Abstract
Although the application of multiple chemotherapy brought revolutionary changes to improve overall survival of osteosarcoma patients, the existence of multidrug resistance (MDR) has become a great challenge for successful osteosarcoma treatment in recent decades. Substantial studies have revealed various underlying mechanisms of MDR in cancers. As for osteosarcoma, evidence has highlighted that microRNAs (miRNAs) can mediate in the processes of DNA damage response, apoptosis avoidance, autophagy induction, activation of cancer stem cells, and signal transduction. Besides, these drug resistance‐related miRNAs showed much promise for serving as candidates for predictive biomarkers of poor outcomes and shorter survival time, and therapeutic targets to reverse drug resistance and overcome treatment refractoriness. This review aims to demonstrate the potential molecular mechanisms of miRNAs‐regulated drug resistance in osteosarcoma, and provide insight in translating basic evidence into therapeutic strategies.
Keywords: biomarker, drug resistance, miRNA, osteosarcoma, therapeutic target
1. INTRODUCTION
Osteosarcoma (OS) with great tumour malignancy, has a predilection for children and adolescents, principally emerging in the metaphysis of long bones.1 The peak age of OS occurrence is approximately 16 years, which was substantiated to have a close association with skeletal growth rate.2 Because of its strong tendency to extensive metastasis and tumour relapse, OS consequently causes high mortality and poses a great threat of life to children and adolescents. With the emergence of next‐generation sequencing, OS was gradually discovered to have a rather complicated genetic background.3 The inactivation of tumour suppressor genes TP53 and/or RB1 was corroborated to remarkably induce OS tumourigenesis.4 The congenital mutations of TP53 and/or RB1 are enough for developing tumour, but the occurrence rate of these congenital mutations was underestimated before.5 Currently, the combination of surgical resection and multiple chemotherapy including neoadjuvant therapy, has been standardized for OS clinical remedy since 1970s. This regimen tremendously ameliorated symptoms and extended overall survival time of OS patients.6 However, there exists a low response to therapeutic drugs in many OS patients, which is responsible for their subsequent aggressive progression and unfavourable outcomes. It is noted that the 5‐year survival rate has remained at the level of 65%‐75% in recent three decades, even with substantial research progress in OS clinical treatment approaches.7, 8, 9 Obviously, distant metastasis, tumour recurrence and drug resistance are three pivotal reasons for the treatment refractoriness of OS. Much attention should be paid to better decipher and understand the underlying molecular mechanisms. Hence, it would be conceivable that molecules implicated in these mechanisms can serve as therapeutic targets for extending survival time of OS patients. Noticeably, extensive evidence has supported the involvement of miRNAs in OS pathogenesis.
In recent years, miRNAs have been explored to have a close connection to the mechanisms of pathogenesis and drug resistance in different cancer types, and establish a competitive endogenous RNA regulatory network that remains to be investigated.10, 11, 12, 13 Inspiringly, miRNAs seem to play an emerging role in OS drug resistance.14, 15 This might provide a brand‐new insight in seeking for promising prognostic biomarkers and therapeutic targets for successful treatment in OS. To our knowledge, a single miRNA can target at least 200 genes involved in one signalling pathway or diverse signalling pathways.16 Therefore, miRNAs might be valuable and effective for treating cancers with inherent heterogeneity and abnormality of multiple genes, among which OS can be taken as a good example.
In this review, we will elaborate on the emerging role of miRNAs in OS drug resistance under the mechanisms of DNA damage response, apoptosis avoidance, autophagy induction, activation of cancer stem cells (CSCs), and alteration in signal pathways. Also, we will provide insight in the potential clinical utility of these miRNAs as promising biomarkers and therapeutic targets to reverse chemoresistance.
2. BIOGENESIS AND BIOLOGICAL FUNCTION OF miRNAs
microRNAs (miRNAs) were first discovered by Victor Ambros et al in 1993, and perceived as endogenous small RNA molecules with biologically regulatory functions.17 They are broadly conserved sequences among species with only 18‐25 nucleotides in length for their mature forms, and have regulatory roles in gene expression at the post‐transcriptional level.18, 19 Through binding to the 3′‐untranslated region (3′‐UTR) perfectly or imperfectly, they consequently contribute to the translational suppression or the degradation of diverse target mRNAs.20 The detailed biogenesis process and functional mechanism of miRNAs have been well‐elucidated (Figure 1). It has been estimated that over 70% of human genome DNA has transcripts. Among them, about 2% transcripts code for protein synthesis and 3% can transcribe endogenous miRNAs. Of note, over 30% of human genes are under the regulation of miRNAs.19, 21 Intriguingly, a single small miRNA can interact with several regions of one or multiple target mRNAs. Conversely, a mRNA can be modulated by a multitude of miRNAs simultaneously, which is a unique advantage of miRNAs for cancer treatment.
Figure 1.
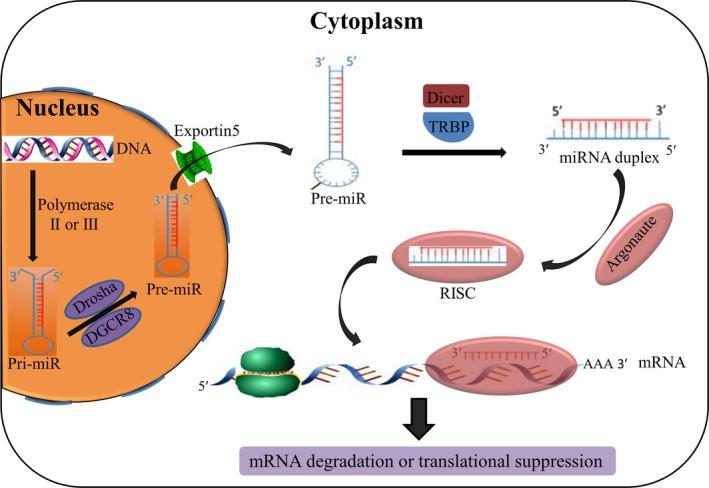
Biogenesis and biological function of miRNAs. First, a specific miRNA gene transcribes into pri‐miRNA through polymerase II or III in the nucleus. Next, Drosha cleaved the hairpin structure of pri‐miRNA to produce pre‐miRNA which is subsequently exported to the cytoplasm by Exportin5. Then, the miRNA duplex is released after the excision of Dicer. After that, a combination of miRNA duplex and Argonaute protein forms a RNA‐induced silencing complex (RISC), in which the passenger strand of miRNA is degraded. Finally, RISC causes mRNA degradation or translational suppression by targeting the 3′‐UTR of mRNA
Evidence has accumulated that miRNAs participate in various biological processes,22, 23 such as development, proliferation, differentiation, apoptosis, cell cycle, and metabolism, together with some human diseases24 including cancer.25 These deregulated miRNAs can be categorized as oncogenic ones and tumour suppressor ones. They play a regulatory function in tumourigenesis, progression, or chemosensitivity of different cancers. Besides, some miRNAs were reported to possess clinical values as predictive factors or therapeutic targets.26, 27, 28 Noticeably, the emerging role of miRNAs in OS chemoresistance has been reported in recent studies, holding promise for improving the quality of life in OS patients.15
3. miRNAs‐MODULATED DRUG RESISTANCE IN CANCER
Although the application of chemotherapeutic agents contributes to effective cancer treatment to a large extent, the occurrence of acquired multidrug resistance (MDR) remains a tough issue that ought to be solved. Substantial studies have discovered several universal mechanisms underlying acquired MDR,29 including drug transport, drug metabolism, aberrant drug targets, DNA damage response, apoptosis evasion, autophagy, epithelial‐to‐mesenchymal transition (EMT), and activation of CSCs.
Drug transport mechanism has been well‐studied in cancer MDR, which is closely associated with up‐regulated drug transport proteins presenting on the surface of cytoplasmic membrane, that is, ATP‐binding cassette (ABC) transporters.30 Drug metabolism is a complicated process of xenobiotics detoxification with the participation of drug metabolism enzymes (DMEs) and the consequent metabolites are transported by ABC transporters.31 As we can see, the concerted efforts of DMEs and ABC transporters finally lead to the decreased drug accumulation in the cytoplasm to reduce drug toxicity. DNA damage response (DDR) is a cellular stress response to DNA damage caused by cytotoxic drugs endogenously or exogenously. It aims to repair existing DNA lesions by arresting cell cycle temporarily, and prevent further or irretrievable damage such as cell senescence and apoptosis.32 Therefore, the enhanced DNA repair can promote cell viability and resistance to cytotoxicity. Programmed cell death, an integrated concept of apoptosis, autophagy, and programmed necrosis, is an intracellular program triggered in the context of adverse conditions to determine the ultimate fate of cells, namely, survival or death. Interestingly, in malignant cells, apoptosis and programmed necrosis are invariably associated with death, while autophagy executes a dual role.33 Furthermore, mechanisms modulated by apoptosis or autophagy have been confirmed to contribute to enhanced drug resistance.
Recently, ever‐growing evidence has shown that exosomes and miRNAs can also play a significant role in drug responsiveness of cancers including OS.15, 34 Extracellular tumour‐derived exosomes can transfer MDR‐related miRNAs through 40‐150 nm vesicles to recipient cells. Of note, miRNAs can modulate all of the above mechanisms of MDR because of their extensive regulation in gene expression in various cancers.35, 36, 37, 38 Therefore, miRNA can be viewed as a pivotal mediator of cancer chemoresistance. In spite of these miRNAs‐modulated drug resistance mechanisms, another challenge we faced is to identify useful targets that can effectively overcome MDR. Given that chemotherapy insensitivity is usually blamed for the rapid growth of local tumours and widespread metastasis to distant organs, it is still an urgent duty to have a thorough understanding of MDR modulated by miRNAs and deeply explore viable methods to reverse drug resistance.
4. THE ROLE OF miRNAs IN OSTEOSARCOMA DRUG RESISTANCE
Clinically, the traditional first‐line chemotherapy regimen for OS patients is a combination of doxorubicin (DOX), cisplatin (CDDP), and methotrexate (MTX). The following resistance to these anticancer drugs is a common phenomenon and contributes to poor clinical outcomes. The underlying mechanisms now have been unveiled.39 With deep investigations of miRNAs in recent years, numerous studies have validated the involvement of miRNAs in OS drug resistance, in addition to tumour initiation and progression.40 These oncogenic or tumour suppressor miRNAs role in chemotherapy sensitivity by the mechanisms of DDR, apoptosis avoidance, autophagy induction, activation of CSCs, and alteration in signal pathways (Figure 2). Besides, they show much promise for predicting clinical outcomes in clinical practice.
Figure 2.
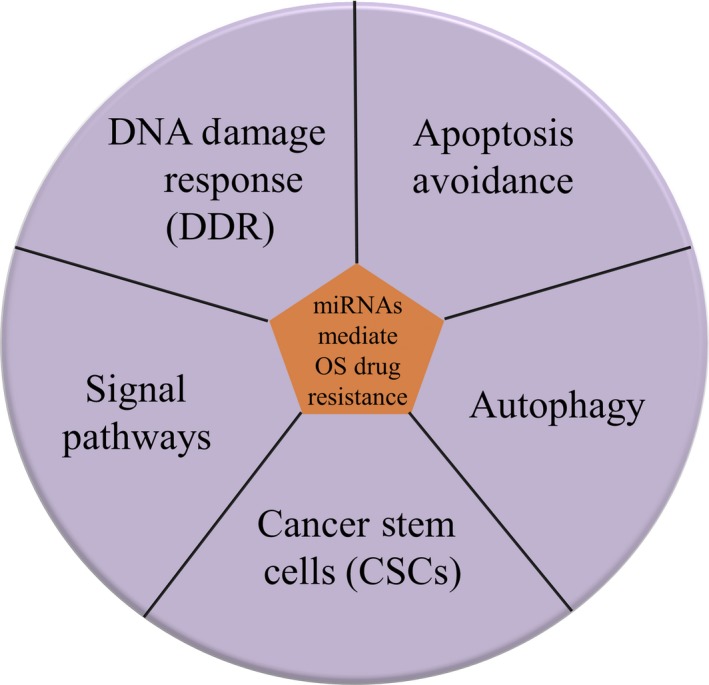
MiRNAs modulate OS drug resistance through several mechanisms
4.1. DNA damage response
Cytotoxic agents can cause cellular DNA damage and initiate a cellular stress response called DDR, which aims to repair existing DNA lesions through temporary cell cycle arrest and protect cells from irreversible damage.32 To our knowledge, the DDR process contains DNA tolerance mechanisms, base excision repair, nucleotide excision repair, mismatch repair, and DNA double‐strand break repair.41 It has been reported that there exists an interplay between DDR genes and noncoding RNAs (ncRNAs) including miRNAs in cancer.42, 43 Several recent studies have shown that miRNAs can be a regulator of OS drug resistance via involving in DDR mechanism (Table 1).
Table 1.
MiRNAs regulate DDR, autophagy, CSCs, and signal pathways
| Mechanism | microRNA | Alteration | Target gene | OS‐derived cell line | Resistant to | References |
|---|---|---|---|---|---|---|
| DNA damage response | miR‐124 | ↓ | ATMIN; PARP1 | U2OS | CPT, VP‐16 and DOX | 46 |
| miR‐15b | ↓ | WEE1 | KHOS, KHOSmr, U2OS, and U2OSmr | DOX | 52 | |
| Autophagy | miR‐101 | ↓ | Not defined | U2OS | DOX | 86 |
| miR‐22 | ↓ | HMGB1 | U2OS and MG63 | DOX and CDDP | 87, 88 | |
| miR‐30a | ↓ | Beclin‐1 | MG63/Dox resistant cell line | DOX | 91 | |
| miR‐199a‐5p | ↓ | Beclin‐1 | MG63 | CDDP | 92 | |
| miR‐155 | ↑ | Not defined | Saos2 and MG63 | DOX and CDDP | 94 | |
| miR‐140‐5p | ↑ | IP3K2 | Saos2 and MG63 | DOX and CDDP | 95 | |
| Cancer stem cells | miR‐143 | ↓ | Not defined | U2OS and Saos2 | DOX | 108 |
| miR‐let‐7 | ↓ | Not defined | KPD, U2OS and Saos2 | Not defined | 110 | |
| miR‐let‐7d | ↓or↑ | Multiple genes | 3AB‐OS CSC line | Not defined | 111 | |
| miR‐29b‐1 | ↓ | Multiple genes | 3AB‐OS CSC line | DOX, CDDP and VP‐16 | 112 | |
| Signal pathways | miR‐34c | ↓ | Notch1; LEF1 | U2OS and MG63 | DOX, CDDP and MTX | 115 |
| miR‐34b | ↓ | PAK1; ABCB1 | MG63/ADM resistant cell line | DOX, GEM and MTX | 116 | |
| miR‐497 | ↓ | VEGFA | Saos2 | CDDP | 117 | |
| miR‐221 | ↑ | PTEN | SOSP‐9607 and MG63 | CDDP | 118 | |
| miR‐146b‐5p | ↑ | ZNRF3 | U2OS and MG63 | DOX, CDDP and MTX | 119 |
ATMIN, ataxia telangiectasia mutated interactor; PARP1, poly (ADP‐ribose) polymerase 1; HMGB1, high‐mobility group box 1; IP3K2, inositol 1,4,5‐trisphosphate kinase 2; LEF1, lymphoid enhancer‐binding factor 1; PAK1, p21‐activated protein kinase 1; ABCB1, ATP‐binding cassette, subfamily B, member 1; VEGFA, vascular endothelial growth factor A; PTEN, phosphatase and tensin homolog; ZNRF3, zinc and ring finger 3; CPT, camptothecin; VP‐16, etoposide; DOX, doxorubicin; CDDP, cisplatin; MTX, methotrexate; GEM, gemcitabine (↑upregulation, ↓downregulation).
MiR‐124 was previously reported to regulate glucocorticoid resistance in haematological malignancies, for which glucocorticoid is common therapeutic drug.44, 45 Up‐regulated miR‐124 was newly shown to enhance cell response to diverse DNA‐damaging drugs by binding to the 3′‐UTR of ATMIN and PARP1 mRNAs in U2OS cells.46 Protein PARP1, an abbreviation of poly (ADP‐ribose) polymerase 1, is well‐known to attract DNA repair proteins for repair through binding to DNA breaks.47 Its inhibitors have been validated to sensitize cancer cells and have an anticancer effect in various cancers.48, 49 Protein ATMIN (ATM interactor) interacts with a significant DNA damage checkpoint kinase, ataxia telangiectasia mutated (ATM), and regulates ATM activity for DNA repair.50 The role of miR‐15b in cancer drug resistance has been reported in the last decade.51 A recent study52 first pointed out a significant decrease in miR‐15b in OS MDR cell lines and identified WEE1 mRNA as its direct target. WEE1 gene codes for a protein kinase to modulate the G2 checkpoint in response to DNA damage. Besides, a restoration of miR‐15b was observed to suppress WEE1 and partially reverse drug resistance in vitro. By establishing a MDR models of OS, Zhenfeng Duan et al discovered an attenuate resistance to DOX after systemic administration of miR‐15b mimics.
As we all know, intracellular genomic instability is an intrinsic hallmark of tumourigenesis and tumour progression. Some cancer cells rely on a limited set of repair mechanisms for survival. Studies have found that disruption of DNA damage repair pathways can be utilized for current anticancer therapies.53, 54 However, it is still obscure in OS chemotherapy and requires deeper exploration of potential mechanisms of miRNAs‐regulated DDR in OS.
4.2. Apoptosis avoidance
Cell apoptosis, characterized by permanent cell cycle arrest, is a complicated prodeath process elicited by activation of a cascade of intracellular caspases.55, 56 It is believed to predict the treatment effect of anticancer drugs. The perturbations in apoptotic process result in uncontrolled cell proliferation, which is an outward manifestation of resistant cancer cells. Previous studies suggest that molecules implicated in apoptotic process can serve as effective targets to reverse cancer drug resistance.57 Inspiringly, recent researches have demonstrated that miRNAs regulate cell apoptosis by affecting apoptosis‐related proteins to obviously influence chemotherapy sensitivity of OS cells (Figure 3, Table 2).
Figure 3.

Aberrant expression of apoptosis‐related miRNAs
Table 2.
MiRNAs involved in apoptosis avoidance
| microRNA | Alteration | Target gene | OS‐derived cell lines | Resistant to | References |
|---|---|---|---|---|---|
| miR‐126 | ↓ | Not defined | U2OS | EGCG | 58 |
| miR‐15a, miR‐16‐1 | ↓ | CCND1 | SOSP‐9607 | Not defined | 59 |
| miR‐217 | ↓ | KRAS | 143B | CDDP | 60 |
| miR‐138 | ↓ | EZH2 | HOS, Saos‐2, MG63, U2OS | CDDP | 61 |
| miR‐382 | ↓ | HIPK3; KLF12 | MNNG/HOS, U2OS and MG63 | DOX, CDDP and MTX | 62 |
| miR‐140 | ↑ | HDAC4 | U2OS | MTX and 5‐FU | 14 |
| miR‐215 | ↑ | DTL | U2OS and MG63 | MTX and tomudex | 63 |
| miR‐301a | ↑ | AMPKα1 | U2OS and MG63 | DOX | 64 |
| miR‐21 | ↑ | Not defined | MG63 | CDDP | 66 |
| miR‐21 | ↑ | Spry2 | U2OS | CDDP | 67 |
| miR‐184 | ↑ | BCL2L1 | U2OS and MG63 | DOX | 68 |
| miR‐367 | ↑ | KLF4 | MG63, U2OS and Saos2 | DOX | 69 |
| miR‐488 | ↑ | Bim | MG63 Saos2 and G293 | DOX | 70 |
| miR‐202 | ↑ | PDCD4 | U2OS and G292 | DOX | 71 |
| miR‐33a | ↑ | TWIST | Saos2 and MG63 | CDDP | 72 |
| miR‐193a‐5p | ↑ | TAp73β | 143B, MNNG/HOS, Saos2, SJSA1, MG63, U2OS and CAL‐72 | CDDP | 73 |
| miR‐34a‐5p | ↑ | AGTR1 | SJSA1 and G292 | CDDP, VP‐16, CDDP and CBP | 75 |
CCND1, Cyclin D1; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; HIPK3, homeodomain interacting protein kinase 3; KLF12, Kruppel‐like factor 12; HDAC4, Histone deacetylase 4; DTL, denticleless protein homolog; Spry2, Sprouty homolog 2; BCL2L1, Bcl‐2‐like protein 1; KLF4, Kruppel‐like factor 4; Bim, Bcl‐2‐interacting mediator of cell death; PDCD4, programmed cell death 4; AGTR1,angiotensin II type 1 receptor; Epigallocatechin‐3‐gallate (EGCG), doxorubicin (DOX), cisplatin (CDDP), methotrexate (MTX), 5‐fluorouracil (5‐FU), etoposide (VP‐16), carboplatin (CBP) (↑upregulation, ↓downregulation).
MiR‐126 is a key regulator in inflammation and angiogenesis. The low expression level of miR‐126 has been commonly reported in cancers. Up‐regulated miR‐126 promoted cell sensitivity to Epigallocatechin‐3‐gallate (EGCG) by enhancing cell apoptosis in U2OS cells.58 Overexpressed miR‐15a and miR‐16‐1 induced apoptosis and cell cycle arrest in SOSP‐9607 cell line and post‐transcriptionally modulated cyclin d1 (CCND1) expression via directly targeting the 3′‐UTR of CCND1.59 CCND1 is a key regulator in the G1 phase, a pivotal cell cycle phase in response to extracellular cues, and is usually up‐regulated in multiple cancers. After adding quercetin drug, there showed enhanced sensitivity to CDDP, the up‐regulation of miR‐217, and down‐regulation of its target KRAS at the level of mRNAs and proteins. This implied that quercetin increased CDDP‐induced cytotoxicity through the miR‐217‐KRAS axis.60 Reduced expression of miR‐138 was assessed in OS tissues and cell lines, and miR‐138 transfection suppressed cell proliferation, induced cell apoptosis, and increased drug responsiveness by binding to EZH2.61 MiR‐382 was detected to decrease in OS specimens with chemoresistance compared to those with chemosensitivity. Further study showed that elevated miR‐382 inhibited cell growth and drug resistance via interacting with KLF12 and HIPK3, respectively. Besides, Meng Xu et al confirmed a relationship between miR‐382 and genes KLF12 and HIPK3 by using a MNNG/HOS xenograft model.62
Expression of miR‐140 is ubiquitous in chondrocyte for bone development during embryonic period. The oncogenic role of miR‐140 in drug resistance relied on the existence of functional wild‐type p53, for which this study was performed in U2OS cells. MiR‐140 inhibited the level of histone deacetylase 4 (HDAC4) and contributed to chemoresistance through G1 and G2 phase arrest and p21 up‐regulation.14 Amplified miR‐215 inhibited cell proliferation through G2 phase arrest and promoted chemotherapy insensitivity to MTX and TDX, accompanied by overexpression of p21 in a p53‐dependent manner.63 Elevated miR‐301a enhanced drug resistance because of apoptosis avoidance by directly targeting AMPKa1.64 It has been identified that miR‐21 mostly exerts oncogenic roles in cancers including OS.65 A study revealed that Bcl‐2 expression had a positive connection with miR‐21 which inhibited apoptosis and induced a resistance to CDDP, while Bcl‐2 siRNA ameliorated miR‐21‐induced resistance.66 Another recent study identified Spry2 as a direct target of miR‐21, and confirmed the positive role of miR‐21in OS drug resistance.67 Time‐dependent expression of miR‐184 was observed in OS cells treated with DOX and up‐regulated miR‐184 caused a poor drug response through targeting bcl‐2‐like protein 1 (BCL2L1).68 MiR‐367 negatively modulated DOX‐induced apoptosis via coupling with KLF4, which could enhance cell apoptosis by regulating Bax and Bcl‐2.69 MiR‐488 was induced by hypoxia because HIF1‐α could interact with the hypoxia response element (HRE) within miR‐488 promoter. Overexpressed miR‐488 resulted in apoptosis avoidance, drug resistance, and promoted proliferation by binding to bcl‐2‐interacting mediator (BIM) of cell death, while an opposite result was obtained via using miR‐488 inhibitor.70 MiR‐202 was found to be up‐regulated in OS tissues and could be induced by TGF‐β1 in OS cells. MiR‐202 mimics transfection led to a significant promotion of chemoresistance together with a decrease in the expression of an apoptosis‐related protein PDCD4, while miR‐202 inhibitor triggered an opposite effect.71 Increased miR‐33a was observed in chemo‐resistant OS and in vitro data showed that miR‐33a enhanced drug resistance by inhibiting CDDP‐induced apoptosis in OS cells with a negative regulation of TWIST. On the contrary, decreased miR‐33a by antagomir‐33a promoted cell apoptosis and increased levels of TWIST mRNA.72 Oncogenic miRNA‐193a‐5p modulated cell viability, colony‐forming capacity, and CDDP‐induced apoptosis in OS cells through targeting TAp73β,73 an isoform of P73 which belongs to the P53‐related transcription factor family and regulates genome stability and chemosensitivity.74 MiR‐34a‐5p was discovered to promote MDR of OS by targeting angiotensin II type 1 receptor (AGTR1) in sensitive (G292) and resistant (SJSA1) OS cells, and function of miR‐34a‐5p in drug resistance was further verified in G292 and SJSA1‐derived xenografts.75
Collectively, these oncogenic or tumour suppressor miRNAs contribute to OS drug resistance by regulating expression of apoptosis‐related genes to avoid cell apoptosis, such as CCND1 and BCL2. Considering that BCL2 is a classic anti‐apoptotic protein that promotes cell survival by inhibiting activation of a caspase cascade, and is associated with several miRNAs in OS chemoresistance, it's presumable that BCL2 might be critical for the reversal of MDR in OS. However, further identification and confirmation of the above miRNAs is needed and great efforts should be invested to translate these findings into clinical applications.
4.3. Autophagy induction
On one hand, autophagy refers to a lysosomal degradation pathway by secluding damaged or excess cellular molecules and organelles within autophagosomes and clearing them to keep cellular homeostasis.76 On the other hand, it's a protective prosurvival pathway by sustaining a balance among the synthesis, degradation, and succeeding recycling of essential molecules in the condition of nutrient deprivation.77 Conversely, autophagy will trigger cell death in the context of excessive loss of proteins, indicating that autophagy can exert paradoxical roles.78 Accumulated evidence has highlighted the participation of autophagy regulation in cancer diseases including OS,79, 80, 81 and revealed the promoted activity of this degradative pathway after administration of cytotoxic drugs to acquire drug resistance.82, 83, 84 Recently, the involvement of autophagy modulated by miRNAs in OS drug resistance has been explored (Table 1).
MiR‐101 is viewed as an important regulator in fibrotic diseases and is used as therapeutic agents.85 But except for that, it is also newly reported in cancer drug resistance. MiR‐101 significantly blocked the expression of autophagy‐related gene in U2OS cells and promoted cell sensitivity to DOX treatment.86 MiR‐22 was reported to couple with high‐mobility group box 1 (HMGB1) and suppress HMGB1‐modulated autophagy in OS cells treated with DOX and CDDP.87, 88 Previous studies imply that HMGB1, a chromatin‐binding nuclear protein, can regulate the balance of autophagy and apoptosis, and promote drug resistance by facilitating autophagy in OS cells with administration of agents.89, 90 It was confirmed that miR‐30a targeting Beclin‐1 reduced chemoresistance to DOX via inhibition of Beclin‐1‐regulated autophagy in vitro.91 MiR‐199a‐5p also bound to Beclin‐1 contributing to blockage of autophagy and CDDP‐induced cytotoxicity in MG63 cells.92 Multifunctional miR‐155 is enriched and important in cellular immune system, and its overexpression is well‐known to result in cancer development and drug resistance. The miR‐155‐based therapy has been commonly considered in cancer treatment.93 A recent study revealed that elevated expression of miR‐155 promoted autophagy induced by anti‐cancer drugs and increased cell viability to modulate drug resistance in OS cells.94 MiR‐140‐5p played a positive role in OS drug resistance through induction of autophagy with a direct interaction with inositol 1,4,5‐trisphosphate kinase 2 (IP3k2).95 Since autophagy is a double‐edged sword in the process of biological degradation, and tight control of autophagy is beneficial for the survival of normal or cancer cells, it would be a considerable notion that manipulation of autophagy can be applied in cancer therapy by inhibiting its protective function and inducing cell death instead.96, 97 This has been studied preclinically in its infancy in OS treatment.98, 99 Therefore, it demands for further investigations in OS to seek for effective therapeutic drugs targeting miRNAs‐modulated autophagy.
4.4. Activation of cancer stem cells
Cancer stem cells (CSCs) refer to a small subpopulation of cells possessing competences of self‐renewal and differentiation, holding malignant potential, showing resistance to therapeutic drugs by expressing ABC transporters, and serving as the source of metastatic and recurrent tumours. Hence, it is universally perceived that an eradication of CSCs is pivotal but challenging for the successful treatment of cancers.100, 101 Ever‐growing evidence indicates that therapeutic approaches targeting CSCs can effectively halt tumour development and ameliorate patient prognosis, which has also been reported in OS.102, 103, 104 Recently, ncRNAs including miRNAs and lncRNAs have been reported to participate in the maintenance of the CSC phenotype, which brought great benefits to better understand CSCs by further exploring CSCs‐related ncRNAs. For example, hypoxia‐inducible factor‐2α promoter upstream transcript (HIF2PUT) was the first lncRNA reported to play a role in OS‐CSCs with expression of CD133.105 Remarkably, several current studies shed light on the involvement of miRNAs in OS‐derived CSCs, which needs much more investigations to have a good understanding of potential mechanisms for their future applications in OS treatment (Table 1).
MiR‐143 is viewed as a novel regulator in type II diabetes, which can specially suppresses insulin‐AKT pathway and causes insulin resistance.106 Besides, chemically modified miR‐143 has been considered as a RNA medicine for treating colorectal tumours.107 A study reported a reduced level of miR‐143 in OS patients with drug treatments, which contributed to enhanced chemoresistance by apoptosis avoidance and activation of autophagy and ALDH1+CD133+ cells.108 It is acknowledged that ALDH1 and CD133 are common cancer stem cell markers for identifying and selecting CSCs.109 Eva Wessel Stratford et al demonstrated that a specific inhibitor of tankyrase JW74 could delay cell cycle progression, induce apoptosis and osteogenic differentiation in OS cells, and up‐regulate miRNA let‐7. MiRNA let‐7 is a main regulator of differentiation and associated with CSC phenotype.110 Notably, the increased level of miRNA let‐7 induced by JW74 triggered poorly differentiated cancer cells to differentiate, implying that tankyrase can modulate a switch between stemness and differentiation through dysregulated miRNAs. Subsequently, a recent study unveiled both tumour suppressor and oncogenic roles of miR‐let‐7d, a member of let‐7 family. MiR‐let‐7d can modulate multiple associated genes in 3AB‐OS cells which is a CSC line derived from MG63 cells.111 A significant decrease in miR‐29b‐1 was detected in 3AB‐OS cells, and miR‐29b‐1 was unveiled to negatively regulate stem cell markers including Oct3/4, Sox2, Nanog, CD133 and N‐Myc, cell cycle‐related markers such as CCND2, E2F1, and E2F2, and anti‐apoptotic markers like Bcl‐2 and IAP‐2. Therefore, elevated miR‐29b‐1 suppressed stemness properties, cell proliferation, self‐renewal, and drug resistance of 3AB‐OS CSCs via direct or indirect interaction with these mRNAs.112 These study findings reveal an internal connection between miRNAs and CSCs in OS, providing a new perspective for the study of CSCs to improve prognosis of OS patients.
4.5. Alteration in signal pathways
Abnormal signal transduction pathways seem to regulate initiation, progression, and chemotherapy sensitivity to anticancer drugs in various cancers. There are several common OS‐associated signal pathways which include Wnt/β‐catenin, PI3K/Akt, IGFIR, Notch, TGF‐β, and so on. Wnt/β‐catenin pathway plays a role in osteoblast differentiation and was reported to be the most important one for OS tumourigenesis.113 PI3K/Akt pathway is another crucial pathway participating in OS pathogenesis, and has been recently confirmed as a key vulnerability for OS treatment.114 Some recent studies have demonstrated that miRNAs could elicit aberrant activities of OS‐associated pathways to affect chemosensitivity (Table 1).
Decreased miR‐34c resulted in OS metastasis and chemoresistance by directly targeting the 3′‐UTR of Notch1 and LEF1.115 Sirolimus was reported to induce cell apoptosis and increase cell sensitivity to therapeutic drugs with an up‐regulation of miR‐34b targeting p21‐activated protein kinase 1 (PAK1) and ABCB1.116 The expression level of miR‐497 was reduced in OS tissues, contributing to enhanced activation of PI3K/Akt signalling and resistance to CDDP through binding to vascular endothelial growth factor A (VEGFA). Further functional confirmation was executed in Saos2 xenograft tumour model.117 MiR‐221 was overexpressed in OS samples. It repressed cell apoptosis, promoted cell survival, and increased CDDP resistance due to its direct interaction with PTEN, which causes the activation of PI3K/Akt pathway.118 Inactivation of PI3K/Akt pathway has been revealed to augment expression of Bcl‐2, CCND1, both of which were under the regulation of miR‐221. In OS tissues treated with anticancer drugs, up‐regulated miR‐146b‐5p was observed to facilitate proliferation, migration, and metastasis by positively regulating MMP‐16, and resistance to chemotherapy via negatively regulating zinc and ring finger 3 (ZNRF3), a molecule inactivating Wnt/β‐catenin signalling pathway.119 Generally, these results provide an appealing strategy to target miRNAs implicated in signal pathways to improve OS therapeutic effectiveness.
5. THE CLINICAL UTILITY OF miRNAs IN OSTEOSARCOMA DRUG RESISTANCE
According to the above preclinical studies, these drug resistance‐related miRNAs are expected to supplement or replace existing biomarkers of diagnosis or prognosis, and serve as promising candidates for therapeutic targets to overcome drug resistance in the coming future.40
Several drug resistance‐related miRNAs were mentioned to have a predictive role in clinical prognosis and survival time of OS patients. Clinically, reduced miR‐382 was correlated with unfavourable prognosis in OS patients, due to its potent effect on chemoresistance to anticancer drugs.62 OS patients with low expression level of miR‐15b had obviously poor prognosis and shorter survival times because of chemotherapy resistance.52 A low expression level of miR‐143 was observed in OS samples, which had a significant connection with poor outcomes and shorter survival of OS patients with chemotherapy.108 Reduced miR‐34b level was perceived as a predictor of unfavourable outcomes of OS patients and associated with MDR, which was reversed by administration of sirolimus in vitro.116 Besides, OS patients had a markedly higher level of serum miR‐21, which was associated with advanced Enneking stage and chemoresistance, and served as an independent prognostic factor for OS patients.120
Noticeably, some miRNAs have been reported to be rather promising therapeutic targets in preclinical or clinical studies in recent years (Table 3). The miR‐34 family including miR‐34a, miR‐34b, and miR‐34c, has been known as a tumour suppressor in cancers including OS and gained extensive attention.121, 122, 123, 124, 125, 126 In substantial preclinical studies, treatment with miR‐34 mimics was viewed as a novel miRNA‐target therapy in cancers.121, 127, 128 Besides, replenishment of miR‐34 encapsulated in lipid nanoparticles was demonstrated to exhibit an anticancer effect in several malignancies in a phase I clinical trail (NCT01829971).129 A recent study revealed that miR‐34 mimics could trigger the perturbation of microtubule network and cell death in OS cells, implicating its possibility as a therapeutic agent in OS.130 Recently, it is noted that miR‐34 mimic brought significant benefits for treatment of metastasis in OS mouse models.131 However, the optimal drug doses require further identification for application. The drug toxicity mentioned in this study was not associated with drug resistance. The loss of let‐7 is a prevalent phenomenon in various cancers, and its restoration obviously suppressed tumour growth and extended survival time in vivo.132 It is indicated that replenishing let‐7 might be a beneficial method in OS treatment, which remains to be investigated. MiR‐155 plays a critical and positive role in diverse cancer types.133 Recently, a study has reported a successful delivery of antimiR‐155 conjugated with a small peptide called pHLIP, and its therapeutic benefits without toxicity in a lymphoma mouse model,134 which requires verification in OS models. Increased level of miR‐221 which targets PTEN is of significance in hepatocellular carcinoma (HCC). A recent study revealed that antimiR‐221 modified with cholesterol significantly inhibited tumour growth and prolonged survival time in a HCC mouse model.135 However, the toxicity of cholesterol‐modified antimiR‐221 was not discussed in this study.
Table 3.
Utility of OS‐related MiRNAs in cancer
| microRNA | Study type | Cancer type | Treatment drug | Therapy effect | References |
|---|---|---|---|---|---|
| miR‐34a | Mouse model | Prostate cancer | Systemically delivered miR‐34a mimics | Inhibited prostate cancer metastasis and extended survival time | 121 |
| Mouse model | Lung cancer | Systemically delivered miR‐34a mimics | A significant decrease in tumor burden | 127 | |
| Mouse model | Pancreatic cancer | A lipid‐based nanoparticle for systemic delivery with miR‐34a | Inhibited tumor growth | 128 | |
| Phase I clinical trial | Advanced solid tumors | A liposomal miR‐34a mimic, MRX34 | Showed evidence of antitumor activity | 129 | |
| Mouse model | Osteosarcoma | Delivery of miR‐34a mimics | Suppressed pulmonary metastases and tumor progression, and improved the overall survival | 131 | |
| miR‐155 | Mouse model | Lymphoma | Delivery with antimiR‐155 conjugated with a small peptide | Showed evidence of antitumor activity | 134 |
| miR‐221 | Mouse model | Hepatocellular carcinoma | Delivery with antimiR‐221 modified with cholesterol | Inhibited tumor growth and prolonged survival time | 135 |
These miRNAs possess a unique advantage of clinical applications in OS treatment. For one thing, a single miRNA can simultaneously target multiple mRNAs implicated in one or several signal pathways. For another thing, OS has a complicated genetic background, and is characterized by inherent heterogeneity and aberrance of multiple genes.3, 16 Therefore, modulating the expression levels of these miRNAs might hold great promise for successful OS treatment and improving survival by effectively reversing chemoresistance. Some well‐studied miRNAs have been investigated in preclinical or clinical trials and exhibited their promising effects. Hence, it remains necessary to further confirm and test the clinical roles of these miRNAs.
6. CONCLUSIONS AND PERSPECTIVE
Drug resistance is the main reason for treatment refractoriness of OS. Recent studies have revealed the emerging roles of miRNAs in OS chemoresistance under the mechanisms of DDR, apoptosis avoidance, autophagy induction, activation of CSCs, and alteration in signal pathways. Perturbed DDR system, a hallmark of cancers, has been targeted to design effective DDR drugs for cancer therapy clinically.136 Autophagy can play both prosurvival and prodeath roles in cancer cells. Hence, researchers considered inhibiting its protective function and inducing cell death to treat cancers.97 The eradication of CSCs has been discussed for successful cancer treatment for years and exosomes were recently reported to have potential capability for targeting CSCs.137 These oncogenic or tumour suppressor miRNAs are expected to serve as promising candidates for prognosis and therapeutic targets, which provides a brand‐new outlook into better clinical treatment in OS. However, it still requires further identification, and more preclinical and clinical evidences in support of their future clinical applications.
Accumulated evidence has emphasized that besides miRNAs, other noncoding RNAs especially lncRNAs could function in OS drug resistance since lncRNAs account for a much bigger percentage than miRNAs in ncRNAs.138, 139 Some recent studies have reported the involvement of lncRNAs in OS chemosensitivity under the mechanisms of mediating MDR associated genes and signal pathways and interacting with miRNAs. LncRNA FOXC2 antisense RNA 1 (FOXC2‐AS1) contributed to poor response to DOX in OS patients by up‐regulating MDR associated proteins involving ABCB1and HIF1A.140 LncRNA OS doxorubicin‐resistance related up‐regulated lncRNA (ODRUL) was revealed to inhibit DOX sensitivity through inducing ABCB1 expression in OS cells.141 Overexpressed lncRNA HOXA transcript at the distal tip (HOTTIP) caused a resistance to CDDP by activating Wnt/β‐catenin signalling pathway.142 Of note, long intergenic noncoding RNA 161 (LINC00161) served as a tumour suppressor lncRNA with respect to OS resistance to CDDP by regulating the miR‐645‐IFIT2 signalling axis,143 which indicates an existence of a competitive endogenous RNA regulatory network. On one hand, a competition for the binding of miRNAs between lncRNAs and another nucleotide sequence or structure could have an effect on the translation of miRNAs’ targets.144 On the other hand, miRNAs might function as an upstream regulator of lncRNAs to regulate their expression levels.145 These interplays among endogenous RNAs are so complicated that demands for more potent evidence supports and deeper exploration of underlying molecular mechanisms. These results provided a new insight into identifying potential therapeutic targets for reversing OS chemoresistance based on the synergetic efforts of lncRNAs and miRNAs.
Of note, researchers have traditionally focused much on DNA, mRNA, and proteins, and viewed them as principal modulators and therapeutic targets. In recent two decades, accumulated evidence has revealed a competitive endogenous RNA regulatory network with the participation of miRNA, lncRNA, and circRNA (Figure 4). MiRNAs have gained increasing attention, and their antagonists or mimics have been designed in cancer therapy to reduce or elevate their previous levels, respectively.146, 147 On one hand, it's acknowledged that a single miRNA simultaneously targets several mRNAs implicated in several signal pathways, which brings great benefits to refractory cancers with genomic heterogeneity. On the other hand, a mRNA can be modulated by multiple miRNAs, implying a therapeutic strategy of applying different miRNA antagonists or mimics to effectively affect the specific target mRNA. However, there exist some disadvantages or challenges with respect to miRNA‐targeted strategy. At first, they may elicit broad effects and unexpected alterations of those unrelated genes targeted by same miRNAs, which obviously break the balance of gene expression profiles in cells. Besides, the existence of off‐target effect for miRNA antagonists cannot be ignored. Furthermore, the quick degradation and cellular delivery are two great challenges ought to be solved. Last but not least, it has been noticed that miRNAs can exert a different effect because of several influence factors such as agents, cell lines, cancer types, and so on. This implies careful and cautious choice of miRNA antagonists or mimics according to different conditions.
Figure 4.
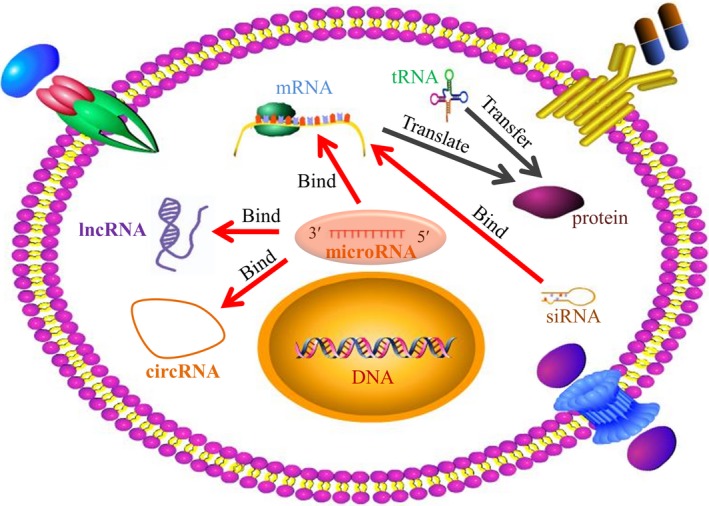
Competitive endogenous RNA regulatory network. Most of human genome DNA has transcripts. About 2% of transcripts code for protein synthesis. The remaining noncoding RNAs include miRNAs, lncRNAs, and cirRNAs. Numerous endogenous RNAs such as mRNAs, lncRNA, and cirRNAs are under the regulation of miRNAs, and they compete for the target binding of miRNAs
To sum up, this review focuses on drug resistance‐related miRNAs in OS through several molecular mechanisms, and provides insight in creating promising therapeutic strategies by targeting these miRNAs to reverse OS chemoresistance.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
This project was supported by NSFC (81502604), Science and Technology Commission of Shanghai Municipality (14140904000), Doctoral Innovation Fund of Shanghai Jiaotong University School of Medicine (No. BXJ201732).
Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Drug resistance‐related microRNAs in osteosarcoma: Translating basic evidence into therapeutic strategies. J Cell Mol Med. 2019;23:2280–2292. 10.1111/jcmm.14064
Ruiling Chen, Gangyang Wang and Ying Zheng contributed equally to this work.
Contributor Information
Gangyang Wang, Email: gangyang_wang@163.com.
Zhengdong Cai, Email: czd856@vip.163.com.
REFERENCES
- 1. Mirabello L, Troisi RJ, Savage SA. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int J Cancer. 2009;125(1):229‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gelberg KH, Fitzgerald EF, Hwang S, Dubrow R. Growth and development and other risk factors for osteosarcoma in children and young adults. Int J Epidemiol. 1997;26(2):272‐278. [DOI] [PubMed] [Google Scholar]
- 3. Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7(1):104‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bousquet M, Noirot C, Accadbled F, et al. Whole‐exome sequencing in osteosarcoma reveals important heterogeneity of genetic alterations. Ann Oncol. 2016;27(4):738‐744. [DOI] [PubMed] [Google Scholar]
- 5. Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336‐2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9(4):422‐441. [DOI] [PubMed] [Google Scholar]
- 7. Eilber F, Giuliano A, Eckardt J, Patterson K, Moseley S, Goodnight J. Adjuvant chemotherapy for osteosarcoma: a randomized prospective trial. J Clin Oncol. 1987;5(1):21‐26. [DOI] [PubMed] [Google Scholar]
- 8. Jaffe N. Osteosarcoma: review of the past, impact on the future. The American experience. Cancer Treat Res. 2009;152:239‐262. [DOI] [PubMed] [Google Scholar]
- 9. Allison DC, Carney SC, Ahlmann ER, et al. A meta‐analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012:704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16(6):257‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861‐874. [DOI] [PubMed] [Google Scholar]
- 12. Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updates. 2013;16(3–5):47‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee JW, Kim N, Park JH, et al. Differential microRNA expression between gastric cancer tissue and non‐cancerous gastric mucosa according to Helicobacter pylori status. J Cancer Prev. 2017;22(1):33‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song B, Wang Y, Xi Y, et al. Mechanism of chemoresistance mediated by miR‐140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28(46):4065‐4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gougelet A, Pissaloux D, Besse A, et al. Micro‐RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int J Cancer. 2011;129(3):680‐690. [DOI] [PubMed] [Google Scholar]
- 16. Jin H, Tuo W, Lian H, Liu Q, Zhu XQ, Gao H. Strategies to identify microRNA targets: new advances. New Biotechnol. 2010;27(6):734‐738. [DOI] [PubMed] [Google Scholar]
- 17. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell. 1993;75(5):843‐854. [DOI] [PubMed] [Google Scholar]
- 18. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522‐531. [DOI] [PubMed] [Google Scholar]
- 19. Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766‐770. [DOI] [PubMed] [Google Scholar]
- 20. Esquela‐Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259‐269. [DOI] [PubMed] [Google Scholar]
- 21. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281‐297. [DOI] [PubMed] [Google Scholar]
- 23. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118‐126. [DOI] [PubMed] [Google Scholar]
- 24. Jiang Q, Wang Y, Hao Y, et al. Liu Y: miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98‐D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong L, Yang Z, Ma J, Fan D. Function of miRNA in controlling drug resistance of human cancers. Curr Drug Targets. 2013;14(10):1118‐1127. [DOI] [PubMed] [Google Scholar]
- 27. Pandima Devi K, Rajavel T, Daglia M, Nabavi SF, Bishayee A, Nabavi SM. Targeting miRNAs by polyphenols: Novel therapeutic strategy for cancer. Semin Cancer Biol. 2017;46:146‐157. [DOI] [PubMed] [Google Scholar]
- 28. Du L, Jiang X, Duan W, et al. Cell‐free microRNA expression signatures in urine serve as novel noninvasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2017;8(25):40832‐40842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kachalaki S, Ebrahimi M, Mohamed Khosroshahi L, Mohammadinejad S, Baradaran B. Cancer chemoresistance; biochemical and molecular aspects: a brief overview. Eur J Pharm Sci. 2016;89:20‐30. [DOI] [PubMed] [Google Scholar]
- 30. Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP‐dependent transporters. Nat Rev Cancer. 2002;2(1):48‐58. [DOI] [PubMed] [Google Scholar]
- 31. Tamasi V, Monostory K, Prough RA, Falus A. Role of xenobiotic metabolism in cancer: involvement of transcriptional and miRNA regulation of P450s. Cell Mol Life Sci. 2011;68(7):1131‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jackson SP, Bartek J. The DNA‐damage response in human biology and disease. Nature. 2009;461(7267):1071‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ouyang L, Shi Z, Zhao S, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45(6):487‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Torreggiani E, Roncuzzi L, Perut F, Zini N, Baldini N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int J Oncol. 2016;49(1):189‐196. [DOI] [PubMed] [Google Scholar]
- 35. Gomes BC, Rueff J, Rodrigues AS. MicroRNAs and cancer drug resistance. Methods Mol Biol. 2016;1395:137‐162. [DOI] [PubMed] [Google Scholar]
- 36. Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13(3):57‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie L, Jing R, Qi J, Lin Z, Ju S. Drug resistance‐related microRNAs in hematological malignancies: translating basic evidence into therapeutic strategies. Blood Rev. 2015;29(1):33‐44. [DOI] [PubMed] [Google Scholar]
- 38. Li F, Mahato RI. MicroRNAs and drug resistance in prostate cancers. Mol Pharm. 2014;11(8):2539‐2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He H, Ni J, Huang J. Molecular mechanisms of chemoresistance in osteosarcoma (Review). Oncol Lett. 2014;7(5):1352‐1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miao J, Wu S, Peng Z, Tania M, Zhang C. MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects. Tumour Biol. 2013;34(4):2093‐2098. [DOI] [PubMed] [Google Scholar]
- 41. Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475‐1485. [DOI] [PubMed] [Google Scholar]
- 42. Chowdhury D, Choi YE, Brault ME. Charity begins at home: non‐coding RNA functions in DNA repair. Nat Rev Mol Cell Biol. 2013;14(3):181‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wouters MD, van Gent DC, Hoeijmakers JH, Pothof J. MicroRNAs, the DNA damage response and cancer. Mutat Res. 2011;717(1–2):54‐66. [DOI] [PubMed] [Google Scholar]
- 44. Liu YX, Wang L, Liu WJ, et al. MiR‐124‐3p/B4GALT1 axis plays an important role in SOCS3‐regulated growth and chemo‐sensitivity of CML. J Hematol Oncol. 2016;9(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ledderose C, Mohnle P, Limbeck E, et al. Corticosteroid resistance in sepsis is influenced by microRNA‐124–induced downregulation of glucocorticoid receptor‐alpha. Crit Care Med. 2012;40(10):2745‐2753. [DOI] [PubMed] [Google Scholar]
- 46. Chen SM, Chou WC, Hu LY, et al. The effect of microRNA‐124 overexpression on anti‐tumor drug sensitivity. PLoS ONE. 2015;10(6):e0128472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. di Fagagna FD, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP‐ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23(1):76‐80. [DOI] [PubMed] [Google Scholar]
- 48. Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol Aspects Med. 2013;34(6):1217‐1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gill SJ, Travers J, Pshenichnaya I, et al. Combinations of PARP inhibitors with temozolomide drive PARP1 trapping and apoptosis in Ewing's Sarcoma. PLoS ONE. 2015;10(10):e0140988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang T, Penicud K, Bruhn C, et al. Competition between NBS1 and ATMIN controls ATM signaling pathway choice. Cell Rep. 2012;2(6):1498‐1504. [DOI] [PubMed] [Google Scholar]
- 51. Xia L, Zhang D, Du R, et al. miR‐15b and miR‐16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123(2):372‐379. [DOI] [PubMed] [Google Scholar]
- 52. Duan Z, Gao Y, Shen J, et al. miR‐15b modulates multidrug resistance in human osteosarcoma in vitro and in vivo. Mol Oncol. 2017;11(2):151‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193‐204. [DOI] [PubMed] [Google Scholar]
- 54. Pearl LH, Schierz AC, Ward SE, Al‐Lazikani B, Pearl FM. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15(3):166‐180. [DOI] [PubMed] [Google Scholar]
- 55. Brentnall M, Rodriguez‐Menocal L, De Guevara RL, Cepero E, Boise LH. Caspase‐9, caspase‐3 and caspase‐7 have distinct roles during intrinsic apoptosis. BMC cell biology. 2013;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tao LJ, Zhou XD, Shen CC, et al. Tetrandrine induces apoptosis and triggers a caspase cascade in U2‐OS and MG‐63 cells through the intrinsic and extrinsic pathways. Mol Med Rep. 2014;9(1):345‐349. [DOI] [PubMed] [Google Scholar]
- 57. Karpel‐Massler G, Shu C, Chau L, et al. Combined inhibition of Bcl‐2/Bcl‐xL and Usp9X/Bag3 overcomes apoptotic resistance in glioblastoma in vitro and in vivo. Oncotarget. 2015;6(16):14507‐14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jiang L, Tao C, He A, He X. Overexpression of miR‐126 sensitizes osteosarcoma cells to apoptosis induced by epigallocatechin‐3‐gallate. World J Surg Oncol. 2014;12:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cai CK, Zhao GY, Tian LY, et al. miR‐15a and miR‐16‐1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep. 2012;28(5):1764‐1770. [DOI] [PubMed] [Google Scholar]
- 60. Zhang X, Guo Q, Chen J, Chen Z. Quercetin enhances Cisplatin sensitivity of human osteosarcoma cells by modulating microRNA‐217‐KRAS axis. Mol Cells. 2015;38(7):638‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR‐138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin‐induced apoptosis in osteosarcoma cells. PLoS ONE. 2016;11(3):e0150026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu M, Jin H, Xu CX, et al. miR‐382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5(19):9472‐9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Song B, Wang Y, Titmus MA, et al. Molecular mechanism of chemoresistance by miR‐215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y, Duan G, Feng S. MicroRNA‐301a modulates doxorubicin resistance in osteosarcoma cells by targeting AMP‐activated protein kinase alpha 1. Biochem Biophys Res Comm. 2015;459(3):367‐373. [DOI] [PubMed] [Google Scholar]
- 65. Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR‐21 in Cancer. Drug Dev Res. 2015;76(6):270‐277. [DOI] [PubMed] [Google Scholar]
- 66. Ziyan W, Yang L. MicroRNA‐21 regulates the sensitivity to cisplatin in a human osteosarcoma cell line. Ir J Med Sci. 2016;185(1):85‐91. [DOI] [PubMed] [Google Scholar]
- 67. Vanas V, Haigl B, Stockhammer V, Sutterluty‐Fall H. MicroRNA‐21 increases proliferation and cisplatin sensitivity of osteosarcoma‐derived cells. PLoS ONE. 2016;11(8):e0161023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin BC, Huang D, Yu CQ, et al. MicroRNA‐184 modulates doxorubicin resistance in osteosarcoma cells by targeting BCL2L1. Med Sci Monit. 2016;22:1761‐1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang GC, He QY, Tong DK, et al. MiR‐367 negatively regulates apoptosis induced by adriamycin in osteosarcoma cells by targeting KLF4. J Bone Oncol. 2016;5(2):51‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhou C, Tan W, Lv H, Gao F, Sun J. Hypoxia‐inducible microRNA‐488 regulates apoptosis by targeting Bim in osteosarcoma. Cell Oncol. 2016;39(5):463‐471. [DOI] [PubMed] [Google Scholar]
- 71. Lin Z, Song D, Wei H, et al. TGF‐beta1‐induced miR‐202 mediates drug resistance by inhibiting apoptosis in human osteosarcoma. J Cancer Res Clin Oncol. 2016;142(1):239‐246. [DOI] [PubMed] [Google Scholar]
- 72. Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR‐33a is up‐regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down‐regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jacques C, Calleja LR, Baud'huin M, et al. miRNA‐193a‐5p repression of p73 controls Cisplatin chemoresistance in primary bone tumors. Oncotarget. 2016;7(34):54503‐54514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ozaki T, Sugimoto H, Nakamura M, et al. Runt‐related transcription factor 2 attenuates the transcriptional activity as well as DNA damage‐mediated induction of pro‐apoptotic TAp73 to regulate chemosensitivity. FEBS J. 2015;282(1):114‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pu Y, Zhao F, Li Y, et al. The miR‐34a‐5p promotes the multi‐chemoresistance of osteosarcoma via repression of the AGTR1 gene. BMC Cancer. 2017;17(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self‐digestion. Nature. 2008;451(7182):1069‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mathew R, Karantza‐Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7(12):961‐967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. White E, DiPaola RS. The double‐edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15(17):5308‐5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen S, Jiang YZ, Huang L, et al. The residual tumor autophagy marker LC3B serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Clin Cancer Res. 2013;19(24):6853‐6862. [DOI] [PubMed] [Google Scholar]
- 82. Liu L, Yang M, Kang R, et al. HMGB1‐induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25(1):23‐31. [DOI] [PubMed] [Google Scholar]
- 83. Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10(9):1533‐1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhao D, Yuan H, Yi F, Meng C, Zhu Q. Autophagy prevents doxorubicininduced apoptosis in osteosarcoma. Mol Med Rep. 2014;9(5):1975‐1981. [DOI] [PubMed] [Google Scholar]
- 85. Pan Z, Sun X, Shan H, et al. MicroRNA‐101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor‐beta1 pathway. Circulation. 2012;126(7):840‐850. [DOI] [PubMed] [Google Scholar]
- 86. Chang Z, Huo L, Li K, Wu Y, Hu Z. Blocked autophagy by miR‐101 enhances osteosarcoma cell chemosensitivity in vitro. ScientificWorldJournal. 2014;2014:794756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Guo S, Bai R, Liu W, et al. miR‐22 inhibits osteosarcoma cell proliferation and migration by targeting HMGB1 and inhibiting HMGB1‐mediated autophagy. Tumour Biol. 2014;35(7):7025‐7034. [DOI] [PubMed] [Google Scholar]
- 88. Li X, Wang S, Chen Y, Liu G, Yang X. miR‐22 targets the 3' UTR of HMGB1 and inhibits the HMGB1‐associated autophagy in osteosarcoma cells during chemotherapy. Tumour Biol. 2014;35(6):6021‐6028. [DOI] [PubMed] [Google Scholar]
- 89. Huang J, Ni J, Liu K, et al. HMGB1 promotes drug resistance in osteosarcoma. Can Res. 2012;72(1):230‐238. [DOI] [PubMed] [Google Scholar]
- 90. Huang J, Liu K, Yu Y, et al. Targeting HMGB1‐mediated autophagy as a novel therapeutic strategy for osteosarcoma. Autophagy. 2012;8(2):275‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu R, Liu S, Chen H, Lao L. MicroRNA‐30a downregulation contributes to chemoresistance of osteosarcoma cells through activating Beclin‐1‐mediated autophagy. Oncol Rep. 2016;35(3):1757‐1763. [DOI] [PubMed] [Google Scholar]
- 92. Li Y, Jiang W, Hu Y, et al. MicroRNA‐199a‐5p inhibits cisplatin‐induced drug resistance via inhibition of autophagy in osteosarcoma cells. Oncol Lett. 2016;12(5):4203‐4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bayraktar R, Van Roosbroeck K. miR‐155 in cancer drug resistance and as target for miRNA‐based therapeutics. Cancer Metastasis Rev. 2018;37(1):33‐44. [DOI] [PubMed] [Google Scholar]
- 94. Chen L, Jiang K, Jiang H, Wei P. miR‐155 mediates drug resistance in osteosarcoma cells via inducing autophagy. Exp Ther Med. 2014;8(2):527‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wei R, Cao G, Deng Z, Su J, Cai L. miR‐140‐5p attenuates chemotherapeutic drug‐induced cell death by regulating autophagy through inositol 1,4,5‐trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells. Biosci Rep. 2016;36(5):e00392‐e392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rosenfeldt MT, O'Prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296‐300. [DOI] [PubMed] [Google Scholar]
- 97. Panda PK, Mukhopadhyay S, Das DN, Sinha N, Naik PP, Bhutia SK. Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin Cell Dev Biol. 2015;39:43‐55. [DOI] [PubMed] [Google Scholar]
- 98. Wu CC, Huang YF, Hsieh CP, Chueh PJ, Chen YL. Combined use of Zoledronic acid augments ursolic acid‐induced apoptosis in human osteosarcoma cells through enhanced oxidative stress and autophagy. Molecules. 2016;21(12):1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang Z, Shao Z, Xiong L, Yang S. Inhibition of autophagy enhances cisplatin‐induced apoptosis in the MG63 human osteosarcoma cell line. Oncol Lett. 2015;10(5):2941‐2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313‐319. [DOI] [PubMed] [Google Scholar]
- 101. Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12(2):133‐143. [DOI] [PubMed] [Google Scholar]
- 102. Subramaniam D, Kaushik G, Dandawate P, Anant S. Targeting cancer stem cells for chemoprevention of pancreatic cancer. Curr Med Chem. 2017;25(22):2585‐2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bekaii‐Saab T, El‐Rayes B. Identifying and targeting cancer stem cells in the treatment of gastric cancer. Cancer. 2017;123(8):1303‐1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Abarrategi A, Tornin J, Martinez‐Cruzado L, et al. Osteosarcoma: cells‐of‐origin, cancer stem cells, and targeted therapies. Stem Cells Int. 2016;2016:3631764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang Y, Yao J, Meng H, et al. A novel long non‐coding RNA, hypoxia‐inducible factor‐2alpha promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol Med Rep. 2015;11(4):2534‐2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li B, Fan J, Chen N. A novel regulator of type ii diabetes: MicroRNA‐143. Trends Endocrinol Metab. 2018;29(6):380‐388. [DOI] [PubMed] [Google Scholar]
- 107. Kitade Y, Akao Y. MicroRNAs and their therapeutic potential for human diseases: microRNAs, miR‐143 and ‐145, function as anti‐oncomirs and the application of chemically modified miR‐143 as an anti‐cancer drug. J Pharmacol Sci. 2010;114(3):276‐280. [DOI] [PubMed] [Google Scholar]
- 108. Zhou J, Wu S, Chen Y, et al. microRNA‐143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med. 2015;240(7):867‐875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Skvortsov S, Debbage P, Skvortsova I. Proteomics of cancer stem cells. Int J Radiat Biol. 2014;90(8):653‐658. [DOI] [PubMed] [Google Scholar]
- 110. Stratford EW, Daffinrud J, Munthe E, et al. The tankyrase‐specific inhibitor JW74 affects cell cycle progression and induces apoptosis and differentiation in osteosarcoma cell lines. Cancer Med. 2014;3(1):36‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Di Fiore R, Drago‐Ferrante R, Pentimalli F, et al. Let‐7d miRNA shows both antioncogenic and oncogenic functions in osteosarcoma‐derived 3AB‐OS cancer stem cells. J Cell Physiol. 2016;231(8):1832‐1841. [DOI] [PubMed] [Google Scholar]
- 112. Di Fiore R, Drago‐Ferrante R, Pentimalli F, et al. MicroRNA‐29b‐1 impairs in vitro cell proliferation, selfrenewal and chemoresistance of human osteosarcoma 3AB‐OS cancer stem cells. Int J Oncol. 2014;45(5):2013‐2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Reimann E, Koks S, Ho XD, Maasalu K, Martson A. Whole exome sequencing of a single osteosarcoma case–integrative analysis with whole transcriptome RNA‐seq data. Hum Genomics. 2014;8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Perry JA, Kiezun A, Tonzi P, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci USA. 2014;111(51):E5564–E5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xu M, Jin H, Xu CX, Bi WZ, Wang Y. MiR‐34c inhibits osteosarcoma metastasis and chemoresistance. Med Oncol. 2014;31(6):972. [DOI] [PubMed] [Google Scholar]
- 116. Zhou Y, Zhao RH, Tseng KF, et al. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA‐34b expression. Acta Pharmacol Sin. 2016;37(4):519‐529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Shao XJ, Miao MH, Xue J, Xue J, Ji XQ, Zhu H. The down‐regulation of microRNA‐497 contributes to cell growth and cisplatin resistance through PI3K/Akt pathway in osteosarcoma. Cell Physiol Biochem. 2015;36(5):2051‐2062. [DOI] [PubMed] [Google Scholar]
- 118. Zhao G, Cai C, Yang T, et al. MicroRNA‐221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS ONE. 2013;8(1):e53906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hu BY, Gu YH, Cao CJ, et al. Reversal effect and mechanism of Ginkgo biloba exocarp extracts in multidrug resistance of mice S180 tumor cells. Exp Ther Med. 2016;12(4):2053‐2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yuan J, Chen L, Chen X, Sun W, Zhou X. Identification of serum microRNA‐21 as a biomarker for chemosensitivity and prognosis in human osteosarcoma. J Int Med Res. 2012;40(6):2090‐2097. [DOI] [PubMed] [Google Scholar]
- 121. Liu C, Kelnar K, Liu B, et al. The microRNA miR‐34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Okada N, Lin CP, Ribeiro MC, et al. A positive feedback between p53 and miR‐34 miRNAs mediates tumor suppression. Genes Dev. 2014;28(5):438‐450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rokavec M, Oner MG, Li H, et al. IL‐6R/STAT3/miR‐34a feedback loop promotes EMT‐mediated colorectal cancer invasion and metastasis. J Clin Investig. 2014;124(4):1853‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bader AG. miR‐34 – a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ghandadi M, Sahebkar A. MicroRNA‐34a and its target genes: key factors in cancer multidrug resistance. Curr Pharm Des. 2016;22(7):933‐939. [DOI] [PubMed] [Google Scholar]
- 127. Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pramanik D, Campbell NR, Karikari C, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10(8):1470‐1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR‐34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35(2):180‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Vetter NS, Kolb EA, Mills CC, Sampson VB. The microtubule network and cell death are regulated by an miR‐34a/Stathmin 1/betaIII‐Tubulin Axis. Mol Cancer Res. 2017;15(7):953‐964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jian C, Tu MJ, Ho PY, et al. Co‐targeting of DNA, RNA, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget. 2017;8(19):30742‐30755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Stahlhut C, Slack FJ. Combinatorial action of microRNAs let‐7 and miR‐34 effectively synergizes with erlotinib to suppress non‐small cell lung cancer cell proliferation. Cell Cycle. 2015;14(13):2171‐2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Tili E, Croce CM, Michaille JJ. miR‐155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28(5):264‐284. [DOI] [PubMed] [Google Scholar]
- 134. Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Park JK, Kogure T, Nuovo GJ, et al. miR‐221 silencing blocks hepatocellular carcinoma and promotes survival. Can Res. 2011;71(24):7608‐7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. O'Sullivan Coyne G, Chen AP, Meehan R, Doroshow JH. PARP inhibitors in reproductive system cancers: current use and developments. Drugs. 2017;77(2):113‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wang J, Zheng Y, Zhao M. Exosome‐based cancer therapy: implication for targeting cancer stem cells. Front Pharmacol. 2016;7:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Chen R, Wang G, Zheng Y, Hua Y, Cai Z. Long non‐coding RNAs in osteosarcoma. Oncotarget. 2017;8(12):20462‐20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhu KP, Zhang CL, Shen GQ, Zhu ZS. Long noncoding RNA expression profiles of the doxorubicin‐resistant human osteosarcoma cell line MG63/DXR and its parental cell line MG63 as ascertained by microarray analysis. Int J Clin Exp Pathol. 2015;8(8):8754‐8773. [PMC free article] [PubMed] [Google Scholar]
- 141. Zhang CL, Zhu KP, Shen GQ, Zhu ZS. A long non‐coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumour Biol. 2016;37(2):2737‐2748. [DOI] [PubMed] [Google Scholar]
- 142. Lim MY, LaMonte G, Lee MC, et al. UDP‐galactose and acetyl‐CoA transporters as plasmodium multidrug resistance genes. Nat Microbiol. 2016;16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang Y, Zhang L, Zheng X, et al. Long non‐coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin‐induced apoptosis by regulating the miR‐645‐IFIT2 axis. Cancer Lett. 2016;382(2):137‐146. [DOI] [PubMed] [Google Scholar]
- 144. Luo W, He H, Xiao W, et al. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7(34):54733‐54743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Al Bshabshe A, Joseph MR, Al Hussein A, Haimour W, Hamid ME. Multidrug resistance Acinetobacter species at the intensive care unit, Aseer Central Hospital, Saudi Arabia: a one year analysis. Asian Pac J Trop Med. 2016;9(9):903‐908. [DOI] [PubMed] [Google Scholar]
- 146. Xue J, Yang J, Luo M, Cho WC, Liu X. MicroRNA‐targeted therapeutics for lung cancer treatment. Expert Opin Drug Discov. 2017;12(2):141‐157. [DOI] [PubMed] [Google Scholar]
- 147. Esposito CL, Nuzzo S, Kumar SA, et al. A combined microRNA‐based targeted therapeutic approach to eradicate glioblastoma stem‐like cells. J Control Release. 2016;238:43‐57. [DOI] [PubMed] [Google Scholar]


