Abstract
Owing to the emerging impact of bioinformatics and computational biology, in this article, we present an overview of the history and current state of the research on this field in Latin America (LA). It will be difficult to cover without inequality all the efforts, initiatives and works that have happened for the past two decades in this vast region (that includes >19 million km2 and >600 million people). Despite the difficulty, we have done an analytical search looking for publications in the field made by researchers from 19 LA countries in the past 25 years. In this way, we find that research in bioinformatics in this region should develop twice to approach the average world scientific production in the field. We also found some of the pioneering scientists who initiated and led bioinformatics in the region and were promoters of this new scientific field. Our analysis also reveals that spin-off began around some specific areas within the biomolecular sciences: studies on genomes (anchored in the new generation of deep sequencing technologies, followed by developments in proteomics) and studies on protein structures (supported by three-dimensional structural determination technologies and their computational advancement). Finally, we show that the contribution to this endeavour of the Iberoamerican Society for Bioinformatics, founded in Mexico in 2009, has been significant, as it is a leading forum to join efforts of many scientists from LA interested in promoting research, training and education in bioinformatics.
Keywords: bioinformatics, computational biology, Latin America, Ibero-America, genomics, SoIBio
Introduction: genome biology, genomics and bioinformatics
As indicated in a report several years ago of the ‘Latin American Genome Initiative’: ‘biological and medical sciences in this large American region were caught unprepared with the explosive development of genome biology with its concomitant use of high-throughput technologies, large scale database management, sophisticated genomic instrumentation and large consortiums collaborative research’ [1]. In contrast, previous biological technologies, such as those developed during the genetic engineering revolution, were readily assimilated and successfully used by laboratories in the region. However, the genomic revolution only became real and effective in Latin America (LA) for research and academia about 15 years ago. Moreover, the omic-wide large-scale data analyses based in the development of original and newly generated bioinformatic tools did not start in a relevant way in LA up to the past decade (showing a lot of differences depending on the areas). Despite this general delay, significant efforts began earlier with the computational studies of some pioneering scientists. In this brief review, we describe the work of some of the research groups that were talented drivers in the first steps of the ‘biocomputational revolution’ in LA. Our analysis is based mainly on the search for scientific publications in the field, carried out by researchers from 19 LA countries in the past 25 years.
Before entering our analysis, we want to indicate that this review is not pretending to ignore other relevant scientific areas related to computational biology (such as ‘theoretical biology’ or ‘mathematical biology’), which have been working and producing interesting studies for many years. The point is that these areas are somewhat out of the focus of our study because we centred our review on the origin of data-driven computational biology and bioinformatics in LA.
Definition of a vast multinational region: Latin- and Ibero-America
To start, it is worth clarifying which is the multinational geographic region that is in the spotlight of our study. The region of the world that we are going to analyse, commonly called ‘Latin America’, is composed of 22 countries: Mexico in North America; Guatemala, Honduras, El Salvador, Nicaragua, Costa Rica and Panama in Central America; Cuba, Dominican Republic, Haiti and Puerto Rico in the Caribbean; and Colombia, Venezuela, Ecuador, Peru, Bolivia, Chile, French Guyana, Brazil, Paraguay, Argentina and Uruguay in South America. In fact, this region is similar to the international area of ‘SoIBio’ (http://www.soibio.org/; Sociedad Iberoamericana de Bioinformática, Iberoamerican Society for Bioinformatics). SoIBio is a scientific and professional society that aims to promote research and development of bioinformatics and computational biology in the international region of Ibero-America (that includes 22 countries). These are 20 countries that are part of LA (i.e. all the ones indicated above, but not Haiti and the French Guyana because they are French-speaking countries), plus two European countries: Spain and Portugal. SoIBio includes Spain and Portugal because they were founders of this scientific society, and several research teams from these countries worked on the development of SoIBio from the beginning. In fact, these two countries are usually considered part of IberoAmerica (‘Ibero-America’) for cultural and historical reasons in multiple international agreements. Despite being full members and founders of SoIBio, in this review, we do not include any data from Spain or Portugal, as we are investigating the development of bioinformatics only in LA.
Analysis of scientific publications on bioinformatics from 1991 to 2016
To have a profile of the scientific records that have been produced in LA in the area of bioinformatics or computational biology over the past 25 years (1991–2016), we have done a search in the reference database Web of Science (previously known as ISI JCR Web of Knowledge, http://webofknowledge.com/). In this analysis, we looked for original scientific articles published by researchers from any of the 19 LA countries included in SoIBio: Argentina, Bolivia, Brazil, Chile, Colombia, Costa Rica, Cuba, Dominican Republic, Ecuador, El Salvador, Guatemala, Honduras, Mexico, Nicaragua, Panama, Paraguay, Peru, Uruguay and Venezuela. As we indicated, we exclude from this investigation Portugal and Spain, as well as Puerto Rico (because it is not an independent country, and in many cases, it cannot be distinguished or separated from the United States).
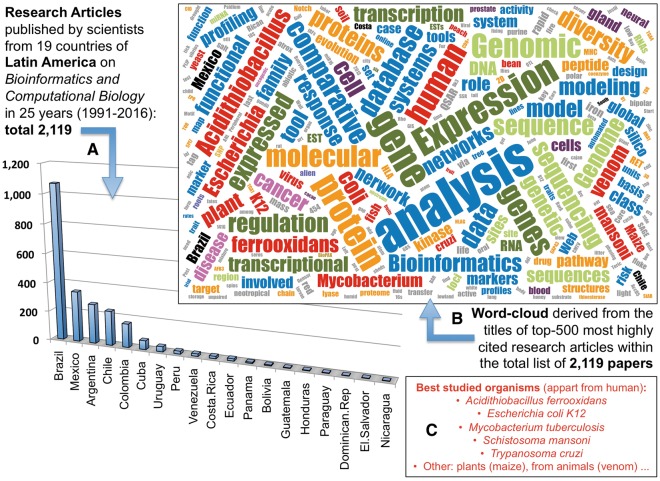
To perform the bibliographic search, we used the following keywords: Bioinformatic* OR Computational Biology* OR Biological Database*. In this way, we found 2119 original research articles published between 1991 and 2016 that are assigned to these countries as shown in Figure 1A. The numbers indicate that there are 1068 articles from Brazil (i.e. articles assigned at least to one author in Brazil), followed by Mexico with 345 articles, Argentina 270, Chile 236, Colombia 162 and Cuba 61. The rest have <50 articles related to bioinformatics in the past 25 years. When we calculated the production per capita (i.e. per million people), the numbers are as follows: Brazil 1068/209.5 = 5.10; Mexico 345/128.6 = 2.68; Argentina 270/43.8 = 6.16; Chile 236/18.1 = 13.04; Colombia 162/48.6 = 3.33; Cuba 61/11.4 = 5.35; Uruguay 35/3.4 = 10.29; Costa Rica 16/4.8 = 3.33; and Panama 8/3.9 = 2.05. The rest of the countries have for this ratio values below 1 (the details can be seen in Supplementary Table S1). All these numbers give a good estimation about the scientific production of LA in the field and show that Brazil is the most prolific producer in absolute numbers. Chile and Uruguay have the best productivity ratio, with a value per capita >10. In comparison with the rest of the world, the total world-wide production that we found in our search was 53 439 scientific articles published. On this figure, LA represents a 3.9% of such production, despite the fact that it represents about an 8.1% of the current world population (600/7400 = 0.08 population ratio). In conclusion, it could be considered that the research on bioinformatics and computational biology in LA should still develop twice to become closer to the general world-wide scientific production in the field.
Figure 1.
(A) Barplot presenting the number of research articles per country published by scientists from 19 countries of LA on bioinformatics and computational biology in 25 years (from 1991 to 2016). The total number found was: 2119. (B) Word cloud derived from the titles of top 500 most highly cited research articles within the total list of 2119 papers. (C) Selection of some of the organisms studied by scientists of LA.
The numbers indicated above are calculated relative to the population of the countries, but it can be illustrative to analyse the productivity also considering the levels of expenditure on research in these LA countries. In this way, new calculations were made to obtain the number of articles published in the field of bioinformatics with respect to the population (per capita), and also relative to each country’s expenditure on Research and Development (R&D). All the results of this analysis are included in Supplementary Table S1 and show a modulation in previous numbers that identifies the top five countries as those that also have the highest productivity in absolute numbers: Brazil, Mexico, Argentina, Chile and Colombia. In this way, Brazil shows to be productive, with a value of 8.11 articles per million people per unit of R&D expenditure. Mexico also shows now a productivity much closer to Brazil (with a value of 7.39). Argentina and Chile show lower productivity (3.52 and 4.56, respectively), but this is because of their higher level of R&D investment. On the contrary, the case of Colombia is interesting because it shows the highest value in this calculation (9.13), which comes from the fact that they have a fairly low level of investment in R&D (only a 0.23% of the gross domestic product, i.e. almost ten times lower than Brazil per capita), and this gives the effect of an apparent higher productivity. A similar anomalous effect also occurs with several small Central America countries (Guatemala, Honduras and Nicaragua; Supplementary Table S1).
To explore the specific and recurrent topics that are behind the literature search done, we performed a word-cloud analysis on the titles of the top 500 most cited articles out of the set of 2119 papers found related to bioinformatics, computational biology or biological databases. The results of this analysis are presented in Figure 1B, which shows the most represented words in these titles (excluding general or ambiguous non-significant words). The cloud shows in blue the most abundant keywords that are the ones related to the search topic: bioinformatics, analysis, data, database, comparative, profiling, modelling, network, system, functional, marker, etc. Apart from these terms that are related to the methodologies used in bioinformatics, there are two other main groups of words that show two thematic tendencies: (i) one in green that represents the terms genomes, genomic, sequences, loci and DNA (in pale green) and gene, genes, expression, expressed, transcription and RNA (in dark green); (ii) another in orange that represents the terms protein, proteins, molecular, peptide, structures, proteome, etc. (Figure 1B). These groups of terms show the over-representation of some thematic trends that we discuss below.
Together to these main subjects, the analysis reveals that the investigations on genes, proteins and bioinformatics in LA have been focused in a group of organisms, mainly microbial or parasites associated to endemic diseases (Figure 1C). In this way, some microorganisms have been more extensively studied with biological and computational approaches: Acidithiobacillus ferrooxidans (studied mainly in Chile by the group of David S. Holmes and collaborators who performed most of the bioinformatic analyses in these studies) [2–6]; Escherichia coli K12 (studied along the past 25 years in Mexico by the group of Julio Collado-Vides and collaborators) [7–13]; Mycobacterium tuberculosis (studied by several groups from Brazil and Colombia, and by an international consortium led by the Institute Pasteur from Guadeluope, France) [14–16]. Most of the articles cited above, associated with studies on microorganisms, correspond to significant contributions on bioinformatics (including new computational methods, tools and developed resources). In fact, some of these publications have received >500 citations in peer-reviewed journals, showing the relevance of the work done.
There are two other organisms featured in our analysis (Figure 1C), which correspond to significant parasites of humans: the trematode Schistosoma mansoni that causes the schistosomiasis (a neglected tropical disease frequent in the Caribbean and in eastern South America), and the protozoan Trypanosoma cruzi that is the causative agent of Chagas disease. Groups from FIOCRUZ in Belo Horizonte (Brazil) and from the University of Sao Paulo (Brazil) participated in the bioinformatic studies on the genome of S.mansoni [17–19]. With respect to T.cruzi, relevant studies using bioinformatics have been performed from the National Universities of San Martin and La Plata (Argentina) [20–23]. Of course, there are other publications and research articles related to human health that involved bioinformatic studies performed by groups in LA, but with the references presented, we have quoted some of the most relevant.
Research interactions and growth of bioinformatics in LA
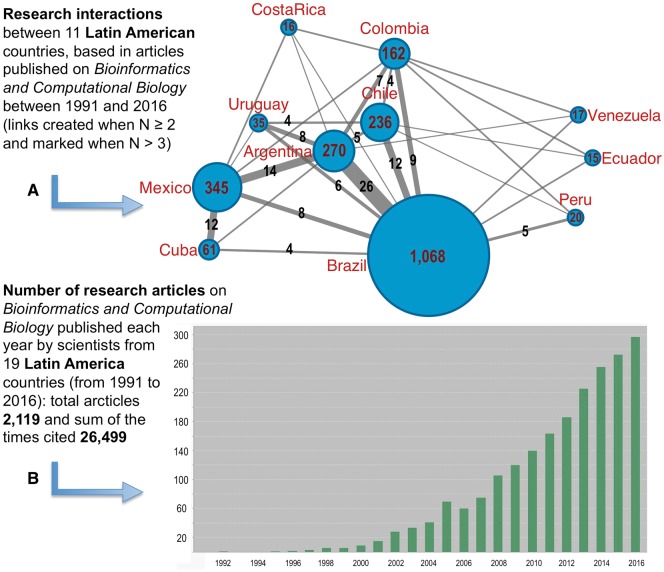
Our investigation based on the analysis of the research publications done by scientists from LA from 1991 to 2016 also reveals the interactions between members of different countries that worked together and collaborated in the same publications. These data are presented in Figure 2A that shows a network built with the number of common publications reported for the 11 most prolific countries. The number of total articles from each country is indicated with a number inside the nodes, and the number of cooperative papers shared by authors from two different countries is indicated with a number on the edges of the network. This analysis shows that the closest collaboration has occurred so far between Argentina and Brazil, followed by Argentina and México. Brazil has links with all the other 10 represented countries (this is expected considering that it is the largest country in LA), but it is interesting to underline a close collaboration triangle Brazil–Argentina–Chile and another close binary interaction Mexico–Cuba.
Figure 2.
(A) Network presenting the research interactions between 11 LA countries, based in articles published on bioinformatics and computational biology between 1991 and 2016 (links created when the number of papers is N ≥ 2 and marked when N > 3). (B) Number of research articles on bioinformatics and computational biology published each year by scientists from 19 LA countries (from 1991 to 2016). The total number of articles was 2119 and the total number of times that have been cited: 26 499.
Regarding the growth of the scientific productivity, Figure 2B shows a continuous increase especially from 2000 to 2016 with an acceleration in the past 4 years (from 2013 to 2016) when the number of articles related to bioinformatics overpass the threshold of 200. As indicated above, LA is still below the average world-wide scientific production in the field, but the growing rate is good and shows a relative improvement in the past 5 years.
SoIBio history and future prospects of bioinformatics in Ibero-America
In 2002, the Red Iberoamericana de Bioinformatica (RIB, Iberoamerican Network for Bioinformatics) was created as a first joint cooperative project in the region. The RIB started as a collaborative effort between several research groups that had some experience working in the field of bioinformatics and computational biology. This project was conducted by Oscar Grau, who was the leader of the EMBnet node in Argentina (http://www.embnet.org/) and who applied for financial support to build the network to the Spanish government, obtaining a grant for 3 years that allowed its formation. The main groups who formed this initiative were from Spain, Portugal and several LA countries (Argentina, Brazil, Chile, Mexico, Uruguay and Venezuela), and they organized a first meeting in Buenos Aires (Argentina) in 2002. Together with this funded network, several members and researchers from the region also discussed at that time the idea of setting up a more stable and broad collaborative initiative considering that ‘regional networks for bioinformatics were going to be highly beneficial for capacity strengthening and cooperation, and for establishing productive interactions between scientists in the fields of biological and informatics sciences’ [24]. In this way, they proposed the creation of a bioinformatics network for LA and the Caribbean (LACBioNet), but this proposal never achieved a real state.
Following the start-up and establishment of the RIB, a series of academic and scientific events were organized by the leaders of this network under the support of the grant that they obtained. In fact, between 2002 and 2008, this network promoted four scientificmeetings on bioinformatics, which took place in Rio de Janeiro (Brazil), Cartagena de Indias (Colombia), Málaga (Spain) and Santiago (Chile). In 2003, the Iberoamerican Science and Technology Cooperation Program CYTED, (http://www.cyted.org/es/) provided official financial support to the RIB for five years, helping to the organization of such meetings and the development of more stable scientific collaborations between scientists from different countries within this region. Some initial examples of these research collaborations can be seen in the first publications on bioinformatics driven by the group of Rosana Chehin in Tucuman, Argentina [25, 26].
As a consequence of all the collaborative work and positive interaction, at the end of the funded period of RIB, the idea of setting up an international scientific society was evolving, and it became mature and formal in 2009 in a specific meeting in Mexico where the SoIBio was founded [27]. SoIBio’s central aim was to promote research and development on bioinformatics and computational biology in the international region of Iberoamerica that includes, as indicated above, 22 countries: Argentina, Bolivia, Brazil, Chile, Colombia, Costa Rica, Cuba, Dominican Republic, Ecuador, El Salvador, Guatemala, Honduras, Mexico, Nicaragua, Panama, Paraguay, Peru, Portugal, Puerto Rico, Spain, Uruguay and Venezuela.
With respect to the current action and future prospects, in the past 6 years, SoIbio has been active in promoting bioinformatics in LA by organizing an annual international meeting that congregates scientists from the region. These meetings help individuals, research groups and national societies who wish to initiate or expand their work in computational biology and bioinformatics, both at academic and professional level. The last three meetings organized by the Society have been in Belo Horizonte (Brazil), Rosario (Argentina) and Riviera Maya (Mexico); and the 2017 meeting will be in Cali (Colombia) (http://ccbcol.org/). In 2016, SoIbio also participated in the organization of the International Society for Computational Biology (ISCB) meeting for LA in Buenos Aires (https://www.iscb.org/iscb-latinamerica2016), and it has a formal agreement with this International Society to be co-organizers of its meeting in LA every 2 years. The Society also has an active Web page (http://www.soibio.org/) with >400 people registered, where most of the researchers are young (i.e. doctoral and postdoctoral) and have submitted their registration in the past 3 years. SoIBio also facilitates transnational exchange, collaboration, formation and development for issues and topics related to bioinformatics and computational biology. A recent example of this effective collaborative research can be seen in the publications derived from the last SoIBio International Conference (http://icmexico2016.soibio.org/) in 2016 [28–30]. SoIBio is also a platform to interact and cooperate with other international societies and networks that work in the same field, and in this way, it is member affiliated to the Global Bioinformatics Network (EMBnet, formerly called European Molecular Biology Network, http://www.embnet.org/) and the ISCB (https://www.iscb.org/). All this collaborative work is reflected also in publications such us the one recently presented in PLoS Computational Biology to explain the roles and active values of a scientific society [31].
Two major thematic trends: genome sequences and protein structures
Focusing now a little more on biological issues, our investigation reveals two major thematic trends that have been drivers and spin-offs for the expansion of bioinformatics in LA. The first thematic trend corresponds to the research focused on the characterization and mapping of new genomes, mostly anchored and supported by the introduction of new-generation high-throughput sequencing technologies (which had from the beginning a great need for computational analysis). Following the genomic studies, more recent work have driven this trend into the field of proteomics, which is a clear complement to the production and analysis of the genome-wide data. The second thematic scenario corresponds to the research focused on the molecular characterization of protein structures, obtained mainly through the application and use of biophysical techniques for the determination of three-dimensional (3D) structures at atomic resolution, such as X-ray crystallography (which also had a high demand for computational power and computational analysis).
To explain and illustrate how these two thematic scenarios were at the origins of bioinformatics, we describe the research trajectory of some scientists and research groups from LA that began to work in these fields. This analysis is not exhaustive and does not attempt to achieve complete coverage of all events, but tries to summarize some of the most referenced scientific papers published by pioneering groups that drove the progression of bioinformatics in the region.
Thematic Trend I: genomes, computational genomics and proteomics
In the first area, related to genomes and genomics, we can mention the work started in Mexico when the National Autonomous University of Mexico (UNAM, Universidad Nacional Autónoma de México) set up in 2004 at the Campus of Cuernavaca, in the state of Morelos, a Center for Genomic Sciences (CCG, Centro de Ciencias Genómicas, http://www.ccg.unam.mx/), transforming a former research institute called Nitrogen Fixation Research Center (CIFN).
In the CCG, it is worth mentioning the work in computational genomics and bioinformatics developed by Julio Collado-Vides who, early in the 90s, used computational methods and tools to study gene regulation [32]. Dr Collado-Vides after a stay in United States moved to Cuernavaca setting up a research group focused on bacterial computational genomics [33–35]. Working in this area, he and colleagues participated in 1997 in the first full-genome sequence of E.coli K-12 [36], and they developed two of the first public biological databases that were established in LA called RegulonDB [7] and EcoCyc [11]. Both databases have been later published in consecutive versions accumulating >1500 citations in scientific publications (Web of Science, http://webofknowledge.com/).
Along with this original work done in Mexico, studies on microbial genomes were also the thematic scenarios for other research groups and institutions in LA that soon began to develop computational biology and bioinformatic studies. Two illustrative examples of this are one institution from Brazil and another from Chile, together with the research groups and scientists who run them. The first is the Laboratory of Bioinformatics within the Laboratório Nacional de Computação Científica (LABINFO, LNCC, http://www.labinfo.lncc.br/), which was created in Brazil in 2000, and incorporated Ana Tereza R. Vasconcelos as director–coordinator. Between 2000 and 2005, this laboratory developed relevant studies on bioinformatics [37–39] mainly applied to microbial genomics, including the sequencing of several new genomes [40]. The second group and institution that we present here is the Center of Bioinformatics and Genome Biology of the Fundación Ciencia and Vida and University of Andres Bello (CienciaVida-UNAB, Chile) that started with this name in 2005. In this institution, one of the most relevant groups working on bioinformatics and computational biology has been the one led by David S. Holmes who, as indicated above, developed important studies on microbial genomes [3–5].
The analysis of the publications of LA groups also indicates that in the past decade, there is a tendency to move from computational genomics into proteomics. This is reflected in the participation of several scientists in important publications in the field of computational proteomics, such us the article of the PRIDE database in 2013 (The PRoteomics IDEntifications database) [41], which has >800 citations, and where several young researchers from LA collaborated. In particular, in previous years, some groups from Cuba and Brazil developed significant bioinformatic software and tools for proteomic analyses. A good example is PatternLab, a tool for differential shotgun proteomics first published in 2008 by the group of Paulo C. Carvalho and collaborators that have received nearly 100 citations [42], or other well-referenced tools also developed by this team from the Federal University of Rio de Janeiro (Brazil): GO-Explorer [43] and SEPro [44]. Another example is the work developed by the group from Gabriel Padron and collaborators of the Center for Genetic Engineering and Biotechnology in La Havana (Cuba), who built bioinformatic applications for the proteomic field such us SCAPE (a tool for the Selective CApture of PEptides in protein identification) [45] and several algorithms for in silico analysis of proteomic data [46, 47].
Thematic Trend II: proteins, 3D structures and structural bioinformatics
In the second thematic area, we can mention the work done by Ramón Latorre de la Cruz (Santiago de Chile, 1941), who is a biochemist from Chile who worked many years in United States (in the Universities of Duke, Chicago and Harvard) and became a member of the US National Academy of Sciences. When Ramón Latorre came back to Chile, he started important biophysical studies mainly on ion channels producing a series of significant publications that included contributions in the field of structural bioinformatics [48–50]. This approach allowed the development of a relevant circle of new scientists interested in the analysis and study of the 3D structures of proteins using computational methods (such as structural dynamic simulations). In this group of researchers, we can find some of the most active scientists in computational biology like Danilo Gonzalez-Nilo who currently is director of the Center for Bioinformatics and Integrative Biology in the Universidad Andres Bello (Chile) [26, 51–53].
Other younger groups in the region have also followed this track of structural bioinformatics and protein modeling, like the one developed by Francisco Melo in the Pontificia Universidad Católica de Chile. He started working in United States [54, 55], but later developed important work in computational structural studies in Chile [56–58]. Finally, other scientists from Argentina have also done relevant work in the field of structural bioinformatics [59–61]. These teams are proactive for the field of bioinformatics participating in the recent development of their national societies, like A2B2C in Argentina (http://www.a2b2c.org.ar/), and also in the interaction with international societies (like ISCB and SoIBio). An interesting review about the initial development of computational biology research in Argentina was published 10 years ago [62], and it presented a view that supports our current view of the field in this country. In the same way, another article in 2007 presented a deep overview of the state and development of computational biology in Brazil [63], describing the history of the field in this country, the origin of the Brazilian Association for Bioinformatics and Computational Biology (AB3C, http://www.ab3c.org), that started in 2004, and the early achievements that were associated in many cases to genome-sequencing projects: like the one of the plant pathogen Xylella fastidiosa [64] or the genomes of several Xanthomonas pathogens [65]. The Xylella genome, published in Nature in 2000, was the first LA genome-sequencing project led by Joao Carlos Setubal and Joao Meidanis of the University of Sao Paulo (Brazil) [64], who also wrote in 1997 a preliminary book entitled ‘Introduction to Computational Molecular Biology’, relevant to the field. This pioneering achievement in bioinformatics linked to genomic studies in Brazil has later continued with other sequencing projects, for example, the one of Anopheles darlingi (the malaria mosquito) published in 2013 [66]. Finally, in relation to the appearance and progress of bioinformatic research in other LA countries, a preliminary publication on the state of the art in Colombia was carried out in 2010 [67]. This article could not present many achievements, because the work done in the field was still preliminary, but it presented a hopeful scenario for bioinformatics in Colombia. As a step in this way, the Colombian Society for Bioinformatics and Computational Biology (Sociedad Colombiana de Bioinformática y Biología Computacional, SC2B2, http://www.sc2b2.org/) was created in Medellín in 2015.
Conclusion
Our analysis presents a revealing silhouette of the historical development of bioinformatics and computational biology in LA over the past two decades, based on the analysis of the original scientific articles published in the field between 1991 and 2016 by research groups of the region. It also shows that the main thematic trends within this research correspond to the production and analysis of genomic data followed by proteomic studies, as well as to structural bioinformatics studies. These research trends are not particularly unique to LA countries, and resemble world-wide growth around new life sciences technologies, but we note that in many cases, they focus on the microbial domain to study relevant pathogenic organisms related with diseases that have high prevalence in the region. Another observation from our analysis is the emergence in the past decade of several scientific societies dedicated to the promotion bioinformatics in LA, starting with SoIBio, that works internationally in most of the region, and followed by three national societies (AB3C in Brazil, A2B2C in Argentina and SC2B2 in Colombia) created in recent years to promote the work in their respective countries.
Taken together, the results are presented as a briefing since it is difficult to write a comprehensive review that may include all the significant achievements on bioinformatics in the vast region of LA. Despite this caution, our studies show information on the current state of the art and allow us to find some relevant trends observed in the literature, which are playing an influential role in the successful advancement of bioinformatics. The figures on publications and the recent creation of multiple scientific societies in the field also reveal a neat positive trend. These indicators of current agile growth do not negate that Latin American publication output in bioinformatics over the last 25 years is approximately half the world average, but they generate optimistic expectations of improvement. An accurate prediction of the future is difficult, but our analyses also indicate that the scientific and academic institutions in the region are rapidly increasing their interest in bioinformatics and computational biology in a similar way to other regions of the world.
Key Points
Scientific publications on bioinformatics and computational biology carried out by researchers from 19 LA countries in the past 25 years reveal state of the art of this field in the region.
Genome studies, proteomics and protein structures have been key topics driving bioinformatics in LA since the 1990s.
Research on bioinformatics in LA should still develop twice to approach the average world scientific production in the field.
SoIBio (Iberoamerican Society for Bioinformatics), founded in Mexico in 2009, is a leading forum for LA scientists interested in promoting research, training and education in bioinformatics.
Three national societies (AB3C in Brazil, A2B2C in Argentina and SC2B2 in Colombia), created in recent years, promote bioinformatics and cooperate actively with SoIBio.
Supplementary Material
Acknowledgements
The authors acknowledge the funding provided to J.D.L.R. group by the Spanish Government, ‘Ministerio de Economia y Competitividad’ (MINECO) with grants of the ‘Instituto de Salud Carlos III’ (ISCiii) co-funded by FEDER (grants AC14/00024 and PI15/00328). The authors also acknowledge a PhD research grant to F.J.C.-L. from JCyL (‘Ayudas a la contratación de Personal Investigador’) provided with the support of the ‘Fondo Social Europeo’ (FSE).
Funding
The publication charges for this article were funded by the research grant PI15/00328, given by the Instituto de Salud Carlos III (ISCiii, MINECO, Spain) and co-funded by the Fondo Europeo de Desarrollo Regional (FEDER, Europe).
Biography
Javier De Las Rivas is PI of the Bioinformatics and Functional Genomics Group of the Cancer Research Center (University of Salamanca and CSIC, Spain). He is the current President of SoIBio and one of its founding members.
Cesar Bonavides-Martinez is an expert in computational genomics, current Treasurer of SoIBio and also one of its founding members in Mexico.
Francisco Jose Campos-Laborie is a bioinformatician working in the group of Dr. De Las Rivas expert in information data analysis.
References
- 1. Ramírez JL, González A, Cantú JM, et al. Latin American genome initiative, the creation of a network and web based resource to aid and nurture genome biology in developing countries. Electronic J Biotechnol 2002;5:3–4. [Google Scholar]
- 2. Barreto M, Jedlicki E, Holmes DS.. Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl Environ Microbiol 2005;71:2902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Quatrini R, Lefimil C, Veloso FA, et al. Bioinformatic prediction and experimental verification of Fur-regulated genes in the extreme acidophile Acidithiobacillus ferrooxidans. Nucleic Acids Res 2007;35:2153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valdés J, Pedroso I, Quatrini R, et al. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 2008;94:180–4. [Google Scholar]
- 5. Quatrini R, Appia-Ayme C, Denis Y, et al. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 2009;10: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esparza M, Cárdenas JP, Bowien B, et al. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans. BMC Microbiol 2010;10:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huerta AM, Salgado H, Thieffry D, et al. RegulonDB: a database on transcriptional regulation in Escherichia coli. Nucleic Acids Res 1998;26:55–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thieffry D, Salgado H, Huerta AM, et al. Prediction of transcriptional regulatory sites in the complete genome sequence of Escherichia coli K-12. Bioinformatics 1998;14:391–400. [DOI] [PubMed] [Google Scholar]
- 9. Salgado H, Santos-Zavaleta A, Gama-Castro S, et al. RegulonDB (version 3.0): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res 2000;28:65–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salgado H, Moreno-Hagelsieb G, Smith TF, et al. Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci USA 2000;97:6652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karp PD, Riley M, Saier M, et al. The EcoCyc database. Nucleic Acids Res 2002;30:56–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gama-Castro S, Salgado H, Santos-Zavaleta A, et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res 2016;44:D133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keseler IM, Mackie A, Santos-Zavaleta A, et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 2017;45:D543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pereira JH, Canduri F, de Oliveira JS, et al. Structural bioinformatics study of EPSP synthase from Mycobacterium tuberculosis. Biochem Biophys Res Commun 2003;312:608–14. [DOI] [PubMed] [Google Scholar]
- 15. Brudey K, Driscoll JR, Rigouts L, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 2006;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Restrepo-Montoya D, Vizcaíno C, Niño LF, et al. Validating subcellular localization prediction tools with mycobacterial proteins. BMC Bioinformatics 2009;10:134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cardoso FC, Pinho JM, Azevedo V, et al. Identification of a new Schistosoma mansoni membrane-bound protein through bioinformatic analysis. Genet Mol Res 2006;5:609–18. [PubMed] [Google Scholar]
- 18. Berriman M, Haas BJ, LoVerde PT, et al. The genome of the blood fluke Schistosoma mansoni. Nature 2009;460:352–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zerlotini A, Aguiar ER, Yu F, et al. SchistoDB: an updated genome resource for the three key schistosomes of humans. Nucleic Acids Res 2013;41:D728–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alvarez VE, Kosec G, Sant'Anna C, et al. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem 2008;283:3454–64. [DOI] [PubMed] [Google Scholar]
- 21. Ackermann AA, Panunzi LG, Cosentino RO, et al. A genomic scale map of genetic diversity in Trypanosoma cruzi. BMC Genomics 2012;13:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cosentino RO, Agüero F.. Genetic profiling of the isoprenoid and sterol biosynthesis pathway genes of Trypanosoma cruzi. PLoS One 2014;9:e96762.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carrea A, Diambra L.. Systems biology approach to model the life cycle of Trypanosoma cruzi. PLoS One 2016;11:e0146947.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Degrave WM, Vargas R, Alvarez F, et al. Towards a bioinformatics network for Latin America and the Caribbean (LACBioNet). Appl Bioinformatics 2002;1:53–6. [PubMed] [Google Scholar]
- 25. Avila CL, Rapisarda VA, Farías RN, et al. Linear array of conserved sequence motifs to discriminate protein subfamilies: study on pyridine nucleotide-disulfide reductases. BMC Bioinformatics 2007;8:96.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avila CL, de Arcuri BF, Gonzalez-Nilo F, et al. Role of electrostatics on membrane binding, aggregation and destabilization induced by NAD(P)H dehydrogenases. Implication in membrane fusion. Biophys Chem 2008;137: 126–32. [DOI] [PubMed] [Google Scholar]
- 27. Lopez-Bojorquez L, Bonavides-Martínez C, Bojorquez C, et al. Report on the International conference and meetings EMBnet-RIBio 2009. EMBnet J 2009;16(Suppl. A):3–5. [Google Scholar]
- 28. Siqueira FM, de Morais GL, Higashi S, et al. Mycoplasma non-coding RNA: identification of small RNAs and targets. BMC Genomics 2016;17(Suppl 8):743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopes KP, Campos-Laborie FJ, Vialle RA, et al. Evolutionary hallmarks of the human proteome: chasing the age and coregulation of protein-coding genes. BMC Genomics 2016; 17(Suppl 8):725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urzúa U, Ampuero S, Roby KF, et al. Dysregulation of mitotic machinery genes precedes genome instability during spontaneous pre-malignant transformation of mouse ovarian surface epithelial cells. BMC Genomics 2016;17(Suppl 8):728.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gaëta BA, De Las Rivas J, Horton P, et al. Ten simple rules for forming a scientific professional society. PLoS Comput Biol 2017;13(3):e1005226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Collado-Vides J. Grammatical model of the regulation of gene expression. Proc Natl Acad Sci USA 1992;89:9405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collado-Vides J. The elements for a classification of units of genetic information with a combinatorial component. J Theor Biol 1993;163:527–48. [DOI] [PubMed] [Google Scholar]
- 34. Rosenblueth DA, Thieffry D, Huerta AM, et al. Syntactic recognition of regulatory regions in Escherichia coli. Comput Appl Biosci 1996;12:415–22. [DOI] [PubMed] [Google Scholar]
- 35. Thieffry D, Rosenblueth DA, Huerta AM, et al. Definite-clause grammars for the analysis of cis-regulatory regions in E. coli. Pac Symp Biocomput 1997;441–52. [PubMed] [Google Scholar]
- 36. Blattner FR, Plunkett G 3rd, Bloch CA, et al. The complete genome sequence of Escherichia coli K-12. Science 1997;277: 1453–62. [DOI] [PubMed] [Google Scholar]
- 37. Vasconcelos AT, Maia MA, de Almeida DF.. Short interrupted palindromes on the extragenic DNA of Escherichia coli K-12, Haemophilus influenzae and Neisseria meningitidis. Bioinformatics 2000;16:968–77. [DOI] [PubMed] [Google Scholar]
- 38. Almeida LG, Paixão R, Souza RC, et al. A new set of bioinformatics tools for genome projects. Genet Mol Res 2004;3:26–52. [PubMed] [Google Scholar]
- 39. Vasconcelos AT, Guimarães AC, Castelletti CH, et al. MamMiBase: a mitochondrial genome database for mammalian phylogenetic studies. Bioinformatics 2005;21:2566–7. [DOI] [PubMed] [Google Scholar]
- 40. Vasconcelos ATR, De Almeida DF, Hungria M, et al. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci USA 2003;100:11660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vizcaíno JA, Côté RG, Csordas A, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 2013;41:D1063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carvalho PC, Fischer JS, Chen EI, et al. PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics 2008;9(1):316.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carvalho PC, Fischer JS, Chen EI, et al. GO Explorer: a gene-ontology tool to aid in the interpretation of shotgun proteomics data. Proteome Sci 2009;7(1):6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carvalho PC, Fischer JS, Xu T, et al. Search engine processor: filtering and organizing peptide spectrum matches. Proteomics 2012;12:944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Betancourt L, Gil J, Besada V, et al. SCAPE: a new tool for the selective capture of peptides in protein identification. J Proteome Res 2005;4:491–6. [DOI] [PubMed] [Google Scholar]
- 46. Perez-Riverol Y, Sánchez A, Ramos Y, et al. In silico analysis of accurate proteomics, complemented by selective isolation of peptides. J Proteomics 2011;74:2071–82. [DOI] [PubMed] [Google Scholar]
- 47. Perez-Riverol Y, Audain E, Millan A, et al. Isoelectric point optimization using peptide descriptors and support vector machines. J Proteomics 2012;75:2269–74. [DOI] [PubMed] [Google Scholar]
- 48. Brauchi S, Orta G, Mascayano C, et al. Dissection of the components for PIP2 activation and thermosensation in TRP channels. Proc Natl Acad Sci USA 2007;104:10246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poblete H, Oyarzún I, Olivero P, et al. Molecular determinants of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) binding to transient receptor potential V1 (TRPV1) channels. J Biol Chem 2015;290:2086–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castillo JP, Sánchez-Rodríguez JE, Hyde HC, et al. β1-subunit-induced structural rearrangements of the Ca2+- and voltage-activated K + (BK) channel. Proc Natl Acad Sci USA 2016;113: E3231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Niemeyer MI, González-Nilo FD, Zúñiga L, et al. Neutralization of a single arginine residue gates open a two-pore domain, alkali-activated K+ channel. Proc Natl Acad Sci USA 2007;104:666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. González W, Riedelsberger J, Morales-Navarro SE, et al. The pH sensor of the plant K+-uptake channel KAT1 is built from a sensory cloud rather than from single key amino acids. Biochem J 2012;442:57–63. [DOI] [PubMed] [Google Scholar]
- 53. Maojo V, Fritts M, Martin-Sanchez F, et al. Nanoinformatics: developing new computing applications for nanomedicine. Comput Sci Eng 2012;94:521–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Melo F, Sánchez R, Sali A.. Statistical potentials for fold assessment. Protein Sci 2002;11:430–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Melo F, Sali A.. Fold assessment for comparative protein structure modeling. Protein Sci 2007;16:2412–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Norambuena T, Melo F.. The protein-DNA interface database. BMC Bioinformatics 2010;11:262.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Slater AW, Castellanos JI, Sippl MJ, et al. Towards the development of standardized methods for comparison, ranking and evaluation of structure alignments. Bioinformatics 2013;29:47–53. [DOI] [PubMed] [Google Scholar]
- 58. Gutiérrez FI, Rodriguez-Valenzuela F, Ibarra IL, et al. Efficient and automated large-scale detection of structural relationships in proteins with a flexible aligner. BMC Bioinformatics 2016;17:20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Juritz EI, Alberti SF, Parisi GD.. PCDB: a database of protein conformational diversity. Nucleic Acids Res 2011;39:D475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Monzon AM, Juritz E, Fornasari MS, et al. CoDNaS: a database of conformational diversity in the native state of proteins. Bioinformatics 2013;29:2512–4. [DOI] [PubMed] [Google Scholar]
- 61. Zea DJ, Monzon AM, Gonzalez C, et al. Disorder transitions and conformational diversity cooperatively modulate biological function in proteins. Protein Sci 2016;25:1138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bassi S, González V, Parisi G.. Computational biology in Argentina. PLoS Comput Biol 2007;3:e257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Neshich G. Computational biology in Brazil. PLoS Comput Biol 2007;3:1845–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Simpson AJ, Reinach FC, Arruda P, et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 2000;406:151–7. [DOI] [PubMed] [Google Scholar]
- 65. Da Silva AC, Ferro JA, Reinach FC, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002;417:459–63. [DOI] [PubMed] [Google Scholar]
- 66. Marinotti O, Cerqueira GC, de Almeida LG, et al. The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res 2013;41:7387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Benítez-Páez A, Cárdenas-Brito S.. Bioinformatics in Colombia: state of the art and perspectives. Biomedica 2010;30:170–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.