Abstract
Inversion polymorphisms are responsible for many ecologically important phenotypes, and are often found under balancing selection. However, the same features that ensure their large role in local adaptation—especially reduced recombination between alternate arrangements—mean that uncovering the precise loci within inversions that control these phenotypes is unachievable using standard mapping approaches. Here we take advantage of long-term balancing selection on a pair of inversions in the mosquito Anopheles gambiae to map desiccation tolerance via pool-GWAS. Two polymorphic inversions on chromosome 2 of this species (denoted 2La and 2Rb) are associated with arid and hot conditions in Africa, and are maintained in spatially and temporally heterogeneous environments. After measuring thousands of wild-caught individuals for survival under desiccation stress, we used phenotypically extreme individuals homozygous for alternative arrangements at the 2La inversion to construct pools for whole-genome sequencing. Genome-wide association mapping using these pools revealed dozens of significant SNPs within both 2La and 2Rb, many of which neighbored genes controlling ion channels or related functions. Our results point to the promise of similar approaches in systems with inversions maintained by balancing selection, and provide a list of candidate genes underlying the specific phenotypes controlled by the two inversions studied here.
Keywords: Balancing selection, recombination, local adaptation, aridity tolerance
Introduction
Mounting evidence suggests that chromosomal inversion polymorphisms play an important role in local adaptation (Hoffmann & Rieseberg 2008; Krimbas & Powell 1992; Wellenreuther & Bernatchez 2018). The pioneering work of Dobzhansky on natural populations of Drosophila showed that selection maintained inversion polymorphisms, as evidenced by stable latitudinal clines and seasonal cycling of inversion frequencies (Dobzhansky 1970). A number of models have been proposed to explain the establishment and maintenance of inversion polymorphisms (Hoffmann & Rieseberg 2008; Kirkpatrick & Barton 2006). These models largely hinge on observations of reduced recombination between inverted versus standard orientations of the chromosomal rearrangement. Through their indirect effect on recombination, inversion polymorphisms preserve sets of locally favored alleles in linkage disequilibrium (LD) and restrict their exchange with other genetic backgrounds, thereby facilitating adaptation to environmental heterogeneities.
Recent studies have implicated chromosomal inversions in environmental adaptation, reproductive isolation, and speciation not only in Drosophila but across taxonomically diverse groups, including a variety of insects, fish, birds, plants, and humans (Wellenreuther & Bernatchez 2018). Yet, despite decades of research, a detailed understanding of the genic targets of selection within inversions is lacking. Reduced recombination inside inversions renders classical genetic mapping (via controlled crosses) an impractical approach for identifying the targets of selection (Roberts 1976; Sturtevant 1917). Importantly, genetic exchange between the alternative arrangements is suppressed but not abrogated altogether. Both gene conversion and double crossovers (jointly termed “gene flux”) can occur and disrupt LD within the inversion, particularly away from the inversion breakpoints (Andolfatto et al. 2001; Navarro et al. 1997). Although recombination events between inverted and standard orientations are individually rare, gene flux has the potential to create a natural (uncontrolled) crossing experiment that in principle could allow the identification of loci under selection within inversions. All that is required is the presence of inversion heterozygotes, a large effective population size, and sufficient time to allow for gene flux.
The Anopheles gambiae species complex is an Afrotropical group of morphologically indistinguishable mosquitoes that radiated both rapidly and recently, within the last 2 million years (Fontaine et al. 2015). Despite retaining substantial amounts of shared ancestral sequence polymorphisms and partial interfertility, these very closely related species have distinguishing physiologies and behaviors that profoundly affect their roles in human malaria (White et al. 2011), a disease that disproportionately strikes Africa and claims the lives of ~400,000 annually (World Health Organisation 2017). To date, neither the relevant phenotypic differences between vector and non-vector species nor the genetic underpinnings of these phenotypes are understood in detail. However, evidence suggests that chromosomal inversion polymorphisms are important, either directly or indirectly. Within the species complex, inversion polymorphisms are abundant, but they are not distributed randomly across chromosomes or among species (Coluzzi et al. 2002; Pombi et al. 2008). Inversions are overrepresented on the second of three chromosomes, and the three major malaria vector species in the group—which are also those whose ranges extend across multiple biomes throughout most of tropical Africa—are the species that carry the vast majority of the common inversion polymorphisms. Non-vector species in the complex, or those whose roles are minor and localized, have much more limited distributions and lower (or no) inversion polymorphism (White et al. 2011). These patterns alone suggest that inversions confer adaptations to environmental heterogeneities, but evidence associated with individual inversions also supports their important role in adaptation.
In the nominal species, the major malaria vector An. gambiae s.s.—the subject of the present study—the frequencies of inversion polymorphisms on chromosome 2 correlate with abiotic variables such as latitude, altitude, seasonality and aridity (Cheng et al. 2012; Coluzzi et al. 1979; Petrarca et al. 1990; Rishikesh et al. 1985; Simard et al. 2009; Toure et al. 1998). Along latitudinal gradients of aridity in West and Central Africa, clines at inversions designated “In(2La)” and “In(2Rb)” are observed in which the frequencies of the inverted orientations are maximal at the northern, arid endpoints, and minimal at the southern, mesic endpoints, suggesting that the inverted orientations confer greater resistance to thermal and arid stress. This hypothesis has been supported by physiological testing in laboratory colonies (Fouet et al. 2012; Gray et al. 2009; Rocca et al. 2009). However, despite transcriptional profiling in the laboratory (Cassone et al. 2011; Cheng et al. 2018) and genome resequencing along a cline (Cheng et al. 2012), the mechanistic basis of this presumably adaptive advantage remains elusive.
In an antecedent study, we performed pooled sequencing of populations (Pool-Seq)(Schlotterer et al. 2014) at opposite ends and at the center of a latitudinal (climatic) cline of In(2La) and In(2Rb) in Cameroon (Cheng et al. 2012). We found significantly elevated differentiation between populations at opposite ends of the cline only in rearranged (not collinear) genomic regions, consistent with spatially varying selection maintaining the inversion cline in the face of unobstructed migration and population connectivity. There was no evidence for adaptive variation outside of In(2La) and In(2Rb). Importantly, nucleotide polymorphisms within the inversions that are fixed or strongly skewed for alternative alleles between endpoint populations showed patterns suggesting large amounts of gene flux between inverted and standard arrangements of In(2La) and In(2Rb) in populations at the center of the cline, where the inversions are at intermediate frequencies and inversion heterozygotes are abundant. Previous whole-genome sequencing of multiple species in the An. gambiae complex revealed that the In(2La) polymorphism originated prior to the species radiation, indicating that it has been polymorphic within populations for more than 2 million years (Fontaine et al. 2015), providing ample time for gene flux to homogenize variants not maintained by selection. Based on divergence outlier analysis we identified candidate genes inside In(2La) and In(2Rb) that might be involved in local adaptation, but could make no direct link with phenotype (Cheng et al. 2012).
Here, we exploit long-term gene flux between opposite orientations of the 22-Mb In(2La) and 7-Mb In(2Rb) rearrangements to fine-scale map genomic regions inside the inversions that contribute to variation in desiccation resistance—an ecologically relevant and fitness-related trait associated with these inversions (Fouet et al. 2012; Gray et al. 2009). Based on a modified Pool-GWAS approach originally developed by Bastide and colleagues (Bastide et al. 2013), we pooled karyotyped mosquitoes with extreme phenotypes and subjected them to Pool-Seq genotyping, in order to estimate allele frequency differences between phenotypic classes of each karyotype. Using replicated pools for both phenotypic extremes and both karyotype classes, we identified allelic variants statistically associated with heightened desiccation resistance across both karyotype classes. These variants are associated with genes whose functions are implicated in physiologies plausibly related to desiccation resistance, suggesting that we have identified changes underlying an adaptive inversion polymorphism.
Materials and Methods
Mosquito Collection
Collections were made between August and October 2012 in Cameroon, near the approximate center of a latitudinal cline of inversions In(2La) and In(2Rb), in a ~25 km2 area surrounding the town of Tibati (6.4703°N, 12.6188°E). In this area, three species within the An. gambiae sensu lato complex are sympatric (An. arabiensis, An. coluzzii, and An. gambiae s.s.), although the latter is the most abundant, reaching ~90% of mixed collections (Simard et al. 2009). At this latitude inversion polymorphism is maximal, ~50% (Simard et al. 2009).
Mosquitoes were collected as larvae by dipping into individual breeding sites. We targeted 3rd-4th instars to limit mortality and artificial selection. Larvae were transferred to the field insectarium facility until pupation, when they were placed into netted cages for adult emergence. Once emerged, adults were maintained without a sugar source to avoid potential bias due to differences in sugar feeding, in conformity with Fouet et al. (2012) and Gray et al. (2009). Adults emerging prior to 22:00h on a given day were discarded. Only those that emerged overnight (between 22:00h and 06:00h) were tested for desiccation resistance, to limit age disparity. In cases where the number of emergences exceeded the maximum that could be tested in one day (N=60), mosquitoes were randomly chosen with respect to breeding sites, locality, size, and gender. In total, we collected 6,148 A. gambiae larvae, and tested 2,850 adult males and females for desiccation tolerance.
Desiccation tolerance assays
To assess acute desiccation tolerance, we followed Fouet et al. (2012). Mosquitoes were individually placed in glass vials plugged with a foam rubber stopper, whose top was positioned at least 2cm below the rim. Once all mosquitoes were placed in vials, a desiccation agent (Drierite®) was added on top of the foam stopper, and the vials were sealed with Parafilm®. This procedure reduced the relative humidity to < 5 % in each vial (Testo™ 435 Multimeter). Our testing started upon sealing of the tubes, at 15:30h, and was terminated after 24 h.
A video recording system was employed to estimate post hoc the time to death, installed in an indoor room dedicated to this purpose. Three CCTV cameras were connected through an EZ Switch® System to a desktop computer equipped with a video recording system (EZ Watch Pro®). Each camera was able to record the behavior of up to 20 mosquitoes with a resolution of 704 × 480 pixels and 24 Hz. The room was continuously lit during the entire experiment. Temperature and relative humidity were continuously logged during the whole study. The time to death of each mosquito was measured after visual inspection of the video recordings. Dead mosquitoes were then individually stored in labeled 1.5 mL tubes with desiccant (Drierite®) until they were transported to the University of Notre Dame for further processing.
Species identification and Inversion 2La karyotyping
All mosquitoes tested in the present study were morphologically identified in the field as belonging to the An. gambiae s.l. complex (Gillies & De Meillon 1968). To determine species identity and inversion status, DNA-based assays were employed. DNA was extracted from one to two legs of each mosquito using CTAB (Morlais et al. 2004). A SINE-based PCR assay (Santolamazza et al. 2008) was used to distinguish An. gambiae from An. coluzzii and An. arabiensis. The PCR assay of White et al. (2007b) provides a reliable tool for molecular karyotyping of In(2La); unfortunately the available PCR assay for In(2Rb) is not sufficiently robust (Lobo et al. 2010). Hence, individual mosquitoes were molecularly karyotyped only with respect to In(2La).
Mosquito body mass
Dry body mass was determined by weighing individual desiccated mosquitoes using a microbalance system, either Mettler Toledo (acc. 0.2 μg) for females (processed at the University of Notre Dame) or a Sartorius-CPA2P (acc. 1 μg) for males (processed at the Institut de Recherche pour le Développement). Damaged mosquitoes were discarded from the analysis.
Identifying individuals to be genotyped
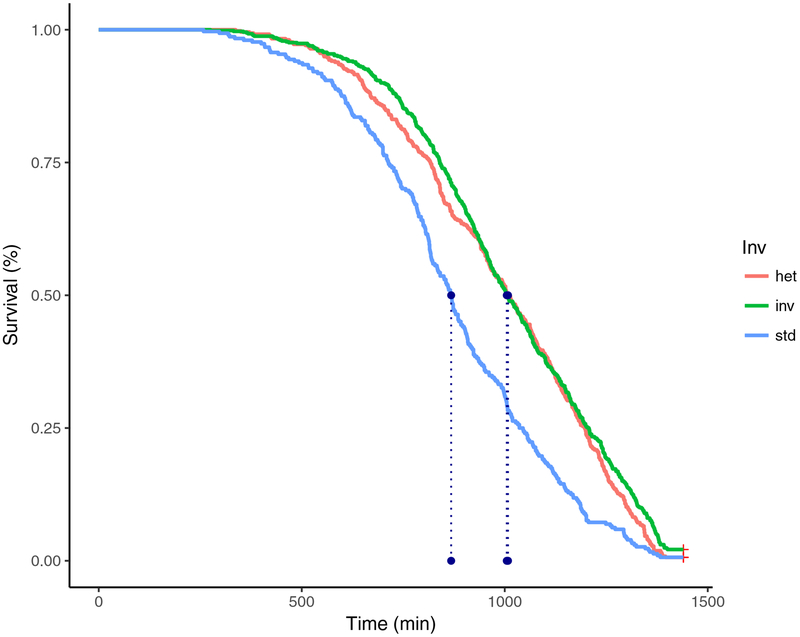
The mapping approach used here genotypes only those individuals with extreme phenotypes (cf. Bastide et al. 2013). As our goal is to map nucleotides associated with desiccation resistance inside In(2La) and In(2Rb), we also need to control for other general factors that may contribute to survival. It is well known from lab colonies that both sex and weight are important factors in desiccation resistance (Fouet et al. 2012), and here we were able to demonstrate these same effects among our wild-caught mosquitoes (Supporting information Figures S1, S2). Because of the large difference in survival between sexes, we chose to conduct mapping using only females; the larger body size of females also ensures enough DNA for genotyping. Survivorship curves of teneral An. gambiae females stratified by In(2La) karyotype are shown in Figure 1. In order to identify individuals with the most extreme resistance phenotypes due only to the karyotype of the 2La inversion they carry, we used the Cox proportional hazard model (Cox 1972) as implemented using the package “coxme” in R (www.r-project.org). We modeled survival using the covariates karyotype, weight (log-transformed), maximum temperature on the day of the experiment, day of experiment, breeding site, and vapor pressure deficit on the day of the experiment; model selection was carried out with the Akaike information criterion (AIC) using the function “aictab” from the “AICcmodavg” package in R. With respect to individuals homozygous for the standard arrangement of In(2La) (“STD”), we identified 50 with the highest survival and 50 with the lowest survival using the residual values from the full model. We similarly identified extreme individuals homozygous for the inverted arrangement (“INV”), 50 with the highest survival and 50 with the lowest survival using residuals.
Figure 1.
Survival of teneral adult female An. gambiae under acute desiccation stress. Data are stratified by In(2La) karyotype. Dashed lines represent 50% survivorship.
Pooled sequencing
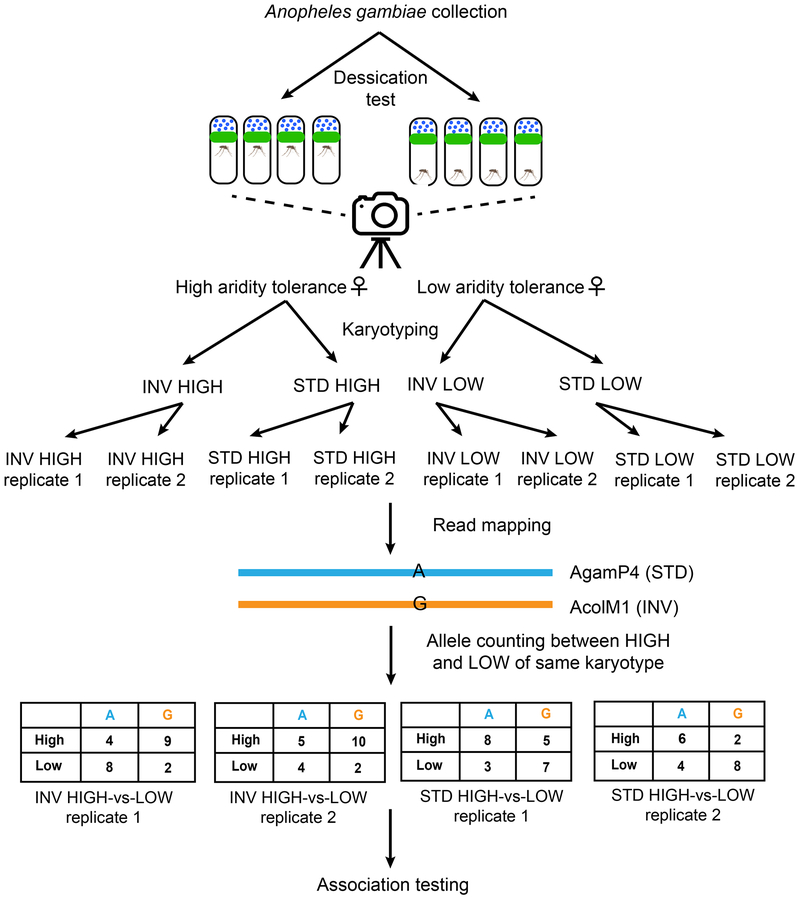
Our survival analysis identified 200 individuals with extreme phenotypes: those homozygous for 2La (INV) with high (n=50) and low (n=50) survival, and those homozygous for 2L+a (STD) with high (n=50) and low (n=50) survival (Figure 2). For each combination of karyotype and phenotype, two biological replicates were used (n=25 individuals in each), yielding 8 pools in total. We refer to each pool by its karyotype followed by phenotype throughout the text. For instance, pools with inverted arrangement and higher resistance to desiccation are referred as INV HIGH. Together, we had two replicate pools each of INV HIGH, INV LOW, STD HIGH, and STD LOW. Each pool was sequenced to approximately 20× coverage using the Illumina HiSeq technology by the BGI at its San Diego sequencing core. Paired-end reads 100 bp in length each (i.e. 2×100 for each cluster) were obtained for each pool. The raw reads were further cleaned of adapters using cutadapt (version 1.7dev; -O 12 –m 35; Martin 2011).
Figure 2.
Experimental design. Mosquitoes collected from nature as larvae were individually phenotyped for dessication tolerance and karyotyped. Pools of female mosquitoes with alternate inversion arrangements and extreme phenotypes were sequenced together in replicate. Reads were mapped to both the reference (standard for In(2La)) and pseudo-reference (inverted for In(2La)) assemblies. Alleles at each polymorphic site are counted to provide estimates of allele frequencies. Between pools with the same karyotype but different phenotypes, contingency tables were created for each polymorphic site. Association mapping using the Cochran-Mantel-Haenszel (CMH) test combines contingency tables across karyotypes and replicates in order to test this global hypothesis at each polymorphic site.
Sequence read mapping
The reference A. gambiae genome assembly (AgamP4, PEST strain) represents the standard arrangements of In(2La) and In(2Rb), which are known to be highly diverged from the inverted arrangements (Cheng et al. 2012; Fontaine et al. 2015), particularly at In(2La). Reads from individuals with inverted karyotypes may be difficult to map unambiguously and accurately if only the reference genome is used (Cheng et al. 2012). To minimize this problem, we simultaneously mapped reads to the AcolM1 assembly which represents the inverted arrangements of In(2La) and In(2Rb), albeit with a high degree of fragmentation. We used BWA (v.0.7.10; -M; Li 2013) to map reads from each pool against AgamP4 and a pseudo-chromosome of AcolM1 representing 2La. The pseudo-chromosome was constructed by extracting the region including In(2La) from the AcolM1 assembly. The mapping results demonstrate that including the pseudo-chromosome captured reads from 2La inverted pools that would otherwise have been missed (Supporting information Figure S3). The chromosomal location and extent of the two inversions are shown in Supporting information Figure S4.
Variant calling
We chose to carry out association mapping using only single nucleotide polymorphisms (SNPs) because of their reliability in pooled sequencing. Allele frequency estimates from pooled sequencing are highly accurate (especially for SNPs with minor allele frequencies greater than 5%) and errors are binomially distributed (Zhu et al. 2012). As our goal is to find SNPs with large frequency differences between pools, the small errors that do exist are less likely to affect our results. To identify SNPs in inverted regions we used two complementary approaches. First, we performed sequence alignments to opposite orientations of In(2La) in AgamP4 and AcolM1 using MUMmer (v3.23; Kurtz et al. 2004). Only unique and optimal alignment blocks were subsequently used, in order to exclude mismatches due to repeats. Nucleotide mismatches resulting from these alignments formed our raw call set (n=116,269 SNPs). We further filtered this set by removing any mismatches between nucleotides and gaps; the alignment for In(2Rb) was highly fragmented due to the incomplete nature of the AcolM1 assembly, and contained many gaps. As a second approach, we used the mapped sequencing reads from the pools directly to identify SNPs. We called variants using three independent programs under default settings: GATK (v3.2; DePristo et al. 2011), FreeBayes (v0.9.18; Garrison & Marth 2012), and SAMtools (v1.0.0; Li 2011). We retained only variable sites with two alleles (n=197,934 SNPs). The final set of SNPs was the intersection obtained from mapping to the reference assemblies and the three call sets from the reads themselves (n=59,381 SNPs).
Association mapping of desiccation tolerance within inversions
We tested for alleles associated with higher desiccation tolerance using the following procedure (Figure 2). For each pool, we first counted the number of alleles at each SNP position (equivalent to the number of reads carrying alternative nucleotides) that matched the inverted or standard alleles for In(2La) or In(2Rb). We only included variable sites that had at least 10 total reads. Next, we created contingency tables for pairs of pools representing the same karyotype but different phenotypes, to compare differences in allelic counts between high and low tolerance pools. We further filtered contingency tables where the sums of both row and column counts were at least 5 (regardless of allelic identity), as these are the minimum counts required to achieve P<0.05. Because each karyotype and phenotype were represented by replicate pools, in total we had four contingency tables for each site. We used the Cochran-Mantel-Haenszel (CMH) test to examine significant associations across tables, an approach more powerful than Fisher’s Exact tests of individual contingency tables (Bastide et al. 2013). We implemented the whole process in a toolkit of publicly available scripts (https://github.com/svm-zhang/poolseq_tk).
Functional classification of SNPs
Significant SNPs were annotated with respect to their predicted functional effects against the An. gambiae reference gene set, AgamP4.9, using SnpEff (v4.1B; Cingolani et al. 2012). SNPs in genes were classified as coding, intronic, splicing, or 5’/3’-UTR. Those within 5 kb of the 5’- and 3’-UTR of a gene were considered “upstream” and “downstream,” respectively, and “intergenic” if the distance upstream or downstream exceeded 5 kb.
Results
Association mapping within the 2La inversion
We identified 729,023 SNPs between opposite orientations of In(2La), corresponding to polymorphisms at 3.4% of all positions within the inversion. After filtering based on coverage in each pool and counts in each contingency table, we tested for associations at the 177,088 SNPs passing our filters using the CMH test. We are particularly interested in alleles that drive higher levels of resistance in the inverted arrangement. Such alleles are expected to be found at higher frequency in the INV HIGH and STD HIGH pools, with the alternative allele (which should be associated with lower resistance) found at higher frequency in the INV LOW and STD LOW pools. We used the odds-ratio across the four 2×2 tables employed for the CMH test to identify the SNPs that vary consistently in this manner, focusing on significant results among this set of sites.
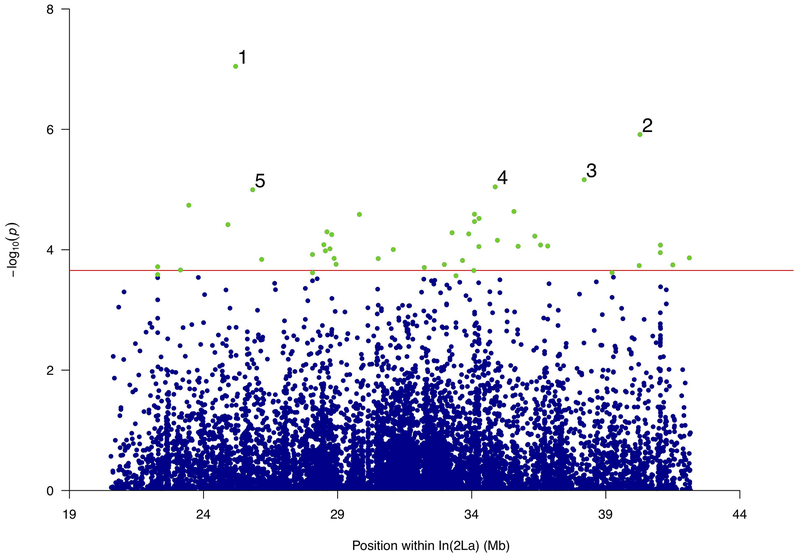
We conducted tests for an association between desiccation tolerance and all 177,088 SNPs within In(2La) passing our filters. To identify “significant” associations in the context of a large number of tests, we controlled for the false discovery rate at a threshold of 5%. In total, we mapped 46 significant SNPs within the 2La inversion that also varied in the manner expected based on their odds-ratios (Figure 3). Notably, these significant SNPs spanned the entire inversion, with little indication of clustering based on visual inspection.
Figure 3.
Manhattan plot of the association P-values for acute desiccation tolerance inside In(2La). Horizontal red line represents the In(2La)-wide significance threshold at FDR of 5% (P < 0.00027). Tested SNPs below that threshold shown as blue dots; green dots represent significant SNPs conferring high tolerance. The SNPs with the lowest P-values are numbered, and correspond to genomic coordinates closest to the following genes: (1) AGAP006026, (2) AGAP006961, AGAP006962, AGAP006963, (3) AGAP006785, AGAP006786, (4) AGAP006633, (5) AGAP006059.
To explore the possible functions among our set of significant associations, we categorized each SNP as coding, intronic, splicing, 5’/3’-UTR, upstream, downstream, and intergenic. We placed 31 significant SNPs in 57 genes (Supporting information Table S1; SNPs can be associated with more than one gene when they are in annotated regions flanking two genes). Most significant SNPs were found in non-coding regions: we found only four synonymous variants among our set of significant sites, and no nonsynonymous variants. Previous studies have found many differences in transcription between inversion arrangements at In(2La) (Cassone et al. 2011; Cheng et al. 2018), allowing us to ask whether the same genes containing significant SNPs also showed evidence for differential expression. We cross-referenced expression profiles from Cheng et al. (2018) with our list of 57 candidate genes and found data for 44 of them. Of these 44 genes, 35 showed significant differences in expression between arrangements of In(2La). This fraction of differentially expressed genes with significant SNPs, 79.5% (35/44), is much higher than the overall fraction of differentially expressed genes within In(2La) between arrangements (~60%; Cheng et al. 2018). Two of our top-ranking candidate genes have presumed roles in response to environmental stimulus. The first (AGAP006026; Supporting information Table S1) encodes an ionotropic glutamate receptor (IR). IRs are commonly associated with chemosensation (Benton et al. 2009) but recent studies in Drosophila have revealed roles in thermosensation and hygrosensation (Enjin 2017; Enjin et al. 2016; Frank et al. 2017; Knecht et al. 2016). The second (AGAP006961; Supporting information Table S1) is located in an area of maximal sequence divergence between alternative In(2La) arrangements (White et al. 2007a) and it encodes a heat shock protein (Hsp90).
Association mapping within the 2Rb inversion
It is important to note that each of our pools was a combination of a phenotype (i.e. high versus low desiccation resistance) and the karyotype of the In(2La) inversion (standard versus inverted). The pooling was blind to the In(2Rb) karyotype, as there is not a 100% reliable DNA marker for this inversion. Although any association test will therefore have much reduced power, if the karyotype at In(2La) is predictive of the karyotype at In(2Rb)—that is, if an individual mosquito homozygous at In(2La) is also likely to be homozygous at In(2Rb) for the same orientation (standard or inverted)—our pools may still allow for an informative test. As an indirect way to examine such linkage disequilibrium (LD), we surveyed the pattern of sequence differentiation at In(2Rb) in our pools. If the inversions are in LD, we expected to observe a similarly elevated level of differentiation in In(2Rb) as in In(2La), although to different degrees (Cheng et al., 2012). We calculated FST along chromosome 2R between the INV HIGH and STD LOW pools. We found a similar pattern as observed for chromosome 2L, with high differentiation inside In(2Rb) compared to the collinear regions outside (Supporting information Figure S5). In addition, we examined patterns of LD between In(2La) and In(2Rb) in mosquitoes previously karyotyped from the same region of Cameroon. Using the “Adamaoua Highands” samples from Simard et al. (2009), we were able to confirm significant linkage disequilibrium between inversions (D’=0.23, P=9.65×10−13, n=737; calculations done using the “genetics” package in R). These independent analyses strongly suggest that the two inversions are in LD (to some degree) with each other in our pools.
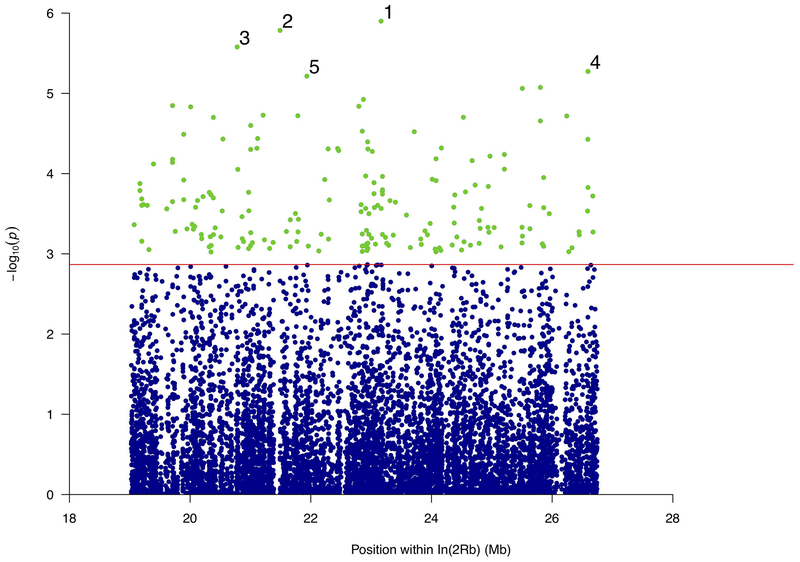
Due to the fragmented nature of our In(2Rb) alignments we identified only 59,381 variable sites. After applying our read-depth and other filters, we tested for associations at 12,993 SNPs, following the same protocol that was used for mapping within In(2La). In total, the CMH test identified 187 significant SNPs within In(2Rb) at an FDR of 5% (Figure 4). Again, the significant SNPs were distributed across the entire inversion, as was the case for In(2La) (Figure 3). Because In(2Rb) is appreciably smaller than In(2La) (7 Mb vs. 22 Mb), it likely has both fewer double-crossovers and more LD within it. These two factors may explain the higher number of significant SNPs in In(2Rb), despite the possible false negatives owing to assembly gaps in In(2Rb).
Figure 4.
Manhattan plot of the association P-values for acute desiccation tolerance inside In(2Rb). Horizontal red line represents the In(2Rb)-wide significance threshold at FDR of 5% (P < 0.00092). Tested SNPs below that threshold shown as blue dots; green dots represent significant SNPs conferring high tolerance. The SNPs with the lowest P-values are numbered, and correspond to genomic coordinates closest to the following genes: (1) AGAP002578, (2) AGAP002444, (3) AGAP002372, (4) AGAP002744, AGAP013160, (5) AGAP002487.
We looked for functions associated with these significant SNPs by performing the same protocol as was done for In(2La). In total, the 187 significant SNPs were assigned to 147 genes (Supporting information Table S2). As with In(2La), the majority of significant SNPs were in non-coding regions. After cross-referencing the expression profiles of these genes using transcription data from (Cheng et al. 2018), we found 132 of the genes with available expression data, 41 of which were reported to exhibit significant differences between mosquitoes homozygous for alternative arrangements of the 2Rb inversion. Similar to In(2La) in which one of two top-ranked candidate genes encodes a ligand-gated transmembrane ion channel, our top-ranking candidate in In(2Rb) (AGAP002578) encodes a voltage-gated calcium channel, whose Drosophila ortholog is named cacophony, a gene involved in male courtship behavior and other neurophysiological processes.
Discussion
In this study, we performed fine-scale association mapping of desiccation tolerance within the 2La and the 2Rb inversions of the mosquito, An. gambiae. We were able to map 46 and 187 SNPs that exhibited significant associations with variation in levels of response to arid environments. Of those located in the vicinity of annotated genes, the vast majority mapped up- or downstream of coding sequence. Between the two inversions only three nonsynonymous SNPs showed significant associations with desiccation tolerance.
We examined the expression profiles of the genes neighboring these non-coding SNPs (Cheng et al. 2018), and found a disproportionate number showing significant differences in expression between mosquitoes carrying alternative arrangements. Indeed, an important finding of Cheng et al. (2018), echoed by other studies of inversions in D. pseudoobscura (Fuller et al. 2016) and D. melanogaster (Lavington & Kern 2017), is that transcriptional profiles are strongly influenced by karyotype, and that genes inside rearranged regions are overrepresented among those differentially expressed genome-wide. Together these findings suggest that gene expression differences preserved by LD inside inversions play an important role in local adaptation (Lavington & Kern 2017; Romero et al. 2012). Nevertheless, important caveats inject caution into the interpretation of our results based on SNP annotations. Not only are An. gambiae gene annotations incomplete and prone to error—especially in putative regulatory regions—but more importantly, it is unlikely that SNPs uncovered in this study as significantly associated with desiccation tolerance are directly causal. Populations of An. gambiae are extraordinarily large and have notoriously high nucleotide diversity (Miles et al. 2016), but our power to detect QTNs instead of merely QTLs was limited by small sample size (including the number of phenotyped mosquitoes, pool size, and pool sequencing coverage). As QTLs, our associated SNPs most likely represent the “low hanging fruit”—loci with relatively strong contributions to the desiccation tolerance phenotype. Low power limits our ability to detect additional QTL of small effect, and strong LD near breakpoints precludes us from detecting QTL in these regions regardless of effect size, as our experimental design depends upon recombination between markers and inversion karyotype. As sequencing costs continue to decline, it may become cost-effective to sequence the genomes of individual mosquitoes instead of pools; such data could allow us to independently assess the effect of SNPs in LD with each other, and possibly the causal role of copy-number variants (CNVs) inside the inversion.
Three of our top-ranked candidate genes are predicted to have functions plausibly connected with desiccation tolerance. These represent a clear starting point for wet-bench experimentation into their roles in resistance to desiccation. Additional candidates inside In(2La) and In(2Rb) have annotations suggesting they are receptors, signal transducers, and transcription factors, but their specific functions have yet to be determined; many other candidates lack functional annotation altogether. Further prioritization of candidate genes might benefit from overlap between gene lists uncovered by multiple independent studies, such as this one and previous studies of In(2La) and In(2Rb) based on complementary approaches, including gene expression profiling in response to thermal or desiccation stress (Cassone et al. 2011; Cheng et al. 2018) and clinal studies of inversion-associated nucleotide divergence (Cheng et al. 2012). Above, we detailed the strong overlap between our candidate In(2La) and In(2Rb) gene lists and those found to be differentially expressed inside these rearrangements by Cheng et al. (2018). We also find some correspondence between our gene lists and the lists of genes overlapping the top 1% windows of FST between northern and southern populations in In(2La) and In(2Rb) found by Cheng et al. (2012). Functional enrichment analyses of those gene lists by Cheng et al. (2012) revealed annotations indicative of cuticle proteins and ion channels/GPCRs to be overrepresented in the In(2La) set, while immunoglobulin-like fold annotations were overrepresented in the In(2Rb) set. Indeed, we find overlap in those functional categories and with individual genes, including our top-ranked ionotropic glutamate channel candidate and two additional ion channel genes in the 2La inversion (Supporting information Table S1). Nevertheless, extensive degrees of overlap between candidates from different studies may not be a realistic expectation, for a variety of reasons. Among the most important, these rearrangements contain hundreds or thousands of genes that likely contribute to multiple complex phenotypes, which may or may not share a common mechanistic basis or a similar transcriptional profile. Here, we studied acute desiccation resistance at the teneral adult stage; previously we studied the transcriptional response to arid acclimation in 8-day old adults (Cheng et al. 2018) or to thermotolerance of fourth instar larvae (Cassone et al. 2011). Our recent results (Cheng et al. 2018) suggest that a more focused examination of the metabolic response, in the framework of energy-limited response to environmental stress, may provide a more synthetic understanding of the role of inversions in adaptation to stressful environments.
Quantitative trait mapping studies generate linkage disequilibrium between marker SNPs and causal mutations using controlled crosses. Such studies depend on recombination to randomize markers across backgrounds, a process that occurs at too low a rate to work efficiently inside inversions. Even standard association mapping, which allows recombination to act over many generations, may not sufficiently break up associations within inversions. Here we have taken advantage of long-term balancing selection at inversions in An. gambiae to carry out association mapping. Despite low levels of gene flux inside inversions, maintenance of these inversions for over two million years (Fontaine et al. 2015) has enabled us to associate individual markers with an ecologically relevant phenotype. While these conditions many not always be available, the regularity of balancing selection on inversions across species likely means that this approach will also be possible in other systems.
Supplementary Material
Acknowledgements
Our gratitude goes to the inhabitants of Tibati and the local authorities for their collaboration in this study. We specially thank Jean-Pierre Agbor and Serge Donfack for their technical support across the field work, and M. Kern for assistance in sample pooling. Funding was provided by the Institut de Recherche pour le Developpement and the National Institutes of Health (grant R01AI076584 to NJB and MWH).
Footnotes
Data accessibility: DNA sequences have been deposited to GenBank under NCBI BioProject ID PRJNA481199.
References
- Andolfatto P, Depaulis F, Navarro A (2001) Inversion polymorphisms and nucleotide variability in Drosophila. Genetical Research, 77, 1–8. [DOI] [PubMed] [Google Scholar]
- Bastide H, Betancourt A, Nolte V, et al. (2013) A genome-wide, fine-scale map of natural pigmentation variation in Drosophila melanogaster. PLoS Genet, 9, e1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell, 136, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone BJ, Molloy MJ, Cheng C, et al. (2011) Divergent transcriptional response to thermal stress by Anopheles gambiae larvae carrying alternative arrangements of inversion 2La. Molecular Ecology, 20, 2567–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Tan JC, Hahn MW, Besansky NJ (2018) A systems genetic analysis of inversion polymorphisms in the malaria mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, White BJ, Kamdem C, et al. (2012) Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics, 190, 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang Le L, et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin), 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Della Torre A, Di Deco MA, Petrarca V (2002) A polytene chromosome analysis of the Anopheles gambiae species complex. Science, 298, 1415–1418. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA (1979) Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Transactions of the Royal Society of Tropical Medicine and Hygiene, 73, 483–497. [DOI] [PubMed] [Google Scholar]
- Cox DR (1972) Regression models and life tables. Journal of the Royal Statistical Society Series B, 34, 187–220. [Google Scholar]
- Depristo MA, Banks E, Poplin R, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics, 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1970) Genetics of the Evolutionary Process Columbia University Press, New York. [Google Scholar]
- Enjin A (2017) Humidity sensing in insects-from ecology to neural processing. Curr Opin Insect Sci, 24, 1–6. [DOI] [PubMed] [Google Scholar]
- Enjin A, Zaharieva EE, Frank DD, et al. (2016) Humidity Sensing in Drosophila. Current Biology, 26, 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, et al. (2015) Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science, 347, 1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C, Gray E, Besansky NJ, Costantini C (2012) Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PloS one, 7, e34841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DD, Enjin A, Jouandet GC, et al. (2017) Early Integration of Temperature and Humidity Stimuli in the Drosophila Brain. Current Biology, 27, 2381–2388 e2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW (2016) Genomics of natural populations: How differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics, 204, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907 [q-bio.GN].
- Gillies MT, De Meillon B (1968) The Anophelinae of Africa South of the Sahara, 2nd edn. South African Institute for Medical Research, Johannesburg. [Google Scholar]
- Gray EM, Rocca KA, Costantini C, Besansky NJ (2009) Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar. J, 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH (2008) Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annual Review of Ecology Evolution and Systematics, 39, 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N (2006) Chromosome inversions, local adaptation and speciation. Genetics, 173, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht ZA, Silbering AF, Ni L, et al. (2016) Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR (1992) Drosophila inversion polymorphism CRC Press, London. [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, et al. (2004) Versatile and open software for comparing large genomes. Genome Biol, 5, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavington E, Kern AD (2017) The effect of common inversion polymorphisms In(2L)t and In(3R)Mo on patterns of transcriptional variation in Drosophila melanogaster. G3 (Bethesda), 7, 3659–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
- Lobo NF, Sangare DM, Regier AA, et al. (2010) Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar J, 9, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal, 17, 10. [Google Scholar]
- Miles A, Harding NJ, Botta G, et al. (2016) Natural diversity of the malaria vector Anopheles gambiae. BioRxiv. [Google Scholar]
- Morlais I, Poncon N, Simard F, Cohuet A, Fontenille D (2004) Intraspecific nucleotide variation in Anopheles gambiae: new insights into the biology of malaria vectors. The American journal of tropical medicine and hygiene, 71, 795–802. [PubMed] [Google Scholar]
- Navarro A, Betran E, Barbadilla A, Ruiz A (1997) Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics, 146, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrarca V, Sabatinelli G, Di Deco MA, Papakay M (1990) The Anopheles gambiae complex in the Federal Islamic Republic of Comoros (Indian Ocean): some cytogenetic and biometric data. Parassitologia, 32, 371–380. [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, et al. (2008) Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC evolutionary biology, 8, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishikesh N, Di Deco MA, Petrarca V, Coluzzi M (1985) Seasonal variations in indoor resting Anopheles gambiae and Anopheles arabiensis in Kaduna, Nigeria. Acta Tropica, 42, 165–170. [PubMed] [Google Scholar]
- Roberts PA (1976) The genetics of chromosome aberration In: The Genetics and Biology of Drosophila (eds. Ashburner M, Novitski E), pp. 67–184. Academic Press, London. [Google Scholar]
- Rocca KA, Gray EM, Costantini C, Besansky NJ (2009) 2La chromosomal inversion enhances thermal tolerance of Anopheles gambiae larvae. Malar J, 8, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y (2012) Comparative studies of gene expression and the evolution of gene regulation. Nature reviews. Genetics, 13, 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Mancini E, Simard F, et al. (2008) Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J, 7, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C, Tobler R, Kofler R, Nolte V (2014) Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nature Reviews Genetics, 15, 749–763. [DOI] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem GC, et al. (2009) Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol, 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH (1917) Genetic factors affecting the strength of linkage in Drosophila. Proceedings of the National Academy of Sciences U S A, 3, 555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure YT, Petrarca V, Traore SF, et al. (1998) The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia, 40, 477–511. [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L (2018) Eco-Evolutionary Genomics of Chromosomal Inversions. Trends Ecol Evol, 33, 427–440. [DOI] [PubMed] [Google Scholar]
- White BJ, Collins FH, Besansky NJ (2011) Evolution of Anopheles gambiae in relation to humans and malaria. Annual Review of Ecology Evolution and Systematics, 42, 111–132. [Google Scholar]
- White BJ, Hahn MW, Pombi M, et al. (2007a) Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae. PLoS Genet, 3, e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Santolamazza F, Kamau L, et al. (2007b) Molecular karyotyping of the 2La inversion in Anopheles gambiae. American Journal of Tropical Medicine and Hygiene, 76, 334–339. [PubMed] [Google Scholar]
- World Health Organisation (2017) World Malaria Report: 2017, http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/.
- Zhu Y, Bergland AO, Gonzalez J, Petrov DA (2012) Empirical validation of pooled whole genome population re-sequencing in Drosophila melanogaster. PloS one, 7, e41901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.