Abstract
The high mobility group A1 (HMGA1) gene plays an important role in numerous malignant cancers. HMGA1 is an oncofoetal gene, and we have a certain understanding of the biological function of HMGA1 based on its activities in various neoplasms. As an architectural transcription factor, HMGA1 remodels the chromatin structure and promotes the interaction between transcriptional regulatory proteins and DNA in different cancers. Through analysis of the molecular mechanism of HMGA1 and clinical studies, emerging evidence indicates that HMGA1 promotes the occurrence and metastasis of cancer. Within a similar location or the same genetic background, the function and role of HMGA1 may have certain similarities. In this paper, to characterize HMGA1 comprehensively, research on various types of tumours is discussed to further understanding of the function and mechanism of HMGA1. The findings provide a more reliable basis for classifying HMGA1 function according to the tumour location. In this review, we summarize recent studies related to HMGA1, including its structure and oncogenic properties, its major functions in each cancer, its upstream and downstream regulation associated with the tumourigenesis and metastasis of cancer, and its potential as a biomarker for clinical diagnosis of cancer.
Keywords: cancer, gene function, HMGA1
1. INTRODUCTION: HMGA1 PROTEINS
High mobility group A (HMGA) proteins are small nuclear proteins with high mobility. The HMGA family consists of four members: three HMGA1 protein isoforms because of alternative splicing, HMGA1a, HMGA1b and HMGA1c, and the fourth member HMGA2. The first three members are located on chromosome 6p21, whereas HMGA2 is transcribed by a separate gene on chromosome 12q15.1 The HMGA family lacks intrinsic transcriptional activity, but it can remodel chromatin structures and later regulate the interaction between the transcriptional regulatory proteins and downstream DNA, the so‐called “architectural transcription factors,” each of which contains three N‐terminal motifs, known as an “AT‐hook.” The HMGA family preferentially binds to other special DNAs, which have AT‐rich sequences and recruit the DNAs to HMGA family binding sites. HMGA proteins also have an acidic C‐terminal, which may be important for protein‐protein interactions or for inducing specific proteins to the enhanceosome.2
It is reported that the high expression of HMGA1 has an essential role in embryonic development. However, in terminal mature differentiation organization, the HMGA1 protein is not detected or is detected at a very low expression. In 1983, Lund et al first discovered HMGA1 expression in aggressive cervical cancer cells.3 Following that discovery, increasing research has provided compelling evidence of elevated HMGA1 expression in malignant cancer,4, 5 regardless of where the neoplasms originated (Table 1), including in epithelial cancers such as breast cancer,6 lung cancer,7, 8 colorectal cancer9, 10 and uterine cancer,11 and mesenchymal tumours such as lipoma/liposarcoma,12 glioma/glioblastoma,13 fibroma/fibrosarcoma,14 leiomyoma15 and osteosarcoma.16 Collectively, these studies reveal that HMGA1 has an important role in tumourigenesis and tumour progression and that the expression level of HMGA1 negatively correlates with clinical prognosis.
Table 1.
The role of HMGA1 in epithelial cancer and in mesenchymal tumours
| Tumour type | Clinical significance | Target gene |
|---|---|---|
| Cancer originated from epithelial tissue | ||
| Thyroid cancer | P, I, M, DB | S100A13, TGF‐β1, HAND1, p53 |
| Gastric cancer | P, I, M, DB | let7 |
| Liver cancer | CP | |
| Cholangiocarcinoma | P, T, DR | |
| Pancreatic cancer | T, DB, DR | COX2, insulin receptor, MMP9, p‐Akt |
| Ovarian carcinomas | P, M, S, DB, DR | ABCG2 |
| Cervical cancer | I, M, DB, DR | MMP2, HPV E6/E7, COX2 |
| Lung cancer | P, I, T, DB, DR | miR222, miR26a, miR26, PPP2R2A, IL24, IL6, CK2,MMP2, p‐Akt |
| Breast cancer | P, M, T, S, CP, EMT, promoting DNA repair | miR625, miR26a, miR181b, Let7a, CBX7,BRCA1, KIT ligand,DNA Ligase IV, CCNE2, TGF‐β1 |
| Colorectal cancer | I, S, DB, DR, chromosome instability | GLUT3, β‐catenin, p53, Sox9, miR137, miR138, miR214 |
| Prostate cancer | P, M, DB, CR, androgen independence | miR296, miR195, miR765, Let7b, MMP2, BCAS2, estrogen receptor β |
| Cancer originated from mesenchymal tissue | ||
| Lipoma/liposarcoma | P, CR | LPP/TPRG1, E2F |
| Leiomyoma | CR | |
| Osteosarcoma | P, I, M | miR142‐3p |
| Hemangioma | CR | TBL1XR1 |
| Medulloblastoma | P, I, M, DB | CRMP1, cdc25A, hsa‐miR124a |
| Glioma/glioblastoma | P, S, CP, DB, DR, angiogenesis | miR1297, miR296‐5p, HIF1A‐AS2, Sox2 |
| Dermatofibroma & dermatofibrosarcoma | DB | |
| Angiomyxoma & angiomyofibroblastoma | CR | |
P, proliferation; I, invasion; M, metastasis; T, tumourigenesis; S, stemness; DB, diagnostic marker; CP, clinical prognosis; DR, drug resistance; CR, chromosomal rearrangement.
As HMGA1 is overexpressed in embryonic tissues, comprehending the role of HMGA1 in cancer is essential for our understanding of HMGA1‐mediated tumourigenesis. HMGA1 functions as an oncogene through transcriptional regulation and protein‐and‐protein interaction. For example, in breast cancer, the expression of HMGA1 protein level indicates the adverse outcome of clinical prognosis. Zhou et al also found that miR‐625 suppresses cell migration and proliferation by decreasing HMGA1 protein expression.17 Overexpression of HMGA1 is correlated with human epidermal growth factor receptor 2 (HER2) and studies show that TGF‐β1 induces HMGA1 expression to promote breast cancer.18 Early studies show that HMGA1 research in colorectal cancer focused on overexpression leading to a worse clinical prognosis.19 Nearly 5 years of research shows that HMGA1 promotes colorectal cancer development, primarily through transcriptional regulation of such targets as the Wnt signalling pathway,20 miR‐13721 and miR‐214.22 The rise of metabolomics in recent years has also observed that HMGA1 can increase glucose uptake, promote aerobic glycolysis and promote the development of colorectal cancer.10, 23 Additionally, in HMGA1 transgenic mice, the faecal metabolome can be used as a non‐invasive diagnostic marker of early colorectal cancer.24
In this article, we discuss the current findings on HMGA1 in tumours as well as recent progress in characterizing the molecular mechanism and function of HMGA1 in the tumourigenesis and malignant progression of numerous cancers. We hope this review will provide a stronger understanding of this small but important oncogene and will inform future studies aimed towards the development of targeted therapy and biomarkers for clinical diagnosis.
2. CLINICAL PROGNOSIS
In all of the literature, HMGA1 is an oncogene in numerous cancers. In this paper, we review cancers classified by location and consider whether HMGA1 serves as a biomarker of clinical prognosis. We systematically classify the expression level and the clinical prognosis of HMGA1 in head and neck cancers, thoracic cancers, abdominal cancers and reproductive system cancers (Table 2). Table 2 shows the specific role of HMGA1 in these cancers.
Table 2.
The systematic classification of the HMGA1 expression level and clinical prognosis in cancer
| Tumour type | Expression level | Clinical prognosis | Ref. |
|---|---|---|---|
| Pituitary tumours | High | Poor | Wang et al (2010) |
| Glioma/Glioblastoma | High | Poor | Donato et al (2004); Pang et al (2012) |
| Thyroid cancer | High (specific) | Czyz et al (2004); Kim et al (2000) | |
| Lung cancer | High (specific) | Poor | Zhang et al (2015) |
| Breast cancer | High | Poor | Sepe et al (2016); Huang et al (2015) |
| Colorectal cancer | High | Poor | Takahashi et al (2013) |
| Hepatobiliary cancer | High | Poor | Chang et al (2005) |
| Pancreatic carcinoma | High (specific) | Poor | Hristov et al (2010) |
| Prostate cancer | High | Poor | Leman et al (2003) |
| Ovarian carcinomas | High | Poor | Zhou et al (2015) |
| Testicular seminomas | High | Chieffi et al (2013) |
Specific: in a specific cancer subtype.
In pituitary tumours, Wang et al reported that an increased expression of HMGA1 in pituitary adenomas, whereas in normal tissues it was negative, and HMGA1 was significantly more expressed in invasive adenomas.25 In gliomas, Pang et al and Donato et al revealed that HMGA1 expression was increased in gliomas compared with normal adjacent tissue; however, expression was lower than in glioblastomas. HMGA1 expression level was correlated with the histological grade in gliomas and in glioblastomas.26, 27 A series of studies have shown that HMGA1 expression is increased in thyroid cancer, especially in follicular carcinoma, but is not detected in normal tissue.28, 29 Zhang et al showed in 2015 that HMGA1 expression was increased in non‐small cell lung cancer (NSCLC), which correlated with clinical prognosis.30 In another thoracic cancer, breast cancer, many studies have confirmed increased HMGA1 expression in cancer tissue and its association with poor clinical prognosis.6, 31 Many articles have reported on HMGA1 in colorectal cancer; for example, Balcerczak et al and Takahashi et al reported an increase in HMGA1 expression in colorectal cancer, especially in small‐sized tumours (<2 cm), and the utility of HMGA1 as an indicator of lymph node metastasis.19 Chang et al revealed HMGA1 as a prognostic marker in hepatocellular carcinoma because its expression is higher in carcinoma tissue than in normal tissue, and its high expression predicts poor prognosis.32 In pancreatic ductal adenocarcinoma, Hristov et al showed a similar result for the role of HMGA1.33 In male reproductive system cancers, Leman et al found evidence that HMGA1 correlates with the clinical prognosis of diagnosed prostate cancer (PCa).34 In female reproductive system cancers, a similar outcome was confirmed. Moreover, Zhou et al showed that urine HMGA1 expression can serve as a non‐invasive prospective diagnostic indicator.35
Overall, it appears that HMGA1 has an important role in cancer, that HMGA1 expression is increased in cancer, and that elevated HMGA1 expression serves as a predictor of poor clinical prognosis. In the following review, we summarize the particular role of HMGA1 in each cancer type.
3. HEAD AND NECK CANCER
According to recent findings, HMGA1 is a potential biomarker for clinical diagnosis in head and neck cancer. Relevant findings primarily focused on pituitary tumours, named for blastoma tumours and thyroid tumours, and we summarize the results for these types of cancer (Figure 1).
Figure 1.
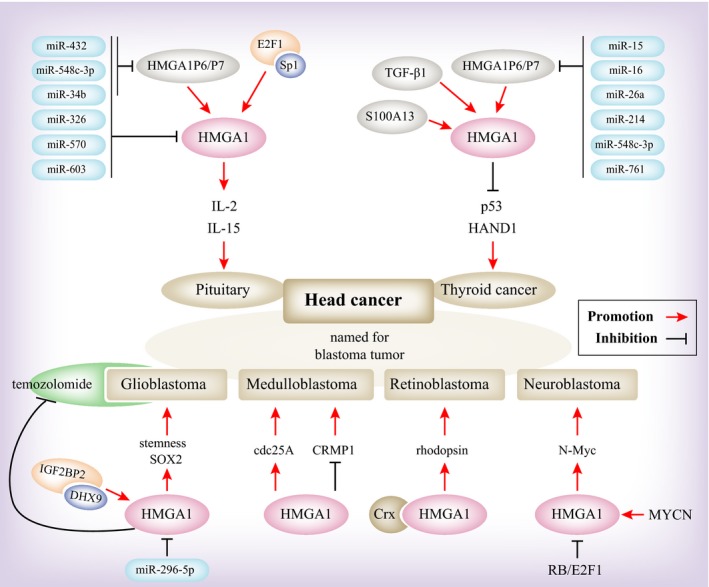
A model depicting the upstream and downstream regulation of HMGA1 in head and neck cancers
In pituitary tumours, HMGA1 transgenic mice tend to develop growth hormone/prolactin cell pituitary adenomas by inducing the expression of IL‐2 and IL‐15 proteins.36 The HMGA1 pseudogenes HMGA1P6 and HMGA1P7 act as competitive endogenous decoys for HMGA1 and contribute to pituitary tumourigenesis by increasing the level of HMGA1.37 The expression profile of microRNA in the pituitary identified a set of miRNAs that are down‐regulated, such as miR‐34b, miR‐326, miR‐432, miR‐548c‐3p, miR‐570 and miR‐603, which target HMGA1, HMGA2 and E2F1. The high expression of these target genes plays an important role in pituitary tumourigenesis.38 These studies show that HMGA1 promotes the progression of pituitary adenomas, but not HMGA1 function. By functional experiments and knockout mouse experiments, it has been observed that Sp1 interacts with E2F1 to promote HMGA1 expression and that deregulation of RB/E2F1 significantly contributes to HMGA1 deregulation in the pituitary.39
This group cancer type is named for blastoma tumours, mainly including glioblastomas, medulloblastomas, retinoblastomas and neuroblastoma. For example, in glioblastoma, the expression of HMGA1 is correlated with malignant progression, histological grade, and time to recurrence,40 while it is interesting that HMGA1 expression is significantly correlated with glioblastoma stem cells (GSCs).41 One potential mechanism is that HMGA1 is regulated by miR296‐5p, but HMGA1 can promote the transcription of SOX2 and thus promote the maintenance of GSCs.42 Another article reported that IGF2BP2 and DHX9 bind to each other and then promote HMGA1‐mediated modulation of GSC responses to hypoxic stress.43 HMGA1 maintained the GSC regulatory property and led to temozolomide resistance.13 In medulloblastomas, it has been reported that HMGA1 can promote the progression of medulloblastoma by promoting cdc25A expression or inhibiting CRMP1 expression.44, 45 In retinoblastomas, HMGA1 expression is correlated with clinical prognosis46 through the same mechanism of RB/E2F1 deregulation by HMGA1.47 In neuroblastoma, HMGA1 has been reported as a biomarker for diagnosis and prognosis.48 Studies have shown that HMGA1 promotes malignant neuroblastoma by inducing expression of the transcription factor N‐Myc.49 Giannini et al revealed that MYCN activated a luciferase reporter expressing the HMGA1 promoter, which contains the first three transcription factor binding sites.48
In thyroid cancer, high HMGA1 expression can be used as a diagnostic marker of thyroid follicular cancer to distinguish between nodular thyroid and thyroid cancer.29 It was reported that HMGA1 has a vital role in thyroid tissue through inhibition of p5350 and HAND151 and induction of TGF‐β152 and S100A13.53 The HMGA1P6 and HMGA1P7 also functions in promoting the malignant progression of thyroid cancer.54
4. THORACIC CANCER
In thoracic cancer, the studies focused primarily on lung cancer and breast cancer. These findings confirmed that HMGA1 could be used as a diagnostic and prognostic biomarker. Research on lung cancer primarily concerns NSCLC, while other research is focused on breast cancer. In this section, we expand on the findings for lung cancer and breast cancer (Figure 2).
Figure 2.
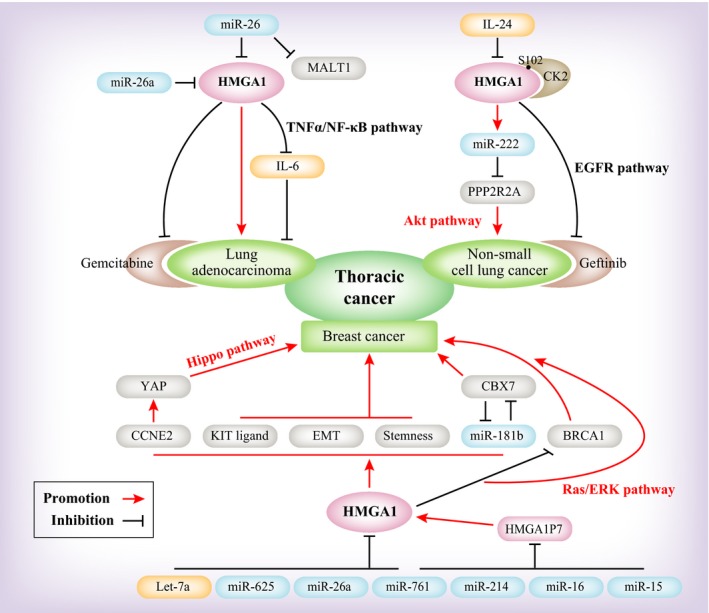
A model depicting the upstream and downstream regulation of HMGA1 in thoracic cancers
In NSCLC, using the protein levels or the circulating blood, expression level of HMGA1 as a biomarker for the diagnosis of NSCLC30 has been studied. The association of SIRT1 expression with HMGA1 expression can be used as a predictor of the prognosis and progression of NSCLC.55 The level of miR‐222 is directly increased by HMGA1 and miR‐222 led to an obvious increase in p‐Akt levels by inhibiting PPP2R2A, thereby promoting the progression of NSCLC.56 Based on these experimental results and recent findings from subsequent studies, IL‐24 inhibits HMGA1 and reduces the expression of miR‐222, inhibiting p‐Akt and thereby suppressing NSCLC.57 Regarding drug resistance, HMGA1 knockdown or mutation restored the efficacy of gefitinib through the reactivation of EGFR signalling in drug‐resistant NSCLC cells.58 Wang et al demonstrated that knockdown of HMGA1 or mutation of S102 of HMGA1, a CK2 phosphorylation site, restored the efficacy of gefitinib through the reactivation of the downstream signalling pathway of EGFR in drug‐resistant NSCLC cells.58
Immunohistochemistry and statistical analysis of HMGA1 in breast cancer showed that its expression is correlated with the histological grade, clinical stage, tumour size, lymph node metastasis, distant metastasis and whether it correlates with triple‐negative breast cancer. These findings indicated that HMGA1 expression could be used as a biomarker of breast cancer.59 Another paper showed that HMGA1 is transcriptionally activated by the KIT ligand (KL) promoter, which implicates serum KL as a diagnostic marker for HMGA1‐positive carcinomas.60 Studies reported down‐regulation of miRNAs in breast cancer, primarily miR‐625, miR‐26a and let‐7a, which inhibit the expression of HMGA1 to suppress breast cancer, thereby elucidating the molecular role of HMGA1 in malignant progression.17, 61 Additionally, the HMGA1 pseudogene HMGA1P7 can competitively bind to endogenous RNA and thus promote the progression of breast cancer. Mansueto et al confirmed that HMGA1 promotes breast cancer progression by promoting miR‐181b and inhibiting CBX7 expression.61 In basal‐like breast cancer and triple‐negative breast cancer, it has been reported that HMGA1 promotes malignant progression and predicts clinical outcome by promoting the transformation of tumour cells into breast stem cells and maintaining stemness.4, 62 Among these breast cancers, HMGA1 can activate YAP through cyclin E2 (CCNE2), promoting nuclear localization and basal‐like breast cancer progression.63 In MCF‐7 breast cancer cells, the data show that HMGA1a increases the activity of the Ras/ERK signalling pathway to promote malignant progression.64 Other findings show that TGF‐β1 induces HMGA1 promoter activity during breast cancer progression.18 With respect to drug resistance, HMGA1 enhances DNA ligase IV activity and influences DNA repair, thereby reducing the killing effect of chemotherapeutic drugs on tumour cells.65 In sporadic breast carcinoma, HMGA1 proteins negatively regulate the BRCA1 gene, which is involved in aggressive mammary carcinomas.66
5. ABDOMINAL CANCER
In addition to head cancer and thoracic cancer, there is a range of abdominal cancers, including gastric cancer, colorectal cancer, liver cancer, cholangiocarcinoma and pancreatic cancer. In this section, we analyse the findings on each of these abdominal cancers (Figure 3). First, the data confirmed that HMGA1 is an oncogene promoting tumourigenesis in abdominal cancer.
Figure 3.
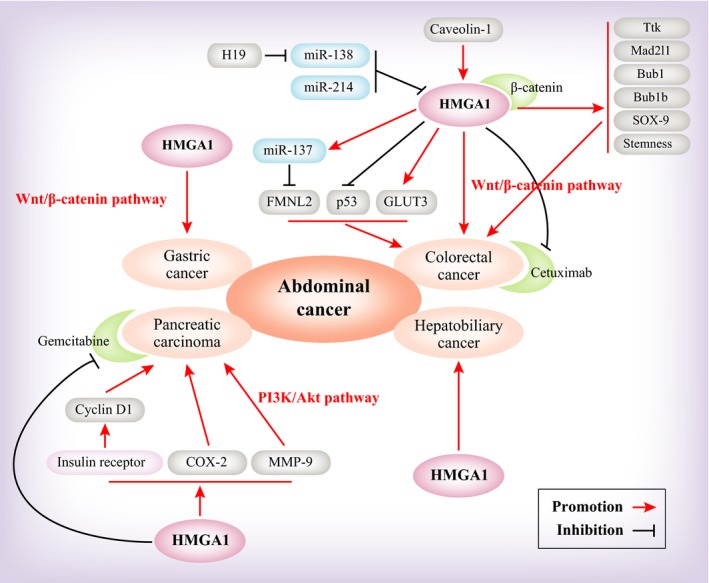
A model depicting the upstream and downstream regulation of HMGA1 in abdominal cancers
In gastric cancer, there is relatively little research on HMGA1. HMGA1 is associated with the malignant phenotype based on immunohistochemical staining of HMGA1 in gastric cancer tissues and relevant normal tissues.67, 68 However, the function of HMGA1 is unknown. Only one paper has reported that HMGA1 increased the expression of the Wnt/beta‐catenin pathway. Cytofunction and transgenic mouse experiments confirmed that HMGA1 maintained cell proliferation and was down‐regulated by beta‐catenin or its downstream factor c‐Myc.69
In colorectal cancer, HMGA1 protein expression in 81 paired tumour tissues and matched, adjacent, non‐malignant tissues indicated that HMGA1 is associated with the tumours, especially advanced tumours and lymph node metastases.9 Another paper showed that in smaller sized (<2 cm) invasive tumours, high HMGA1 expression can increase lymph node metastasis.19 The faecal metabolome reveals that the expression of the HMGA1 protein is associated with abnormal proliferation in the Hmga1 transgenic mouse intestinal epithelium. This finding is notable because faecal metabolomic analysis can serve as a non‐invasive screening tool in the early precursor lesions of colorectal cancer.24 In the regulation of metabolites, HMGA1 contributes to CRC by inducing fatty acid synthesis and aerobic glycolysis.10, 23 Above all, we are certain that HMGA1 can be used as a diagnostic indicator of CRC. To determine the mechanism of HMGA1 function, we reviewed the findings from recent years. First, HMGA1 can induce chromosomal instability in CRC by the regulation of spindle assembly checkpoint genes, such as Ttk, Mad2l1, Bub1 and Bub1b.70 Second, miRNAs, such as miR‐137, miR‐138 and miR‐214, have important roles in inhibiting the function of HMGA1 and suppressing CRC progression.21, 22, 71 Third, Wnt/β‐catenin signalling is a classical pathway to promote tumourigenesis in colorectal cancer. The data show that HMGA1 can regulate Wnt signalling to promote CRC progression and “build” an intestinal stem cell niche through inducing SOX9, interacting with β‐catenin and specifically binding to the β‐catenin/TCF‐4 complex.20 Lastly, HMGA1 regulates the symmetric/asymmetric cell division ratio and self‐renewal in CSCs through transcriptional regulation of p53.72 However, HMGA1‐relevant small molecule compounds or drugs are rarely reported. Only one paper suggests that HMGA1 proteins led to chemoresistance against drugs such as cetuximab and 5‐fluorouracil.73
In hepatobiliary cancer, there are few relevant studies. These studies were mainly focused on HMGA1 expression and correlated with worse clinical outcome.32, 74 HMGA1 has negative expression in hepatocellular carcinoma and different degrees of positive expression in intrahepatic cholangiocarcinoma and metastatic adenocarcinoma to the liver, especially in the metastatic lesions from pancreatic carcinoma, where it shows 100% positive expression.75 Another paper shows that HMGA1, a predictive marker in hepatocellular carcinoma, is involved intrahepatic metastasis, rather than non‐intrahepatic metastasis.76 There is only one paper on cholangiocarcinoma. Studies show that HMGA1 enhances tumourigenicity and confers resistance to therapy. This paper indicates that HMGA1 promotes colony formation, cell proliferation and resistance to gemcitabine treatment.77
The quantitative immunohistochemical analysis in HMGA1 transgenic mice aged 5, 11 and 15 months showed that HMGA1 expression is significantly increased in pancreatic intraepithelial neoplasia.78 HMGA1 serves as a potential diagnostic molecular marker in intraductal papillary mucinous tumours and pancreatic duct cell carcinomas.79 A series of findings may have determined the molecular function of HMGA1. At the transcriptional level, HMGA1 transcription induces the expression of cyclooxygenase 2 (COX‐2) or regulates the insulin receptor to increase cyclin D1 translation and later promotes malignant progression.80, 81 The roles of the phosphatidylinositol 3‐kinase (PI3‐K)/Akt signalling pathway are very important in HMGA1 inducing pancreatic carcinoma. Relevant studies note that HMGA1 led to cellular invasiveness and metastasis in a PI3‐K/Akt‐dependent mechanism through Akt phosphorylation at Ser (473).82 Based on the preclinical models of drug resistance, chemosensitivity is increased when HMGA1 expression is suppressed,83 and HMGA1 promotes the chemoresistance of pancreatic carcinoma to gemcitabine.84 Studies indicate that the PI3‐K/Akt pathway has an important role in the specifics of HMGA1 mediated chemoresistance.85 Li et al report that metformin up‐regulated the expression of miRNA, such as let‐7c, miR‐26a and miR‐192, in a dose‐dependent manner. These miRNAs can inhibit pancreatic cancer proliferation. One of these miRNAs, miR‐26a directly targets HMGA1 to inhibit pancreatic cancer progression. Therefore, we can conclude that metformin has a role in the treatment pancreatic cancer.86
6. REPRODUCTIVE SYSTEM CANCER
There are many papers that reveal that HMGA1 induces malignant progression and serves as a potential clinical biomarker in reproductive system cancer. PCa is the most investigated cancer in the male genitourinary system, while in the female reproductive system, ovarian carcinoma, uterine cancer, cervical cancer and endometrial cancer studies have been reported in recent decades. We review these studies individually (Figure 4).
Figure 4.
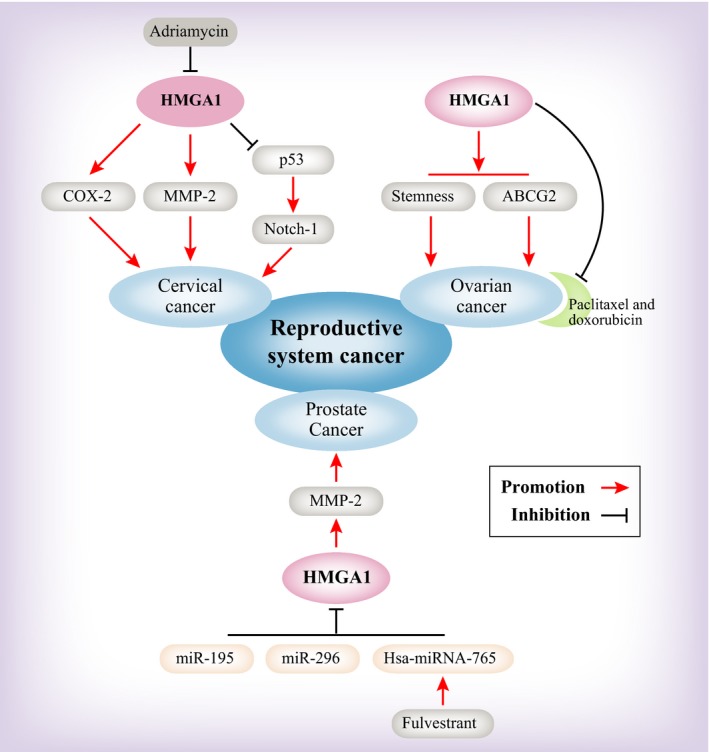
A model depicting the upstream and downstream regulation of HMGA1 in reproductive system cancers
In PCa, HMGA1 has been shown to be associated with the nuclear matrix. In examining the HMGA1 protein level during the progression from normal to prostate neoplasia in HMGA1 transgenic mice, the data show that HMGA1 expression is correlated with malignant properties in PCa.34 Because the prostate belongs to the male internal genital organ and it can be influenced by androgen, HMGA1 could transform PCa cells from androgen‐sensitive to androgen‐insensitive and could play a vital role in the cell proliferation of androgen‐independent PCa cells.87 It is well known that HMGA1 is highly expressed in embryogenesis, as well as in most malignant neoplasms, and it has been found in post‐pubertal testicular germ cell tumours, including in seminomas and embryonal carcinomas. Chieffi et al reported that HMGA1 has high expression in testicular seminomas.88 They also revealed that HMGA1 interacts with estrogen receptor β (ERβ) in nuclear of germ cells; however, such interaction is impaired by the absence of ERβ in testicular seminomas.89 Additionally, HMGA1 represents a valid diagnostic marker by immunohistochemistry analysis. How HMGA1 performs its function is subsequently reviewed. The miRNA‐microarray analysis in PCa patients has identified several candidate miRNAs (let‐7 family, miR‐181b, ‐515‐3p/5p, ‐361 and ‐146b) with differential expression, especially let‐7b, which targets HMGA1 to inhibit PCa.90 Other miRNAs, including miR‐195 and miR‐296, perform a similar function to inhibit the expression of HMGA1.91, 92 Importantly, fulvestrant treats a novel PCa pathway through ERβ‐mediated transcriptional up‐regulation of hsa‐miR‐765.
Studies note that HMGA1 serves as a useful diagnostic biomarker in ovarian carcinomas. HMGA1 has high expression in malignant cancer, as determined through immunohistochemistry, when comparing normal tissue to malignant ovarian cancer.93 Liu et al indicates that using short/small hairpin RNAs of HMGA1 led to decreased growth and metastasis potential of ovarian cancer.94 In a non‐invasive urinary detection test, HMGA1 is correlated with the degree of malignancy of ovarian cancer. It has been verified that HMGA1 served as a non‐invasive biomarker in ovarian diagnosis.35 How HMGA1 performs its function in ovarian cancer is subsequently reviewed. Studies indicated that overexpression of HMGA1 can elevate Spheroid‐forming cancer stem cells, increasing stemness‐related gene expression, such as ALDH, SOX2, ABCB1, ABCG2 and KLF4. At the same time, ovarian cancer showed resistance to chemotherapeutic agents, such as doxorubicin and paclitaxel.95
Studies show that HMGA1 is an oncoprotein in cervical cancer; however, the molecular underpinnings of malignant progression remain poorly understood. Human papilloma virus (HPV) infection is an important factor in cervical cancer. Studies have shown that HMGA1 expression is sustained by HPV E6/E7 proteins, and a positive autoregulatory loop has been established between these proteins in cervical cancer.96 By generating transgenic mice with HMGA1a, the following research shows that the HMGA1a protein level is increased in high‐grade cancer but not in normal uterine tissue. Additionally, these studies found that HMGA1a binds directly to the COX‐2 promoter based on chromatin immunoprecipitation.97 At the same time, Di Cello et al also demonstrated that COX‐2 inhibitors can rescue the role of HMGA1 in cervical cancer.98 Cervical cancer growth is impaired in an matrix metalloproteinase‐2 (MMP‐2) deficient background in HMGA1a transgenic mice. This finding indicated that HMGA1 positively regulated MMP‐2 to induce cervical cancer.11
7. CONCLUSIONS
HMGA1 is an oncofoetal protein that participates in tumourigenesis and tumour progression. As an oncogene, HMGA1 is up‐regulated in many different tumours, including epithelial and mesenchymal tissue‐originated tumours, as shown in this review. HMGA1 overexpression is correlated with poor clinical outcome, distant metastasis and advanced tumour stage in many cancers. As an architectural transcription factor, HMGA1 is involved in many biological pathways, such as the TNF‐α/NF‐κB pathway, EGFR pathway, Hippo pathway, Ras/ERK pathway, Akt pathway, Wnt/beta‐catenin pathway and PI3‐K/Akt pathway. In these biological pathways, HMGA1 targets different downstream genes. However, most of the studies on HMGA1 upstream genes are miRNAs. Most of the studies reported that miRNAs, such as miR‐15, miR‐16, miR‐26, miR‐214, miR‐296, miR‐761 and Let‐7a, can inhibit the expression of HMGA1, thereby suppressing tumour progression. Additionally, Zhang et al reported that HMGA1 can promote the development of NSCLC by promoting miR‐222 expression. There is emerging evidence that HMGA1 is a vital tumourigenic factor in the tumours of every functional system, but most of the relevant studies are in breast cancer and colorectal cancer. HMGA1 as an oncogene has been able to provide certain evidence for clinical diagnosis. Further studies of the targets of the HMGA1 gene, such as small molecules or drug interventions, will be useful for the clinical treatment of tumours. It has been reported that HMGA1 leads to tumour resistance to chemotherapeutic drugs by maintaining the stemness property of the tumour cells. Akhter et al found that Adriamycin can interact with HMGA1 and is involved in the inhibition of HMGA1 and its role in cervical cancer. Other studies on small molecules or drugs targeting HMGA1 are few, and more studies are needed for further discovery.
DISCLOSURE
No potential conflicts of interest were disclosed.
AUTHORS’ CONTRIBUTION
YW, LG conceptualized and designed the study; also written, reviewed and revised the manuscript. YW, LH developed the methodology. YW, LH, YZ analysed and interpreted the data. LG supervised the study.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (no. 31700778) and the China Postdoctoral Science Foundation funded project (no. 7131702818). We thank Wiley for the English language editing of the article.
Wang Y, Hu L, Zheng Y, Guo L. HMGA1 in cancer: Cancer classification by location. J Cell Mol Med. 2019;23:2293–2302. 10.1111/jcmm.14082
Yuhong Wang and Lin Hu contributed equally to this article.
REFERENCES
- 1. Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899‐910. [DOI] [PubMed] [Google Scholar]
- 2. Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63‐81. [DOI] [PubMed] [Google Scholar]
- 3. Lund T, Holtlund J, Fredriksen M, Laland SG. On the presence of two new high mobility group‐like proteins in HeLa S3 cells. FEBS Lett. 1983;152:163‐167. [DOI] [PubMed] [Google Scholar]
- 4. Shah SN, Cope L, Poh W, et al. HMGA1: a master regulator of tumor progression in triple‐negative breast cancer cells. PLoS ONE. 2013;8:e63419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belton A, Gabrovsky A, Bae YK, et al. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS ONE. 2012;7:e30034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang R, Huang D, Dai W, Yang F. Overexpression of HMGA1 correlates with the malignant status and prognosis of breast cancer. Mol Cell Biochem. 2015;404:251‐257. [DOI] [PubMed] [Google Scholar]
- 7. Kettunen E, Anttila S, Seppanen JK, et al. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer‐related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149:98‐106. [DOI] [PubMed] [Google Scholar]
- 8. Hillion J, Wood LJ, Mukherjee M, et al. Upregulation of MMP‐2 by HMGA1 promotes transformation in undifferentiated, large‐cell lung cancer. Mol Cancer Res. 2009;7:1803‐1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balcerczak M, Pasz‐Walczak G, Balcerczak E, Wojtylak M, Kordek R, Mirowski M. HMGI(Y) gene expression in colorectal cancer: comparison with some histological typing, grading, and clinical staging. Pathol Res Pract. 2003;199:641‐646. [DOI] [PubMed] [Google Scholar]
- 10. Williams MD, Zhang X, Belton AS, et al. HMGA1 drives metabolic reprogramming of intestinal epithelium during hyperproliferation, polyposis, and colorectal carcinogenesis. J Proteome Res. 2015;14:1420‐1431. [DOI] [PubMed] [Google Scholar]
- 11. Hillion J, Roy S, Heydarian M, et al. The high mobility group A1 (HMGA1) gene is highly overexpressed in human uterine serous carcinomas and carcinosarcomas and drives Matrix Metalloproteinase‐2 (MMP‐2) in a subset of tumors. Gynecol Oncol. 2016;141:580‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Zamolyi RQ, Zhang H, et al. Fusion of HMGA1 to the LPP/TPRG1 intergenic region in a lipoma identified by mapping paraffin‐embedded tissues. Cancer Genet Cytogenet. 2010;196:64‐67. [DOI] [PubMed] [Google Scholar]
- 13. Colamaio M, Tosti N, Puca F, et al. HMGA1 silencing reduces stemness and temozolomide resistance in glioblastoma stem cells. Expert Opin Ther Targets. 2016;20:1169‐1179. [DOI] [PubMed] [Google Scholar]
- 14. Li N, McNiff J, Hui P, Manfioletti G, Tallini G. Differential expression of HMGA1 and HMGA2 in dermatofibroma and dermatofibrosarcoma protuberans: potential diagnostic applications, and comparison with histologic findings, CD34, and factor XIIIa immunoreactivity. Am J Dermatopathol. 2004;26:267‐272. [DOI] [PubMed] [Google Scholar]
- 15. Nezhad MH, Drieschner N, Helms S, et al. 6p21 rearrangements in uterine leiomyomas targeting HMGA1. Cancer Genet Cytogenet. 2010;203:247‐252. [DOI] [PubMed] [Google Scholar]
- 16. Xu G, Wang J, Jia Y, Shen F, Han W, Kang Y. MiR‐142‐3p functions as a potential tumor suppressor in human osteosarcoma by targeting HMGA1. Cell Physiol Biochem. 2014;33:1329‐1339. [DOI] [PubMed] [Google Scholar]
- 17. Zhou WB, Zhong CN, Luo XP, et al. miR‐625 suppresses cell proliferation and migration by targeting HMGA1 in breast cancer. Biochem Biophys Res Commun. 2016;470:838‐844. [DOI] [PubMed] [Google Scholar]
- 18. Zu X, Zhong J, Tan J, et al. TGF‐beta1 induces HMGA1 expression in human breast cancer cells: implications of the involvement of HMGA1 in TGF‐beta signaling. Int J Mol Med. 2015;35:693‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi Y, Sawada G, Sato T, et al. Microarray analysis reveals that high mobility group A1 is involved in colorectal cancer metastasis. Oncol Rep. 2013;30:1488‐1496. [DOI] [PubMed] [Google Scholar]
- 20. Xian L, Georgess D, Huso T, et al. HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche. Nat Commun. 2017;8:15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang L, Li X, Zhang X, et al. MicroRNA‐137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology. 2013;144:624–635 e624. [DOI] [PubMed] [Google Scholar]
- 22. Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. MicroRNA‐214 suppresses growth, migration and invasion through a novel target, high mobility group AT‐hook 1, in human cervical and colorectal cancer cells. Br J Cancer. 2016;115:741‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ha TK, Her NG, Lee MG, et al. Caveolin‐1 increases aerobic glycolysis in colorectal cancers by stimulating HMGA1‐mediated GLUT3 transcription. Cancer Res. 2012;72:4097‐4109. [DOI] [PubMed] [Google Scholar]
- 24. Williams MD, Xian L, Huso T, et al. Fecal metabolome in Hmga1 transgenic mice with polyposis: evidence for potential screen for early detection of precursor lesions in colorectal cancer. J Proteome Res. 2016;15:4176‐4187. [DOI] [PubMed] [Google Scholar]
- 25. Wang EL, Qian ZR, Rahman MM, et al. Increased expression of HMGA1 correlates with tumour invasiveness and proliferation in human pituitary adenomas. Histopathology. 2010;56:501‐509. [DOI] [PubMed] [Google Scholar]
- 26. Pang B, Fan H, Zhang IY, et al. HMGA1 expression in human gliomas and its correlation with tumor proliferation, invasion and angiogenesis. J Neurooncol. 2012;106:543‐549. [DOI] [PubMed] [Google Scholar]
- 27. Donato G, Martinez Hoyos J, Amorosi A, et al. High mobility group A1 expression correlates with the histological grade of human glial tumors. Oncol Rep. 2004;11:1209‐1213. [PubMed] [Google Scholar]
- 28. Czyz W, Balcerczak E, Jakubiak M, Pasieka Z, Kuzdak K, Mirowski M. HMGI(Y) gene expression as a potential marker of thyroid follicular carcinoma. Langenbecks Arch Surg. 2004;389:193‐197. [DOI] [PubMed] [Google Scholar]
- 29. Kim SJ, Ryu JW, Choi DS. The expression of the high mobility group I(Y) mRNA in thyroid cancers: useful tool of differential diagnosis of thyroid nodules. Korean J Intern Med. 2000;15:71‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Z, Wang Q, Chen F, Liu J. Elevated expression of HMGA1 correlates with the malignant status and prognosis of non‐small cell lung cancer. Tumour Biol. 2015;36:1213‐1219. [DOI] [PubMed] [Google Scholar]
- 31. Chiappetta G, Botti G, Monaco M, et al. HMGA1 protein overexpression in human breast carcinomas: correlation with ErbB2 expression. Clin Cancer Res. 2004;10:7637‐7644. [DOI] [PubMed] [Google Scholar]
- 32. Chang ZG, Yang LY, Wang W, et al. Determination of high mobility group A1 (HMGA1) expression in hepatocellular carcinoma: a potential prognostic marker. Dig Dis Sci. 2005;50:1764‐1770. [DOI] [PubMed] [Google Scholar]
- 33. Hristov AC, Cope L, Di Cello F, et al. HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod Pathol. 2010;23:98‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leman ES, Madigan MC, Brunagel G, Takaha N, Coffey DS, Getzenberg RH. Nuclear matrix localization of high mobility group protein I(Y) in a transgenic mouse model for prostate cancer. J Cell Biochem. 2003;88:599‐608. [DOI] [PubMed] [Google Scholar]
- 35. Zhou J, Xie M, He H, et al. Increases urinary HMGA1 in serous epithelial ovarian cancer patients. Cancer Biomark. 2015;15:325‐331. [DOI] [PubMed] [Google Scholar]
- 36. Fedele M, Pentimalli F, Baldassarre G, et al. Transgenic mice overexpressing the wild‐type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene. 2005;24:3427‐3435. [DOI] [PubMed] [Google Scholar]
- 37. Esposito F, De Martino M, D'Angelo D, et al. HMGA1‐pseudogene expression is induced in human pituitary tumors. Cell Cycle. 2015;14:1471‐1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. D'Angelo D, Palmieri D, Mussnich P, et al. Altered microRNA expression profile in human pituitary GH adenomas: down‐regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab. 2012;97:E1128–E1138. [DOI] [PubMed] [Google Scholar]
- 39. Massimi I, Guerrieri F, Petroni M, et al. The HMGA1 protoncogene frequently deregulated in cancer is a transcriptional target of E2F1. Mol Carcinog. 2013;52:526‐534. [DOI] [PubMed] [Google Scholar]
- 40. Liu B, Pang B, Liu H, et al. High mobility group A1 expression shows negative correlation with recurrence time in patients with glioblastoma multiforme. Pathol Res Pract. 2015;211:596‐600. [DOI] [PubMed] [Google Scholar]
- 41. Fan H, Guo H, Zhang IY, et al. The different HMGA1 expression of total population of glioblastoma cell line U251 and glioma stem cells isolated from U251. Brain Res. 2011;1384:9‐14. [DOI] [PubMed] [Google Scholar]
- 42. Lopez‐Bertoni H, Lal B, Michelson N, et al. Epigenetic modulation of a miR‐296‐5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene. 2016;35:4903‐4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mineo M, Ricklefs F, Rooj AK, et al. The long non‐coding RNA HIF1A‐AS2 facilitates the maintenance of mesenchymal glioblastoma stem‐like cells in hypoxic niches. Cell Rep. 2016;15:2500‐2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lau KM, Chan QK, Pang JC, et al. Overexpression of HMGA1 deregulates tumor growth via cdc25A and alters migration/invasion through a cdc25A‐independent pathway in medulloblastoma. Acta Neuropathol. 2012;123:553‐571. [DOI] [PubMed] [Google Scholar]
- 45. Li KK, Qi Y, Xia T, et al. CRMP1 inhibits proliferation of medulloblastoma and is regulated by HMGA1. PLoS ONE. 2015;10:e0127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mu G, Liu H, Zhou F, et al. Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let‐7 down‐regulation in retinoblastomas. Hum Pathol. 2010;41:493‐502. [DOI] [PubMed] [Google Scholar]
- 47. Ueda Y, Watanabe S, Tei S, Saitoh N, Kuratsu J, Nakao M. High mobility group protein HMGA1 inhibits retinoblastoma protein‐mediated cellular G0 arrest. Cancer Sci. 2007;98:1893‐1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Giannini G, Cerignoli F, Mellone M, et al. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res. 2005;65:8308‐8316. [DOI] [PubMed] [Google Scholar]
- 49. Yanagita T, Manabe T, Okuda H, et al. Possible involvement of the expression and phosphorylation of N‐Myc in the induction of HMGA1a by hypoxia in the human neuroblastoma cell line. Neurosci Lett. 2005;374:47‐52. [DOI] [PubMed] [Google Scholar]
- 50. Frasca F, Rustighi A, Malaguarnera R, et al. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Res. 2006;66:2980‐2989. [DOI] [PubMed] [Google Scholar]
- 51. Martinez Hoyos J, Ferraro A, Sacchetti S, et al. HAND1 gene expression is negatively regulated by the high mobility group A1 proteins and is drastically reduced in human thyroid carcinomas. Oncogene. 2009;28:876‐885. [DOI] [PubMed] [Google Scholar]
- 52. Zhong J, Liu C, Zhang QH, et al. TGF‐beta1 induces HMGA1 expression: the role of HMGA1 in thyroid cancer proliferation and invasion. Int J Oncol. 2017;50:1567‐1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhong J, Liu C, Chen YJ, et al. The association between S100A13 and HMGA1 in the modulation of thyroid cancer proliferation and invasion. J Transl Med. 2016;14:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Esposito F, De Martino M, Petti MG, et al. HMGA1 pseudogenes as candidate proto‐oncogenic competitive endogenous RNAs. Oncotarget. 2014;5:8341‐8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin SY, Peng F. Association of SIRT1 and HMGA1 expression in non‐small cell lung cancer. Oncol Lett. 2016;11:782‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Y, Ma T, Yang S, et al. High‐mobility group A1 proteins enhance the expression of the oncogenic miR‐222 in lung cancer cells. Mol Cell Biochem. 2011;357:363‐371. [DOI] [PubMed] [Google Scholar]
- 57. Panneerselvam J, Srivastava A, Muralidharan R, et al. IL‐24 modulates the high mobility group (HMG) A1/miR222 /AKT signaling in lung cancer cells. Oncotarget. 2016;7:70247‐70263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang YT, Pan SH, Tsai CF, et al. Phosphoproteomics reveals HMGA1, a CK2 substrate, as a drug‐resistant target in non‐small cell lung cancer. Sci Rep. 2017;7:44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dolde CE, Mukherjee M, Cho C, Resar LM. HMG‐I/Y in human breast cancer cell lines. Breast Cancer Res Treat. 2002;71:181‐191. [DOI] [PubMed] [Google Scholar]
- 60. Treff NR, Dement GA, Adair JE, et al. Human KIT ligand promoter is positively regulated by HMGA1 in breast and ovarian cancer cells. Oncogene. 2004;23:8557‐8562. [DOI] [PubMed] [Google Scholar]
- 61. Mansueto G, Forzati F, Ferraro A, et al. Identification of a new pathway for tumor progression: microRNA‐181b up‐regulation and CBX7 down‐regulation by HMGA1 protein. Genes Cancer. 2010;1:210‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pegoraro S, Ros G, Piazza S, et al. HMGA1 promotes metastatic processes in basal‐like breast cancer regulating EMT and stemness. Oncotarget. 2013;4:1293‐1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pegoraro S, Ros G, Ciani Y, Sgarra R, Piazza S, Manfioletti G. A novel HMGA1‐CCNE2‐YAP axis regulates breast cancer aggressiveness. Oncotarget. 2015;6:19087‐19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Treff NR, Pouchnik D, Dement GA, Britt RL, Reeves R. High‐mobility group A1a protein regulates Ras/ERK signaling in MCF‐7 human breast cancer cells. Oncogene. 2004;23:777‐785. [DOI] [PubMed] [Google Scholar]
- 65. Pellarin I, Arnoldo L, Costantini S, et al. The architectural chromatin factor high mobility group A1 enhances DNA ligase IV activity influencing DNA repair. PLoS ONE. 2016;11:e0164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Baldassarre G, Battista S, Belletti B, et al. Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol. 2003;23:2225‐2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nam ES, Kim DH, Cho SJ, et al. Expression of HMGI(Y) associated with malignant phenotype of human gastric tissue. Histopathology. 2003;42:466‐471. [DOI] [PubMed] [Google Scholar]
- 68. Rahman MM, Qian ZR, Wang EL, et al. Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let‐7 downregulation. Br J Cancer. 2009;100:501‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Akaboshi S, Watanabe S, Hino Y, et al. HMGA1 is induced by Wnt/beta‐catenin pathway and maintains cell proliferation in gastric cancer. Am J Pathol. 2009;175:1675‐1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pierantoni GM, Conte A, Rinaldo C, et al. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget. 2015;6:17342‐17353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging miR‐138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50:1801‐1809. [DOI] [PubMed] [Google Scholar]
- 72. Puca F, Colamaio M, Federico A, et al. HMGA1 silencing restores normal stem cell characteristics in colon cancer stem cells by increasing p53 levels. Oncotarget. 2014;5:3234‐3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. D'Angelo D, Mussnich P, Rosa R, Bianco R, Tortora G, Fusco A. High mobility group A1 protein expression reduces the sensitivity of colon and thyroid cancer cells to antineoplastic drugs. BMC Cancer. 2014;14:851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Andreozzi M, Quintavalle C, Benz D, et al. HMGA1 expression in human hepatocellular carcinoma correlates with poor prognosis and promotes tumor growth and migration in in vitro models. Neoplasia. 2016;18:724‐731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Abe N, Watanabe T, Izumisato Y, et al. High mobility group A1 is expressed in metastatic adenocarcinoma to the liver and intrahepatic cholangiocarcinoma, but not in hepatocellular carcinoma: its potential use in the diagnosis of liver neoplasms. J Gastroenterol. 2003;38:1144‐1149. [DOI] [PubMed] [Google Scholar]
- 76. Chuma M, Saeki N, Yamamoto Y, et al. Expression profiling in hepatocellular carcinoma with intrahepatic metastasis: identification of high‐mobility group I(Y) protein as a molecular marker of hepatocellular carcinoma metastasis. Keio J Med. 2004;53:90‐97. [DOI] [PubMed] [Google Scholar]
- 77. Quintavalle C, Burmeister K, Piscuoglio S, et al. High mobility group A1 enhances tumorigenicity of human cholangiocarcinoma and confers resistance to therapy. Mol Carcinog. 2017;56:2146‐2157. [DOI] [PubMed] [Google Scholar]
- 78. Veite‐Schmahl MJ, Joesten WC, Kennedy MA. HMGA1 expression levels are elevated in pancreatic intraepithelial neoplasia cells in the Ptf1a‐Cre; LSL‐KrasG12D transgenic mouse model of pancreatic cancer. Br J Cancer. 2017;117:639‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Abe N, Watanabe T, Masaki T, et al. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000;60:3117‐3122. [PubMed] [Google Scholar]
- 80. Kolb S, Fritsch R, Saur D, Reichert M, Schmid RM, Schneider G. HMGA1 controls transcription of insulin receptor to regulate cyclin D1 translation in pancreatic cancer cells. Cancer Res. 2007;67:4679‐4686. [DOI] [PubMed] [Google Scholar]
- 81. Hillion J, Smail SS, Di Cello F, et al. The HMGA1‐COX‐2 axis: a key molecular pathway and potential target in pancreatic adenocarcinoma. Pancreatology. 2012;12:372‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liau SS, Jazag A, Whang EE. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66:11613‐11622. [DOI] [PubMed] [Google Scholar]
- 83. Trapasso F, Sarti M, Cesari R, et al. Therapy of human pancreatic carcinoma based on suppression of HMGA1 protein synthesis in preclinical models. Cancer Gene Ther. 2004;11:633‐641. [DOI] [PubMed] [Google Scholar]
- 84. Liau SS, Ashley SW, Whang EE. Lentivirus‐mediated RNA interference of HMGA1 promotes chemosensitivity to gemcitabine in pancreatic adenocarcinoma. J Gastrointest Surg, 2006;10:1254–1262; discussion 1263. [DOI] [PubMed] [Google Scholar]
- 85. Liau SS, Jazag A, Ito K, Whang EE. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer. 2007;96:993‐1000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86. Li W, Yuan Y, Huang L, Qiao M, Zhang Y. Metformin alters the expression profiles of microRNAs in human pancreatic cancer cells. Diabetes Res Clin Pract. 2012;96:187‐195. [DOI] [PubMed] [Google Scholar]
- 87. Takeuchi I, Takaha N, Nakamura T, et al. High mobility group protein AT‐hook 1 (HMGA1) is associated with the development of androgen independence in prostate cancer cells. Prostate. 2012;72:1124‐1132. [DOI] [PubMed] [Google Scholar]
- 88. Chieffi P, Chieffi S. Molecular biomarkers as potential targets for therapeutic strategies in human testicular germ cell tumors: an overview. J Cell Physiol. 2013;228:1641‐1646. [DOI] [PubMed] [Google Scholar]
- 89. Esposito F, Boscia F, Gigantino V, et al. The high‐mobility group A1‐estrogen receptor beta nuclear interaction is impaired in human testicular seminomas. J Cell Physiol. 2012;227:3749‐3755. [DOI] [PubMed] [Google Scholar]
- 90. Schubert M, Spahn M, Kneitz S, et al. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let‐7 as prognostic marker in high‐risk prostate cancer. PLoS ONE. 2013;8:e65064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wei JJ, Wu X, Peng Y, et al. Regulation of HMGA1 expression by microRNA‐296 affects prostate cancer growth and invasion. Clin Cancer Res. 2011;17:1297‐1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang X, Tao T, Liu C, et al. Downregulation of miR‐195 promotes prostate cancer progression by targeting HMGA1. Oncol Rep. 2016;36:376‐382. [DOI] [PubMed] [Google Scholar]
- 93. Masciullo V, Baldassarre G, Pentimalli F, et al. HMGA1 protein over‐expression is a frequent feature of epithelial ovarian carcinomas. Carcinogenesis. 2003;24:1191‐1198. [DOI] [PubMed] [Google Scholar]
- 94. Liu Y, Wang Y, Zhang Y, Fu J, Zhang G. Knockdown of HMGA1 expression by short/small hairpin RNA inhibits growth of ovarian carcinoma cells. Biotechnol Appl Biochem. 2012;59:1‐5. [DOI] [PubMed] [Google Scholar]
- 95. Kim DK, Seo EJ, Choi EJ, et al. Crucial role of HMGA1 in the self‐renewal and drug resistance of ovarian cancer stem cells. Exp Mol Med. 2016;48:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mellone M, Rinaldi C, Massimi I, et al. Human papilloma virus‐dependent HMGA1 expression is a relevant step in cervical carcinogenesis. Neoplasia. 2008;10:773‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tesfaye A, Di Cello F, Hillion J, et al. The high‐mobility group A1 gene up‐regulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res. 2007;67:3998‐4004. [DOI] [PubMed] [Google Scholar]
- 98. Di Cello F, Hillion J, Kowalski J, et al. Cyclooxygenase inhibitors block uterine tumorigenesis in HMGA1a transgenic mice and human xenografts. Mol Cancer Ther. 2008;7:2090‐2095. [DOI] [PMC free article] [PubMed] [Google Scholar]


