Abstract
The ability to collect, store and analyze massive amounts of molecular and clinical data is fundamentally transforming the scientific method and its application in translational medicine. Collecting observations has always been a prerequisite for discovery, and great leaps in scientific understanding are accompanied by an expansion of this ability. Particle physics, astronomy and climate science, for example, have all greatly benefited from the development of new technologies enabling the collection of larger and more diverse data. Unlike medicine, however, each of these fields also has a mature theoretical framework on which new data can be evaluated and incorporated—to say it another way, there are no ‘first principals’ from which a healthy human could be analytically derived. The worry, and it is a valid concern, is that, without a strong theoretical underpinning, the inundation of data will cause medical research to devolve into a haphazard enterprise without discipline or rigor. The Age of Big Data harbors tremendous opportunity for biomedical advances, but will also be treacherous and demanding on future scientists.
Keywords: translational bioinformatics, data mining, observational analysis, translational medicine, biomedical informatics, Big Data
Introduction
Medicine is a uniquely complex scientific enterprise. Today, we think of medical research as a quantitative science, guided by statistics and models, driven forward by rigorously conducted clinical trials. However, medicine as a practice is stubbornly qualitative. Often decisions are made based on personal experience and anecdotes. These may well be the ‘right’ decisions for the patients, but they are often taken without much scientific evidence. It is the complexity of medicine that produces this gulf between what is acceptable research and acceptable practice.
Unlike other quantitative sciences, medicine is not founded on first principals from which a healthy human can be derived. The natural laws governing molecules and submolecules that have been so successful in describing physics, environmental science and chemistry simply do not translate to the medical scale. The order of interactions of molecules in biomedicine is so high that there is no model now, or for the foreseeable future, and that will be able to fully explain the human condition.
That is not to say that we know little about medicine. On the contrary, the past 60 years have been transformative. In less than a human lifespan, we have gone from discovering the structure of DNA to the description of 3712 disease-causing genetic variants [1]. We have evolved the simple ‘Central Dogma of Biology’ into one that includes feedback regulation, gene silencing and alternative methods of inheritance outside of DNA. We have cured diseases and extended human life expectancy. We have transformed debilitating diagnoses into manageable chronic conditions [2].
The collection, storage and analysis of ‘Big Data’, data that are either too massive or too complex for single-machine analysis, are beginning to change how the science of medicine advances. A genetic study of >300 000 individuals revealed variants associated with major depressive disorder that we were previously not powered to detect [3]. A reevaluation of the clinical data for 11 000 patients using a new mathematical model called topological data analysis revealed subgroups that would have remained hidden [4]. Deep neural networks trained on nearly 130 000 images of skin legions can now detect malignancies as well as—and faster than—experts [5]. Recently, we used 1.8 million adverse event reports and 1.6 million electrocardiograms to discover a previously unknown arrhythmiogenic drug–drug interaction [6, 7], an interaction that would have not otherwise been suspected.
Data are changing all aspects of the scientific method, but the most profound changes are in the type, complexity and scale of the hypotheses that we can generate. Large data sets allow for the exploration of biomedical hypotheses that, simply, would never have been feasible to consider using a traditional, knowledge-driven approach. This shifting of the source of the hypothesis is uncomfortable and fraught with challenges. However, the advantages Big Data represent can vastly outnumber their shortcomings. Here, I explore how data are changing the fundamental practice of translational research and their impact on the scientific method, highlighting notable works.
Data and the source of the hypothesis
Observation is the starting point of scientific discovery. Making observations informs new hypotheses. When Charles Darwin sailed around the Galapagos Islands, it was the careful and diligent recording of his observations that led to the formation of biology’s most elegant theory. This, of course, applies to medicine as well. For example, William McBride and Widukind Lenz’s observation of extreme birth defects in babies born to women exposed to thalidamide largely forms the basis of the pharmacovigilance discipline as we know it.
The tools that we use to make observations are advancing. The first scientists used manual techniques and their natural senses to record and analyze their observations. Modern technology has drastically changed the scale and type of data that are being collected. Instead of the bytes and kilobytes of data collected by those like Darwin, McBride and Lenz, we are working with terabytes and petabytes worth of data every day. Instead of observing biomedical phenomenon at a human scale, we are dealing with systems containing thousands of invisible molecules representing millions of interactions. Technologies like next-generation sequencing, high-throughput chemical screening and mass spectroscopy and meta-databases thereof have all contributed to the changes in how biomedical data are collected and stored. It is not unreasonable to admit that the human mind, by itself, is not capable of processing the scale and complexity of these massive resources. This is where data mining—the automated discovery of relationships in large data sets—comes into play.
Data mining is about making the tools of analyzing data catchup with our ability to make and record new observations. In other words, data mining transfers the computing burden of generating new hypotheses from the scientist’s mind to computer hardware, freeing up the scientist to consider a broader set of hypotheses. Data mining implementations can take many different forms, from simple models like statistical correlation to the more complex, such as genetic interaction networks or systems of differential equations. Generally, however, the distinguishing feature of this approach to science is that a class of hypotheses is defined with each member of that class being evaluated simultaneously. For example, a pharmacovigilance scientist, based on their knowledge, may come up with the single hypothesis, ‘rofecoxib increases the risk of heart attack’, where a data-pharmacovigilance scientist would consider all members of the class, ‘[drug] increases the risk of heart attack’. This switch, from a single-hypothesis instance to a hypothesis class, is what makes data mining powerful and allows for the emergence of previously unconsidered hypotheses. However, this is also the source of much of the discomfort and is often derided, by those outside the field, as a ‘fishing expedition’. These criticisms are the result of a misconception of data mining’s role in the scientific method and common misuses of data mining methods in research.
Miscommunication about the role data play in science has led to distrust by those outside the field as well as misuse by those within. Data can be used in both the hypothesis generating and the hypothesis testing phases of the scientific method, and it is the blurring of the lines between them that results in this miscommunication. It is the data scientist’s responsibility to draw this line distinctly and clearly. There are additional responsibilities in the way we, as data scientists, must communicate the design and the results of our studies, especially when exploring hypotheses. Avoiding the common pitfalls of ‘Big Data’ will improve the rigor of our analyses, the reproducibility of our results and the adoption of our approach to science. In the following section, I enumerate some of these major challenges and pitfalls.
The pitfalls and traps of Big Data
There are challenges to using large data sets at every stage of the scientific method. Here, we focus specifically on those pertaining to hypothesis generation, as they are the most often misused and misconstrued. The data that are being mined for new hypotheses are often observational in nature, meaning that they will suffer from missingness and noise, they will have known and unknown covariances and they will contain systematic errors that introduce bias. Data mining in this environment is challenging, with model-free approaches being especially sensitive to these issues. Some of the major misuses include overemphasis of the P-value, improper validation of findings and the role of replication, miscommunication to the scientific community and the public and the use of model-free approaches in a Big Data setting.
The curse of big N
One of the most dramatic changes in this era of Big Data has been the role that the P-value plays in reporting research results. The P-value describes the probability of observing an effect by chance. When effect estimates are noisy (e.g. when the sample size is small) then significance analysis through examination of the P-value is paramount to protect against affirmation biases. However, when sample sizes (i.e. N) are large, it is often trivial to achieve so-called ‘significant’ P-values even when correcting for multiple hypothesis testing. It may be that these studies are overpowered and can detect small—essentially meaningless—effect sizes with statistical significance. This makes it more important than ever to focus on the effect estimates rather than just their significance. Statistical significance is necessary, but not sufficient when performing hypothesis generation.
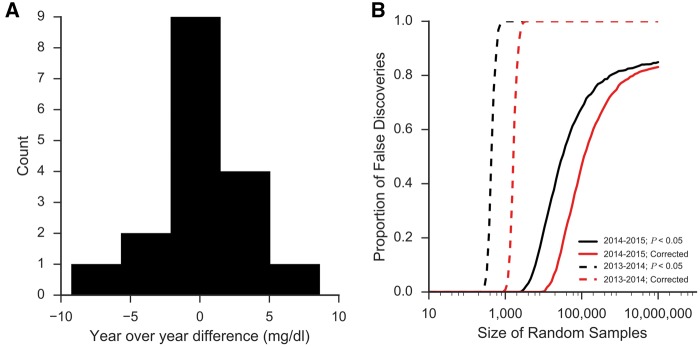
A more concerning, and common, source of trivial P-values in observational analysis is when the findings are significant because the data do not conform to the assumptions of the statistical model being used. Consider the situation where you are studying the effects on blood glucose between two drugs approved in adjacent years. At Columbia University Medical Center/New York-Presbyterian Hospital (CUMC/NYP), we have run >10 million random blood glucoses over the past 15 years. The average difference in glucose values measured year-over-year has been 0.05 mg/dl over this period—<0.05% of the average measured value (Figure 1A). However, in certain years, like 2013–14, we see as much as a 9.3 mg/dl (6%) change in the average measured value. The reason behind such a large change is not clear and, often, impossible to determine. It could be that there were unique characteristics of the patient population those years or perhaps slight changes in practice patterns. In any case, the implications for our hypothetical drug study are profound. Any drug approved and used in 2013 will appear to have a different effect than one approved in 2014, regardless of the actual effects of these two drugs. Even a clinically insignificant difference, like that between 2014 and 2015 of 1.1 mg/dl (0.7%), will confound the analysis and lead to high false discovery rates (Figure 1B).
Figure 1.
The triviality of P-values. When ‘big data’ do not conform to the assumptions of statistical tests being performed on them, P-values become meaningless. (A) CUMC/NYP has run >10 million blood glucose tests over the past 15 years and over that time that year-over-year difference in the average value has barely changed (0.05 mg/dl, 0.05%). However, between some years, larger differences in average glucose are observed. Between 2013 and 2014, for example, the difference in the average glucose measurement was 9.3 mg/dl or 6%—a difference that is likely explained by slight differences in patient population or practice patterns. (B) These insignificant changes can confound analyses and produce trivially significant P-values with cohorts of ≥ 1000 showing significant differences, regardless of what is being studied. Even years with smaller differences (1.1 mg/dl between 2014 and 2015) will produce false discoveries at extremely high rates.
If the sources of the biases are known and measured, then a simple epidemiological approach, like propensity score matching or stratification, may be sufficient to mitigate their effects [8, 9]. However, this is rarely the case. We, and others, have worked on systematic methods to identify and correct for confounding in large data sets [10–12]. These methods work by using the internal covariances in the data to identify better (i.e. less biased) controls. Application of these methods has led to significantly improved performance when identifying known drug side effects and drug–drug interactions [10] from the FDA’s Adverse Event Reporting System. There is no easy solution to correcting for systematic errors in observational data mining. Data scientists must exercise discipline and rigor when conducting these studies and maintain a robust skepticism when evaluating their findings.
Invalid validation
Machine learning is used heavily in translational bioinformatics and biomedical data mining with almost no area of medicine remaining untouched. Most commonly used are logistic regression, random forests and deep neural networks. Once these models are trained, it is essential that an honest evaluation of their performance is conducted. There are many tools for this evaluation including cross, hold-out, leave-one-out and out-of-bag validation depending on the model being used. The accuracy of these methods at estimating the performance is dependent on an assumption of the independence of the training examples, which is commonly violated in large biomedical data sets.
Consider the task of using existing data to identify a new indication for an existing, approved drug—so-called drug ‘repositioning’. There are numerous studies that have applied machine learning for this purpose using drug target data [13], pathway data [14], genome-wide association study data [15], gene-expression data [16, 17] and others [18]. The assumption of independence between training examples made by machine learning validation strategies is broken in this case. The features that are available for each drug are dependent on the use of that drug in clinical medicine—which cell lines are used for gene expression experiments, which targets are assayed for binding affinity, and which population is studied in a genome wide association study (GWAS) to name a few. Evaluation strategies naive to these confounding biases will produce inflated estimates of performance [19, 20]. Permutation testing that preserves the confounding covariance in the data will reveal this bias by producing models with seemingly good performance even when trained on null—or randomized—data. Setting up such an experiment requires deep knowledge of the study domain, and generalized solutions are not yet available.
Instead, data scientists should consider independent experiments that replicate or corroborate the findings of their data-driven analyses. The more independent the analysis and data are, the more robust the discovery will be. For example, in previous work, we used an integrative translational bioinformatics approach to identify causal relationships between drug exposures and adverse reactions [7, 21]. In the most recent case, we mined the FDA’s data and discovered that ceftriaxone, a common antibiotic, and lansoprazole, an over-the-counter proton pump inhibitor, were associated with arrhythmia risk when taken concomitantly [6]. This finding alone is too fragile to be acted on. Therefore, in a follow-up analysis, we used an independent data source, the electrocardiogram reports from our electronic health records, to corroborate this finding. We found that patients exposed to these two drugs also had significantly longer QT intervals—a risk factor for arrhythmia. Finally, we used a prospective cell line experiment to prove a causal relationship between combination drug exposure and changes in the molecular activity of important cardiac ion channels [7]. In response to its publication, additional reports of this drug–drug interaction in patients have surfaced [22]. Notably, others in translational bioinformatics are also integrating prospective experiments into their pipelines [16, 17, 23]. The application of this rigorous approach of corroboration and replication focuses data-driven science toward the most meaningful and valid hypotheses.
Data miseducation
Data science and ‘big data’ analyses are not well communicated to the public. An average interested and informed citizen does not have the time to fully understand the scientific methods behind each important research study. It is important, however, that they can discern the quality of the study to identify those they can trust. Whether intentional, through the communication and reporting of research progress, the scientific community has provided some guidelines of trust to help the public distinguish the quality of research studies. Thus far, the primary guideline of trust that we have provided the public is sample size. This is intuitive considering that since its conception, biomedical science has been constrained primarily by what data are available. As we know, this is no longer the case.
The implications of this public miseducation struck me while giving a lecture at the New York Genome Center as part of a series on Big Data analytics. I presented our work using the medical records of >1.7 million patients to identify seasonal risk factors of disease at birth [24]. There are many caveats that go along with our approach which I, naturally, presented alongside our findings, including that this was a single-site analysis, the environmental factors are not directly identified and changes in disease prevalence and birth trends over time could affect our results. I was stunned when asked by one of the attendees how any of my findings could be wrong with a sample size of nearly 2 million patients. I realized then that data are not just transforming the way that we conduct the scientific method but also will change how the public evaluates and thinks about scientific results. As data scientists, we have an obligation to educate the public on issues specific to large data, including bias, error, noise and missingness—nuances that, I am confident, the most educated public in history [25] is eager to learn.
The model-free trap
In what I imagine to be an elegantly delivered lecture at New York University in 1959, Eugene Wigner presents his assertion that ‘mathematics plays an unreasonably important role in physics [26]’. Wigner extols the virtues of mathematics and its ability to accurately describe physical phenomenon even given limited information. Fifty years later, three researchers from Google write an opinion asserting the ‘unreasonable effectiveness of data’ and argue for a diminished role for modeling [27]. Their argument, using semantic extraction from a massive corpus of text as an example, is that with enough data the underlying models will reveal themselves—there is no need for them to be predefined. This knowledge-free approach is alluring to the burgeoning data scientist, who is often armed with technical mastery but has limited domain experience. If enough data can be collected, then application of state-of-the-art learning algorithms will reveal the underlying models—this is the ‘model-free trap’. We have discovered that it is not sufficient to have a massive amount of data; you must also have the right data. I will admit that, for the context the authors presented—semantic text processing—the data do appear unreasonably effective. Nonetheless, it is poetic that one of the most often cited misuses of this model-free approach is Google Flu Trends (GFT). Early in 2013, it became apparent that the errors in GFT were getting out of control. An autopsy of the algorithm by Lazer et al. [28] identified common mistakes in Big Data analysis (e.g. overfitting a large number of data to a small number of cases) as the likely culprits and warned against ‘big data hubris’. Lazer accurately identifies some of the major issues with this approach but stops short of generalizing what this means more broadly for the practice of data science. GFT’s failures are a consequence of conflating multiple steps of the scientific method. Instead of using their data mining to identify hypotheses of which lexical concepts will predict flu and then evaluating each of these hypotheses rigorously, the researchers moved directly into building and deploying an unvalidated model.
Fear of the model-free approach is widespread, as evidenced by recent articles calling out ‘research parasites [29]’. However, being a good translational data scientist means knowing when models are needed and when they are not, understanding the provenance of the data collected and the correct application of technology to biomedical challenges.
Big Data having an impact
The role that data are playing in shaping the conduct and progress of science can be felt across disciplines. Of particular note are rigorously conducted observational analyses with broad-reaching impact. The following is a selection of three exemplary studies from human genetics to clinical practice.
The Exome Aggregation Consortium (ExAC) has had a swift and profound impact on the field of medical genetics. Over 5 years, the first release of ExAC amalgamated >60 000 exomes and released aggregate results publicly. Despite the inherent biases that come with integrating data from multiple studies of different populations and diseases, the availability of these much genetic data has fundamentally changed rare disease genetics and, in several cases, corrected erroneous conclusions about the role of some genetic variants in disease [30]. Most notably, previously thought pathogenic variants in the prion protein gene (PRNP) were found to occur at a much higher frequency that should have been possible, reversing some of the prior conclusions on these variants [31].
Massive collection of data can overcome inherent sources noise. For example, the consumer genetics company, 23andMe, has been using user-provided data collected online in through their mobile applications to run GWAS on a wide range of traits. This type of survey has many known limitations, including self-selection biases and erroneous data entry [32]. However, using self-reported data on depression from >300 000 individuals, 15 genetic loci were significantly identified. Previously associated variants that were discovered only occurred at low frequencies and explained only a small portion of the variance in depression [3].
Data are transforming the practice of medicine in addition to its research. A landmark report from Frankovich and colleagues [33] presents a case of electronic health record-assisted clinical decision-making. Faced with a pediatric patient presenting with lupus, the providers were concerned about the potential for blood clots, but there is no reliable evidence on whether to put these patients on anticoagulants. The authors mined the electronic health records of 98 patients with similar presentations and used their treatment outcomes to inform their decisions. This informatics-enabled and data-derived approach is the premier example of a new way to practice medicine. However, even though this case was successful, the hospital has since prohibited others from following suite citing concerns that the systems to code, organize and search the data are not mature enough to be used in clinical care. The procedural and ethical considerations of using historical records in this way have yet to be worked out.
A deluge of opportunity
Massive expansions in our capability to collect, store and analyze observations of our natural world are affecting all scientific disciplines. In a field as complex as translational medicine, where disease is as unique as the patient it affects, data have the potential to be truly transformative. Extreme phenotyping, prognostic forecasting and precision treatment are all within reach because of the availability of these new data. In addition, there is an opportunity to use simpler, more general, models to characterize our observations. When data are large with sufficient coverage, we can make fewer assumptions and use models with fewer caveats. This shift could be profound and has the potential to drastically increase the rate of discovery in translational medicine. A traditional approach may discover the role of a single protein in a particular disease, for example. Models learned from Big Data, however, can reveal something fundamental about how disease manifests across modalities. This potential, however, is paralleled by an equal amount of peril—errors, noise, nonrandom missingness and unknown biases [34] threaten the validity of Big Data methods. It is up to us, as translational data scientists, to exercise the rigor and discipline necessary to produce research results that are robust, and to communicate fully the limitations of our analyses as well as the results. This will engender trust in the scientific community and encourage the adoption of our new approach to science. The role of the translational data scientist has never been more important than it is today.
Key Points
The ability to collect massive amounts of data is changing the way biomedical research is conducted.
The most profound impact data are having is the ability to explore novel and unexpected hypotheses.
Misunderstanding and misuse of the hypothesis-free approach to science lead to skepticism by those outside of data science.
The challenges and pitfalls of working with large data can be foreseen and are avoidable.
Data scientists can lead the next wave of innovation in translational medicine, but must exercise discipline and rigor in their studies and effectively communicate their work to the scientific community and the public.
Acknowledgements
The author would like to express his sincere gratitude to those who contributing ideas, literature and opinions to this work, including Rami Vanguri, Yun Hao, Phyllis Thangaraj, Mary Regina Boland, Tal Lorberbaum, Theresa Koleck and Konrad Karczewski.
Biography
Nicholas Tatonetti, PhD, is Herbert Irving Assistant Professor of Biomedical Informatics at Columbia University. He has dedicated his research to the use of observational data to discover and explain adverse drug effects and drug–drug interactions.
Funding
The author is supported by R01 GM107145, OT3 TR002027, and an award from the Herbert and Florence Irving Foundation.
References
- 1. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University Baltimore, MD. Online Mendelian Inheritance in Man. OMIM®.
- 2. Deeks SG, Lewin SR, Havlir DV.. The end of AIDS: HIV infection as a chronic disease. Lancet 2013;382:1525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hyde CL, Nagle MW, Tian C, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 2016;48:1031–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li L, Cheng WY, Glicksberg BS, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 2015;7:311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lorberbaum T, Sampson KJ, Woosley RL, et al. An integrative data science pipeline to identify novel drug interactions that prolong the QT interval. Drug Saf 2016; 39:433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lorberbaum T, Sampson KJ, Chang JB, et al. Coupling data mining and laboratory experiments to discover drug interactions causing QT prolongation. J Am Coll Cardiol 2016;68:1756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pattanayak CW, Rubin DB, Zell ER.. Propensity score methods for creating covariate balance in observational studies. Rev Esp Cardiol 2011;64:897–903. [DOI] [PubMed] [Google Scholar]
- 9. Cook EF, Goldman L.. Performance of tests of significance based on stratification by a multivariate confounder score or by a propensity score. J Clin Epidemiol 1989;42:317–24. [DOI] [PubMed] [Google Scholar]
- 10. Tatonetti NP, Ye PP, Daneshjou R, et al. Data-driven prediction of drug effects and interactions. Sci Transl Med 2012; 4:125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simpson SE, Madigan D, Zorych I, et al. Multiple self-controlled case series for large-scale longitudinal observational databases. Biometrics 2013;69:893–902 [DOI] [PubMed] [Google Scholar]
- 12. Schneeweiss S, Rassen JA, Glynn RJ, et al. High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology 2009;20:512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng F, Liu C, Jiang J, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol 2012;8:e1002503.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J, Lu Z.. Pathway-based drug repositioning using causal inference. BMC Bioinform 2013;14:S3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanseau P, Agarwal P, Barnes MR, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol 2012;30:317–20. [DOI] [PubMed] [Google Scholar]
- 16. Dudley JT, Sirota M, Shenoy M, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med 2011;3:96ra76.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med 2011;3:96ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J, Zheng S, Chen B, et al. A survey of current trends in computational drug repositioning. Brief Bioinform 2016;17:2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pahikkala T, Airola A, Pietilä S, et al. Toward more realistic drug-target interaction predictions. Brief Bioinform 2015;16:325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peón A, Naulaerts S, Ballester PJ.. Predicting the reliability of drug-target interaction predictions with maximum coverage of target space. Sci Rep 2017;7:3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011;90:133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lazzerini PE, Bertolozzi I, Rossi M.. Combination therapy with ceftriaxone and lansoprazole, acquired long QT syndrome, and torsades de pointes risk. J Am Coll Cardiol 2017;69:1876–77. [DOI] [PubMed] [Google Scholar]
- 23. Effective combination therapies for b-cell lymphoma predicted by a virtual disease model. Cancer Res 2017;77:1818–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boland MR, Shahn Z, Madigan D, et al. Birth month affects lifetime disease risk: a phenome-wide method. J Am Med Inform Assoc 2015;22:1042–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barro RJ, Lee JW.. A new data set of educational attainment in the world, 1950–2010. J Dev Econ 2013;104:184–98. [Google Scholar]
- 26. Wigner EP. The unreasonable effectiveness of mathematics in the natural sciences. Richard courant lecture in mathematical sciences delivered at New York University, May 11, 1959. Commun Pure Appl Math 1960;13:1–14. [Google Scholar]
- 27. Halevy A, Norvig P, Pereira F.. The unreasonable effectiveness of data. IEEE Intell Syst 2009; 24:8–12. [Google Scholar]
- 28. Lazer D, Kennedy R, King G, et al. The parable of Google Flu: traps in big data analysis. Science 2014;343:1203–5. [DOI] [PubMed] [Google Scholar]
- 29. Longo DL, Drazen JM.. Data sharing. N Engl J Med 2016;374:276–77. [DOI] [PubMed] [Google Scholar]
- 30. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Minikel EV, Vallabh SM, Lek M, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med 2016;8:322ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Groves RM. Research on survey data quality. Public Opin Q 1987;51:S156. [Google Scholar]
- 33. Frankovich J, Longhurst CA, Sutherland SM.. Evidence-based medicine in the EMR era. N Engl J Med 2011;365:1758–9. [DOI] [PubMed] [Google Scholar]
- 34. Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2012;20:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]