). Alcohol and tobacco are frequently co-abused (16, 17). Cigarette smoke exposure promotes alcoholic cirrhosis in patients (18) and enhances experimental alcoholic fatty liver in mice (19). Nicotine, a major stimulant and addiction-forming alkaloid in tobacco smoke, can also enhance alcoholic fatty liver in mice but it didn’t enhance alcohol-induced liver inflammation (20). Recently we found that cotinine, a major metabolite of nicotine can also enhance alcoholic fatty liver, and nicotine- and cotinine-enhanced alcoholic fatty liver were not observed in CYP2A5 knockout (KO) mice while the fatty liver was enhanced in CYP2A5 wild type (WT) mice, suggesting a pivotal role of CYP2A5 (21).
AALD is associated with impaired liver regeneration (22). The liver regeneration is inhibited in alcoholic cirrhotic patients (23,24). Steatosis has an impact on inhibited liver regeneration, but steatosis is not enough to inhibit liver regeneration (25, 26). Oxidative stress is identified as a mechanism involved in the inhibited regeneration of fatty livers (25). During nicotine and cotinine metabolism, CYP2A5-dependent reactive oxygen species (ROS) generation and lipid peroxidation (LPO) might contribute to the CYP2A5-dependent enhancing effects (21). In this study, we performed liver differential gene expression analyses from RNA-seq data. We found that Cdkn1a gene encoding P21, a cell proliferation inhibiting protein, was upregulated by both nicotine and cotinine in WT mice but not in KO mice. Consistent with the upregulation of P21, as indicated by Ki67 and proliferating cell nuclear antigen (PCNA), hepatocyte proliferation was inhibited by both nicotine and cotinine in WT mice but not in KO mice. LPO end product 4-Hydroxynonenal (HNE) was reported to be able to increase P21 expression and inhibit cell proliferation (27). Consistently, we observed an increased formation of HNE by nicotine and cotinine in ethanol-fed WT mice but not in KO mice.
METHODS
Animals
Female, 8–10 weeks old CYP2A5 KO mice and WT mice were selected for experiment. All the mice were housed in temperature-controlled animal facilities with 12-hour light/dark cycles and were permitted consumption of tap water and Purina standard chow ad libitum. The mice received humane care, and experiments were carried out according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals. The animal studies were approved by University Committee on Animal Care at East Tennessee State University. Before experiments, the mice were fed the control liquid dextrose diet (Bio-Serv, Frenchtown, NJ, USA) for 3 days to acclimate them to the liquid diet. Then these mice were either fed the liquid ethanol diet (Bio-Serv, Frenchtown, NJ, USA) or fed the liquid dextrose diet as a control. Nicotine hydrogen tartrate salt was mixed in the liquid ethanol diet at 30 mg/L and cotinine was mixed in the liquid ethanol diet at 9 mg/L. The content of ethanol was gradually increased every 3 days from 10% to15%, 20%, 25%, 30%, and finally 35% of total calories. After 18 days of ethanol feeding, the mice were sacrificed after a 6 h of fast. The liver aliquots from same lobes of different mice were put in RNAlater for extracting RNA for RNA-seq analysis, or put in neutral Formalin buffer for preparing paraffin sections for immunohistochemistry staining (IHC).
RNA Sequencing
Liver samples in RNALater were sent to Macrogen (Rockville, MD, USA) for RNA extraction, RNA quality control, cDNA library construction and sequencing. Bioinformatics quality control Differential gene expression analysis was performed by Yotta Biomed, LLC (Potomac, MD, USA) using FastQC, version 0.11.7. The raw reads were aligned to mm10 reference genome using STAR, version 2.6.1a. The number of reads was mapped to genes using htseq, version 0.11.0. Differential gene expression analysis was performed using DESeq2, version 1.20.0 with the cutoff of 0.05 on False Discovery Rate (FDR) with Independent Hypothesis Weighting. R version 3.5.1 (2018-07-02) was used, and Bioconductor version 3.7 with BiocInstaller version 1.30.0 were used.
Immunohistochemistry
Liver paraffin sections were used for IHC. ADRP/Plin2, P21, PCNA and Ki67 were detected by IHC using anti-ADRP (Abcam, Cat#: ab52356), anti-P21 (Abcam, Cat#: ab188224), anti-PCNA (Santa Cruz Biotechnology, Cat#: sc-32757), anti-Ki67 (Santa Cruz Biotechnology, Cat#: sc-23900) and anti-HNE (R&D Systems, Cat# MAB3249SP) followed by a Broad Spectrum IHC Select® HRP/DAB kit (from EMD Millipore, Cat#: DAB150). ADRP is mainly located around lipid droplets in the liver sections. HNE is mainly located around central veins. Quantification for HNE was made by calculating the percentage of positive staining in all central veins. P21, PCNA, and Ki67 are detected in cell nuclei. Quantification for P21, PCNA, and Ki67 was made by counting the positively stained nuclei in 40x bright field under microscope.
Western Blotting analysis
Liver homogenates were prepared and SDS-PAGE and chemiluminescence imaging was carried out as described before (7–10). ADRP was detected in whole cell tissue protein and P21 was detected in nuclear protein. Liver nuclear protein was extracted using NE-PER Nucear and Cytoplasmic Extraction Reagent (Thermo Scientific, Prod# 78835). β-actin and lamin B were detected in whole cell tissue protein and nuclear protein as loading controls, respectively.
Statistical analysis
Results are expressed as mean ± S.D. Statistical evaluation was carried out by using one-way analysis of variance with subsequent the Student-Newman-Keuls post hoc test. P<0.05 was considered as statistical significance.
RESULTS
Both nicotine and cotinine up-regulates Plin2 and Cdkn1a genes in ethanol-fed WT mice but not in ethanol-fed KO mice
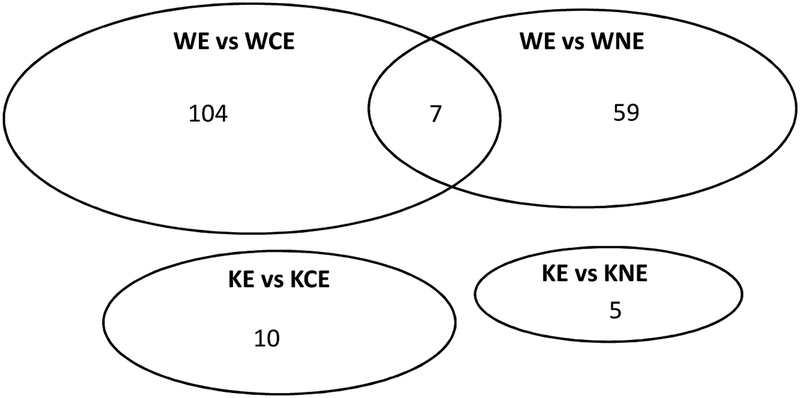
Previously we reported that nicotine and cotinine enhanced alcoholic fatty liver in WT mice but not in KO mice as evaluated by Hematoxylin & Eosin staining, Oil Red O staining, and liver triglyceride (TG) biochemical measurement (21). To further identify the mechanisms by which nicotine and cotinine enhance alcoholic fatty liver, differential gene expression analysis was performed using the RNA sequencing data. When alcohol/nicotine and alcohol/cotinine were compared with alcohol alone, 38 and 43 genes were up-regulated and 21 and 61 genes were down-regulated in WT mice; but only 4 and 5 genes were up-regulated and 1 and 5 genes were down-regulated in KO mice, respectively (Table 1). Among those 38 and 43 up-regulated genes in WT mice, 7 genes were up-regulated by both nicotine and cotinine in ethanol-fed WT mice but not in KO mice (Fig. 1, Table 2). In ethanol-fed WT mice, no gene was down-regulated by both nicotine and cotinine. Considering that both nicotine and cotinine can enhance alcoholic fatty liver in WT mice but not in KO mice, it is plausible that one or more of the 7 genes play a pivotal role in the nicotine- and cotinine-enhancing effect on alcoholic fatty liver. Among those 7 genes, Plin 2 and Cdkn1a were selected for further study. Plin2 encodes adipose differentiation related protein (ADRP), a marker of high fat diet-induced fatty liver (28). Actually, Plin2 mRNA was elevated by ethanol alone in both WT mice and KO mice (data not shown), but ethanol combined with nicotine or cotinine further increased Plin2 in WT mice but not in KO mice (Table 2). Upregulation of Plin2 gene was confirmed by increased hepatic expression of ADRP by nicotine and cotinine in WT mice but not in KO mice (Fig. 2 A). IHC showed that many sinusoid cells in all groups were positively stained with ADRP, which was evenly distributed in cytoplasm (Fig. 2B). These positively stained sinusoidal cells are probably hepatic stellate cells which contain small lipid droplets storing retinol (29). In nicotine/alcohol and cotinine/alcohol groups of WT mice, lipid droplets were surrounded by ADRP in hepatocytes (Fig. 2B). These results suggest that ADRP is also a marker of nicotine- and cotinine-enhanced alcoholic fatty liver.
Table 1.
The number of genes up-regulated and down-regulated in Nicotine/Ethanol and Cotinine/Ethanol groups when compared with Ethanol alone
| Comparison | Up-regulated | Down-regulated | Total |
|---|---|---|---|
| WT EthanoI vs WT Nicotine/Ethanol | 38 | 21 | 59 |
| WT Ethanol vs WT Cotinine/Ethanol | 43 | 61 | 104 |
| KO Ethanol vs KO Nicotine/Ethanol | 4 | 1 | 5 |
| KO Ethanol vs KO Cotinine/Ethanol | 5 | 5 | 10 |
Figure 1.
The number of genes that were up-regulated or down-regulated in Nicotine/Ethanol and Cotinine/Ethanol groups when compared with Ethanol groups in WT mice and KO mice. WE, WT Ethanol; WNE, WT Nicotine+Ethanol; WCE, WT Cotinine+Ethanol; KE, KO Ethanol; KNE, KO Nicotine+Ethanol; KCE, KO Cotinine+Ethanol.
Table 2.
The 7 genes up - regulated in Nicotine/Ethanol and Cotinine/Ethanol groups were only observed in WT mice but not in KO mice
| Gene | Ethanol vs Nicotine/Ethanol | Ethanol vs Cotinine/Ethanol | ||
|---|---|---|---|---|
| log2FoldChange | padj | log2FoldChange | padj | |
| Plin2 | −0.990068449 | 0.042329495 | −0.818311894 | 0.000184317 |
| Cdkn1a | −2.036431864 | 0.033638921 | −2.913107961 | 0.036949565 |
| Haus8 | −1.89789859 | 0.02491449 | −1.367318125 | 0.008570789 |
| Chchd10 | −0.711896364 | 0.002858506 | −0.526557686 | 0.00169264 |
| Slc18a1 | −2.140699664 | 6.82E-05 | −1.684747193 | 0.004863641 |
| Ifit3 | −1.018860029 | 0.044612117 | −0.938051923 | 0.007718369 |
| 2010003K11Rik | −1.707657595 | 6.82E-05 | −1.254458756 | 0.005462912 |
Padj, adjusted P value.
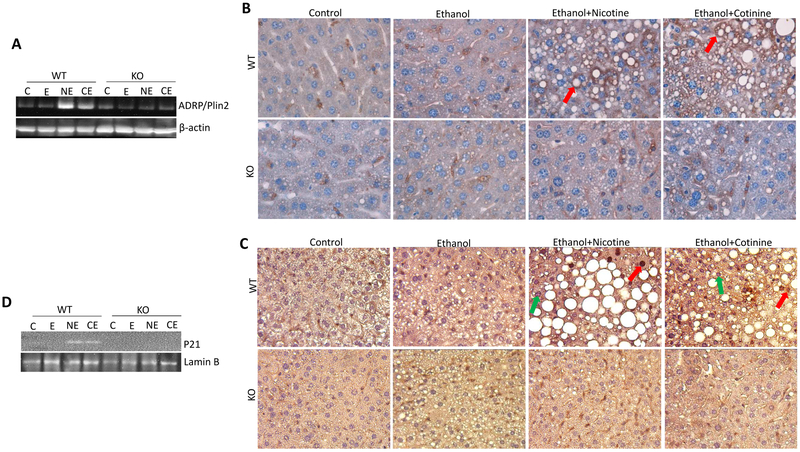
Figure 2.
ADRP/Plin2 and P21 were upregulated by nicotine and cotinine in ethanol-fed WT mice but not in KO mice. (A) Liver expression of ADRP/Plin2 detected by Western blotting analysis. (B) IHC staining for ADRP/Plin2. Red arrows show stained ADRP/Plin2 surrounding the lipid droplets. (C) IHC staining for P21. Red arrows show positively stained nuclei and green arrows show negatively stained nuclei. (D) Nuclear P21 content detected by Western blotting analysis.
Cdkn1a encodes P21. Nuclear positive staining for P21 in hepatocytes was only observed in nicotine/alcohol and cotinine/alcohol groups of WT mice (Fig. 2C), Consistently, Western blotting analysis showed that P21 was only detected in nuclear protein from nicotine/alcohol and cotinine/alcohol groups of WT mice (Fig. 2D). These results suggest that P21 was induced by nicotine and cotinine in combination with ethanol in WT mice but not in KO mice.
Both nicotine and cotinine inhibit liver regeneration in ethanol-fed WT mice but not in KO mice
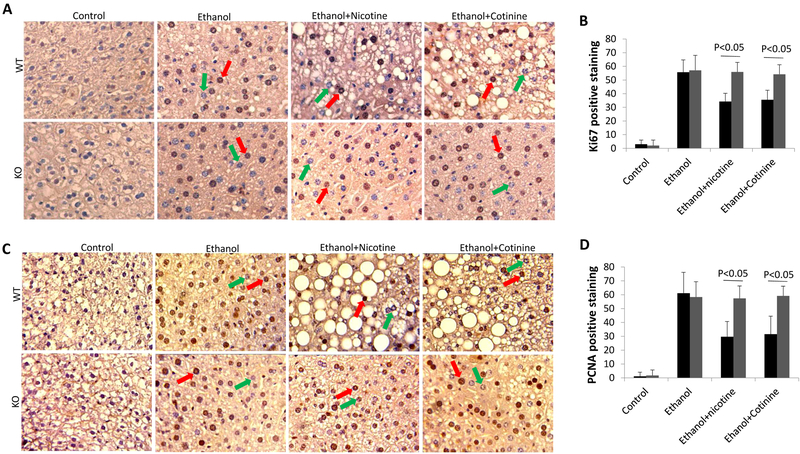
P21 is an inhibitor of cell proliferation (30,31). In nuclei, P21 directly binds with PCNA to inhibit DNA replication (32,33). Up-regulation of P21 may inhibit liver regeneration. Here, Ki67 and PCNA were detected as markers of hepatocyte proliferation. As shown in Fig. 3, nuclear Ki67 staining and PCNA staining were increased by ethanol feeding in both WT mice and KO mice, but nicotine and cotinine blunted the increased Ki67 and PCNA staining in WT mice but not in KO mice, suggesting that nicotine and cotinine inhibit liver regeneration in ethanol-fed WT mice but not in ethanol-fed KO mice.
Figure 3.
Liver regeneration was inhibited by nicotine and cotinine in combination with ethanol in WT mice but not in KO mice. (A) IHC staining for Ki67. Red arrows show positively stained nuclei and green arrows show negatively stained nuclei. (B) Quantification of Ki67 staining. Black bar indicating WT mice, and gray bar indicating KO mice. (C) IHC staining for PCNA. Red arrows show positively stained nuclei and green arrows show negatively stained nuclei. (D) Quantification of PCNA staining. Black bar indicating WT mice, and gray bar indicating KO mice.
Both nicotine and cotinine enhance HNE formation in ethanol-fed WT mice but not in KO mice
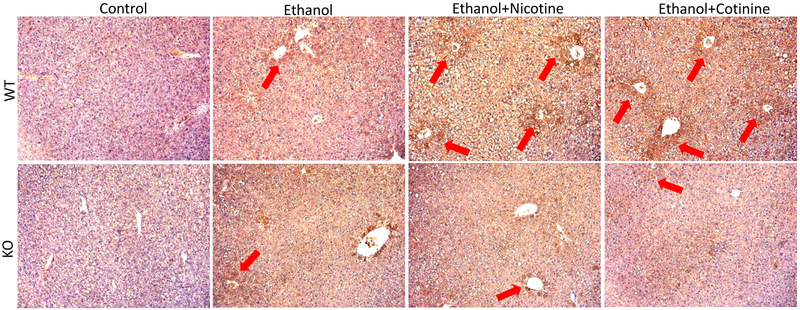
LPO end product HNE inhibits cell proliferation (34,35). Under metabolism by CYP2A5, both nicotine and cotinine can produce ROS and enhance formation of MDA, another LPO end product, which was observed in WT mice but not in KO mice (21). Here we further detected HNE. As shown in Fig. 4A, HNE was induced by ethanol feeding in both WT mice and KO mice, and about half of central vein areas were positively stained by HNE. When ethanol is in combination with nicotine or cotinine, almost all the central vein areas were positively stained by HNE in WT mice; but in KO mice, it was still about half of central vein areas positively stained by HNE. These results suggest that HNE formation was enhanced by nicotine and cotinine in WT mice but not in KO mice.
Figure 4.
IHC staining for HNE in ethanol and ethanol in combination with nicotine and cotinine groups. Red arrows show positive HNE staining.
DISCUSSION
It is well known that liver regeneration is inhibited in AALD (22). In this study we found that impaired liver regeneration is also associated with nicotine- and cotinine-enhanced alcoholic fatty liver, which is probably through elevating P21, a cell cycle inhibitor. LPO end product HNE is a P21 inducer (27) and nicotine was reported to inhibit cell growth by increasing expression of P21 in oral keratinocytes (36). Previously we reported that metabolism of nicotine and cotinine by CYP2A5 can produce ROS and induce LPO (21). In this study HNE was detected to be increased by nicotine and cotinine in combination with ethanol in WT mice but not in KO mice. Consistently, P21 was also increased by nicotine and cotinine in WT mice but not in KO mice. Therefore, nicotine and cotinine suppressed liver regeneration probably via a ROS-HNEP21 pathway.
TG is synthesized in endoplasmic reticulum (ER). In the ER of the liver, synthesized TG binds to ADRP/Plin2 and form steatosis as we showed in Fig. 2 B. CYP2A5 is located in ER, so ROS is also produced in ER during CYP2A5-mediated nicotine metabolism. Thus, ROS in ER may somehow make TG bind to upregulated ADRP/Plin2 and enhance fatty liver. P21 has an antioxidant function to protect cells against oxidative damage. In response to oxidative stress, p21 is upregulated to promote cell survival through activation of the Nrf2 signaling pathway (37). Therefore, p21 might be a compensation in response to ROS to protect against fatty liver, which probably occurs in cytoplasm. P21 may also translocate into nuclei to inhibit cell proliferation and cause cell senescence (30, 31). Hepatocyte senescence predicts progression of fatty liver disease (38, 39). However, cause-effect relationship between P21 and fatty liver disease needs further studies using p21 KO mice.
In conclusion, nicotine and cotinine enhance alcoholic fatty liver in a CYP2A5-dependent manner. During the process of CYP2A5-dependent metabolism of nicotine and cotinine, generated ROS induces LPO resulting in increased HNE, which in turn increases P21 and cause impaired hepatocyte proliferation. However, cause-effect relationship between liver regeneration and the nicotine– and cotinine-enhanced alcoholic fatty liver still need a further study.
Supplementary Material
7 genes were increased by nicotine and cotinine and Plin 2 and Cdkn1a were focused;
Plin 2 encoded ADRP was increased and mainly surrounded lipid droplets;
Cdkn1a encoded P21, a cell cycle inhibitor, was confirmed to be elevated in nuclei;
LPO end product HNE was detected to be increased by nicotine and cotinine
A ROS-HNE-P21 pathway might be involved in the impaired liver regeneration.
Acknowledgement
We thank Dr. Xinxin Ding for the CYP2A5 KO mice.
This work was supported by the National Institutes of Health [grant numbers R01AA024723, C06RR0306551].
Abbrieviation:
- AALD
alcohol-associated liver disease
- ADRP
adipose differentiation-related protein
- ER
endoplasmic reticulum
- HNE
4-hydroxynonenal
- IHC
immunohistochemistry staining
- LPO
lipid peroxidation
- PCNA
proliferating cell nuclear antigen
- ROS
reactive oxygen species
- TG
triglyceride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
REFERENCES
- 1.Purohit V, Gao B, Song BJ. Molecular mechanisms of alcoholic fatty liver. Alcohol Clin Exp Res. 2009. February;33(2):191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacSween RNM, Burt AD. Histologic spectrum of alcoholic liver disease. Semin. Liver Dis, 6 (1986), pp. 221–232 [DOI] [PubMed] [Google Scholar]
- 3.Miller AM, Horiguchi N, Jeong WI, Radaeva S, Gao B. Molecular mechanisms of alcoholic liver disease: innate immunity and cytokines. Alcohol Clin Exp Res. 2011. May;35(5):787–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber CS. (2005). Metabolism of alcohol. Clin Liver Dis. 9, 1–35. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Cederbaum AI. (2008). CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 44, 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cederbaum AI, Lu Y, Wu D. Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol. 2009. June;83(6):519–48 [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Zhuge J, Wang X, Bai J, Cederbaum AI. Cytochrome P450 2E1 contributes to ethanol-induced fatty liver in mice. Hepatology. 2008. May;47(5):1483–94 [DOI] [PubMed] [Google Scholar]
- 8.Lu Y, Wu D, Wang X, Ward SC, Cederbaum AI. Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in humanized cytochrome P4502E1 knock-in mice. Free Radic Biol Med. 2010. November 15;49(9):1406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Zhuge J, Wu D, Cederbaum AI. (2011). Ethanol induction of CYP2A5: permissive role for CYP2E1. Drug Metab Dispos. 39, 330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Zhang XH, Cederbaum AI. (2012). Ethanol induction of CYP2A5: role of CYP2E1-ROS-Nrf2 pathway. Toxicol Sci. 128, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matta SG, Balfour DJ, Benowitz NL, et al. (2007). Guidelines on nicotine dose selection for in vivo research. Psychopharmacology. 190, 269–319. [DOI] [PubMed] [Google Scholar]
- 12.Messina ES, Tyndale RF, Sellers EM. (1997). A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 282, 1608–1614. [PubMed] [Google Scholar]
- 13.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, Funae Y, Shimada N, Kamataki T, Kuroiwa Y. (1996). Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 24, 1212–1217. [PubMed] [Google Scholar]
- 14.Raunio H, Pokela N, Puhakainen K, Rahnasto M, Mauriala T, Auriola S, Juvonen RO. (2008). Nicotine metabolism and urinary elimination in mouse: in vitro and in vivo. Xenobiotica. 38, 34–47. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X, Zhuo X, Xie F, Kluetzman K, Shu YZ, Humphreys WG, Ding X. (2010). Role of CYP2A5 in the clearance of nicotine and cotinine: insights from studies on a Cyp2a5-null mouse model. J Pharmacol Exp Ther. 332, 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sellers EM, Tyndale RF, Fernandes LC. (2003). Decreasing smoking behavior and risk through CYP2A6 inhibition. Drug Discov Today. 8, 487–493. [DOI] [PubMed] [Google Scholar]
- 17.Schoedel KA, Tyndale RF. (2003). Induction of nicotine-metabolizing CYP2B1 by ethanol and ethanol-metabolizing CYP2E1 by nicotine: summary and implications. Biochim Biophys Acta. 1619, 283–290. [DOI] [PubMed] [Google Scholar]
- 18.Klatsky AL, Armstrong MA. (1992). Alcohol, smoking, coffee, and cirrhosis. Am J Epidemiol. 136, 1248–1257. [DOI] [PubMed] [Google Scholar]
- 19.Bailey SM, Mantena SK, Millender-Swain T, Cakir Y, Jhala NC, Chhieng D, Pinkerton KE, Ballinger SW. (2009). Ethanol and tobacco smoke increase hepatic steatosis and hypoxia in the hypercholesterolemic apoE(−/−) mouse: implications for a “multihit” hypothesis of fatty liver disease. Free Radic Biol Med. 46, 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y, Ward SC, Cederbaum AI. Nicotine enhances ethanol-induced fat accumulation and collagen deposition but not inflammation in mouse liver. Alcohol. 2013. August;47(5):353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Owoseni E, Salamat J, Cederbaum AI, Lu Y. Nicotine enhances alcoholic fatty liver in mice: Role of CYP2A5. Arch Biochem Biophys. 2018. November 1;657:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French SW, Liao G, Li J, Vitocruz E, Liu H, Tillman B, French BA, Mendoza AS. What are the mechanisms of regeneration inhibition in alcoholic hepatitis? Exp Mol Pathol. 2016. June;100(3):502–5 [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Bao H, Sawyer S, Kunos G, Gao B. Effects of short and long term ethanol on the activation of signal transducer and activator transcription factor 3 in normal and regenerating liver. Biochem Biophys Res Commun. 1997. October 29;239(3):666–9. [DOI] [PubMed] [Google Scholar]
- 24.Horiguchi N1, Ishac EJ, Gao B. Liver regeneration is suppressed in alcoholic cirrhosis: correlation with decreased STAT3 activation. Alcohol. 2007. June;41(4):271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allaire M, Gilgenkrantz H. The impact of steatosis on liver regeneration. Horm Mol Biol Clin Investig. 2018. November 21 pii: /j/hmbci.ahead-of-print/hmbci-2018-0050/hmbci-2018-0050.xml. doi: 10.1515/hmbci-2018-0050. [Epub ahead of print] Review. [DOI] [PubMed] [Google Scholar]
- 26.Picard C, Lambotte L, Starkel P, Sempoux C, Saliez A, Van den Berge V, et al. Steatosis is not sufficient to cause an impaired regenerative response after partial hepatectomy in rats. J Hepatol. 2002;36:645–52. [DOI] [PubMed] [Google Scholar]
- 27.Laurora S, Tamagno E, Briatore F, Bardini P, Pizzimenti S, Toaldo C, Reffo P, Costelli P, Dianzani MU, Danni O, Barrera G. 4-Hydroxynonenal modulation of p53 family gene expression in the SK-N-BE neuroblastoma cell line. Free Radic Biol Med. 2005. January 15;38(2):215–25. [DOI] [PubMed] [Google Scholar]
- 28.Motomura W, Inoue M, Ohtake T, Takahashi N, Nagamine M, Tanno S, Kohgo Y, Okumura T. Up-regulation of ADRP in fatty liver in human and liver steatosis in mice fed with high fat diet. Biochem Biophys Res Commun. 2006. February 24;340(4):1111–8. [DOI] [PubMed] [Google Scholar]
- 29.Shmarakov IO, Jiang H, Liu J, Fernandez EJ, Blaner WS. Hepatic stellate cell activation: A source for bioactive lipids. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. February 5;1864(5):629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998. November 20;282(5393):1497–501. [DOI] [PubMed] [Google Scholar]
- 31.Smits VA, Klompmaker R, Vallenius T, Rijksen G, Mäkela TP, Medema RH. p21 inhibits Thr161 phosphorylation of Cdc2 to enforce the G2 DNA damage checkpoint. J Biol Chem. 2000. September 29;275(39):30638–43. [DOI] [PubMed] [Google Scholar]
- 32.Flores-Rozas H, et al. (1994). “Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme”. Proceedings of the National Academy of Sciences. 91(18): 8655–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waga S, et al. (1994). “The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA”. Nature. 369 (6481): 574–578 [DOI] [PubMed] [Google Scholar]
- 34.Barrera G, Pizzimenti S, Laurora S, Briatore F, Toaldo C, Dianzani MU. 4-hydroxynonenal and cell cycle. Biofactors. 2005;24(1–4):151–7. [DOI] [PubMed] [Google Scholar]
- 35.Barrera G, Pizzimenti S, Dianzani MU. 4-hydroxynonenal and regulation of cell cycle: effects on the pRb/E2F pathway. Free Radic Biol Med. 2004. September 1;37(5):597–606. [DOI] [PubMed] [Google Scholar]
- 36.Lee HJ, Guo HY, Lee SK, Jeon BH, Jun CD, Lee SK, Park MH, Kim EC. Effects of nicotine on proliferation, cell cycle, and differentiation in immortalized and malignant oral keratinocytes. J Oral Pathol Med. 2005. August; 34(7):436–43. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009. June 26;34(6):663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aravinthan A, Scarpini C, Tachtatzis P, Verma S, Penrhyn-Lowe S, Harvey R, Davies SE, Allison M, Coleman N, Alexander G. Hepatocyte senescence predicts progression in non-alcohol-related fatty liver disease. J Hepatol. 2013. March;58(3):549–56. [DOI] [PubMed] [Google Scholar]
- 39.Aravinthan A, Pietrosi G, Hoare M, Jupp J, Marshall A, Verrill C, Davies S, Bateman A, Sheron N, Allison M, Alexander GJ. Hepatocyte expression of the senescence marker p21 is linked to fibrosis and an adverse liver-related outcome in alcohol-related liver disease. PLoS One. 2013. September 23;8(9):e72904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.