Abstract
Maize is monecious, with separate male and female inflorescences. Maize flowers are initially bisexual but achieve separate sexual identities through organ arrest. Loss-of-function mutants in the jasmonic acid (JA) pathway have only female flowers due to failure to abort silks in the tassel. Tasselseed5 (Ts5) shares this phenotype but is dominant. Positional cloning and transcriptomics of tassels identified an ectopically expressed gene in the CYP94B subfamily, Ts5 (ZmCYP94B1). CYP94B enzymes are wound inducible and inactivate bioactive jasmonoyl-L-isoleucine (JA-Ile). Consistent with this result, tassels and wounded leaves of Ts5 mutants displayed lower JA and JA-lle precursors and higher 12OH-JA-lle product than the wild type. Furthermore, many wounding and jasmonate pathway genes were differentially expressed in Ts5 tassels. We propose that the Ts5 phenotype results from the interruption of JA signaling during sexual differentiation via the upregulation of ZmCYP94B1 and that its proper expression maintains maize monoecy.
China Lunde et al. used cloning, transcriptomics, and metabolomics to identify the Tasselseed5 (Ts5) gene in maize, and its role in the jasmonate metabolism. They find that the phenotype of the Ts5 mutants is the result of interrupted jasmonic acid signaling during sexual differentiation.
Introduction
Most plants produce hermaphroditic flowers with the male organs, stamens, surrounding an inner whorl of female organs, the pistils. Fertilization occurs when pollen from the stamens successfully reaches the ovule within the pistil. While this arrangement assures successful seed production, it may also lead to inbreeding and reduced fitness. Plants have evolved several mechanisms to avoid inbreeding. One mechanism is genetic self-incompatibility, in which distinct alleles in the male pollen and female pistil are required for successful seed set1. Another mechanism is physical separation, which involves the development of unisexual flowers. In monoecy, male and female flowers are on the same plant2. It is estimated that 30% of the plant species produce some unisexual flowers3, including maize. In contrast, diecious plants, such as Cannabis, Corylus, or Asparagus, have separate male and female plants.
Maize produces separate staminate (male) and pistillate (female) inflorescences called the tassel and the ear, respectively. All grasses contain flowers (referred to as florets) flanked by sterile bracts called glumes within spikelets, units of the grass inflorescence. During early floral development, the two florets in both the tassel and the ear spikelets are hermaphroditic, and monoecy is conferred by the selective abortion of pistillate organs in tassel florets and the arrest of staminate organs in the ear floret4,5.
A large number of sex-determination mutants have been identified in maize, given the easy visibility of tasselseed mutants in which silks (pistils) and kernels (seeds) are found in the normally male tassel. Recessive mutants, ts1 and ts2, and dominant mutants, Ts3 and Ts5, fail to abort carpels in tassels and also fail to abort the lower floret in ears6–8. ts1 encodes a lipoxygenase, also called ZmLox8, that acts in jasmonic acid (JA) biosynthesis9,10, and ts2 encodes a monocot-specific3 short-chain alcohol dehydrogenase11. Both ts1 and ts2 have a reduced plant height8. Another tasselseed mutant was created by knocking out the duplicated orthologs of OPR3, a major OPR (12-oxo-phytodienoic acid reductase) gene in Arabidopsis that acts in JA biosynthesis12,13. The resulting maize opr7opr8 mutants are phenotypically similar to ts1 and ts2 mutants14,15.
Other mutants with silks in the tassels include ts4, which encodes an miR172 microRNA, and Ts6, which has a mutation in the ts4-binding site16. Brassinosteroid biosynthetic mutants17,18 and epigenetic mutants such as required to maintain repression6 (rmr6)19 also show feminized tassels. Clearly, many modes of regulation are necessary to keep hormone action balanced for proper sex determination20.
Here, we use positional cloning, transcriptomics, and metabolomics to identify the Ts5 gene and its role in jasmonate metabolism. The mechanism of Ts5 function reveals a role for jasmonate catabolism through the ω-oxidation pathway in both attenuating response to wounding and specification of sexual identity.
Results
Ts5 tassels are feminized and ear florets fail to abort
Maize bears a terminal staminate inflorescence (tassel) and lateral pistillate inflorescences (ears), which arise in the axils of leaves. Early development of ear and tassel is similar except that the tassel initiates branches prior to initiating spikelets. Both male and female spikelets initiate two floral meristems inside the sterile glumes. In the tassel spikelet, the pistils abort producing two male florets. In the ear, the stamens arrest and the pistil of the lower floret aborts, leaving a single pistil to grow out and receive pollen in the female spikelet5.
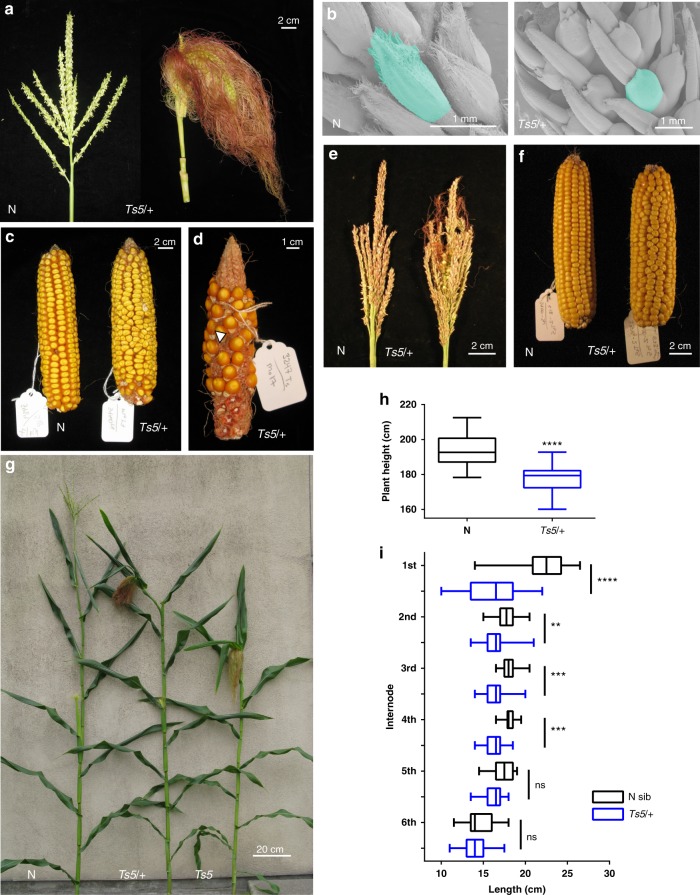
Ts5 tassels are feminized; the pistil fails to abort and, consequently, stamen development is arrested in affected florets. Although covered in silks, the Ts5 tassel remains branched (Fig. 1a–e) as previously described6–8. Scanning electron micrographs reveal that in developing tassels of Ts5/+ , glumes are short and glabrous (Fig. 1b), resembling those of ear glumes, a phenotype shared with ts211, and indicating that the entire spikelet is feminized in the Mo17 background. In contrast, tassel glumes of normal siblings are long and produce trichomes (Fig. 1b). Ts5/ + ears are also abnormal; they fail to abort the lower floret, leading to disordered vertical kernel files (Fig. 1c–f)6,7. In cases of reduced seed set, unfilled pericarps are visible, appressed to the kernel of the upper floret (Fig. 1d); these are present but hidden by filled kernels in fully pollinated ears as in Fig. 1c, f.
Fig. 1.
Inflorescence and vegetative phenotype of Ts5. a Normal and Ts5/ + mature tassels in Mo17. b SEM micrograph of central spike of 4 cm tassel of normal and Ts5/ + in Mo17. c Normal and Ts5/ + ears in Mo17. d A poorly pollinated Ts5/ + ear in Mo17 showing empty pericarps. Arrow: appressed empty pericarp of the lower floret. e Normal and Ts5/ + mature tassels in B73. f Normal and Ts5/ + ears in B73. g Mature plants of normal, Ts5/ + , and Ts5/Ts5 in A188. h Plant height (cm) of normal siblings (n = 30) and Ts5/ + (n = 31) in A188. Height was significantly different by two-tailed unpaired t test, P < 0.0001 (t = 6.778, df = 59; 95% CI −20.21 to −11.00). Bar, mean; whiskers, SD. i Graph of upper internode lengths of the same plants graphed in panel (h), normal siblings (n = 30) and Ts5/ + (n = 31) in A188. The entry 1st internode is measured from the last tassel branch to the first subtending node. Means of the first four internodes measured were significantly different, one-way ANOVA with Sidak’s multiple comparison test adjusted for multiple comparisons: 1st, t = 13.96, P < 0.0001, 95% CI = 4.778–7.0137; 2nd, t = 3.3874, 95% CI = 0.3131771–2.548113, P = 0.0047; 3rd, t = 4.162688, 95% CI = 0.6405964–2.875533, t = 4.287441, 95% CI = 0.6932846–2.928221, P = 0.0002; 4th, P = 0.0001, global DF = 353, ns = non-significant. Bar, mean; whiskers, SD. SEM, scanning electron microscope
In crossing Ts5 to different inbred backgrounds, we observed strong differences in expressivity. In the Mo17 background, the tassel is completely feminized and produces no pollen (Fig. 1a). In B73 tassels, Ts5 displays a mild phenotype and is male fertile (Fig. 1e). Spikelets in B73 Ts5 tassels are a mix of either male or female identity, but female florets can be found inside male glumes. The weak expressivity seen in B73 tassels, however, is not true for the ear because lower floret abortion still fails and the rows are uneven (Fig. 1f). The phenotype of Ts5 in A188 is intermediate between that of Mo17 and B73. The tassel is highly feminized, but still produces pollen. We quantified tassel-feminization traits by measuring two ratios, the number of feminized branches/total branch number (FBN/TBN), and the length of the main spike that was feminized/spike length (FSL/SL) and found that feminization increases with increase in copies of Ts5. In A188, homozygotes have more feminization along the main rachis and heterozygotes have an intermediate phenotype (Supplementary Fig. 1). Raw data used to create each graph are available in Supplementary Data 4.
Ts5 plants in A188 are noticeably shorter. Plants heterozygous for the dominant Ts5 allele have intermediate heights and Ts5 homozygotes are the shortest (Fig. 1g). We quantified height differences between Ts5/ + and wild type (A188, BC4) and found that mutants were nearly 16 cm shorter (Fig. 1h). Using these same individuals, we measured the internode length to determine the cause of the height difference and found a reduction in elongation of the four internodes immediately subtending the tassel (Fig. 1i). Genotype did not affect the lengths of the lower internodes.
Ts5 maps to ZmCYP94B1 and has JA-deficient phenotypes
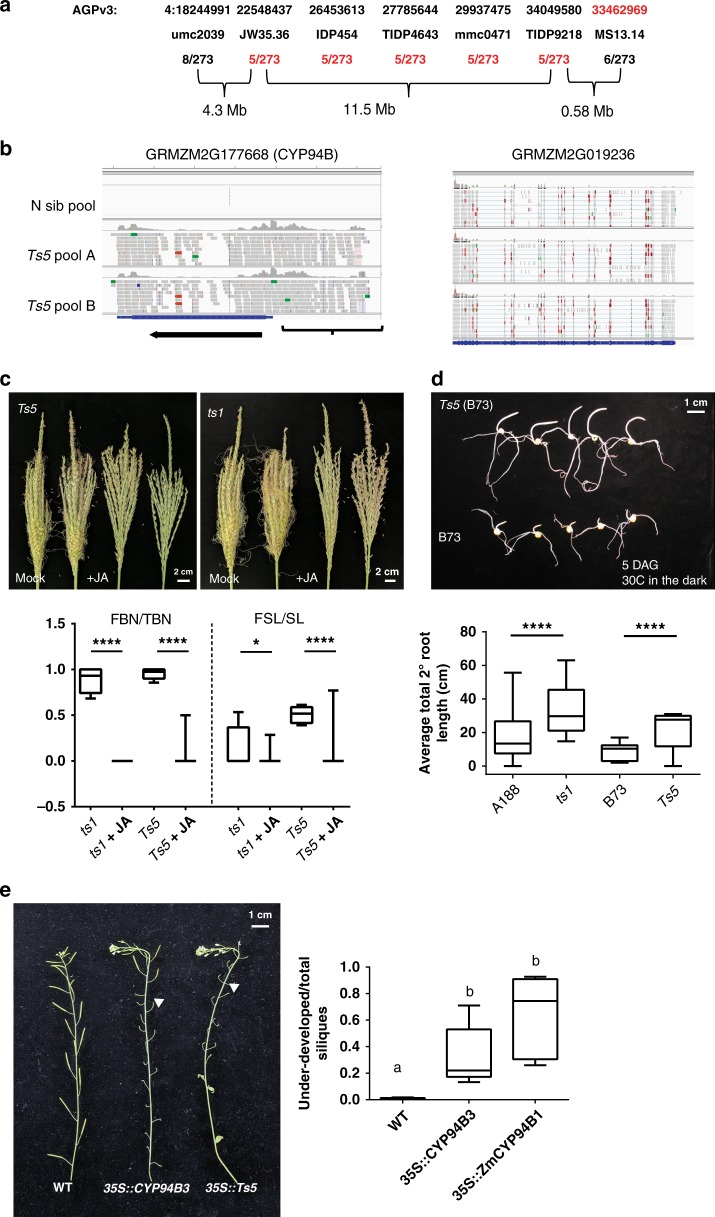
Ts5 appears on a genetic linkage map published by R.A. Emerson21. Current genetic mapping data placed the Ts5 mutation within bin 4.03 (www.maizegdb.org)22. Using molecular markers, we found a lack of recombination between umc2039 and five adjacent markers. In our mapping population, TIDP9218 was distal to MS13.14, compared to its reported location in B73 AGPv3. We fine-mapped Ts5 to a 15 Mb interval containing 65 genes between umc2039 and our custom indel marker JW35.36 (Fig. 2a; for primer sequences, see Supplementary Data 1).
Fig. 2.
Ts5 maps to a CYP94B that catabolizes JA, is corrected by JA application, and causes under-developed siliques when overexpressed in Arabidopsis. a Schematic of the Ts5 mapping locus. Red text indicates loci lacking recombination or marker order inconsistent with B73 AGPv3. b A CYP94B (GRMZM2G177668) is upregulated in Ts5/ + tassels relative to normal siblings in the Mo17 background visualized in IGV. Ectopic 5′ reads are indicated by brackets. A linked gene (GRMZM2G019236) has similar read counts in each pool. c Mature tassel phenotypes after the exogenous application of JA to developing tassels. Graphs of the ratios of feminized branch number to total branch number (FBN/TBN) and feminized spike length to total spike length (FSL/SL) of ts1 (n = 11), ts1 + JA (n = 8), Ts5 (n = 12), and Ts5 + JA (n = 11). Bar, mean, whiskers, SD. Treated mean of FBN/TBN and FSL/SL for ts1 and Ts5 are significantly less than untreated using an unpaired, one-tailed Student’s t test with Welch’s correction, ts1 FBN/TBN treated vs untreated t = 24.0708, df = 10.0000, P < 0.0001, 95% CI = −0.980591 to −0.814434; ts1 FSL/SL treated vs untreated t = 1.799 df = 15.18, P = 0.0459, 95% CI = −0.2855 to 0.02397; Ts5 FBN/TBN treated vs untreated, t = 18.8039, df = 12.4881, P < 0.0001, 95% CI = −1.01137 to −0.802148, Ts5 FSL/SL treated vs untreated, t = 5.94347, df = 12.3037, P < 0.0001, 95% CI = −0.600572 to −0.279006. d Photo of seedling phenotypes after growing in the dark for 5 days (30 °C) and graph of mean total secondary root length of ts1 in the A188 background (n = 12), A188 (n = 10), Ts5 homozygotes in the B73 background (n = 12), and B73 (n = 11). Mean root lengths of ts1 and Ts5 were significantly longer than those of wild type by two-tailed unpaired Student’s t test: for ts1, P = 0.0419, t = 2.173557, df = 20, 95% CI = 0.6021773 to 29.28449; for Ts5, P = 0.0036, t = 3.272358, df = 21, 95% CI = 4.680919 to 21.00393. e Photo of mature Arabidopsis lines Col-O, CYP94B3-OE, and ZmCYP94B1-OE showing under-developed siliques in both overexpression lines. Graph of the mean ratio of under-developed to normal siliques in each line is shown. Letters denote that wild-type plants are significantly distinct using two-tailed Student’s t test with Welch’s correction: WT vs 35S::CYP94B3, t = 3.123, P = 0.0353, 95% CI = 0.03581 –0.6044, df = 4.009; WT vs 35S::ZmCYP94B1, t = 4.538, P = 0.0105, 95% CI = 0.2446 –1.015, df = 4.005; 35S::CYP94B3 vs 35S::ZmCYP94B1, P = 0.1136. Bar, mean; whiskers, SD
To identify the potential mutations within these genes, we performed RNA sequencing (RNA-Seq) using 9–11 mm developing tassels from Ts5/ + and normal siblings in the phenotypically expressive Mo17 background. This approach enriched for potential transcripts causal for the phenotype and excluded secondary transcripts simply related to ectopic silk production. Plants were genotyped by polymerase chain reaction (PCR) using flanking markers. A CYP94B gene GRMZM2G177668 was not expressed in the normal sibling pool but was highly expressed in two Ts5/ + pools when comparing mapped reads (logFC 11.69) (Fig. 2b; Supplementary Data 2). In addition to an increase in read number, ectopic 5′ reads were detected, mapping to an area annotated as being 5′ of the canonical transcriptional start site of GRMZM2G177668. These 5′ reads are unique to Ts5 and were not found in other published transcriptomes10,23,24, suggesting that the Ts5 phenotype is the result of a novel transcript at this locus. Because Ts5 behaves as a dominant mutation, the expression of a novel transcript or the overexpression of a normal transcript could be causal. The sequence reads of adjacent genes were similar in the Ts5/ + pools compared to the normal pool (Fig. 2b), implying that although the order of two markers (MS13.14 and TIDP9218) is reversed in the mapping population, a gross rearrangement affecting linked genes had not occurred. Two other genes within the mapping interval were slightly upregulated in Ts5 mutant pools. GRMZM2G079452 (logFC 1.35) is annotated as a hypothetical protein whereas GRMZM2G112795 (logFC 1.07) is annotated as a putative uncharacterized protein. Otherwise, the expression of nearby genes was unchanged between Ts5 and normal plants, suggesting that the chromosomal rearrangement did not broadly impact regional transcription, and that the increase in GRMZM2G177668 expression is most likely the specific cause of Ts5 phenotypes.
We sought to confirm that Ts5 is in the JA pathway given that GRMZM2G177668 encodes a paralog of an enzyme known to catabolize bioactive jasmonate25,26 and Ts5 shares phenotypes with jasmonic acid biosynthetic mutants14,15. We tested if an exogenous JA application could block the growth of silks in homozygous Ts5 mutant tassels and suppress the mutant phenotype (Fig. 2c). We used tasselseed1 (ts1), a mutation in a JA biosynthetic enzyme, as a control. The feminization of that mutant was previously shown to be reversed by the exogenous application of JA9. JA or a mock solution was applied directly into the whorls of 4-week-old plants every 2 days for 2 weeks at a concentration of 1 mM as they transitioned from vegetative to inflorescence development. FBN/TBN and FSL/SL were quantified and compared to the reduction, after treatment, of ts1 mutants (Fig. 2c). Ts5 tassels responded similarly to ts1 tassels, revealing that high concentrations of exogenous JA can rescue the feminization of the Ts5 phenotype. We applied JA at a concentration of 1 mM, but endogenous levels are much lower. This finding is consistent with Ts5 encoding an enzyme that catabolizes JA, though it is unable to metabolize an excess of JA applied directly to the developing tassel. Similar cases have been reported with transgenic lines overexpressing enzymes in the JA catabolic pathways26–28.
Since Ts5 could be corrected by the addition of JA, we measured root traits, known to be affected in other JA biosynthetic mutants such as opr7opr8 in maize14 or allene oxide cyclase mutants in rice29. In addition, the overexpression of either CYP94B1 or CYP94B3 in Arabidopsis has been shown to display a similar long root phenotype26,27. Dark-grown Ts5 homozygous (produced after seven backcrosses to B73 (BC7)) seedlings have prematurely elongated coleoptiles and longer roots than B73 (Fig. 2d), similar to those found in opr7opr8 double mutants. This suggests that Ts5 plants have reduced bioactive jasmonate and that bioactive jasmonate negatively regulates root growth, as previously reported14,15,29. To assess the similarity in the function of ZmCYP94B1 and CYP94B3, we overexpressed the maize gene in Arabidopsis. ZmCYP94B1-OE lines had a similar ratio of under-developed to developed siliques as CYP94B3-OE lines (Fig. 2e). Arabidopsis CYP94B1-OE lines have been reported to display similar flower-development defects as CYP94B3-OE26. From this experiment, we conclude that ZmCYP94B1, like CYP94B1 or CYP94B3, functions to hydrolyze JA-Ile to control flower and fruit development in planta.
Ts5 mutants accumulate metabolites of jasmonate ω-oxidation
We profiled jasmonate metabolites in the developing tassels of Ts5 homozygous plants (B73 BC7) and B73 wild-type controls using liquid chromatography–mass spectrometry to test whether they are predictably altered. Arabidopsis CYP94B126 and CYP94B327 are known to convert JA-Ile, the most bioactive form of jasmonate30–33 to the inactive form 12OH-JA-Ile during oxidative catabolism of JA34,35 as summarized in Fig. 3a. Comparison of CYP94B protein sequences from maize and Arabidopsis did not distinguish whether GRMZM2G177668 is more similar to Arabidopsis CYP94B3 or to CYP94B1 (Fig. 3b). Both the Arabidopsis proteins, however, oxidize JA-Ile to 12OH-JA-Ile semi-redundantly leading to less-bioactive jasmonate26,27,34,36. Based on the phylogeny and evidence presented in this paper, we named GRMZM2G177668 ZmCYP94B1. Our phylogenic analysis also revealed that GRMZM2G164074 is a close paralog, although not differentially expressed in Ts5 heterozygous tassels (Supplementary Data 2).
Fig. 3.
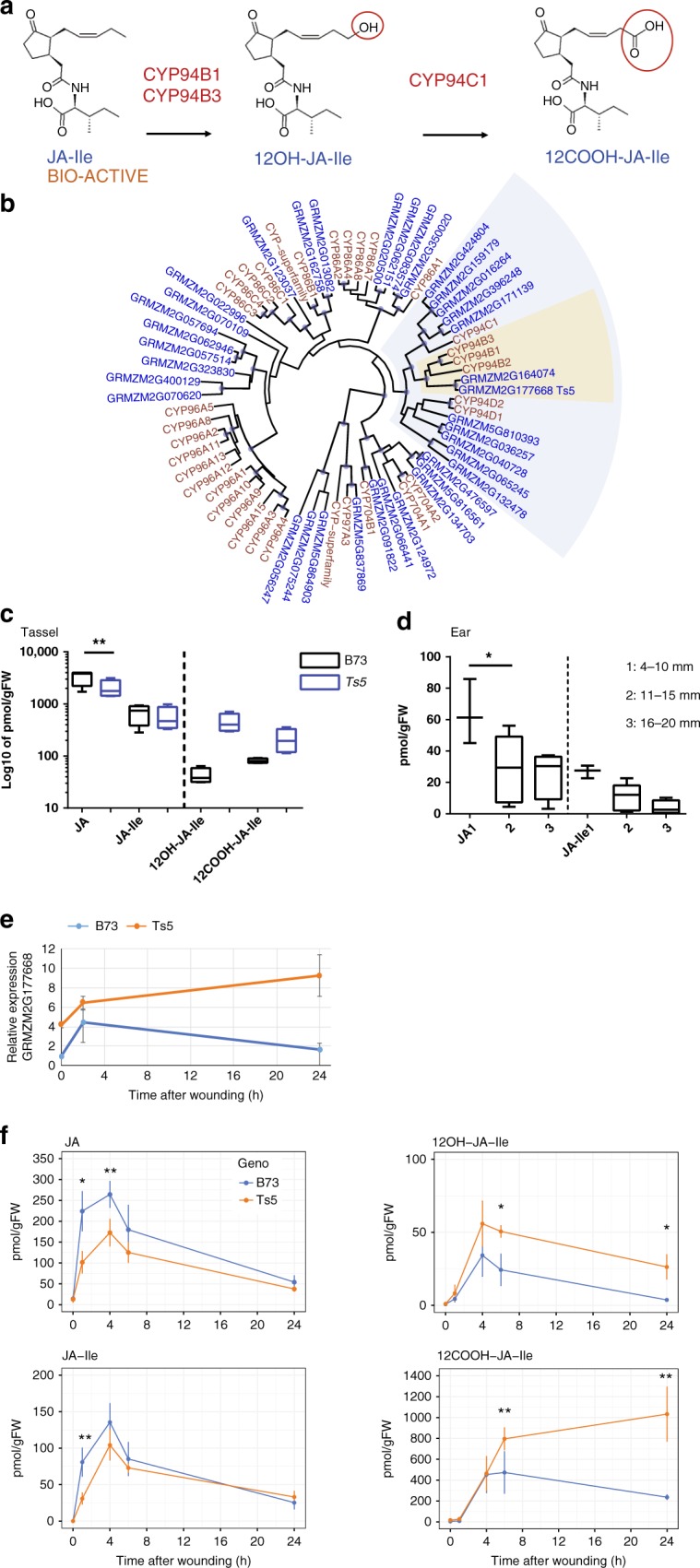
A wound-inducible CYP94B is upregulated in Ts5. a A diagram of steps of JA-Ile catabolism via the ω-oxidation pathway. b A phylogenetic tree of CYP genes from maize (blue) and Arabidopsis (brown). CYP94 genes (blue-gray) and CYP94B genes (yellow) are shaded. A blue dot at a node indicates branches with > 95% support. c Graph of jasmonate levels in developing tassels (2 cm) in B73 (black) and Ts5 homozygotes (blue). The amount of 12OH-JA-Ile is significantly higher in Ts5 than in B73 using a one-tailed Student’s t test with Welch’s correction, P = 0.0109, t = 4.348524 df = 3.037953, 95% CI = 112.0289 to 707.7894. d JA and JA-Ile levels in three sizes of B73 ears during early development. The amount of JA in 11–15 mm ears is significantly less than that in 4–10 mm ears by one-way ANOVA; F = 7.078, P = 0.008, DF = 18. Bar, mean; whiskers, SD. e Relative expression of GRMZM2G177668 measured by qRT-PCR at 0, 2, and 24 h after wounding in pooled (n = 4) second leaves of Ts5 homozygotes (BC7 B73) and B73. Graph depicts means of three technical replicates. Error bars, SD. f Graphs depicting a time-course of LC–MS outputs of JA, JA-Ile, 12OH-JA-Ile, and 12COOH-JA-Ile accumulation (pmol/gFW) in wounded leaves of Ts5 homozygotes (B73) and B73 at 0, 1, 4, 6, and 24 h. Ts5 leaves had significantly less JA at 1 h (t = 4.567575, P = 0.0038, df = 6, 95% CI = −188.1638 to −56.88620) and 4 h (t = 3.548059, P = 0.0238, df = 4, 95% CI = −163.2358 to −19.91499) after wounding than B73. Ts5 leaves had significantly less JA-Ile at 1 h (t = 4.802854, P = 0.0030, df = 6, 95% CI = −75.41736 to −24.50798) post-wounding than B73 leaves. Ts5 leaves had significantly more 12OH-JA-Ile than B73 at 6 h (t = 3.961804, P = 0.0107, df = 5, 95% CI = 9.232525–43.35088) and at 24 h (t = 9.473349, P = 0.0002, df = 5, 95% CI = 18.97077–33.10019) post-wounding than B73 leaves. Ts5 leaves had significantly more 12COOH-JA-Ile at 6 h (t = 5.007114, P = 0.0075, df = 4, 95% CI = 184.6060–644.1580) and at 24 h (t = 8.622493, P = 0.0003, df = 5, 95% CI = 626.7214–1159.130) post-wounding than B73 leaves. f Error bars, SD of four biological replicates. LC–MS, liquid chromatography–mass spectrometry
Compared to B73 wild type, steady-state levels of JA and JA-Ile are lower in Ts5 tassels, whereas steady-state levels of 12OH-JA-Ile, the product of JA-Ile oxidation by Arabidopsis CYP94B1 and CYP94B3, are higher in Ts5 mutants (Fig. 3c). This result is consistent with the increased expression of GRMZM2G177668 promoting catalysis of active jasmonates in Ts5 tassels. In Arabidopsis, the prolonged catabolism of JA-Ile provides more substrate for CYP94C1, which preferentially catalyzes carboxy-derivative formation, explaining the increased levels of its downstream product 12COOH-JA-Ile34, also seen in Fig. 3c. 12OH-JA is a wound-induced jasmonate, also known as tuberonic acid37, that results either from the hydrolytic cleavage of 12OH-JA-Ile by amidohydrolases38,39 or from the direct oxidation of JA40,41. We also profiled wild-type B73 ear jasmonates and found that the levels of all tested metabolites (JA, JA-Ile, 12OH-JA, 12OH-JA-Ile, and 12COOH-JA-Ile) were much lower in ears than in tassels. Moreover, the level of JA-Ile decreased significantly as wild-type ears grew from 6 mm (comprised of spikelet and spikelet pair meristems) to 20 mm (comprised of floral meristems) (Fig. 3d).
The CYP94B3 enzyme is known to be wound inducible in Arabidopsis27,35. To evaluate our hypothesis that Ts5 is a maize functional homolog of CYP94B, we assayed its gene expression and profiled JA metabolites via a wounding time-course experiment. We found that GRMZM2G177668 is expressed at higher levels in Ts5 homozygous (B73 BC7) leaves even before wounding (Fig. 3e), consistent with the increase seen in our RNA-Seq experiments in Ts5 tassels (Fig. 2b). Post-wounding, GRMZM2G177668 levels increased within 2 h in both wild-type and Ts5 plants. In wild type, GRMZM2G177668 expression levels dropped to pre-wounding levels within 24 h, yet GRMZM2G177668 expression levels remained high in Ts5 (Fig. 3e). These results suggest that the regulation of the mutant allele is altered in Ts5, causing enhanced ZmCYP94B1 transcript accumulation.
The JA metabolite profile in wounded Ts5 homozygous leaves, in the B73 genetic background, is consistent with the prolonged and increased expression of ZmCYP94B1. JA-lle and JA levels were lower in Ts5, but 12OH-JA-lle and 12COOH-JA-lle levels are increased (Fig. 3f). Similar to reports in Arabidopsis27, the jasmonate catabolites showed a slight time lag behind the increase in JA and JA-Ile. One hour post-wounding, JA and JA-Ile had already increased by 30–60% of their highest levels, but 12OH-JA-Ile and 12COOH-JA-Ile had only increased by 5–10%. Importantly, JA and JA-Ile levels were lower in Ts5 compared to B73 at–4-6 h post-wounding, whereas oxidized jasmonate levels were significantly higher in Ts5, consistent with the enhanced metabolism via the ω-oxidation pathway. Twenty-four hours post-wounding, the JA catabolites in wild-type plants, especially 12COOH-JA-Ile, were lowered to near starting levels, but remained high in the mutant. Taken together, these results support the hypothesis that Ts5 is altered in JA metabolism due to the overexpression of GRMZM2G177668 because metabolites in the JA pathway are affected in Ts5 mutants in a specific manner consistent with higher CYP94B enzymatic activity.
Ts5 differentially expresses JA signaling genes
Of the 231 significantly differentially expressed genes detected in our RNA-Seq analysis (false discovery rate (FDR) < 0.05), 89% were downregulated. Eight percent (11/131) of the annotated differentially expressed genes are involved in JA signaling (Supplementary Data 2; Supplementary Data 3), including lipoxygenase genes (LOX1, LOX2, and LOX5), an allene oxide synthase (AOS or CYP74 A), an allene oxide cyclase (AOC3), and 12-oxophytodienoate reductase (OPR1). This finding is consistent with these JA-Ile-inducible genes being downregulated due to the increased turnover of JA-Ile in Ts5 mutants. ts2, which encodes a dehydrogenase implicated in JA biosynthesis11, was also differentially expressed in Ts5 tassels although we did not detect any difference in JA-Ile, nor several other jasmonates in ts2 mutants compared to their non-mutant siblings following wounding (Supplementary Figure 2).
Ts5 interacts genetically with ts2 and sk1
Given the down-regulation of ts2 in Ts5 mutants, we made a double mutant to assay the potential dosage effects between the mutant loci. The crosses were carried out in A188, which is a moderately expressive background for both mutations. ts2 is known to be completely recessive, and its genetic interaction with Ts5 in a heterogeneous genetic background has been previously described8. Spikes were completely feminized in plants homozygous for ts2, independent of Ts5 dosage (Fig. 4a). Plants with an FSL/SL ratio of 0 are males and those with a ratio of 1 are completely feminized. Surprisingly, lowering the dose of TS2 increased the feminization of Ts5 tassels. Ts5/ + ; + / + ; and Ts5/Ts5; + / + (n = 8) plants had variable FSL/SL ratios with a mean of less than 0.20 while double heterozygotes, Ts5/ + ; ts2/ + ; (n = 5), had a mean ratio of almost 0.60 (Fig. 4a). These results suggest different possible routes for a feminization switch. That switch can be reached by mutating ts2 or overexpressing Ts5, or by a combination of both.
Fig. 4.
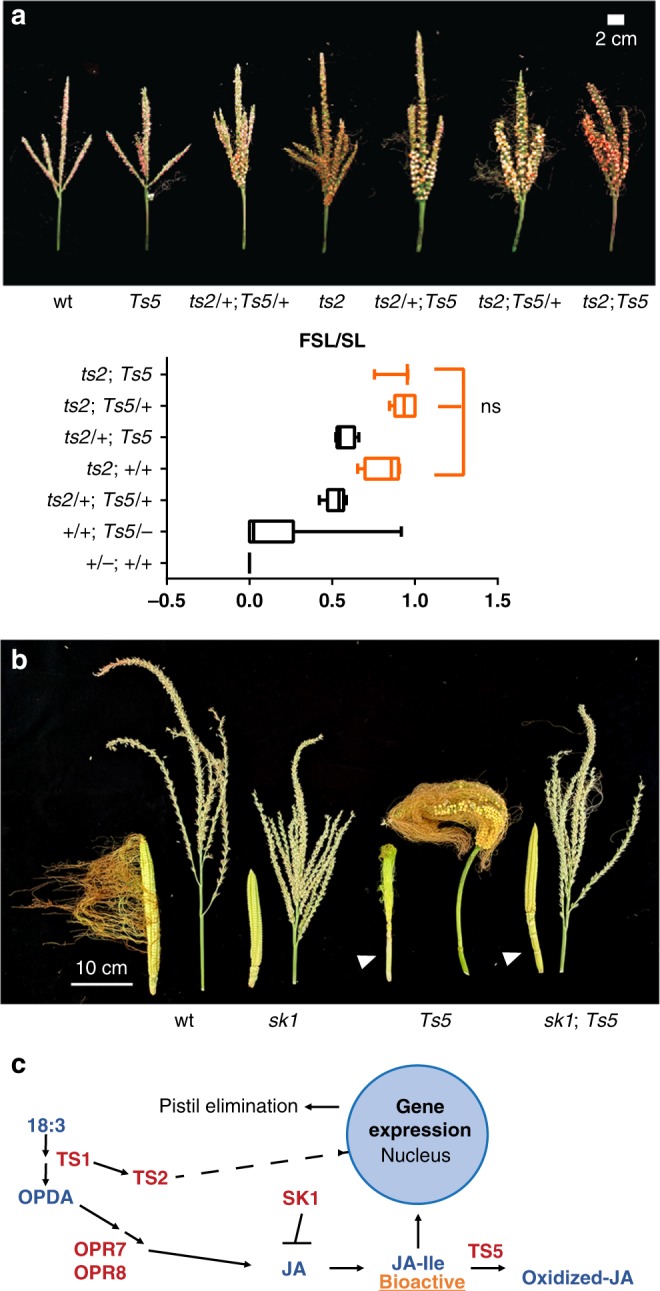
Genetic dissection of Ts5. a Analysis of ts2;Ts5 double mutants in the A188 background and their effect on feminized spike length over spike length (FSL/SL). Photos are of mature tassels showing feminization, silks, and kernels in genotypes in which they are formed. In graph, measurements of individual plants (double homozygous mutants (n = 3), ts2:Ts5/ + (n = 6), ts2/ + ;Ts5 (n = 5), ts2: + / + (n = 4), ts2/ + ;Ts5/ + (n = 5), + / + ; Ts5/− (n = 8), and wild type ( + /−; + / + ) (n = 6)) are shown. Boxes for genetic classes with completely feminized tassels are orange. These classes were not significantly different (ns) by one-way ANOVA with Tukey’s multiple comparison test, P < 0.05. For ts2; + / + vs ts2/ + ; Ts5, q = 3.26787, multiplicity-adjusted P = 0.3376, 95% CI = −0.105368 to 0.575774, for ts2; + / + vs. ts2; Ts5/ + , q = 1.60441, multiplicity adjusted P = 0.9314, 95% CI = −0.456047 to 0.225095, for ts2; + / + vs. ts2; Ts5, q = 0.825390, multiplicity adjusted P = 0.9977, 95% CI = −0.473261 to 0.332678. Dashes represent pooled classes wherein the allele may be mutant or wild type at the locus. Bars, mean; whiskers, SD. b Unpollinated ears and mature tassels of sk1;Ts5 double mutants in the Mo17 background. White arrowheads indicate elongated ear shanks. c A model for the actions of known gene products of the maize JA pathway.
We also crossed Ts5 to silkless1 (sk1), which is in the same genetic pathway with ts2 and ts18,42,43. sk1 is a uridine diphosphate glycosyltransferase that blocks the production of JA44. Ts5;sk1 double homozygous mutants in the Mo17 background were silkless in the ear and had just a few silks formed in the tassel, indicating that sk1 is epistatic (Fig. 4b). Because Ts5 functions later in the pathway to catabolize JA-Ile and SK1 functions earlier to block the production of JA-Ile, we reason that sufficient JA-Ile is still present to prevent silks in both the tassel and ear of the double mutant.
Interestingly, we observed elongated ear shanks in both Ts5 and double mutants in this family (Fig. 4b). This phenotype was described in opr7opr8 mutants and is seen in teosinte branch1 (tb1) mutants45,46, a gene which is downregulated in Ts5 (Supplementary Data 2). The elongated internodes of the ear shank contrast with the shortened internodes below the terminal inflorescence. Perhaps this differential response is due to the differences in JA metabolism or signaling in these two organs, or results from the differences in the amounts of any other hormone.
Discussion
Maize plants have separate male and female flowers (are monecious) due to pistil abortion in the tassel and stamen arrest in the ear. In addition, maize ears contain straight rows of kernels due to the abortion of the lower floret. In Ts5, pistil abortion is blocked with the result that the tassel is feminized and kernel rows in the ear are irregular. These phenotypes are shared by ts1, ts2, and opr7opr8, as well as by lines that overexpress SK1. TS1 is a lipoxygenase needed for the biosynthesis of JA, and TS2 is a short-chain alcohol dehydrogenase purported to be in the JA pathway9,11. The study of its substrate specificity, however, suggests that it more likely affects steroid compounds such as brassinosteroids and may not play a direct role in the jasmonate signaling pathway47. SK encodes a uridine diphosphate glycosyltransferase that blocks the production of JA44. Thus, JA biosynthesis and metabolism are critical for pistil abortion in the tassel and abortion of the lower floret in the ear.
Here, we present evidence that the dominant Ts5 mutant is due to the misexpression of a maize homolog of Arabidopsis, CYP94Bs. In Arabidopsis, CYP94B126 and CYP94B327 catabolize JA-Ile, the active jasmonate metabolite that binds to the nuclear localized jasmonate co-receptor complex32. Transcript levels of ZmCYP94B1 are dramatically upregulated in Ts5 mutant tassels, and jasmonate metabolites are altered consistently with the increased catabolism of JA-Ile through the ω-oxidation pathway25. Ts5 leaves have ~ 4-fold more ZmCYP94B1 transcripts prior to wounding, and transcript levels continue to rise over time, failing to return to non-wounded levels even a day after wounding. Indeed, the catabolic products of JA-Ile remain high in wounded Ts5 leaves, unlike those of wild types. The increase in ZmCYP94B1 transcripts in Ts5 tassel and leaf, along with changes in JA metabolism, support the hypothesis that the feminization of Ts5 tassels is due to reduced JA-Ile levels, similar to the observations of ts1 or opr7opr89,14.
The genetic lesion of Ts5 is unknown. The first mention of Ts5 is on a genetic map in 1932 without further description21. Given the novel 5′ transcripts found by RNA-Seq and the reduction in recombination around the locus, it is likely to be a genomic rearrangement. Reduced recombination can be found in heterozygous genomic regions of differing retrotransposon haplotypes48. Our mapping population, made by backcrossing a Ts5/+ parent in an unknown progenitor background to Mo17, may indeed have a structural variation that affects genetic distances by contracting or expanding physical and genetic distances between markers48. Moreover, we were unable to amplify the 5′-end of the gene using PCR, possibly due to the presence of a large insertion such as a transposon. Unique dominant alleles often result from large rearrangements that alter cis regulatory sequences, such as Abphyl2, which is a transposition49, Kn1-O, which is a tandem duplication50 or Wab1-R, which is ectopically expressed in the leaf due to unknown cis regulatory changes51.
If higher jasmonate levels lead to pistil abortion in the tassel, what keeps it from eliminating the pistils in the ear? This quandary was first investigated in 1925 in the analysis of silkless1 (sk1)42,52. Tassels of sk1 mutants are normal, but ears lack silks, although they retain other female features such as stamen suppression and short glumes. The double mutant with ts2 restores the silks to the ear, while the tassel remains mostly staminate8,43. Although the exact substrate of SK1 is unknown, its overexpression leads to a tasselseed phenotype and reduces 12-oxo-phytodienoic acid (OPDA, a precursor of JA) and JA-Ile44. The authors hypothesize that SK1 inactivates JA or a precursor in a tissue-specific manner. The rescue of silks in the sk1;ts2 double mutant occurs because the loss of SK1 has no effect in the ts2 mutant background43. SK1 is expressed at very low levels, but is most highly expressed in ears, consistent with its function to protect the silks of ears. The Ts5;sk1 double mutant is different from ts2;sk1 in that no silks form in the ear and very few in the tassel, plus lower floret abortion still occurs in ears. The absence of SK1 is expected to cause an increase in JA and, consequently, its downstream product JA-Ile. Overexpression of ZmCYP94B1 in Ts5 catabolizes JA-Ile, but the turnover effect may be somewhat compensated by the higher levels of JA-Ile. This result is similar to the rescue of Ts5 mutants by exogenous JA. The increased catabolism from Ts5 is unable to compensate for the extra jasmonate predicted in the sk1 mutant.
Although sex determination in maize is dependent upon jasmonate suppression of pistils, regulating pistil outgrowth does not explain why tasselseed florets have pistils and no stamens. The function of jasmonate in promoting stamen development is well established in Arabidopsis. Mutants that are deficient in jasmonate biosynthesis, perception, or signaling are also defective in stamen differentiation12,32,53. In fact, the overexpression of CYP94B1 or CYP94B3 in Arabidopsis leads to a partial loss of fertility. The stigma is extended, the anther filament is shortened, and pollen viability is reduced27. We also saw a similar phenotype when overexpressing Ts5 in Arabidopsis. The high levels of JA and JA-Ile in wild-type tassels compared to ears and the repression of stamens in JA maize mutants suggest that jasmonate is needed in both maize and Arabidopsis for proper stamen development.
Additional jasmonate-deficient phenotypes are described in rice that are consistent with sex-determination phenotypes in maize. The allene oxide cyclase mutants of rice have elongated sterile lemmas and, occasionally, longer palea or additional bract-like organs47 similar to tassel glumes. The eg1 mutant, which encodes a plastid-targeted lipase, a homolog of DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1) of Arabidopsis, and the eg2 mutant, defective in JAZMONATE ZIM-DOMAIN (JAZ) gene, have extra glume-like organs and altered floral organ numbers54. In Sorghum, pedicellate spikelets are normally sterile but not in multiseeded (msd) mutants that produce extra spikelets. The increase in fertile spikelets in msd1 mutants is lost with the addition of JA55. Clearly, jasmonate plays a role in blocking the growth of pistils, leaf-life organs, and spikelets in addition to its well-established role in stamen fertility.
Our analysis of Ts5 adds to the growing understanding of how jasmonates are involved in maize sex determination and that not only its biosynthesis but also its turnover plays a role by contributing to its homeostasis (Fig. 4c). The lipoxygenase TS1 putatively converts α-linolenic acid (18:3) to an intermediary and then to cis-( + )-12-oxophytodienoic acid (OPDA)9, a precursor to JA-Ile that is further metabolized to JA via the function of OPR7 and OPR8, which are partially and functionally redundant14. The activation of the short-chain dehydrogenase/reductase TS2 requires TS143. Although, its exact substrates are unknown47, TS2 promotes JA production. Once the bioactive jasmonate, JA-Ile, is made, it relieves transcription factors from JAZ repression32, leading to the activation of several pathways, including those necessary for floral development12,32,53. ZmCYP94B1 then acts to inactivate JA-Ile by oxidation.
In conclusion, study of the gain-of-function mutant, Ts5, has revealed that the proper expression of ZmCYP94B1 is necessary to maintain maize monoecy. The conversion of JA-Ile to 12OH-JA-Ile via the ZmCYP94B1 enzyme is an important regulatory mechanism for sex determination in normal ear and tassel development. Given that this enzyme also has a role in ameliorating wound-induced JA response in leaves, it provides an elegant example of a gene product having multiple tissue-specific functions.
Methods
Plant materials and growth conditions
The Ts5-ref allele was obtained from the Maize Cooperative Center and backcrossed at least seven times to B73 and Mo17 and four times to A188. Mapping was performed after backcrossing seven times into Mo17 population. The ts2-ref and ts1-ref alleles were obtained from the Maize Cooperative Center. Genetic analysis of Ts5;ts2 double mutants was performed in the A188 background after three backcrosses and genotyping of all possible alleles. Introgressed stocks of sk1-R in Mo17 were a gift from Erik Vollbrecht (Iowa State). Genetic analysis of Ts5;sk1 double mutants was performed after crossing sk1-R in Mo17 to Ts5 in Mo17 and self-pollinating the F1 to create an F2 population. Ecotype Col-0 was used for all Arabidopsis thaliana experiments, and plants were grown under standard greenhouse conditions. Primers CL 682 and CL 758 (Supplementary Data 1) were used to amplify Ts5 (GRMZM2G177668), which has no introns, from B73 genomic DNA. The resulting amplicon was cloned into the pENTR/D-TOPO Vector using the manufacturer’s instructions (Thermo Fisher Scientific) to make a Gateway compatible entry vector. pEarleyGate10356 was linearized with EcoRV and recombined with the Ts5 entry clone via LR Clonase™ II Reaction. The resulting clone places the Ts5 ORF under control of the cauliflower mosaic virus 35S promoter. Plants were transformed using the floral dip method57. Transformants were also genotyped and transgene expression was confirmed with primers CL 682 and CL 758 (Supplementary Data 1). CYP94B3-OE lines have been described27.
Plant treatments
Plants for the rescue of tassel feminization were treated with JA as in Yan et al.14.
For the root-growth assay, plants were grown on filter paper in the dark at 30 °C for 5 days or grown in the medium vermiculite for 10 days under normal greenhouse conditions.
Genetic mapping of the Ts5 locus
We fine mapped Ts5 to a 15 Mb interval flanked by custom indel markers CL589.590 and MS13.14 in a backcross population of Ts5 to Mo17 consisting of 273 plants. This was further narrowed to the interval flanked by umc2039 and TIDP9218. DNA extraction was performed using standard protocols (Lunde, 2018)58 using the primers listed in Supplementary Data 1.
RNA-expression analysis
RNA-Seq libraries were constructed as in Tsuda et al.59 except that 6 µg of total RNA was used rather than 3 µg. Three libraries were made of four pooled 9–11 mm tassels: two mutants and one wild type. Libraries were sequenced on a NextSeq Illumina platform with 75 bp paired-end reads. Raw reads were aligned to the maize B73 genome AGPv3.30 using HISAT260 and counted to AGPv3.30 gene models using HTSeq-counts61 using the cyberinfrastructure provided by Cyverse Atmosphere62. Reads were visualized using the Integrative Genomics Viewer (Broad Institute)63. Counted reads were tested for differential expression with edgeR using a generalized linear model (GLM) approach on transcripts with a raw count greater than 5 in at least one condition and FDR significance threshold of 0.0564,65. Differentially expressed genes (FDR ≤ 0.05) between Ts5 and WT siblings were separated by -log(fold-change) into upregulated and downregulated differentially expressed gene lists. Gene accessions from each list were tested for Gene Ontology term enrichment by singular Gene Ontology term enrichment analysis (SEA) with agriGO v2.066. Quantitative reverse transcription PCR was performed as in Bolduc et al.67.
Phylogenetic analysis of maize CYP94 genes
The amino-acid sequence of GRMZM2G177668_P01 was blasted against Zea mays AGPv3.30 and against A. thaliana Araport11 genomes68–70. Canonical protein isoforms with blastp bit scores > 100 were run in an ETE371 pipeline that included alignment by Clustal Omega72 phylogeny model evaluation using PhyML73 and tree branchpoint evaluation using 100 bootstraps. Trees were visualized and annotated in R74 with the ggtree package75.
Analytical methods and chemicals
Hormone extraction from tissues was according to a previously described method with minor modifications31 for the quantification of jasmonate by mass spectrometry. Frozen leaf tissues and inflorescences (100‒200 mg) were pulverized using metal beads in TissueLyser II (Qiagen) and extracted multiple times with 3 ml of ethyl acetate (0.5% acetic acid) containing a mixture of dihydro-JA (dhJA) and [13C6]-JA-Ile as an internal standard. The combined extracts were evaporated under a stream of nitrogen gas, and the dried residue was reconstituted in 0.2 ml of 70% methanol/water/acetic acid (v/v/v = 70:29.5:0.5). The resulting tissue extract was cleared by centrifugation at 18,000 × g for 30 min in 4 °C. Analysis and quantification of jasmonate were carried out based on methods described previously26 using an electrospray ionization triple quadrupole mass spectrometer (Xevo T-QS, Waters) interfaced with an ultra-performance liquid chromatography (ACUITY H-class, Waters). Characteristic mass spectrometry transitions detected under multiple reaction monitoring in electrospray ionization-negative mode were for JA (m/z 209 > 59), dhJA (211 > 59), 12OH-JA (225 > 59), JA-Ile (322 > 130), [13C6]-JA-Ile (328 > 136), 12OH-JA-Ile (338 > 130), and 12COOH-JA-Ile (352 > 130). MassLynx 4.1 and TargetLynx (Waters) were used to analyze the data.
Statistical analysis and plotting
Student’s t tests, one-way analysis of variance, and graphs were made using GraphPad Prism software76, with the exception of graphs in 3F, which were designed in core R packages74. Raw data used to create graphs are available in Supplementary Data 4.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Many thanks to Nathalie Bolduc, Katsu Tsuda, and Jazmin Abraham-Juarez for help with RNA-expression analysis. Alleles of ts1-R, ts2-R, Ts5-R, and sk1-R were provided by Maize Genetics Coop. Junyi Wu, Daniele Paschoal, and Monica Sadhu designed the molecular markers for mapping. Introgressed stocks of sk1-R in Mo17 were a gift from Erik Vollbrecht (Iowa State). Nathan Springer (University of Minnesota) provided haplotype information for the undefined Ts5-R allele, useful for marker design. We thank the members of Hake Lab for comments on the manuscript and Kjel Johnson for photos in Fig. 4a. We also thank Dr. Jacob Brunkard, Thai Dao and Michael Busche for technical advice on Arabidopsis transformation. The work was supported by grants from the National Science Foundation (IOS-1557439 to A.J.K., IOS-1238202 and CRIS 2030-21000-052-00D to S.H., and IOS-1612268 to S.L.).
Author contributions
C.L., S.H. and A.J.K. designed the experiments. C.L. and A.K. performed the experiments. C.L., A.K. and S.L. analyzed the data. C.L. and S.L. prepared the figures. C.L. and S.H. wrote the manuscript. A.J.K. and S.L. edited the manuscript.
Data availability
Transcriptomic data for 10 mm tassel RNA-Seq of Ts5/ + and normal siblings are available at the NCBI sequence read archive (SRA) under the accession code PRJNA495059.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0354-1.
References
- 1.Fujii S, Kubo KI, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants. 2016;2:16130. doi: 10.1038/nplants.2016.130. [DOI] [PubMed] [Google Scholar]
- 2.Pannell JR. Plant sex determination. Curr. Biol. 2017;27:R191–R197. doi: 10.1016/j.cub.2017.01.052. [DOI] [PubMed] [Google Scholar]
- 3.Malcomber ST, Kellogg EA. Evolution of unisexual flowers in grasses (Poaceae) and the putative sex-determination gene, TASSELSEED2 (TS2) New Phytol. 2006;170:885–899. doi: 10.1111/j.1469-8137.2006.01726.x. [DOI] [PubMed] [Google Scholar]
- 4.Bonnett OT. Development of the staminate and pistillate inflorescences of sweet corn. J. Agric. Res. 1940;60:25–37. [Google Scholar]
- 5.Cheng PC, Greyson RI, Walden DB. Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am. J. Bot. 1983;70:450–462. doi: 10.1002/j.1537-2197.1983.tb06411.x. [DOI] [Google Scholar]
- 6.Emerson RA. Heritable characters in maize. II. Pistillate flowered maize plants. J. Hered. 1920;11:65–76. doi: 10.1093/oxfordjournals.jhered.a101971. [DOI] [Google Scholar]
- 7.Nickerson NH, Dale EE. Tassel modifications in Zea mays. Ann. Mo. Bot. Gard. 1955;42:195–211. doi: 10.2307/2394655. [DOI] [Google Scholar]
- 8.Irish EE, Langdale JA, Nelson TM. Interactions between tassel seed genes and other sex determining genes in maize. Dev. Genet. 1994;15:155–171. doi: 10.1002/dvg.1020150206. [DOI] [Google Scholar]
- 9.Acosta IF, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science (80-). 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 10.Eveland AL, et al. Regulatory modules controlling maize inflorescence architecture. Genome Res. 2014;24:431–443. doi: 10.1101/gr.166397.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong A, Calderon-Urrea A, Dellaporta SL. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell. 1993;74:757–768. doi: 10.1016/0092-8674(93)90522-R. [DOI] [PubMed] [Google Scholar]
- 12.Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U S A. 2000;97:10625–10630. doi: 10.1073/pnas.190264497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaller F, Biesgen C, Müssig C, Altmann T, Weiler EW. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 2000;210:979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- 14.Yan Y, et al. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell. 2012;24:1420–1436. doi: 10.1105/tpc.111.094151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Y, Huang PC, Borrego E, Kolomiets M. New perspectives into jasmonate roles in maize. Plant Signal. Behav. 2014;9:1–5. doi: 10.4161/15592316.2014.970442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 2007;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- 17.Hartwig T, et al. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA. 2011;108:19814–19819. doi: 10.1073/pnas.1108359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Best, N. B. et al. nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis protein Dwarf1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol. 10.1104/pp.16.00399 (2016). [DOI] [PMC free article] [PubMed]
- 19.Parkinson SE, Gross SM, Hollick JB. Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 2007;308:462–473. doi: 10.1016/j.ydbio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Boualem A, Bendahmane A, Ming R. Genomics of sex determination. Curr. Opin. Plant. Biol. 2014;18:110–116. doi: 10.1016/j.pbi.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Emerson, R. A. The present status of maize genetics. In Proc. Sixth International Congress of Genetics, 141–152 (Brooklyn Botanical Garden, Brooklyn, NY, USA, 1932).
- 22.Andorf CM, et al. MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 2016;44:D1195–D1201. doi: 10.1093/nar/gkv1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosa, M. et al. The maize mid-complementing activity homolog cell number regulator13/narrow odd dwarf coordinates organ growth and tissue patterning. Plant Cell. 10.1105/tpc.16.00878 (2017). [DOI] [PMC free article] [PubMed]
- 24.Zhang, D. et al. GRF-interactingfactor1 (gif1) regulates shoot architecture and meristem determinacy in maize. Plant Cell. 10.1105/tpc.17.00791 (2018). [DOI] [PMC free article] [PubMed]
- 25.Koo AJK, Howe GA. Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 2012;3:19. doi: 10.3389/fpls.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo AJ, et al. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-L-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J. Biol. Chem. 2014;289:29728–29738. doi: 10.1074/jbc.M114.603084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo AJK, Cooke TF, Howe G. a. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. US A. 2011;108:9298–9303. doi: 10.1073/pnas.1103542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T, et al. Hormone crosstalk in wound stress response: Woundinducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2016;67:2107–2120. doi: 10.1093/jxb/erv521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riemann M, et al. Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 2013;74:226–238. doi: 10.1111/tpj.12115. [DOI] [PubMed] [Google Scholar]
- 30.Staswick PE. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung HS, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thines B, et al. JAZ repressor proteins are targets of the SCFCOI1complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 33.Fonseca S, et al. ( + )-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 34.Heitz T, et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012;287:6296–6306. doi: 10.1074/jbc.M111.316364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitaoka N, et al. Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 2011;52:1757–1765. doi: 10.1093/pcp/pcr110. [DOI] [PubMed] [Google Scholar]
- 36.Poudel AN, et al. Mutations in jasmonoyl-L-isoleucine-12-hydroxylases suppress multiple JA-dependent wound responses in Arabidopsis thaliana. Biochim. Biophys. Acta. 2016;1861:1396–1408. doi: 10.1016/j.bbalip.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 2008;177:114–127. doi: 10.1111/j.1469-8137.2007.02252.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Li X, Yu R, Han M, Wu Z. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family. Mol. Genet. Genomics. 2015;290:1849–1858. doi: 10.1007/s00438-015-1042-6. [DOI] [PubMed] [Google Scholar]
- 39.Widemann E, et al. Sequential oxidation of Jasmonoyl-Phenylalanine and Jasmonoyl-Isoleucine by multiple cytochrome P450 of the CYP94 family through newly identified aldehyde intermediates. Phytochemistry. 2015;117:388–399. doi: 10.1016/j.phytochem.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 40.Caarls L, et al. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc. Natl. Acad. Sci. USA. 2017;114:6388–6393. doi: 10.1073/pnas.1701101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smirnova E, et al. Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against botrytis cinereaInfection. Mol. Plant. 2017;10:1159–1173. doi: 10.1016/j.molp.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Jones DF. The interaction of specific genes determining sex in dioecious maize. In Proc. Sixth International Congress of Genetics. 1932;2:104–107. [Google Scholar]
- 43.Calderon-Urrea A, Dellaporta SL. Cell death and cell protection genes determine the fate of pistils in maize. Development. 1999;126:435–441. doi: 10.1242/dev.126.3.435. [DOI] [PubMed] [Google Scholar]
- 44.Hayward AP, et al. Control of sexuality by the sk1-encoded UDP-glycosyltransferase of maize. Sci. Adv. 2016;2:e1600991–e1600991. doi: 10.1126/sciadv.1600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doebley J, Stec A, Gustus C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, et al. Biochemical characterization of TASSELSEED 2, an essential plant short-chain dehydrogenase/reductase with broad spectrum activities. FEBS J. 2007;274:1172–1182. doi: 10.1111/j.1742-4658.2007.05642.x. [DOI] [PubMed] [Google Scholar]
- 48.Dooner HK, He L. Maize genome structure variation: interplay between retrotransposon polymorphisms and genic recombination. Pant Cell. 2008;20:249–258. doi: 10.1105/tpc.107.057596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang F, et al. A maize glutaredoxin gene, abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell. 2015;27:121–131. doi: 10.1105/tpc.114.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veit B, Vollbrecht E, Mathern J, Hake S. A tandem duplication causes the Kn1-O allele of knotted, a dominant morphological mutant of maize. Genetics. 1990;125:623–631. doi: 10.1093/genetics/125.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis MW, et al. Gene regulatory interactions at lateral organ boundaries in maize. Development. 2014;141:4590–4597. doi: 10.1242/dev.111955. [DOI] [PubMed] [Google Scholar]
- 52.Jones DF. Heritable characters of maize. XXIII–Silkless. J. Hered. 1925;16:339–342. doi: 10.1093/oxfordjournals.jhered.a102629. [DOI] [Google Scholar]
- 53.Feys B. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai Q, et al. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014;5:3476. doi: 10.1038/ncomms4476. [DOI] [PubMed] [Google Scholar]
- 55.Jiao Y, et al. MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat. Commun. 2018;9:822. doi: 10.1038/s41467-018-03238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Earley, K. W. et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 10.1111/j.1365-313X.2005.02617.x (2006). [DOI] [PubMed]
- 57.Clough, S. J. & Bent, A. F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 10.1046/j.1365-313X.1998.00343.x (1998). [DOI] [PubMed]
- 58.Lunde C. Small-scale DNA extraction method for maize and other plants. Bio Protoc. 2018;101:e2782. [Google Scholar]
- 59.Tsuda K, Kurata N, Ohyanagi H, Hake S. Genome-wide study of knox regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell. 2014;26:1–14. doi: 10.1105/tpc.114.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goff SA, et al. The iPlant Collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2011;2:1–16. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian T, et al. AgriGOv2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolduc N, et al. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012;26:1685–1690. doi: 10.1101/gad.193433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng CY, et al. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- 69.Cannon EKS, et al. POPcorn: An Online resource providing access to distributed and diverse maize project data. Int. J. Plant Genomics. 2011;2011:923035. doi: 10.1155/2011/923035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berardini TZ, et al. The arabidopsis information resource: Making and mining the ‘gold standard’ annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2014;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 74.R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2018).
- 75.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 76.Miller JM. GraphPad PRISM. Analysis. 2003;52:2–3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic data for 10 mm tassel RNA-Seq of Ts5/ + and normal siblings are available at the NCBI sequence read archive (SRA) under the accession code PRJNA495059.