Abstract
Background and purpose
Dioscin shows potent effects against cancers. We aimed to elucidate its pharmacological effects and mechanisms of action on hepatocellular carcinoma (HCC) in vivo and in vitro.
Experimental approach
Effects of dioscin were investigated in SMMC7721 and HepG2 cells, diethylnitrosamine‐induced primary liver cancer in rats, and cell xenografts in nude mice. Isobaric tags for relative and absolution quantitation (iTRAQ)‐based proteomics was used to find dioscin's targets and investigate its mechanism.
Key results
In SMMC7721 and HepG2 cells dioscin markedly inhibited cell proliferation and migration, induced apoptosis, autophagy, and DNA damage. It inhibited DEN‐induced primary liver cancer in rats, markedly changed body weights and restored levels of α fetoprotein, alanine transaminase, aspartate transaminase, γ‐glutamyltransferase, alkaline phosphatase, and Ki67. It also inhibited growth of xenografts in mice. In SMMC7721 cells, 191 differentially expressed proteins were found after dioscin, based on iTRAQ‐based assay. TP53‐inducible glycolysis and apoptosis regulator (TIGAR) was identified as being significantly down‐regulated by dioscin. Dioscin induced cell apoptosis, autophagy, and DNA damage via increasing expression levels of p53, cleaved PARP, Bax, cleaved caspase‐3/9, Beclin‐1, and LC3 and suppressing those of Bcl‐2, p‐Akt, p‐mammalian target of rapamycin (mTOR), CDK5, p‐ataxia telangiectasia‐mutated gene (ATM). The transfection of TIGAR siRNA into SMMC7721 cells and xenografts in nude mice further confirmed that the potent activity of dioscin against HCC is evoked by adjusting TIGAR‐mediated inhibition of p53, Akt/mTOR, and CDK5/ATM pathways.
Conclusions and implications
The data suggest that dioscin has potential as a therapeutic, and TIGAR as a drug target for treating HCC.
Abbreviations
- AFP
α fetoprotein
- ALP
alkaline phosphatase
- ALT
alanine transaminase
- AST
aspartate transaminase
- ATM
ataxia telangiectasia‐mutated gene
- Bcl‐2
B‐cell CLL/lymphoma 2
- CDK5
cyclin‐dependent kinases‐5
- CMC‐Na
sodium carboxymethyl cellulose
- CQ
chloroquine
- DEN
diethylnitrosamine
- HCC
hepatocellular carcinoma
- iTRAQ
isobaric tags for relative and absolution quantitation
- mTOR
mammalian target of rapamycin
- p53
tumour protein 53
- TIGAR
TP53‐inducible glycolysis and apoptosis regulator (fructose‐2,6‐bisphosphatase)
- γ‐GT
γ‐glutamyltransferase
1. INTRODUCTION
Hepatocellular carcinoma (HCC), the third leading cause of cancer‐related mortality worldwide, can cause over 6,000,000 deaths every year (Polina, Lubov, & Timchenko, 2011). At present, surgery and nonsurgical strategies have been used for the treatment of HCC. Surgical treatment including liver resection, percutaneous ablation, and liver transplantation is one common therapeutic option (Qian et al., 2015). Up to now, some biological methods including molecular‐targeted therapy, immunotherapy, and gene therapy have shown potential as treatments for HCC (Greten, Xin, & Korangy, 2015; Marquardt, Galle, & Teufel, 2012). Apart from these, drugs including doxorubicin, cisplatin, and 5‐fluorouracil (5‐Fu) have achieved survival benefits against HCC (Gao, Zhen, Liao, Zhuang, & Guo, 2018). However, side effects of these drugs, including cardiotoxicity and neurotoxicity, limit their clinical application. Thus, it is necessary to develop potent therapeutic agents with high efficiency and low toxicity against HCC.
Some biological processes including apoptosis, autophagy, and DNA damage play critical roles in regulating HCC (Faridah, Ataollahi, & Asmah, 2014; Gong & Li, 2011; Liu et al., 2018; Yu et al., 2017). Excessive accumulation of intracellular ROS can trigger a series of mitochondria‐associated events, and regulating apoptosis, autophagy, and DNA damage can be considered as one important target for the development of anticancer drugs (Lv et al., 2013). Nowadays, large‐scale in depth quantitative proteomic analysis has been widely used to find biomarkers, drug targets, molecular mechanisms, and elucidate pathways affected by drugs against HCC (Yin et al., 2017; Zhang, Xu, et al., 2015). Isobaric tags for relative and absolution quantitation (iTRAQ), combined with multidimensional LC and tandem MS assay, is one powerful quantitative proteomic method, which has been widely used to identify biomarkers and drug targets (Chen et al., 2014).
Traditional Chinese medicines have recently being attracting more and more attention. Some natural products including curcumin, matrine, and resveratrol from medicinal plants have anti‐HCC activities (Jain et al., 2015). Therefore, the exploration of effective natural products from medicinal plants to treat HCC is reasonable. Dioscin (Figure S1), one such natural product, has been shown to have anti‐inflammatory, antifungal, antivirus, and antihepatic fibrosis activities (Cho, 2013; Liu et al., 2013; Lu et al., 2012; Wang et al., 2010; Zhao et al., 2012). In addition, dioscin shows potent effects against colon cancer, lung cancer, laryngeal cancer, and glioblastoma multiforme (Si et al., 2016; Wei et al., 2013). Moreover, dioscin can induce apoptosis and autophagy in Huh‐7 cells (Xu et al., 2018), suppress cell proliferation in BEL‐7402 cells (Zhang et al., 2016), and reverses multidrug resistance in human hepatoma HepG2/adriamycin cells (Sun et al., 2011). However, the effects of dioscin on SMMC7721 and HepG2 cells have not been reported. Moreover, the effects of dioscin on diethylnitrosamine (DEN)‐induced hepatocarcinogenesis and its mechanism and target are also unclear.
Therefore, the aim of the present work was to elucidate the pharmacological effects and molecular mechanisms of dioscin against HCC in vitro and in vivo.
2. METHODS
2.1. Cell culture
The human cell lines including Huh‐7 (RRID: CVCL_0336), SMMC7721 (RRID: CVCL_0534), HepG2 (RRID: CVCL_0027, Problematic cell line: Misidentified), BEL7402 (RRID: CVCL_5492), and L‐02 (RRID: CVCL_6926) were obtained from China Centre of Type Culture Collection (Wuhan, China). Huh‐7, SMMC7721, and L‐02 cells were cultured in RPMI1640, while the HepG2 and BEL7402 cells were cultured in DMEM medium, supplemented with 10% FBS, 100 U·ml−1 of penicillin and 100 U·ml−1 of streptomycin, and cultured at 37°C in a humidified atmosphere of 5% CO2. The medium was changed every other day.
2.2. MTT assay
The cells were seeded into 96‐well plates (1 × 105 cells per well) and incubated overnight. Then, the medium was removed, and 100 μl of sample solution with various concentrations of dioscin (0.7, 1.4, 2.9, 5.8, and 11.8 μM) was added under different treatment times for 12, 24, and 36 hr. After 10 ml of MTT stock solution (5 mg·ml−1) was added, the plates were incubated for another 4 hr at 37°C and DMSO (100 ml per well) was added to dissolve formazan crystals. We measured the absorbance with a microplate reader (Thermo, USA) at 490 nm, and the results were normalized to control for unwanted sources of variation, and the cell morphology was imaged with a phase contrast microscope (Nikon, Japan).
2.3. Acridine orange/Ethidium bromide and DAPI staining
SMMC7721 and HepG2 cells at a density of 2 × 105 cells per well were plated in 6‐well plate and incubated for 24 hr, then treated with dioscin (1.4, 2.9, and 5.8 μM) for 24 hr. After incubation, the solution was removed and the cells were washed with PBS twice. Then, 20 μl of solution containing the same volume of AO (diluted in PBS for 1.0 mg·ml−1) and EB (diluted in PBS for 1.0 mg·ml−1) was added. The cells were observed under a fluorescence microscope (OLYMPUS, Japan). DAPI staining was carried out as mentioned above. Finally, the images were obtained by using a fluorescence microscope (OLYMPUS, Japan).
2.4. Plate colony‐forming assay
SMMC7721 and HepG2 cells were harvested, and seeded (500 cells per well) into six‐well plates. After overnight incubation, the cells were treated with dioscin (1.4, 2.9, and 5.8 μM) once every 3 days, and the medium was replaced every 3 days. The process lasted for 2 weeks. Finally, the colonies were stained with crystal violet solution for 10 min at room temperature, and the colonies containing more than 50 cells were counted.
2.5. Scratch assay
SMMC7721 and HepG2 cells were plated into six‐well plates at 2 × 105 cells per well. Wounds were scratched with a sterile micropipette tip and washed with PBS to remove the floating cells in the serum‐free medium. Next, the cells were treated with different concentrations of dioscin (1.1, 1.4, and 1.7 μM) for 24 hr. Finally, the images of the wound gap were observed with an inverted microscope (Nikon, Japan).
2.6. Transwell migration assay
The migration and invasion of SMMC7721 and HepG2 cells were measured according to the manufacturer's instruction with sterile transwell chambers (Corning Incorporated, Corning, NY, USA). Total of 5 × 104 cells in serum‐free medium (200 μl) were added into the upper chambers while the lower chamber was filled with 500 μl of medium containing 10% FBS. After incubation with dioscin (1.1, 1.4, and 1.7 μM) for 24 hr, the cells were fixed with 4% formaldehyde for 20 min and stained with haematoxylin for 20 min. The cells in five randomly selected fields were counted under an inverted phase‐contrast microscope (Nikon, Japan).
2.7. Transmission electron microscopy
SMMC7721 and HepG2 cells were seeded into six‐well plates (3.5 × 105 cells per well) and treated with dioscin for 24 hr. The cells were then fixed in 2% glutaraldehyde at 4°C for 24 hr, postfixed in 1% OSO4 dehydrated in graded ethanol, and then embedded in epoxy resin. The images were acquired using a transmission electron microscope (JEM‐2000, JEDL, Japan).
2.8. Single cell gel electrophoresis assay
SMMC7721 and HepG2 cells at a density of 1.5 × 105cells per well were incubated in six‐well plates. After being treated with different concentrations of dioscin (1.4, 2.9, and 5.8 μM) for 24 hr, single cell gel electrophoresis assay was determined by the kit according to the manufacturer's instructions. Images of the cells were obtained by a fluorescence microscope (OLYMPUS, Osaka, Japan); 200 randomly selected cells were analysed by the Comet Assay Software Project.
2.9. Animal model and experimental protocol
Male Wistar rats (RRID: RGD_10044) weighing 120–150 g and 4‐week‐old BALB/c nude mice (RRID: MGI: 5649767) weighing 18–22 g were provided by the Experimental Animal Centre of Dalian Medical University, Dalian, China. All experimental procedures were performed strictly complied with Legislation Regarding the Use and Care of Laboratory Animals of China, and all experiments involving animals were approved by the Animal Care and Use Committee of Dalian Medical University. Animals were housed in wire mesh floor cages under 12 h light/dark cycles, a controlled temperature of 22–24°C and a relative humidity of 60 ± 10%, which were sustained on standard rat chow.
Primary liver tumours in rats were induced by DEN. The animals were randomly distributed into six groups (n = 16), including a control group, model group (DEN), DEN + high dose of dioscin (60 mg·kg−1) group, DEN + medium dose of dioscin (30 mg·kg−1) group, DEN + low dose of dioscin (15 mg·kg−1) group, and dioscin group (60 mg·kg−1). The rats in the model and DEN+dioscin groups received 0.01% DEN for 18 weeks and other rats were given PBS (Barajas et al., 2001). The body weights of the rats were measured once a week during the process. Animals were killed under deep anesthesia, and blood samples were collected from abdominal aorta to produce the serum and the liver samples were fixed in 10% buffered formalin and embedded in paraffin.
Xenograft models in nude mice were established on male BALB/c nude mice. SMMC7721 and HepG2 cells at a density of 1.5 × 106 suspended in 100 μl of PBS were injected s.c. into the right flanks of mice. Fifty mice were randomly divided into five groups (n = 10). The mice in control group were i.p. administered with 0.5% CMC‐Na as vehicle group. The mice in positive group were injected with 5‐fluorouracil at the dose of 20 mg·kg−1·day−1 (Qian et al., 2015). The animals in the dioscin‐treated groups were treated with 20, 40, and 80 mg·kg−1 of dioscin. The drug was administered by gavage once daily for 27 days, and the tumour volumes were calculated according to the following formula: V = (max diameter) × (min diameter)2/2. At the end of the test, the animals were killed, and the tumours were then photographed and weighed. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology.
2.10. Biochemical assays
The serum levels of α fetoprotein (AFP), alkaline phosphatase (ALP), alanine transaminase (ALT), aspartate transaminase (AST), and γ‐glutamyltransferase (γ‐GT) in rats were measured using the detection kits based on the manufacturer's instruction.
2.11. Histological and immunohistochemical assays
Formalin‐fixed livers and tumour tissues were embedded in paraffin and cut into 5‐μm sections. The rat liver samples were stained with haematoxylin and eosin (H&E), Masson, Sirius red, and Ki67 according to the manufacturer's instruction. The tumour sections of nude mice were stained with H&E and Ki67. The images of the stained sections were obtained using a light microscope (Nikon Eclipse TE2000‐U, Japan) with 200× magnification. The immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology.
2.12. TUNEL and DNA fragmentation assays
Apoptosis detection in cells and liver tissues was carried out using the kit based on the manufacturer's protocol. The cells and liver tissues were treated with or without dioscin before the fluorescein (green)‐labelled dUTP solution was added. The images were taken using fluorescence microscopy (Nikon Eclipse TE2000‐U, NIKON, Japan) with 200× magnification. DNA samples from SMMC7721 cells were extracted using the kit, and then the concentration and purity of extracted DNA samples were determined. For the laddering assay, an equal amount of DNA samples from treated and control groups were mixed with 1× tracking dye (bromophenol blue) solution and then loaded for electrophoretic separation on 1.5% agarose gel. A standard 100 base pair DNA marker was also loaded. After electrophoresis, DNA was stained with 20 mg·ml−1 of ethidium bromide and the gel was visualized under UV light and photographed using a Bio‐Spectrum Gel imaging System (UVP, Upland, CA, USA).
2.13. Protein preparation and iTRAQ labelling
SMM7721 cells of control and dioscin (5.8 μM)‐treated groups were washed with ice‐cold PBS and lysed with radioimmunoprecipitation assay lysis buffer (50‐Mm Tris–HCl, 1% SDC, 150‐mM NaCl, 0.1% Triton X‐100, pH 8.0), which was supplemented with 1 mM PMSF. The samples were homogenized by sonication for three times after being vortexed and incubated in boiling water for 5 min. Next, protein concentration was quantified by BCA Protein Assay kit. The iTRAQ labelling was carried out based on the manufacturer's instructions. The samples of dioscin‐treated group were labelled with iTRAQ 115, 116, and 117, and the samples of control group were labelled with iTRAQ 118, 119, and 121. All tagged samples were prepared for 1 hr at room temperature and then pooled together and lyophilized to powder.
2.14. Protein identification and bioinformatics analysis
The peptide data were analysed by Protein Pilot Software 4.5, and the identified proteins were classified according to annotations from the UniProt (RRID: SCR_002380) knowledge base, which were further analysed using Web Gene Ontology Annotation Plot and enrichment analysis. The gene ontology database was used to elucidate biological process (BP), molecular function (MF), and cellular components. The fold change was the ratio of protein differently produced in dioscin‐induced cells relative to those of in control group. A fold change cut‐off of 1.5 was set to identify molecules whose expression was significantly affected in both duplicate experiments, and P < 0.05 was taken as a significant screening.
2.15. Immunofluorescence assay
The paraffin‐embedded sections of liver and tumour tissues were prepared for examining the expressions levels of TIGAR. The tissue slices or formalin‐fixed cells were incubated with anti‐TIGAR antibody (Cat# ab62533, RRID: AB_2066497) in a humidified box at 4°C overnight, then incubated with a fluorescein‐labelled secondary antibody for 1 hr, and cell nuclei were stained with DAPI (5 μg·ml−1). The samples were imaged using fluorescence microscopy (OLYMPUS, Japan).
2.16. Western blotting assay
Total proteins from SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice were isolated using the kit based on the manufacturer's instructions. Protein concentration was measured using a BCA assay kit. Then, the protein samples were loaded onto SDS‐PAGE gels, and transferred onto PVDF membrane (Millipore, USA). Membranes were incubated overnight at 4°C with the primary antibodies (listed in Table S1). After being blocked with 5% dried skimmed milk for 3 hr at a room temperature, the membranes were incubated with HRP‐conjugated antibody at room temperature for 2 hr. Finally, the proteins were detected using an enhanced chemiluminescence method and imaged by a Bio‐Spectrum Gel imaging System (UVP, Upland, CA, USA). Intensity values of the relative protein levels were normalized to GAPDH.
2.17. Real‐time PCR assay
Total RNA samples were obtained from SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice using RNA iso Plus Reagent following the manufacturer's protocol (the primer sequences of TIGAR are listed in Table S2). Each RNA sample was reverse‐transcribed into cDNA using the TransStart Top Green qPCR SuperMix kit. Among the data from each sample, the Ct value of the target gene was normalized to that of GAPDH. The unknown template in our study was calculated using the standard curve for quantitative analysis.
2.18. TIGAR siRNA transfection experiments in vitro
In vitro transfection experiments were performed on SMMC7721 and HepG2 cells. The TIGAR‐targeted siRNA (siRNA1, siRNA2, and siRNA3) and control siRNA were dissolved in Opti‐MEM. The solutions were equilibrated for 5 min at room temperature. Then, the TIGAR‐targeted siRNA and control siRNA were mixed gently with transfection regent‐lipofectamine 2000 for 20 min to form siRNA liposomes. Cell apoptosis, proliferation, migration, and invasion were detected after 24 hr of transfection. In addition, DNA damage and the expression levels of TIGAR, tumour protein 53 (p53), Akt, p‐Akt, mammalian target of rapamycin (mTOR), p‐mTOR, cyclin‐dependent kinases‐5 (CDK5), ataxia telangiectasia‐mutated gene (ATM), and p‐ATM were measured after 24 hr of transfection.
2.19. TIGAR siRNA transfection experiments in vivo
In vivo transfection experiments, SMMC7721 cells (1.5 × 106) suspended in 100 μl of PBS were injected s.c. into the right flanks of nude mice. Fifty mice were then randomly divided into five groups (n = 10) including control group, control‐siRNA group, 80 mg·kg−1 of dioscin group, TIGAR‐siRNA (200 nmol·kg−1) group, and TIGAR‐siRNA+dioscin group. TIGAR‐siRNA was diluted in PBS and injected into the tummours of the mice once every 3 days. The tumour volumes were measured once every 3 days and calculated using the indicated formula. The tumour burden did not exceed the recommended maximum diameter (1.5 cm in therapeutic studies). After scarification, the tumours were weighed and photographed. Then, a TUNEL assay was performed, and immunofluorescence staining for TIGAR was also carried out. In addition, the expression levels of TIGAR, p53, Akt, p‐Akt, mTOR, p‐mTOR, CDK5, ATM, and p‐ATM were measured.
2.20. Statistical analysis
All data are expressed as the mean ± SD. Statistical analysis was performed with GraphaPad Prism 5.0 software (San Diego, CA, USA) and one‐way ANOVA followed by Tukey's post hoc test when comparing multiple independent groups. Comparisons between two groups were performed by use of Student's unpaired t‐test. P < 0.05 were considered to be significant. The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018).
2.21. Chemicals and materials
Dioscin purified in our laboratory was dissolved in DMSO for cell experiments or in 0.5% sodium carboxymethyl cellulose (CMC‐Na) solution for in vivo tests. Diethylnitrosamine (DEN) and chloroquine (CQ) were obtained from Sigma (St. Louis, MO, USA). The detection kits of aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), γ‐glutamyl transpeptidase (γ‐GT) and cells apoptosis, and DNA Ladder Extraction kit were obtained from Nanjing Jiancheng Institute of Biotechnology (Nanjing, China). TransZolTM, TransScript® All‐in‐One First‐Strand cDNA Synthesis SuperMix for qPCR (One‐Step gDNA Removal), and TransStart® Top Green qPCR SuperMix were purchased from Beijing TransGen Biotech Co., Ltd (Beijing, China). Agarose gel, ethidium bromide, and bromophenol blue were obtained from TransGen Institute of Biotechnology (Beijing, China). The rat α fetoprotein (AFP) ELISA kit was purchased from Shanghai Langdun Institute of Biotechnology (Shanghai, China). We got the DMEM and FBS from Gibco (California, USA). Tissue Protein Extraction Kit was obtained from KEYGEN Biotech. Co., Ltd. (Nanjing, China). The bincinchoninic acid (BCA) Protein Assay Kit was purchased from Beyotime Institute of Biotechnology (Jiangsu, China). 3‐(4, 5‐Dimethylthiazol‐2‐yl)‐2, 5‐diphenyl terazolium bromide (MTT) was obtained from Roche Diagnostics (Basel, Switzerland). Acridine orange (AO) and ethidum bromide (EB) fluorescent dyes, DAPI, tris (hydroxymethyl) aminomethane (Tris), SDS, and CMC‐Na were purchased from Sigma. Comet assay kit was purchased from Cell Biolabs, Inc. (San Diego, USA). Primary and secondary antibodies were purchased from Proteintech™ (Wuhan, China) and Bioworld Technology, Inc. (Nanjing, China). Lipofectamin2000 and TP53‐induced glycolysis and apoptosis regulator (TIGAR) siRNA was purchased from RiboBio. Co., Ltd. (Guangzhou, China). The TUNEL assay kit was performed using the In Situ Cell Death Detection Kit (TMR Red, Roche, NJ, USA).
2.22. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander, Fabbro et al., 2017; Alexander, Kelly, et al., 2017).
3. RESULTS
3.1. Cytotoxicity of dioscin on HCC cells
SMMC7721, HepG2, Huh‐7, and BEL‐7402 cell lines were used to investigate the effects of dioscin against HCC in vitro. The results shown in Figure 1a revealed that dioscin substantially inhibited cell viabilities in a time and dose‐dependent manner (the IC50 values for the cell lines are listed in Table S3), especially to SMMC7721 and HepG2 cells with the IC50 values under 24‐hr treatment at 2.55 and 3.24 μM, which were more sensitive than the effect of the compound on L0–2 cells. As shown in Figure S2, dioscin obviously caused the death of SMMC7721 and HepG2 cells, and the apoptotic cells with orange fluorescence and condensed nucleus were increased with the increased concentrations of dioscin.
Figure 1.
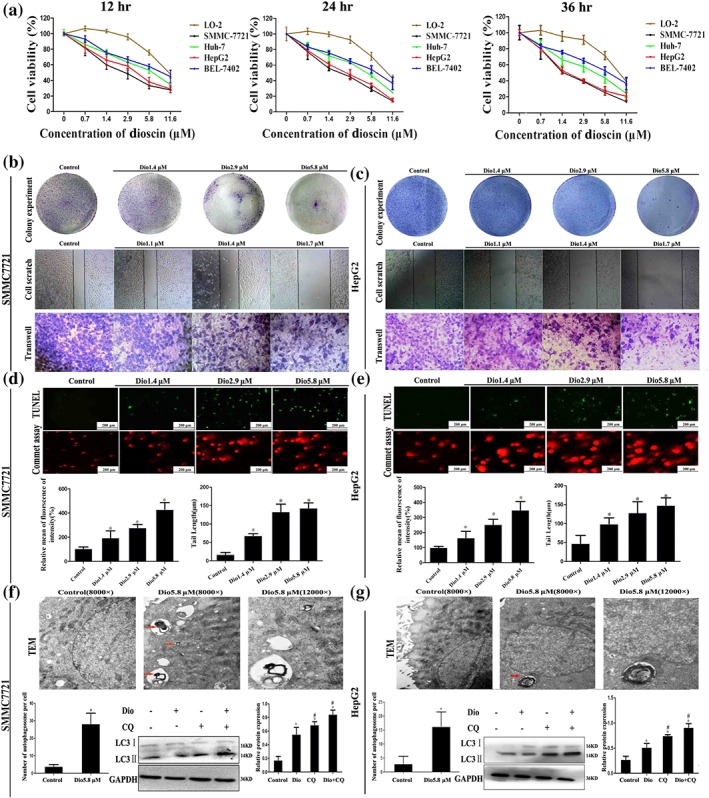
Effects of dioscin on HCC cells. (a) Effects of dioscin on viabilities of Huh‐7, SMMC7721, HepG2, BEL7402, and L‐02 cells. The cells were seeded into 96‐well plates at a density of 1 × 104 cells per well and treated with different concentrations of dioscin for 24 hr. The percentages of viable cells were determined using MTT assay. (b–c) Effects of dioscin on colony formation and motility in SMMC7721 and HepG2 cells. SMMC7721 and HepG2 cells were seeded (500 cells per well) into six‐well plates and treated by dioscin (1.4, 2.9, and 5.8 μM). The colonies were stained with haematoxylin solution and then counted. SMMC7721 and HepG2 cells were seeded into six well and treated by dioscin (0, 1.1, 1.4, and 1.7 μM) for 24 hr, and then cell migration was measured by wound‐healing assay. After incubation with dioscin for 24 hr, the invasive properties of SMMC7721 and HepG2 cells were tested in transwell plates. (d–e) Effects of dioscin on cell apoptosis and DNA damage in SMMC7721 and HepG2 cells under 24 hr treatment based on TUNEL and commet assays. (f–g) Effects of dioscin on autophagy and autophagic flux in SMMC7721 and HepG2 cells. The red arrows indicated the individual autophagosomes, and complete autophagic flux in the cancer cells was detected by determining the expression levels of LC‐3 II in the presence of CQ. Data are presented mean ± SD (n = 5). *P < 0.05 compared with control group; # P < 0.05 compared with dioscin group
3.2. Effects of dioscin on wound, invasion, and migration in SMMC7721 and HepG2 cells
As shown in Figure 1b–c, dioscin at the concentrations of 1.4, 2.9, and 5.8 μM significantly caused the decrease in colony formation on SMMC7721 and HepG2 cells. In addition, the capacities of wound healing on SMMC7721 and HepG2 cells were obviously attenuated by low concentrations of dioscin (1.1, 1.4, and 1.7 μM) in a dose‐dependent manner. Cell migration and invasion assays indicated that dioscin suppressed the invasive and migratory capabilities of the cancer cells compared with control groups. In a word, dioscin significantly inhibited the proliferation, migration, and invasion of SMMC7721 and HepG2 cells.
3.3. Dioscin causes apoptosis, autophagy, and DNA damage in cells
The data in Figure 1d–e showed that TUNEL‐positive cells were markedly increased by dioscin compared with control groups. In the commet assay, the nuclei in control groups were intact, round in shape and DNA remaining in nuclear matrix. However, in dioscin‐treated cells, DNA fragment migration formed smears with a smack head and big tail, suggesting that the lengths of DNA migration smear (comet tail) were significantly increased and the contents of head DNA were markedly decreased by dioscin. As shown in Figure S3, stronger DNA ladders were obviously found in dioscin‐treated groups compared with control groups. In addition, as shown in Figure 1f–g, treatment with 5.8 μM of dioscin for 24 hr significantly affected the morphological changes of SMMC7721 and HepG2 cells based on Transmission electron microscopy assay. Furthermore, the increased autophagosome formations in dioscin‐treated groups compared with control groups were found. Furthermore, CQ up‐regulated the expression levels of LC3‐II in dioscin‐treated cells, indicating that dioscin induced the complete autophagic flux in SMMC7721 and HepG2 cells. These findings demonstrated that dioscin significantly caused apoptosis, autophagy, and DNA damage in SMMC7721 and HepG2 cells.
3.4. Dioscin inhibited DEN‐induced primary liver cancer in rats
As shown in Figure 2a–b, the livers in DEN group were puffy and stiff, and enlarged livers and more numbers of nodules compared with control group were found, which were all ameliorated by dioscin. The body weights of rats in model group were lower than those of in control group (Figure 2c). However, dioscin significantly prevented the decrease in body weights compared with model group from 12 to 18 weeks (P < 0.05). In addition, the serum levels of AFP, ALT, AST, ALP, and γ‐GT were markedly increased in model group, which were also significantly reversed by dioscin (Figure 2d). H&E, Masson, Sirius red, Ki67, and α‐SMA staining indicated that the livers of the animals in control and dioscin‐treated groups showed normal architecture, whereas the livers in model group were destroyed with obvious liver fibrosis. Dioscin significantly reduced the extent of collagen deposition and improved hepatocellular architecture compared with DEN group (Figure 2e and Figure S4). In addition, TUNEL‐positive cells in dioscin groups were obvious compared with model group (Figure 2e). Taken together, dioscin showed active effects against DEN‐induced primary liver cancer in rats.
Figure 2.
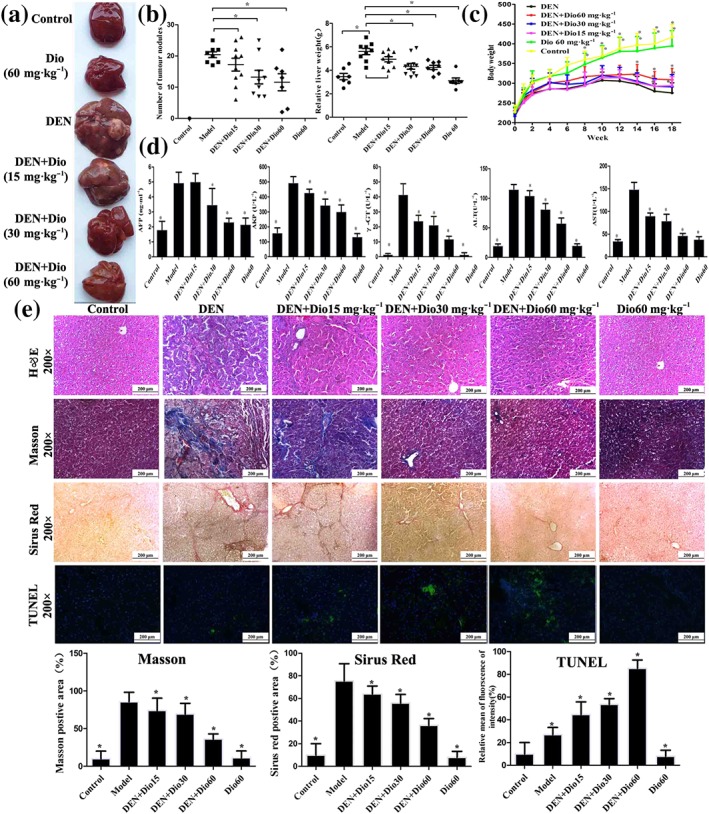
Effects of dioscin on primary liver cancer induced by DEN in rats. (a–b) Effects of dioscin on hepatic morphology of rats induced by DEN. The animals were randomly distributed into control group, model group (DEN), DEN+dioscin (60, 30, and 15 mg·kg−1) groups and dioscin group (60 mg·kg−1). The rats in model and DEN+dioscin groups received 0.01% DEN for 18 weeks, and other animals were given PBS. (c) Effects of dioscin on body weights of rats treated with DEN. The body weights of rats were measured once a week during the process. (d) Effects of dioscin on the serum levels of AFP, ALT, AST, ALP, and γ‐GT of rats induced by DEN. After 18 weeks of DEN administration, blood samples were collected and then the serum levels of AFP, ALT, AST, ALP, and GT were detected. (e) Effects of dioscin on primary liver cancer based on H&E, Masson, Sirius red, and TUNEL staining. All data are expressed as mean ± SD (n = 8). *P < 0.05 versus DEN model group
3.5. Dioscin inhibited the growth of HCC xenografts in nude mice
To evaluate the in vivo anticancer of dioscin, SMMC7721 and HepG2 cells tumour xenograft model were used and 5‐FU, which is a fluorinated pyrimidine analogue that acts as an antimetabolic agent, inhibiting thymidylate synthase and interfering with RNA synthesis and representing the standard first‐line chemotherapy to treat cancer (Suehiro et al., 2018), was applied as the positive drug. As shown in Figure 3a, the significant differences in tumour volumes were found based on tumour images after 27 days of treatment. The tumour volumes in 80 mg·kg−1 of dioscin‐treated group and 5‐FU‐treated group were decreased by 75.1% and 36.3% in nude mice transplanted with SMMC7721 cells and by 70.3% and 35.0% in nude mice transplanted with HepG2 cells respectively. The tumour weights were also significantly decreased. As shown in Figure 3b, H&E, Ki67, and TUNEL staining suggested that cell injuries were obviously found in dioscin‐treated groups compared with control groups. Taken together, dioscin showed potent effects against SMMC7721 and HepG2 cell xenografts in nude mice.
Figure 3.
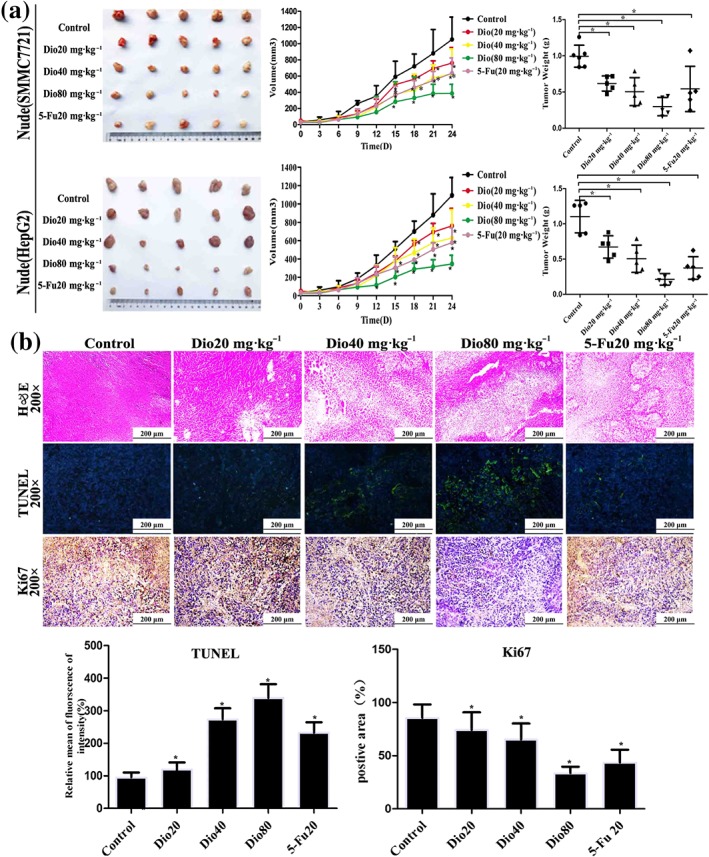
Effects of dioscin on xenografts in nude mice. (a) Effects of dioscin on tumour volumes and tumour weights of xenografts in nude mice. HepG2 and SMCC7721 cells were s.c. injected into the right flanks of nude mice to establish xenograft models. The mice were treated with dioscin (20, 40, and 80 mg·kg−1·day−1) or 5‐FU (20 mg·kg−1·day−1) for 24 days. After treatment, all mice were killed and the tumour volume was measured and calculated using the formula: V = A × B2/2. (b) Effects of dioscin on xenografts in nude mice based on H&E, TUNEL, and Ki67 staining. All data are expressed as mean ± SD (n = 5). *P < 0.05 compared with control group
3.6. Differentially expressed proteins in SMMC7721 cells caused by dioscin
The differentially expressed proteins caused by dioscin in SMMC7721 cells were identified by iTRAQ assay. A total of 5,189 proteins were quantified, and the isoelectric point distribution, protein mass distribution, and peptide number distribution were assayed in Figure S5A. Total of 190 differentially expressed proteins with the fold changes >1.5 and P < 0.05 were found, in which 117 proteins were up‐regulated (Table 1) and 73 proteins were down‐regulated (Table 2) by dioscin compared with control group. Then, the differentially expressed proteins were classified according to BP, cell component, and MF by gene ontology categories. As shown in Figure S5B, the differentially expressed proteins involved in MF included ion binding (38%), RNA binding (38%), enzyme binding (22%), oxidoreductase activity (21%), DNA binding (19%), cytoskeletal protein binding (16%), lipid binding (13%), transmembrane transporter activity (11%), and enzyme regulator activity (10%). A classification based on BP revealed that these proteins were mainly involved in biosynthetic process, anatomical structure development, cellular nitrogen compound metabolic process, and signal transduction. Importantly, the biological processes of the down‐regulated proteins associated with cell death, autophagy, and DNA metabolic process suggested that dioscin should induce apoptosis, autophagy, and DNA damage on SMMC7721 cells.
Table 1.
The differentially up‐regulated proteins in SMMC7721 cells caused by dioscin
| Protein name | Gene symbol | Fold change |
|---|---|---|
| Hydroxymethylglutaryl‐CoA synthase, cytoplasmic | HMGCS1 | 4.467876 |
| Acyl carrier protein, mitochondrial | NDUFAB1 | 4.270876 |
| Squalene synthase | FDFT1 | 4.137811 |
| Keratin, type I cytoskeletal 10 | KRT10 | 4.08447 |
| Isopentenyl‐diphosphate 𝛿‐isomerase 1 | IDI1 | 2.954908 |
| LDL receptor (fragment) | LDLR | 2.897039 |
| Keratin, type II cytoskeletal 1 | KRT1 | 2.742402 |
| Complement decay‐accelerating factor | CD55l | 2.63388 |
| Fatty acid‐binding protein, heart (fragment) | FABP3 | 2.547575 |
| Lanosterol 14‐α demethylase | CYP51A1 | 2.493801 |
| Keratin, type II cytoskeletal 2 epidermal | KRT2 | 2.481374 |
| 14‐3‐3 protein σ | SFN | 2.449948 |
| SOD | SOD2 | 2.370577 |
| Signal peptidase complex subunit 2 | SPCS2 | 2.313942 |
| Long‐chain‐fatty‐acid‐CoA ligase 4 | ACSL4 | 2.309261 |
| 𝛿(24)‐sterol reductase | DHCR24 | 2.303789 |
| 7‐dehydrocholesterol reductase | DHCR7 | 2.237158 |
| Annexin A1 | ANXA1 | 2.218028 |
| Protein S100‐P | S100P | 2.205782 |
| ATPase inhibitor, mitochondrial | ATPIF1 | 2.17796 |
| Microsomal GSH S‐transferase 3 | MGST3 | 2.152152 |
| Forkhead box protein K1 | FOXK1 | 2.03466 |
| PG G/H synthase 2 | PTGS2 | 1.977949 |
| Myristoylated alanine‐rich C‐kinase substrate | MARCKS | 1.94862 |
| Gamma‐IFN‐inducible protein 16 | IFI16 | 1.941166 |
| Peroxiredoxin‐5, mitochondrial | PRDX5 | 1.936759 |
| Enoyl‐CoA hydratase, mitochondrial | ECHS1 | 1.933657 |
| Proprotein convertase subtilisin/kexin type 9 | PCSK9 | 1.932362 |
| Protein TBRG4 | TBRG4 | 1.92955 |
| OCIA domain‐containing protein 2 | OCIAD2 | 1.923373 |
| RNA exonuclease 4 | REXO4 | 1.921324 |
| Ras‐related GTP‐binding protein C | RRAGC | 1.918965 |
| Ras‐related protein Rab‐5B | RAB5B | 1.894384 |
| Translocator protein | TSPO | 1.887573 |
| CD97 antigen | CD97 | 1.884349 |
| Niemann‐Pick C1 protein | NPC1 | 1.855988 |
| Prelamin‐A/C | LMNA | 1.846607 |
| Integrin α‐2 | ITGA2 | 1.845733 |
| Cytochrome c oxidase subunit 7C, mitochondrial | COX7C | 1.84376 |
| Farnesyl pyrophosphate synthase | FDPS | 1.83849 |
| V‐type proton ATPase subunit d 1 | ATP6V0D1 | 1.835165 |
| Proliferation‐associated protein 2G4 | PA2G4 | 1.826637 |
| Translation machinery‐associated protein 7 | TMA7 | 1.81405 |
| Endoplasmin | HSP90B1 | 1.808616 |
| Mitochondrial carrier homolog 2 | MTCH2 | 1.790893 |
| Urokinase plasminogen activator surface receptor | PLAUR | 1.789592 |
| 3‐ketoacyl‐CoA thiolase, mitochondrial | ACAA2 | 1.781226 |
| Heterogeneous nuclear ribonucleoprotein F | HNRNPF | 1.780622 |
| High mobility group protein B3 (fragment) | HMGB3 | 1.773229 |
| Dual specificity tyrosine‐phosphorylation‐regulated kinase 1A | DYRK1A | 1.768352 |
| Vimentin | VIM | 1.762179 |
| Acyl‐CoA‐binding protein | DBI | 1.734179 |
| Ceramide synthase 2 | CERS2 | 1.730889 |
| ORM1‐like protein 2 | ORMDL2 | 1.722456 |
| Disks large‐associated protein 4 | DLGAP4 | 1.721456 |
| Protein S100‐A13 | S100A13 | 1.717785 |
| Basic leucine zipper and W2 domain‐containing protein 1 | BZW1 | 1.712186 |
| Insulin‐like growth factor‐binding protein 1 | IGFBP1 | 1.710111 |
| Perilipin‐3 | PLIN3 | 1.696373 |
| Voltage‐dependent anion‐selective channel protein 3 | VDAC3 | 1.682508 |
| Nicalin | NCLN | 1.665872 |
| NADH dehydrogenase (Ubiquinone) flavoprotein 1, 51 kDa, isoform CRA_c | NDUFV1 | 1.657116 |
| Myeloid‐derived growth factor | MYDGF | 1.653477 |
| Integrin α‐5 | ITGA5 | 1.651647 |
| Plectin | PLEC | 1.649441 |
| Isoleucine–tRNA ligase, mitochondrial | IARS2 | 1.637101 |
| Cytochrome c oxidase subunit 6C | COX6C | 1.631336 |
| ATP synthase F(0) complex subunit B1, mitochondrial | ATP5F1 | 1.627885 |
| Cytochrome c1, haem protein, mitochondrial | CYC1 | 1.620842 |
| Transformer‐2 protein homolog α | TRA2A | 1.610992 |
| 28S ribosomal protein S35, mitochondrial | MRPS35 | 1.607127 |
| Sorting nexin‐9 | SNX9 | 1.605466 |
| Lamin‐B1 | LMNB1 | 1.605438 |
| Reticulon‐3 | RTN3 | 1.605057 |
| Dolichyl‐diphosphooligosaccharide‐protein glycosyltransferase subunit 1 | RPN1 | 1.599851 |
| Protein FAM3C | FAM3C | 1.585922 |
| Transcription factor MafK | MAFK | 1.580469 |
| Alcohol dehydrogenase class‐3 | ADH5 | 1.579261 |
| PG E synthase | PTGES | 1.578501 |
| Minor histocompatibility antigen H13 | HM13 | 1.577779 |
| Cytochrome c oxidase copper chaperone | COX17 | 1.573968 |
| Soluble calcium‐activated nucleotidase 1 | CANT1 | 1.570252 |
| NADH dehydrogenase [ubiquinone] iron–sulfur protein 3, mitochondrial | NDUFS3 | 1.569836 |
| Obg‐like ATPase 1 | OLA1 | 1.569714 |
| Thiosulfate sulfur transferase | TST | 1.56659 |
| Acidic leucine‐rich nuclear phosphoprotein 32 family member A | ANP32A | 1.566353 |
| Acetyl‐CoA acetyltransferase, mitochondrial | ACAT1 | 1.564878 |
| Purine nucleoside phosphorylase | PNP | 1.563292 |
| Prefoldin subunit 3 | VBP1 | 1.562049 |
| Prohibitin‐2 | PHB2 | 1.560065 |
| Actin‐related protein 2/3 complex subunit 5 | ARPC5 | 1.559988 |
| ATP‐dependent RNA helicase DDX3X | DDX3X | 1.553437 |
| Calpain small subunit 1 | CAPNS1 | 1.549143 |
| Sulfide:quinone oxidoreductase, mitochondrial | SQRDL | 1.548049 |
| Amyloid β A4 protein | APP | 1.543608 |
| Kinesin light chain 2 | KLC2 | 1.543524 |
| Barrier‐to‐autointegration factor | BANF1 | 1.538895 |
| Lanosterol synthase | LSS | 1.537363 |
| Mitochondrial import receptor subunit TOM5 homolog | TOMM5 | 1.536781 |
| SWI/SNF complex subunit SMARCC2S | MARCC2 | 1.534996 |
| Hydroxyacyl‐CoA dehydrogenase, mitochondrial | HADH | 1.531318 |
| Syntenin‐1 | SDCBP | 1.527044 |
| Acidic leucine‐rich nuclear phosphoprotein 32 family member E | ANP32E | 1.520728 |
| E3 ubiquitin‐protein ligase RNF149 | RNF149 | 1.515388 |
| ATP synthase subunit O, mitochondrial | ATP5O | 1.515363 |
| Metal transporter CNNM2 | CNNM2 | 1.514863 |
| Ubiquitin‐40S ribosomal protein S27a | RPS27A | 1.514685 |
| Serine palmitoyltransferase 1 | SPTLC1 | 1.513388 |
| Carbamoyl‐phosphate synthase [ammonia], mitochondrial | CPS1‐ | 1.512657 |
| Integrin β‐1 | ITGB1 | 1.512505 |
| Uroporphyrinogen decarboxylase | UROD | 1.512451 |
| All‐trans‐retinol 13,14‐reductase | RETSAT | 1.512367 |
| Keratin, type II cytoskeletal 5 | KRT5 | 1.510872 |
| NADPH–cytochrome P450 reductase | POR | 1.505179 |
| Beta‐2‐microglobulin | B2M | 1.504594 |
| Vacuolar protein sorting‐associated protein 35 | VPS35 | 1.503595 |
Table 2.
The differentially down‐regulated proteins in SMMC7721 cells caused by dioscin
| Protein name | Gene symbol | Fold change |
|---|---|---|
| Plakophilin‐3 | PKP3 | 0.667412 |
| Ribosomal biogenesis protein LAS1L | LAS1L | 0.666549 |
| Ras GTPase‐activating‐like protein IQGAP2 | IQGAP2 | 0.666464 |
| Alkaline phosphatase, placental‐like | ALPPL2 | 0.66389 |
| 40S ribosomal protein S28 | RPS28 | 0.663475 |
| Protein arginine N‐methyltransferase 7 | PRMT7 | 0.659268 |
| OTU domain‐containing protein 7B | OTUD7B | 0.658997 |
| Fructose‐2,6‐bisphosphatase | TIGAR | 0.657632 |
| Brain‐specific angiogenesis inhibitor 1‐associated protein 2 | BAIAP2 | 0.656172 |
| DNA‐directed RNA polymerase I subunit RPA34 | CD3EAP | 0.653801 |
| Multidrug resistance‐associated protein 4 | ABCC4 | 0.65079 |
| IL‐1α | IL1A | 0.650269 |
| Catenin β‐1 | CTNNB1 | 0.64775 |
| Nuclease‐sensitive element‐binding protein 1 | YBX1 | 0.646298 |
| Aryl hydrocarbon receptor nuclear translocator | ARNT | 0.644848 |
| Kinase D‐interacting substrate of 220 kDa | KIDINS220 | 0.644789 |
| 40S ribosomal protein S23 | RPS23 | 0.642951 |
| Histone‐lysine N‐methyltransferase | EHMT1 | 0.642702 |
| Ras‐related protein Rab‐5C | RAB5C | 0.638376 |
| Choline transporter‐like protein 2 | SLC44A2 | 0.638143 |
| Heterogeneous nuclear ribonucleoprotein Q | SYNCRIP | 0.636071 |
| Programmed cell death protein 2 | PDCD2 | 0.635812 |
| Alkaline phosphatase, placental type | ALPP | 0.630986 |
| Rap1 GTPase‐GDP dissociation stimulator 1 | RAP1GDS1 | 0.629451 |
| Thrombospondin‐1 | THBS1 | 0.625131 |
| Serine/arginine‐rich splicing factor 6 | SRSF6 | 0.624837 |
| tRNA (guanine(26)‐N(2))‐dimethyltransferase | TRMT1 | 0.623349 |
| Importin subunit α‐3 | KPNA4 | 0.62148 |
| Keratin, type II cytoskeletal 80 | KRT80 | 0.621039 |
| 60S ribosomal protein L28 | RPL28 | 0.619835 |
| Vacuolar protein sorting‐associated protein 13A | VPS13A | 0.619703 |
| GSH peroxidase 1 | GPX1 | 0.619061 |
| Vacuolar protein sorting‐associated protein 13C | VPS13C | 0.617022 |
| Ribosomal L1 domain‐containing protein 1 | RSL1D1 | 0.614332 |
| Calpain‐7 | CAPN7 | 0.612377 |
| Protein CYR61 | CYR61 | 0.611353 |
| Multidrug resistance‐associated protein 1 | ABCC1 | 0.605965 |
| NEDD8 ultimate buster 1 | NUB1 | 0.603901 |
| F‐box‐like/WD repeat‐containing protein TBL1XR1 | TBL1XR1 | 0.596918 |
| Protein CutA | CUTA | 0.586437 |
| LIM and calponin homology domains‐containing protein 1 | LIMCH1 | 0.584768 |
| Protein PRRC2B | PRRC2B | 0.574416 |
| Periplakin | PPL | 0.566886 |
| Eukaryotic translation initiation factor 4B | EIF4B | 0.563664 |
| Dystonin | DST | 0.562774 |
| Protein HGH1 homolog | HGH1 | 0.55638 |
| Nucleoredoxin | NXN | 0.552367 |
| Polymerase I and transcript release factor | PTRF | 0.552291 |
| Histone H2B type 1‐D | HIST1H2BD | 0.549395 |
| Drebrin | DBN1 | 0.54526 |
| Basal cell adhesion molecule | BCAM | 0.543613 |
| Protein SCAF11 | SCAF11 | 0.541997 |
| Cyclin‐dependent kinase 6 | CDK6 | 0.536053 |
| Coiled‐coil domain‐containing protein 85C | CCDC85C | 0.534299 |
| Tropomyosin α‐1 chain | TPM1 | 0.526925 |
| Palladin | PALLD | 0.525911 |
| GSH peroxidase | GPX4 | 0.525253 |
| Eukaryotic translation initiation factor 5A‐1 | EIF5A | 0.518732 |
| CD276 antigen | CD276 | 0.517228 |
| Disabled homolog 2 | DAB2 | 0.505867 |
| PGF2 receptor negative regulator | PTGFRN | 0.490684 |
| Radixin | RDX | 0.488652 |
| Nuclear ubiquitous casein and cyclin‐dependent kinase substrate 1 | NUCKS1 | 0.465746 |
| mRNA‐capping enzyme | RNGTT | 0.439002 |
| Nesprin‐1 | SYNE1 | 0.41975 |
| Probable methyltransferase TARBP1 | TARBP1 | 0.396494 |
| Transferrin receptor protein 1 | TFRC | 0.34983 |
| Tropomyosin β chain | TPM2 | 0.34593 |
| YTH domain‐containing protein 1 | YTHDC1 | 0.330133 |
| Serine/threonine‐protein kinase | ULK4 | 0.016407 |
3.7. Dioscin decreased the expression levels of TIGAR in vivo and in vitro
Among the differentially expressed proteins, the expression level of one target protein named TIGAR was markedly inhibited by dioscin compared with control group. As a novel p53‐inducible gene, TIGAR's association with p53, Akt/mTOR, and other signal pathways was verified based on KEGG pathway database (Figure S6). Hence, TIGAR was selected as the target protein for further understanding the mechanisms of dioscin against HCC. As expected, dioscin significantly suppressed the expression levels of TIGAR in SMMC7721 cells, HepG2 cells, rats, and mice based on immunofluorescence assay (Figure S7). In addition, the results in Figure 4a–b indicated that the mRNA and protein levels of TIGAR in SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice were markedly decreased by dioscin compared with control groups based on real‐time PCR and Western blotting assays.
Figure 4.
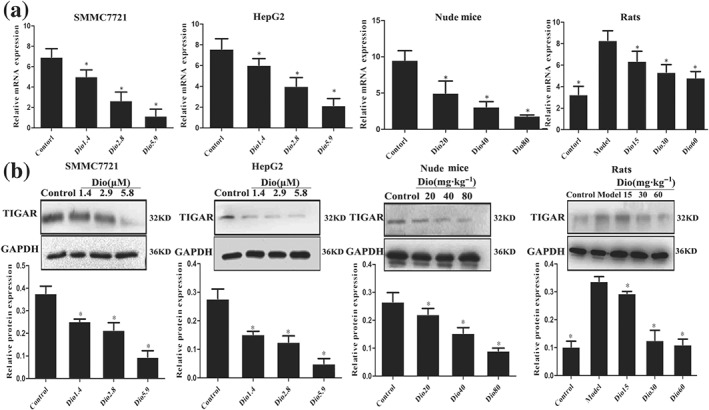
Dioscin decreased the expression levels of TIGAR in vitro and in vivo. (a) Effects of dioscin on the mRNA levels of TIGAR in vitro and in vivo based on real‐time PCR assay. Total RNA samples were obtained from SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice. (b) Effects of dioscin on the protein levels of TIGAR in vitro and in vivo based on Western blotting assay. Total proteins from SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice were isolated. After separation on SDS‐PAGE gels, the proteins were detected and imaged. All data are expressed as mean ± SD (n = 5). *P < 0.05 versus control group
3.8. Dioscin affected TIGAR‐mediated apoptosis, autophagy, and DNA damage
As shown in Figure 5a, the results indicated that the expression levels of p53, cleaved caspase‐3, cleaved caspase‐9, cleaved PARP, and Bax were markedly increased, and the levels of Bcl‐2 were decreased compared with control groups in SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice. As shown in Figure 5b, dioscin significantly down‐regulated the expression levels of p‐Akt and p‐mTOR and up‐regulated the expression levels of LC3 and Beclin1 in SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice. As shown in Figure 5c, dioscin markedly restrained the expression levels of CDK5 and p‐ATM in vitro and in vivo to adjust DNA damage (details of fold changes and significances of these proteins in are listed in Figure S8).
Figure 5.
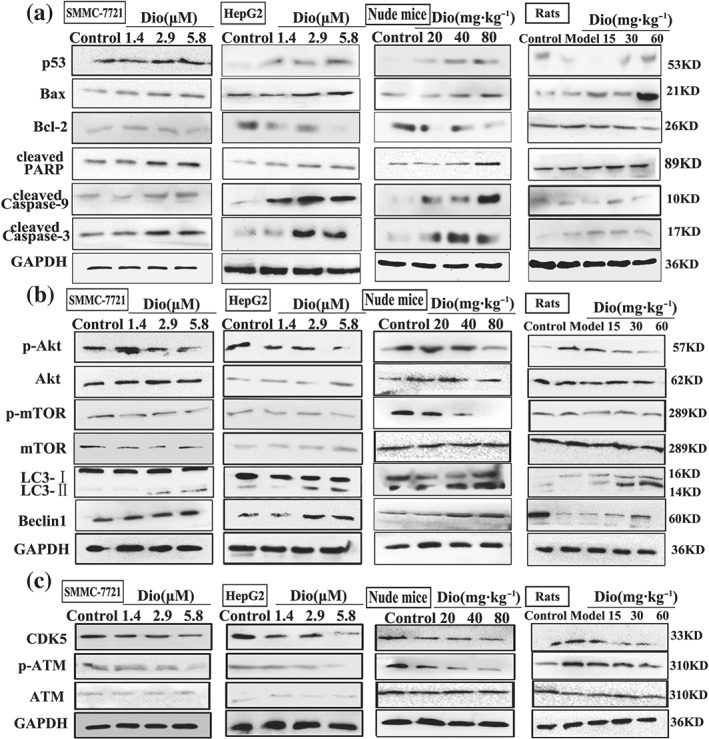
Dioscin adjusted TIGAR‐mediated apoptosis, autophagy, and DNA damage. (a) Effects of dioscin on the expression levels of p53, Bax, Bcl‐2, cleaved PAPR, and cleaved caspase‐3/9 in vitro and in vivo. (b) Effects of dioscin on the expression levels of p‐Akt/Akt, p‐mTOR/mTOR, Beclin‐1, and LC3 in vitro and in vivo. (c) Effects of dioscin on the expression levels of CDK5 and p‐ATM/ATM in vitro and in vivo. Total proteins from SMMC7721 and HepG2 cells, livers of rats, and tumour tissues of nude mice were isolated using the kit. After separation on SDS‐PAGE gels, the proteins were detected and imaged. Intensity values of the relative protein levels were normalized to GAPDH
3.9. TIGAR siRNA aggravated the effects of dioscin on TIGAR signalling in vitro
To explore the role of TIGAR in anticancer activity of dioscin, TIGAR siRNA transfection approach in vitro was tested. Knockdown of TIGAR was performed with three siRNAs (si‐h‐TIGAR1, GTATGACCTACAGGATCA, si‐h‐TIGAR2, CAGCGGTATTCCAGGATTA, si‐h‐TIGAR3, GATGAACCTCTTTCAGAAA) and ideally si‐h‐TIGAR1 was selected for the transfection (Figure S9). As shown in Figure 6a and Figure S10, effects of dioscin on colony formation, motility, apoptosis, and DNA damage in SMMC7721 cells were all aggravated by TIGAR siRNA. Moreover, no significant differences in the colony formation, wound healing, cell migration, apoptosis, and DNA damage between TIGAR siRNA group and dioscin‐treated group after transfection of TIGAR siRNA were found (P > 0.05). In addition, after transfecting with TIGAR siRNA in the presence or absence of dioscin, the expression levels of TIGAR, p‐Akt, p‐mTOR CDK5, and p‐ATM were notably decreased, and the expression levels of p53 were markedly increased compared with control group (Figure 6b).
Figure 6.
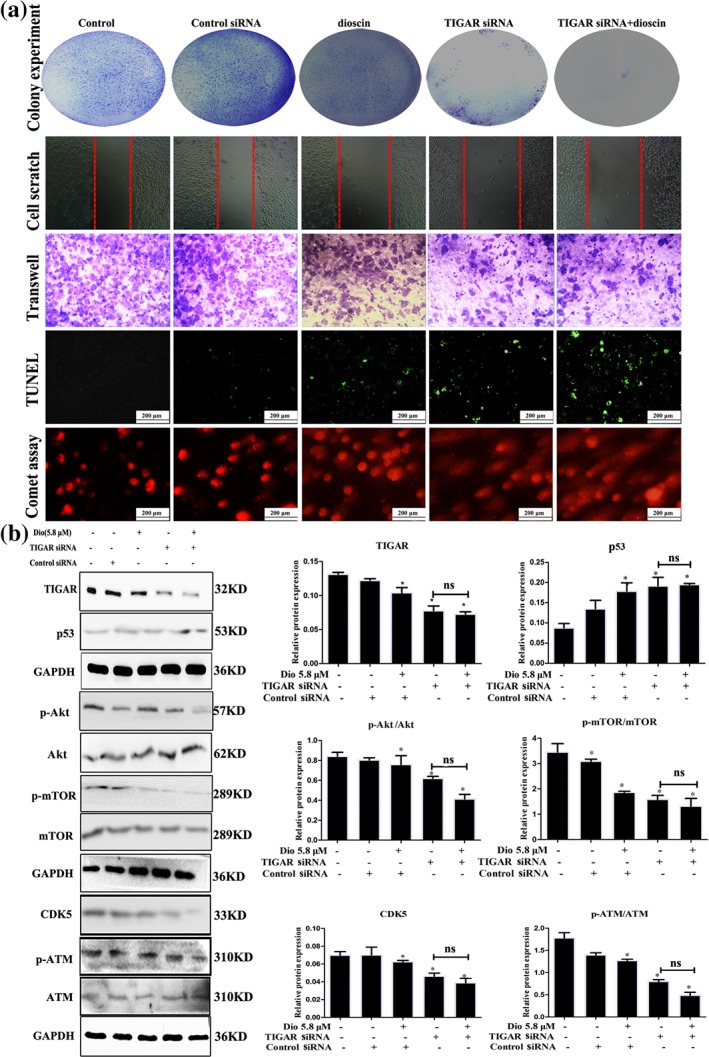
TIGAR siRNA aggravated the effects of dioscin on TIGAR‐mediated signal in vitro. (a) Effects of dioscin on colony formation, motility, cell apoptosis, and DNA damage after TIGAR‐siRNA transfection in SMMC7721 cells. SMMC7721 cells were cultured in six‐well plates and transfected with the siRNA liposomes in antibiotic‐free cell medium. Colony formation, motility, cell apoptosis, and DNA damage were detected after 24 hr of transfection in the absence or presence of dioscin (5.6 μM). (b) Effects of dioscin on the expression levels of TIGAR, p53, p‐Akt/Akt, p‐mTOR/mTOR, CDK5, and p‐ATM/ATM in SMMC7721 cells after TIGAR‐siRNA transfection in SMMC7721 cells. All data are expressed as mean ± SD (n = 5). *P < 0.05 versus control group; n.s: no significance
3.10. TIGAR siRNA aggravated the effects of dioscin on TIGAR signalling in vivo
As shown in Figure 7a–c, the results indicated that dioscin notably decreased tumour volume and tumour weight on SMMC7721 cell tumour xenograft in nude mice. Moreover, TIGAR siRNA transfection decreased tumour volume and weight with or without dioscin, suggesting that TIGAR siRNA aggravated the inhibitory effects of dioscin on tumour growth in vivo. The results of immunofluorescence and TUNEL assays indicated that transfection with TIGAR siRNA markedly decreased the expression level of TIGAR and induced cell apoptosis (Figure 7d and Figure S11). In addition, as shown in Figure 7e, the expression levels of TIGAR, p‐mTOR, p‐Akt, CDK5, and p‐ATM were decreased, and the expression levels of p53 were markedly increased compared with control group. These data revealed that inhibiting TIGAR aggravated the action of dioscin against HCC, suggesting that dioscin reduced TIGAR expression to exert active effects against HCC.
Figure 7.
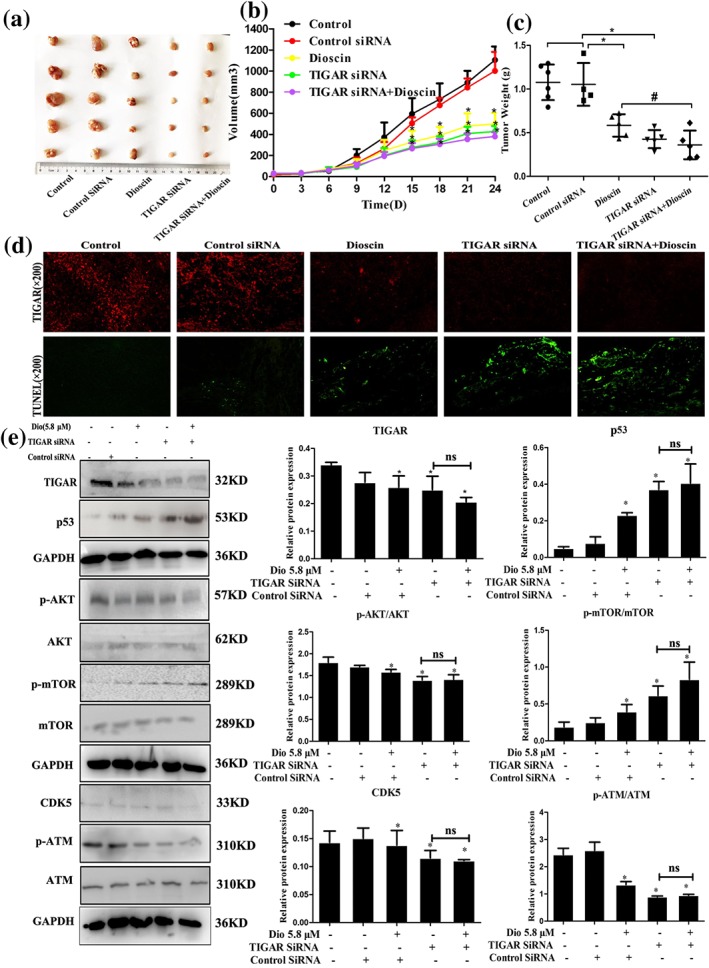
TIGAR siRNA exacerbated the effects of dioscin on TIGAR‐mediated signalling in mice. (a) Effects of dioscin on SMMC7721 cell tumour xenograft in nude mice after TIGAR‐siRNA transfection. The mice were randomly divided into the indicated groups, and dioscin was administered by gavage once daily for 24 days. Chemically modified TIGAR‐siRNA was diluted in PBS and intratumoural injected into the mice once every 3 days. After administration, the tumours were collected and imaged. (b–c) Effects of dioscin on tumour volume (*P < 0.05 vs. control group) and tumour weight (# P < 0.05 vs. dioscin group) on SMMC7721 cell tumour xenograft in nude mice after TIGAR‐siRNA transfection. (d) Effects of dioscin on TIGAR expression level and cell apoptosis in tumour tissues of SMMC7721 cell tumour xenograft in nude mice based on immunofluorescence and TUNEL assays after TIGAR‐siRNA transfection. (e) Effects of dioscin on the expression levels of TIGAR, p53, p‐Akt/Akt, p‐mTOR/mTOR, CDK5, and p‐ATM/ATM in tumour tissues of SMMC7721 cell tumour xenograft in nude mice based on Western blotting assay after TIGAR‐siRNA transfection (*P < 0.05 vs. control group; n.s: no significance). All data are expressed as mean ± SD (n = 5)
4. DISCUSSION
Natural products from medicinal plants may be the new and ideal sources for discovery of anti‐HCC drugs (Yuan, Wang, Wang, Xiao, & Liu, 2014). Dioscin, a natural product, has beneficial effects against colon cancer, glioblastoma multiforme, and lung cancer (Lv et al., 2013; Wei et al., 2013; Zhang, Han, et al., 2015). In our previous studies, dioscin has excellent protective effects against ethanol‐induced liver injury (Xu et al., 2014). In the present work, the results demonstrated that dioscin notably inhibited the viability, colony formation, migration, and invasion on SMMC7721 and HepG2 cells. DEN, one of the most significant environmental carcinogens, can induce carcinoma in animals and humans. Reportedly, overproduction of ROS can cause oxidative stress and cellular injury (Zhang, Zeng, Zhao, & Xie, 2015). A large amounts of 8‐hydroxy‐2‐deoxyguanosine produced by DEN in rat liver can exert its carcinogenic effects. Thus, DEN‐induced hepatocarcinoma animal model has been considered as one of the most accepted experimental models to evaluate the chemoprotective effects of drugs or chemicals in a variety of studies. Hence, DEN‐induced hepatocarcinoma model in rats was used to evaluate the chemoprotective effects of dioscin, and the results showed that dioscin significantly reduced the numbers and sizes of tumour nodules and improved histopathological damage and serum biochemical indices against primary liver cancer in rats. Furthermore, dioscin at the dose of 80 mg·kg−1 significantly suppressed tumour growth of SMMC7721 and HepG2 cell xenografts in nude mice. In general, the above data indicated that dioscin showed potent effects against HCC in vitro and in vivo via guiding cell apoptosis, autophagy, and DNA damage.
Up to now, proteomic methods are powerful tools for identification of biological markers (Li, Zhang, Wang, Liu, & Lu 2014). The proteomic method of stable‐isotope labelling by amino acids has been used to find that dioscin can target integrin α5 to suppress collagen synthesis against liver fibrosis (Xu et al., 2017). iTRAQ‐based proteomic method has been used to discover the molecular targets of dioscin on human HCT‐116 colon cancer cells (Chen et al., 2014). In the present work, we used iTRAQ method to obtain valuable evidences on proteomic changes to understand the mechanisms of dioscin against HCC, and the differentially expressed protein named TIGAR associated with apoptosis, autophagy, and DNA damage was selected as the drug target for mechanism investigation.
TIGAR, a novel p53‐inducible gene, functions as a fructose‐2,6‐bisphosphatase that inhibits cellular glycolysis and plays a crucial role in apoptosis (Green & Chipuk, 2006). High expression levels of TIGAR in glioblastoma, breast cancer, and colorectal cancer have been reported (Kim, Choi, Won, & Lim, 2016; Li, Sun, & Cao, 2014; Zhou et al., 2016). Previous studies have also demonstrated that knockdown of TIGAR can induce apoptosis and autophagy in tumour cells (Ye et al., 2013). In general, TIGAR as one p53‐inducible gene should be up‐regulated since p53 is induced in multiple cell lines. However, decreased ROS levels in response to the expression of TIGAR has also been reported to play a role of p53 for protecting cells against genomic damage, which is considered another relationship between p53 and the metabolic changes in cancer (Bensaad et al., 2006). In addition, the increased expression of TIGAR through down‐regulated p53 expression can decrease cellular sensitivity to ROS‐associated apoptosis (Kim et al., 2016). In the present work, we found that dioscin significantly decreased the expression levels of TIGAR in vitro and in vivo, suggesting that the anticancer effect of dioscin against HCC may be through adjusting TIGAR. As one p53‐targeted gene associated with low levels of stress, Bensaad et al. have also demonstrated that high expression of p53 can be produced by low expression of TIGAR in human breast cancers (Bensaad et al., 2006). In this study, we observed that dioscin significantly increased the expression levels of the proteins including p53, Bcl‐2 associated X protein, cleaved cysteinylaspartate specific proteinase 3/9, cleaved PARP, and decreased Bcl‐2 expression level to induce cell apoptosis in vivo and in vitro. Further transfection tests of TIGAR‐siRNA in vitro also showed that TIGAR‐siRNA aggravated the effects of dioscin on SMMC7721 cell colony formation, apoptosis, and migration. In addition, in vivo TIGAR‐siRNA transfection in mice further showed that the decreased tumour volume and weight were found. The similar results were obviously observed in the expression levels of TIGAR and p53 in vitro and in vivo, indicating that dioscin exerted anticancer effect against HCC via regulating TIGAR‐mediated p53 signal.
Autophagy, one conserved mechanism for degradating intracellular substances, is responsible for the recycle of metabolic substances and the maintenance of intracellular stability (Xie et al., 2014). To further understand the mechanism of dioscin on affecting autophagy, some experiments of detecting the expression levels of autophagy‐related proteins and determining autophagy flux by employing autophagy flux inhibitor CQ caused by dioscin were carried out. The results revealed that dioscin induced autophagy against HCC in vivo and in vitro. The signal of Akt/mTOR is important in regulating intracellular homeostasis, and it is reported that activation of Akt/mTOR signal is essential in autophagy (Yan et al., 2018). Interestingly, recent studies have highlighted that TIGAR knockdown can enhance epriubicin‐induced autophagy by inhibiting mTOR pathway, suggesting the protective effect of autophagy on the survival of cancer cells when TIGAR is knocked down (Kumar, Iqbal, Singh, & Bamezai, 2015). In the present study, we found that dioscin significantly decreased the expression levels of p‐Akt/Akt and p‐mTOR/mTOR in vitro and in vivo. TIGAR‐siRNA transfection tests in vitro and in vivo showed that dioscin altered the expression levels of p‐Akt/Akt and p‐mTOR/mTOR. Knockdown of TIGAR showed no significant effect on the expression levels of Akt and mTOR but decreased the phosphorylation levels of Akt and mTOR, suggesting that dioscin showed potent effects against HCC via regulating TIGAR‐mediated Akt/mTOR signal pathway.
DNA damage response results in DNA repair associated with cell survival or cell death (Driscoll & Chowdhury, 2012). TIGAR functions to lower level of fructose‐2,6‐bisphosphate, resulting in higher intracellular NADPH level and lower ROS level (Yu et al., 2015). ROS can induce DNA damages including base oxidation, sugar fragmentation, and single strand DNA breaks (Camins et al., 2009). Some studies have explored the role of TIGAR in DNA damage response, and more serious DNA damage after TIGAR knockdown has been found, suggesting the protective role of TIGAR on DNA damage (Lee, Kim, Lee, & Kim, 2007). Our present study found that only DNA damage induced by dioscin based on comet assay was found at the very beginning. Consistent with previous studies, the present results showed that down‐regulated TIGAR by dioscin reduced the expression levels of CDK5 as well as phosphorylated ATM (Tian, Yang, & Mao, 2009; Yu et al., 2015). The above data showed that dioscin showed potent effects against HCC via adjusting TIGAR‐mediated CDK5/ATM signal pathway.
Therefore, our findings in this paper provided novel insights into the molecular mechanisms of dioscin against HCC (Figure S12). Dioscin should be developed as an efficient candidate, and TIGAR should also be considered as one drug target to treat liver cancer in clinical in the future. However, the proteomic data showed that there have some other differently expressed proteins caused by dioscin which may exert the effects associated with TIGAR. In the present work, we only selected TIGAR as the targeted gene for the study. Besides, the anticancer effects of dioscin against HCC are complex, and one gene cannot fully demonstrate the actions of the compound. Network regulation associated with multiple genes, multiple signal pathways, and multiple biological processed should be suitable for the study and we will try our best to fully elucidate the molecular mechanisms of dioscin against HCC in our future work.
AUTHOR CONTRIBUTIONS
Z.M., X.H. and J.P. designed the experiments and wrote the manuscript. Z.M., D.C. and Y.X. performed the animal and cell experiments. Z.M., L.Y. and L.X. performed the quantitative real‐time PCR and western blotting assays. Z.M., H.S. and Y.Q. performed the gene transfection experiments. L.F., K.L. and J.P. edited the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation, and as recommended by funding agencies, publishers, and other organizations engaged with supporting research.
Supporting information
Data S1. Supporting information
ACKNOWLEDGEMENTS
This study was financially supported by the Key Research and Development Project of Liaoning Province (2017225090), the Special Grant for Translational Medicine, Dalian Medical University (2015004), and the Basic Scientific Research Projects of Liaoning University (LF2017010).
Mao Z, Han X, Chen D, et al. Potent effects of dioscin against hepatocellular carcinoma through regulating TP53‐induced glycolysis and apoptosis regulator (TIGAR)‐mediated apoptosis, autophagy, and DNA damage. Br J Pharmacol. 2019;176:919–937. 10.1111/bph.14594
REFERENCES
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , … CGTP Collaborators (2017). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. British Journal of Pharmacology, 174(S1), S272–S359. 10.1111/bph.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , Harding, S. D. , … CGTP Collaborators (2017). The Concise Guide to PHARMACOLOGY 2017/18: Overview. British Journal of Pharmacology, 174(S1), S1–S16. 10.1111/bph.13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas, M. , Mazzolini, G. , Genove, G. , Bilbao, R. , Narvaiza, I. , Schmitz, V. , … Prieto, J. (2001). Gene therapy of orthotopic hepatocellular carcinoma in rats using adenovirus coding for interleukin 12. Hepatology, 33, 52–61. 10.1053/jhep.2001.20796 [DOI] [PubMed] [Google Scholar]
- Bensaad, K. , Tsuruta, A. , Selak, M. A. , Vidal, M. N. , Nakano, K. , Bartrons, R. , … Vousden, K. H. (2006). TIGAR, a p53‐inducible regulator of glycolysis and apoptosis. Cell, 126, 107–120. 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- Camins, A. , Crespo‐Biel, N. , Junyent, F. , Verdaguer, E. , Canudas, A. M. , & Pallàs, M. (2009). Calpains as a target for therapy of neurodegen‐erative diseases: Putative role of lithium. Current Drug Metabolism, 10, 433–447. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Xu, L. , Yin, L. , Xu, Y. , Han, X. , Qi, Y. , … Peng, J. (2014). iTRAQ‐based proteomic analysis of dioscin on human HCT‐116 colon cancer cells. Proteomics, 14, 51–73. 10.1002/pmic.201300101 [DOI] [PubMed] [Google Scholar]
- Cho, J. Y. (2013). The antifungal activity and membrane‐disruptive action of dioscin extracted from Dioscorea nipponica . Biochimica et Biophysica Acta, 1828, 1153–1158. 10.1016/j.bbamem.2012.12.010 [DOI] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175(7), 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, J. J. , & Chowdhury, R. D. (2012). Molecular crosstalk between the proteasome, aggresomes and autophagy: Translational potential and clinical implications. Cancer Letters, 325, 147–154. 10.1016/j.canlet.2012.06.016 [DOI] [PubMed] [Google Scholar]
- Faridah, H. , Ataollahi, E. S. , & Asmah, R. (2014). Thecentella asiaticajuice effects on DNA damage, apoptosis and gene expression in hepatocellular carcinoma (HCC). BMC Complementary and Alternative Medicine, 14, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Zhen, R. , Liao, H. , Zhuang, W. , & Guo, W. (2018). Pharmacokinetics of continuous transarterial infusion of 5‐fluorouracil in patients with advanced hepatocellular carcinoma. Oncology Letters, 15, 7175–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, K. , & Li, W. (2011). Shikonin, a Chinese plant‐derived naphthoquinone, induces apoptosis in hepatocellular carcinoma cells through reactive oxygen species: A potential new treatment for hepatocellular carcinoma. Free Radical Biology & Medicine, 51, 2259–2271. [DOI] [PubMed] [Google Scholar]
- Green, D. R. , & Chipuk, J. E. (2006). p53 and metabolism: Inside the TIGAR. Cell, 126, 30–32. 10.1016/j.cell.2006.06.032 [DOI] [PubMed] [Google Scholar]
- Greten, T. F. , Xin, W. W. , & Korangy, F. (2015). Current concepts of immune based treatments for patients with HCC: From basic science to novel treatment approaches. Gut, 64, 842–848. 10.1136/gutjnl-2014-307990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR (2018). The IUPHAR/BPS guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46, D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, A. , Samykutty, A. , Jackson, C. , Browning, D. , Bollag, W. B. , Thangaraju, M. , … Singh, S. R. (2015). Curcumin inhibits PhIP induced cytotoxicity in breast epithelial cells through multiple molecular targets. Cancer Letters, 365, 122–131. 10.1016/j.canlet.2015.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. 10.1111/j.1476-5381.2010.00872.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. H. , Choi, S. I. , Won, K. Y. , & Lim, S. J. (2016). Distinctive interrelation of p53 with SCO2, COX, and TIGAR in human gastric cancer. Pathology, Research and Practice, 212, 904–910. 10.1016/j.prp.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Kumar, B. , Iqbal, M. A. , Singh, R. K. , & Bamezai, R. N. (2015). Resveratrol inhibits TIGAR to promote ROS induced apoptosis and autophagy. Biochimie, 118, 26–35. 10.1016/j.biochi.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Lee, J. H. , Kim, H. S. , Lee, S. J. , & Kim, K. T. (2007). Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death. Journal of Cell Science, 120, 2259–2271. 10.1242/jcs.03468 [DOI] [PubMed] [Google Scholar]
- Li, M. , Sun, M. , & Cao, L. (2014). A TIGAR‐regulated metabolic pathway is critical for protection of brain ischemia. The Journal of Neuroscience, 34, 7458–7471. 10.1523/JNEUROSCI.4655-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. Z. , Zhang, S. N. , Wang, K. X. , Liu, S. M. , & Lu, F. (2014). iTRAQ‐based quantitative proteomics study on the neuroprotective effects of extract of Acanthopanax senticosus harm on SH‐SY5Y cells overexpressing A53T mutant α‐synuclein. Neurochemistry International, 72, 37–47. 10.1016/j.neuint.2014.04.012 [DOI] [PubMed] [Google Scholar]
- Liu, C. , Wang, Y. , Wu, C. , Pei, R. , Song, J. , Chen, S. , & Chen, X. (2013). Dioscin's antiviral effect in vitro. Virus Research, 172, 9–14. 10.1016/j.virusres.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Yu, H. , Zhang, X. , Wang, Y. , Song, Z. , Zhao, J. , … Zhang, L. W. (2018). The protective role of autophagy in nephrotoxicity induced by bismuth nanoparticles through AMPK/mTOR pathway. Nanotoxicology, 6, 1–16. [DOI] [PubMed] [Google Scholar]
- Lu, B. , Xu, Y. , Xu, L. , Cong, X. , Yin, L. , Li, H. , & Peng, J. (2012). Mechanism investigation of dioscin against CCl4‐induced acute liver damage in mice. Environmental Toxicology and Pharmacology, 34, 127–135. 10.1016/j.etap.2012.03.010 [DOI] [PubMed] [Google Scholar]
- Lv, L. , Zheng, L. , Dong, D. , Xu, L. , Yin, L. , Xu, Y. , … Peng, J. (2013). Dioscin, a natural steroid saponin, induces apoptosis and DNA damage through reactive oxygen species: A potential new drug for treatment of glioblastoma multiforme. Food and Chemical Toxicology, 59, 657–669. 10.1016/j.fct.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Marquardt, J. U. , Galle, P. R. , & Teufel, A. (2012). Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): An emerging field for advanced technologies. Journal of Hepatology, 56, 267–275. 10.1016/j.jhep.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Polina, I. , Lubov, T. , & Timchenko, N. A. (2011). Intracellular signaling and hepatocellular carcinoma. Seminars in Cancer Biology, 21, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, L. , Liu, Y. , Xu, Y. , Ji, W. , Wu, Q. , Liu, Y. , … Su, C. (2015). Matrine derivative WM130 inhibits hepatocellular carcinoma by suppressing EGFR/ERK/MMP‐2 and PTEN/Akt signaling pathways. Cancer Letters, 368, 126–134. 10.1016/j.canlet.2015.07.035 [DOI] [PubMed] [Google Scholar]
- Si, L. , Zheng, L. , Xu, L. , Yi, L. , Han, X. , Qi, Y. , … Peng, J. (2016). Dioscin suppresses human laryngeal cancer cells growth via induction of cell‐cycle arrest and MAPK‐mediated mitochondrial‐derived apoptosis and inhibition of tumor invasion. European Journal of Pharmacology, 774, 105–117. 10.1016/j.ejphar.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Suehiro, Y. , Takemoto, Y. , Nishimoto, A. , Ueno, K. , Shirasawa, B. , Tanaka, T. , … Hamano, K. (2018). Dclk1 inhibition cancels 5‐FU‐induced cell‐cycle arrest and decreases cell survival in colorectal cancer. Anticancer Research, 38, 6225–6230. 10.21873/anticanres.12977 [DOI] [PubMed] [Google Scholar]
- Sun, B. T. , Zheng, L. H. , Bao, Y. L. , Yu, C. L. , Wu, Y. , Meng, X. Y. , & Li, Y. X. (2011). Reversal effect of dioscin on multidrug resistance in human hepatoma HepG2/adriamycin cells. European Journal of Pharmacology, 654, 129–134. 10.1016/j.ejphar.2010.12.018 [DOI] [PubMed] [Google Scholar]
- Tian, B. , Yang, Q. , & Mao, Z. (2009). Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nature Cell Biology, 11, 211–218. 10.1038/ncb1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. X. , Han, J. , Zhao, L. W. , Jiang, D. X. , Liu, Y. T. , & Liu, X. L. (2010). Anthelmintic activity of steroidal saponins from Paris polyphylla. Phytomedicine, 17, 1102–1105. 10.1016/j.phymed.2010.04.012 [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Xu, Y. , Han, X. , Qi, Y. , Xu, L. , Xu, Y. , … Peng, J. (2013). Anti‐cancer effects of dioscin on three kinds of human lung cancer cell lines through inducing DNA damage and activating mitochondrial signal pathway. Food and Chemical Toxicology, 59, 118–128. 10.1016/j.fct.2013.05.054 [DOI] [PubMed] [Google Scholar]
- Xie, J. M. , Li, B. , Yu, H. P. , Gao, Q. G. , Li, W. , Wu, H. R. , & Qin, Z. H. (2014). TIGAR has a dual role in cancer cell survival through regulating apoptosis and autophagy. Cancer Research, 74, 5127–5138. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Yin, L. , Tao, X. , Qi, Y. , Han, X. , Xu, Y. , … Peng, J. (2017). Dioscin, a potent ITGA5 inhibitor, reduces the synthesis of collagen against liver fibrosis: Insights from SILAC‐based proteomics analysis. Food and Chemical Toxicology, 107, 318–328. 10.1016/j.fct.2017.07.014 [DOI] [PubMed] [Google Scholar]
- Xu, T. , Zheng, L. , Xu, L. , Yin, L. , Qi, Y. , Xu, Y. , … Peng, J. (2014). Protective effects of dioscin against alcohol‐induced liver injury. Archives of Toxicology, 88, 739–753. [DOI] [PubMed] [Google Scholar]
- Xu, W. , Song, F. , Wang, B. , Li, K. , Tian, M. , Yu, M. , … Jiang, H. (2018). The effect of and mechanism underlying autophagy in hepatocellular carcinoma induced by CH12, a monoclonal antibody directed against epidermal growth factor receptor variant III. Cellular Physiology and Biochemistry, 46, 226–237. 10.1159/000488425 [DOI] [PubMed] [Google Scholar]
- Yan, X. T. , Sun, Y. S. , Ren, S. , Zhao, L. C. , Liu, W. C. , Chen, C. , … Li, W. (2018). Dietary α‐mangostin provides protective effects against acetaminophen‐induced hepatotoxicity in mice via Akt/mTOR‐mediated inhibition of autophagy and apoptosis. International Journal of Molecular Sciences, 19, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L. , Zhao, X. , Lu, J. , Qian, G. , Zheng, J. C. , & Ge, S. (2013). Knockdown of TIGAR by RNA interference induces apoptosis and autophagy in HepG2 hepatocellular carcinoma cells. Biochemical and Biophysical Research Communications, 437, 300–306. 10.1016/j.bbrc.2013.06.072 [DOI] [PubMed] [Google Scholar]
- Yin, L. , Qi, Y. , Xu, Y. , Xu, L. , Han, X. , Tao, X. , … Peng, J. (2017). Dioscin inhibits HSC‐T6 cell migration via adjusting SDC‐4 expression: Insights from iTRAQ‐based quantitative proteomics. Frontiers in Pharmacology, 8, 665 10.3389/fphar.2017.00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. P. , Xie, J. M. , Li, B. , Sun, Y. H. , Gao, Q. G. , Ding, Z. H. , … Qin, Z. H. (2015). TIGAR regulates DNA damage and repair through pentosephosphate pathway and Cdk5‐ATM pathway. Scientific Reports, 5, 9853 10.1038/srep09853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S. , Wang, Y. , Jing, L. , Claret, F. X. , Li, Q. , Tian, T. , … Guo, H. (2017). Autophagy in the “inflammation‐carcinogenesis” pathway of liver and HCC immunotherapy. Cancer Letters, 411, 82–89. 10.1016/j.canlet.2017.09.049 [DOI] [PubMed] [Google Scholar]
- Yuan, L. , Wang, Y. , Wang, J. , Xiao, H. , & Liu, X. (2014). Additive effect of zinc oxide nanoparticles and isoorientin on apoptosis in human hepatoma cell line. Toxicology Letters, 225, 294–304. 10.1016/j.toxlet.2013.12.015 [DOI] [PubMed] [Google Scholar]
- Zhang, C. L. , Zeng, T. , Zhao, X. L. , & Xie, K. Q. (2015). Garlic oil suppressed nitrosodiethylamine‐induced hepatocarcinoma in rats by inhibiting PI3K‐AKT‐NFκB pathway. International Journal of Biological Sciences, 11, 643–651. 10.7150/ijbs.10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Zeng, X. , Zhang, R. , Liu, J. , Zhang, W. , Zhao, Y. , … du, B. (2016). Dioscin suppresses hepatocellular carcinoma tumor growth by inducing apoptosis and regulation of TP53, BAX, BCL2 and cleaved CASP3. Phytomedicine, 23, 1329–1336. 10.1016/j.phymed.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Han, X. , Yin, L. , Xu, L. , Qi, Y. , Xu, Y. , … Peng, J. (2015). Potent effects of dioscin against liver fibrosis. Scientific Reports, 5, 9713 10.1038/srep09713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Xu, L. , Yin, L. , Qi, Y. , Xu, Y. , Han, X. , & Peng, J. (2015). Quantitative chemical proteomics for investigating the biomarkers of dioscin against liver fibrosis caused by CCl4 in rats. Chemical Communications, 51, 11064–11067. 10.1039/C4CC09160D [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Cong, X. , Zheng, L. , Xu, L. , Yin, L. , & Peng, J. (2012). Dioscin, a natural steroid saponin, shows remarkable protective effect against acetaminophen‐induced liver damage in vitro and in vivo. Toxicology Letters, 214, 69–80. 10.1016/j.toxlet.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Zhou, J. H. , Zhang, T. T. , Song, D. D. , Xia, Y. F. , Qin, Z. H. , & Sheng, R. (2016). TIGAR contributes to ischemic tolerance induced by cerebral preconditioning through scavenging of reactive oxygen species and inhibition of apoptosis. Scientific Reports, 6, 27096 10.1038/srep27096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information


