Abstract
Some early-stage clear cell renal cell carcinomas (ccRCCs) of ≤7 cm are associated with a poor clinical outcome. In this study, we investigated molecular biomarkers associated with aggressive clinical T1 stage ccRCCs of ≤7 cm, which were used to develop a risk prediction tool toward guiding the decision of treatment. Among 1069 nephrectomies performed for ccRCC of ≤7 cm conducted between January 2008 and December 2014, 177 cases with available formalin-fixed paraffin-embedded tissue were evaluated. An aggressive tumor was defined as a tumor exhibiting synchronous metastasis, recurrence, or leading to cancer-specific death. Expression levels of six genes (FOXC2, CLIP4, PBRM1, BAP1, SETD2, and KDM5C) were measured by reverse-transcription polymerase chain reaction (qRT-PCR) and their relation to clinical outcomes was investigated. Immunohistochemistry was performed to validate the expression profiles of selected genes significantly associated with clinical outcomes in multivariate analysis. Using these genes, we developed a prediction model of aggressive ccRCC based on logistic regression and deep-learning methods. FOXC2, PBRM1, and BAP1 expression levels were significantly lower in aggressive ccRCC than non-aggressive ccRCC both in univariate and multivariate analysis. The immunohistochemistry result demonstrated the significant downregulation of FOXC2, PBRM1, and BAP1 expression in aggressive ccRCC. Adding immunohistochemical staining results to qRT-PCR, the aggressive ccRCC prediction models had the area under the curve (AUC) of 0.760 and 0.796 and accuracy of 0.759 and 0.852 using the logistic regression method and deep-learning method, respectively. Use of these biomarkers and the developed prediction model can help stratify patients with clinical T1 stage ccRCC.
Keywords: Renal cell cancer, Biomarker, Prediction model
Abbreviations: BAP1, BRCA1 associated protein-1; BMI, Body mass index; ccRCC, Clear cell renal cell carcinoma; CLIP4, CAP-Gly, cytoskeleton-associated protein-glycine rich domain-containing linker protein family member 4; DNN, Deep neural network; EDTA, Ethylenediaminetetraacetic acid; FFPE, Formalin-fixed paraffin-embedded; FOXC2, Forkhead box protein C2; KDM5C, Lysine-specific demethylase 5C; MSKCC, Memorial Sloan Kettering Cancer Center; PBRM1, Polybromo 1; PBS, Phosphate-buffered saline; qRT-PCR, Quantitative reverse transcription-polymerase chain reaction; SETD2, SET domain-containing 2; TNM, Tumor-node-metastasis
Graphical Abstract
1. Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common subtype of kidney cancer, and its detection and diagnosis have been continuously increasing in high-income countries worldwide over the last few decades [1,2]. In particular, this increase is largely attributed to advances in diagnostic imaging techniques, including cross-sectional imaging, allowing for the detection of clinical T1 stage with a tumor of 7 cm or smaller, which have improved identification of incidental renal masses that are suspicious of malignancy [3,4].
Once clinical T1 stage ccRCC has been identified, clinicians are faced with various treatment options ranging from surgical resection to non-surgical approaches such as cryoablation, radiofrequency ablation, and active surveillance, which are deemed to be particularly appropriate for patients of older age, harboring a single kidney, or those who have comorbidities and/or are reluctant to undergo a major surgery [[5], [6], [7]]. These clinical guidelines on the management of renal masses have been accepted by the American Urological Association, European Association of Urology, and National Comprehensive Cancer Network [[6], [7], [8]].
However, the tumor biology of clinical T1 stage ccRCC remains poorly understood. Approximately 30% of patients treated for localized ccRCC ultimately relapse, and 15% of these cases exhibit metastatic potential, which can potentially lead to death in certain cases [[8], [9], [10]]. Many factors have been reported to be associated with these inferior oncological outcomes, and several systems have been suggested for prognosis incorporating the tumor-node-metastasis (TNM) staging system with clinical and pathological features established by multiple institutions, including the Mayo clinic score, the University of California Los Angeles Integrated Staging System, and the Memorial Sloan Kettering Cancer Center (MSKCC) postoperative nomogram [[11], [12], [13], [14]]. However, some of the factors used in these prognostic models are time-dependent, such as tumor growth and changes in radiographic images, and other parameters such as tumor grade, tumor necrosis, and patient performance status are subject to inter-observer variability [8,15].
As the alternative, multigene assays have been shown to provide prognostic information beyond that possible with traditional approaches, and are beginning to be included in standard treatment guidelines for some tumors [15]. For example, previous studies on RCC discovered novel, prevalent genomic mutations of polybromo 1 (PBRM1), BRCA1 associated protein-1 (BAP1), SET domain-containing 2 (SETD2), and lysine-specific demethylase 5C (KDM5C) in patients with poor oncological outcomes [8,16]. Moreover, we reported that forkhead box protein C2 (FOXC2) and cytoskeleton-associated protein-glycine rich (CAP-Gly) domain-containing linker protein family member 4 (CLIP4) mutations were associated with clinical T1 stage ccRCC with synchronous metastasis [10]. Thus, the aim of the present study was to investigate whether these molecular biomarkers are associated with aggressive clinical T1 stage ccRCCs and develop a risk prediction tool to improve prognostic prediction in ccRCC and serve as a guide for future mechanistic research on this type of tumor.
2. Materials and Methods
2.1. Sample and Data Collection
For this retrospective study, we used data from 1069 patients with ccRCC (≤7 cm) who underwent radical and partial nephrectomy between January 2008 and December 2014 at our institution. The study protocol was approved by the Institutional Review Board of the Yonsei University Health System (project no: 4–2013-0742). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was not required for the purposes of this study, as the study was based on retrospective anonymous patient data and did not involve patient intervention or the use of human tissue samples.
Inclusion criteria were patients with ccRCC (≤7 cm) treated with nephrectomy alone and with available formalin-fixed paraffin-embedded (FFPE) tumor tissue. Exclusion criteria were those that had received neoadjuvant or adjuvant systemic therapy to rule out any possible effect on the risk of recurrence, along with patients with a history of inherited von Hippel-Lindau disease or synchronous or metachronous bilateral RCC because they might not be representative of sporadic ccRCC. Moreover, cases in which no or very little tumor (<5% of the area occupied by invasive cancer cells) tissue was available, or insufficient RNA (<1000 ng) or inadequate RNA quality measured by standard methods for quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis were excluded from analysis. In addition, cases that recurred within 6 months after the surgery in the absence of adequate imaging were excluded to rule out the possibility of undetected metastasis.
Data on clinical features [age, sex, height, weight, body mass index (BMI), tumor size, presence of metastasis] were recorded for each patient. The diameters of the primary tumors were obtained from the imaging modalities. Data on histological subtype was assessed according to the 2004 WHO Renal Neoplasms guidelines [17] and data on Fuhrman grade, invasion (perinephric/sinus fat or microscopic vascular invasion), and lymph node involvement were assessed according to the 2010 American Joint Committee on Cancer system [18]. Grading was based on a standardized four-tier system [19]. An aggressive tumor was defined as a tumor exhibiting synchronous metastasis, recurrence, or cancer-specific death, and synchronous metastasis was defined as metastasis detected at or within three months of the primary RCC diagnosis [20].
2.2. Sample Preparation
FFPE sections from ccRCC patients were obtained from the archives of the Department of Pathology at the Yonsei University College of Medicine (Seoul, Korea). All cases were reviewed and classified by a urologic pathologist (N.H.C.). Non-tumor elements were identified based on hematoxylin and eosin-stained slides by a urologic pathologist (N.H.C) after manual microdissection before being transferred to the extraction tube.
2.3. qRT-PCR
Total RNA was extracted from microdissected FFPE samples using TRIzol® reagent (Ambion, Life technologies, USA), and 1 μg of total RNA was reverse-transcribed into first-strand cDNA using a iNtRon Maxime RT PreMix (Intronbio. Cat No. 25081) according to the manufacturer's protocol. qRT-PCR was performed with Power SYBR® Green Master Mix (Thermo Fisher, Cat No. A25742, USA) in a 10-μl reaction volume comprising 5 μl of SYBR® Green master PCR mix, 1 μl each forward and reverse primers (10 pmol), 1 μl of diluted cDNA template, and sterile distilled water. Conditions for the amplification of genes were as follows: initial denaturation at 95 °C for 10 min; 45 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 60 s, and elongation at 72 °C for 60 s; and final elongation at 72 °C for 5 min. qRT-PCR was performed on the ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). All quantifications were performed with GAPDH as a reference gene for standardization of relative expression levels. The PCR primer sequences were as follows: FOXC2, 5′-GAT CAC CTT GAA CGG CAT CT-3′ (sense) and 5′-ACC TTG ACG AAG CAC TCG TT-3′ (antisense); CLIP4, 5′-GCA TCA TGC CAG GAA ATT CT-3′ (sense) and 5′-TTT GTT GGA CCT GAG GAA CC-3′ (antisense); PBRM1, 5′-TGA TGG CCA ACA AGT ACC AA-3′ (sense) and 5′-AGA TCA AAG ACT CCG GCT CA-3′ (antisense); BAP1, 5′-GCC TGA GGA GTC CAA GTC AG-3′ (sense) and 5′-CTG GAG GCT TCA CCA CTA GC-3′ (antisense); SETD2, 5′-TCA CAA GGC AGA CTC AGT GG -3′ (sense) and 5′-CTG CTG TCT TGG GCT TTT TC-3′ (antisense); KDM5C, 5′-GTC ATT TGC AAC CCC TGA GT-3′ (sense) and 5′-AAT GGG ATG AGG GGT AAA GG-3′ (antisense); GAPDH, 5′-CAG CCT CAA GAT CAT CAG CA-3′ (sense) and 5′-GGT GCT AAG CAG TTG GTG GT-3′ (antisense). Relative gene expression was analyzed using the 2–ΔΔCT method, and the results are expressed as the extent of change with respect to control values. qRT-PCR experiments were replicated at least three times. qRT-PCR analysis was performed by an investigator who was masked to the clinical data.
2.4. Immunohistochemistry
We have performed immunochemistry for the genes that show significant differences between aggressive and non-aggressive ccRCC. Rabbit anti-human FOXC2 (1:100 dilution; Thermo Fisher Scientific), rabbit anti-human BAF1 (1:2000 dilution; Abcam), and rabbit anti-human BAP1 (1:200 dilution; Abcam) were used for immunohistochemical investigations. FFPE specimens were cut into 4-μm-thick paraffin sections and were placed on Superfrost Plus microscope slides (Thermo Fisher Scientific). Slides were immunostained with Benchmark automated system (Ventana Medical System, Tucson, Arizona, USA). Sections were deparaffinized with EZ Prep (Ventana) for 8 min at 75 °C. Antigen retrieval was performed in Cell Conditioning Solution (high pH CC1 standard) for 60 min at 100 °C. The DAB inhibitor (3% H2O2 endogenous peroxidase) was blocked for 4 min at 37 °C. The slides were incubated with the respective primary antibodies for 32 min at 37 °C, followed by incubation with the secondary antibody (Universal HRP Multimer) for 8 min at 37 °C. Subsequently, the slides were treated with the DAB + H2O2 substrate for 8 min followed by hematoxylin II and the bluing reagent counterstain at 37 °C. The reaction buffer (pH 7.6 Tris buffer) was used as a wash solution. Finally, the slides were evaluated by light microscopy at 100× to 400× to document the staining intensity and categorized tumors as positive-expression for presence of PBRM1 and BAP1 nuclear tissue staining and FOXC2 cytoplasm tissue staining and negative-expression for absence of PBRM1 and BAP1 nuclear tissue staining and FOXC2 cytoplasm tissue staining, respectively.
2.5. Outcomes
The primary outcome of interest was to predict aggressive clinical T1 stage ccRCC (≤7 cm) that exhibit (1) synchronous distant metastasis by imaging, (2) recurrence (local or distant metastases identified by imaging, biopsy, or physical examination), or (3) cancer-specific death.
2.6. Statistical Analyses
The results are reported as the mean ± standard deviation for continuous variables and as a percentage for categorical variables. For the univariate analysis, the t-test was used to compare continuous variables. Multivariate analysis was based on logistic regression, including all risk factors that were significantly associated with clinical outcome in the univariate analysis. According to the univariate results, we developed a multiple logistic regression model that included gene expression levels of FOXC2, PBRM1, and BAP1. Moreover, we have added immunohistochemical staining of FOXC2, PBRM1, and BAP1 in addition to gene expression levels of FOXC2, PBRM1, and BAP1. Diagnostic indicators such as accuracy, and area under the receiver operating characteristic curve (AUC) were evaluated with the same validation group to assess the performance of the deep-learning model. SPSS software version 23.0 (IBM Corp., Armonk, NY) was used for all statistical analyses. All statistical tests were two-tailed, and a P value <.05 was considered statistically significant.
2.7. Deep Learning
All variables, including FOXC2, PBRM1, and BAP1 expression levels, used for development of the logistic regression model were normalized, and each value was changed into a range of variables from “0” to “1” using the following equation: {Zi = [xi − min(x)]/[max(x) − min(x)]}. The dataset was divided randomly into two independent training and validation groups to test for internal validation. The training group, comprising 70% of the dataset (123 subjects, including 28 with aggressive ccRCC), was used to construct the prediction models. The validation group, comprising 30% of the dataset (54 subjects, including 12 with aggressive ccRCC), was used to assess the performance of the model for aggressive ccRCC prediction. Receiver operating characteristic curves and AUC analyses were executed to verify the performance of each prediction model for aggressive ccRCC.
The main algorithms conventionally used for deep-learning approaches are deep neural networks (DNN), deep convolutional neural networks, deep belief networks, and recurrent neural networks. We selected DNN using the python library Keras (version 2.2.0) with TensorFlow (version 1.8.0) backend. The scikit-learn library (http://scikitlearn.org/) was used for data management and preprocessing. In this study, we used a two-layer DNN network with a 30% dropout rate to handle the overfitting problem. The models were optimized using the Adam optimizer with a loss function of binary cross entropy. Neuron activation functions were set as sigmoid for the first layer and as rectified linear unit for the second layer. We selected 500 epochs and a batch size of 30 for the DNN model.
3. Results
The clinical and pathological characteristics of the study population (n = 177) are listed in Table 1. The majority of the patients were male (71.8%). A total of 40 patients (22.6%) were defined as having aggressive ccRCC based on the presence of synchronous metastasis, recurrence, or cancer-specific death.
Table 1.
Baseline characteristics of patients with clinical T1 stage clear cell renal cell cancers (ccRCC) (≤7-cm) included in this study.
| Clinical T1 stage ccRCC (≤7-cm) (n = 177) | |
|---|---|
| Gender, n (%) | |
| Male | 127 (71.8%) |
| Female | 50 (28.2%) |
| Age (years) | 58.5 ± 11.7 |
| BMI (kg/m2) | 24.8 ± 3.6 |
| Radical surgery, n (%) | 94 (53.1%) |
| Tumor size (cm) | 4.1 ± 1.6 |
| Fuhrman grade | |
| 1 | 8 (4.5%) |
| 2 | 79 (44.6%) |
| 3 | 81 (45.8%) |
| 4 | 9 (5.1%) |
| Invasion (perinephric/sinus fat/vascular), n (%) | 11 (6.2%) |
| Positive nodal status, n (%) | 1 (1.0%) |
| Synchronous metastasis, n (%) | 19 (10.7%) |
| Recurrences, n (%) | 23 (13.0%) |
| Cancer-specific death, n (%) | 30 (16.9%) |
| Gene expressions | |
| FOXC2 | 0.0065 ± 0.0152 |
| CLIP4 | 0.0077 ± 0.0152 |
| PBRM1 | 0.0041 ± 0.0111 |
| SETD2 | 0.0004 ± 0.0006 |
| BAP1 | 0.0002 ± 0.0005 |
| KDM5C | 0.0027 ± 0.0083 |
Data are shown as mean ± SD or number of subjects (%)
BMI, body mass index
As shown in Table 2, there were no differences between patients with and without aggressive characteristics of ccRCC according to gender; age; BMI; tumor size, invasion to the perinephric, sinus fat, or vessel; and positive nodal status. However, patients with aggressive ccRCC had a significantly higher rate of radical surgery than those without aggressive characteristics. Aggressive tumors also showed significantly higher percentages of higher Fuhrman grade (3–4) both in univariate and multivariate analysis. With regards to gene expression, FOXC2, CLIP4, PBRM1, and BAP1 were expressed at significantly lower levels in aggressive ccRCC than non-aggressive ccRCC in the univariate analysis. However, in multivariate analysis, only FOXC2, PBRM1, and BAP1 were still independently significantly associated with the aggressiveness of ccRCC. Immunohistochemical staining of FOXC2, PBRM1, and BAP1 were significantly lower in aggressive ccRCC, both in univariate and multivariate analysis. Among aggressive RCC patients, there was no significant difference between whether it was performed with radical or partial nephrectomy (p = 0.816) in recurrence (23 patients). Similarly, there was also no significant difference between radical or partial nephrectomy in cancer-specific death (30 patients) (p = 0.838).
Table 2.
Comparison of clinical T1 stage clear cell renal cell cancers (ccRCC) (≤7-cm) with or without aggressive characteristics (metastasis, recurrence, or cancer-specific death).
| RCC with aggressive characteristics (n = 40) | RCC without aggressive characteristics (n = 137) | Pa | Pb | |
|---|---|---|---|---|
| Gender, n (%) | ||||
| Male | 26 (65.0%) | 101 (73.7%) | 0.281 | |
| Female | 14 (35.0%) | 36 (26.3%) | ||
| Age (years) | 58.0 ± 11.3 | 58.7 ± 11.8 | 0.755 | |
| BMI (kg/m2) | 25.2 ± 4.2 | 24.8 ± 3.4 | 0.520 | |
| Radical surgery, n (%) | 29 (72.5%) | 65 (47.4%) | 0.005 | 0.053 |
| Tumor size (cm) | 4.4 ± 1.5 | 4.1 ± 1.7 | 0.241 | |
| Fuhrman grade | ||||
| 1 | 1 (2.5%) | 7 (5.1%) | 0.023 | |
| 2 | 12 (30.0%) | 67 (48.9%) | ||
| 3 | 22 (55.0%) | 59 (43.1%) | ||
| 4 | 5 (12.5%) | 4 (2.9%) | ||
| 3–4, n (%) vs 1–2 | 27 (67.5%) | 63 (46.0%) | 0.017 | 0.015 |
| Invasion (perinephric/sinus fat/vascular), n (%) | 1 (2.5%) | 10 (7.3%) | 0.269 | |
| Positive nodal status, n (%) | 1 (2.5%) | 0 (0.0%) | 0.063 | |
| Gene expressions | ||||
| FOXC2 | 0.0033 ± 0.0057 | 0.0074 ± 0.0169 | 0.018 | 0.031 |
| CLIP4 | 0.0042 ± 0.0055 | 0.0088 ± 0.0169 | 0.008 | 0.781 |
| PBRM1 | 0.0008 ± 0.0014 | 0.0051 ± 0.0124 | <0.001 | 0.035 |
| SETD2 | 0.0003 ± 0.0003 | 0.0004 ± 0.0007 | 0.092 | |
| BAP1 | 0.0001 ± 0.0001 | 0.0002 ± 0.0005 | 0.004 | 0.049 |
| KDM5C | 0.0016 ± 0.0015 | 0.0030 ± 0.0094 | 0.350 | |
| Immunohistochemical staining | ||||
| FOXC2 (positive), n (%) | 24 (60.0%) | 110 (80.3%) | 0.008 | 0.011 |
| PBRM1 (positive), n (%) | 10 (25.0%) | 83 (60.6%) | <0.001 | 0.009 |
| BAP1 (positive), n (%) | 25 (62.5%) | 123 (89.8%) | <0.001 | 0.037 |
Data are shown as mean ± SD or number of subjects (%)
BMI body mass index
P-value calculated using t-test for continuous variables and chi-square test for categorical variables
P-value calculated using logistic regression for multivariate analysis
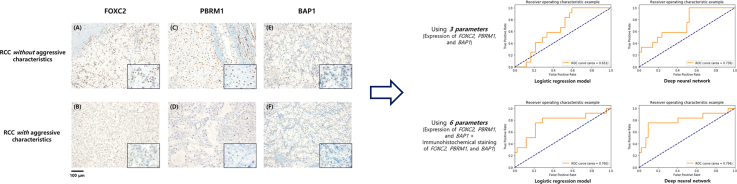
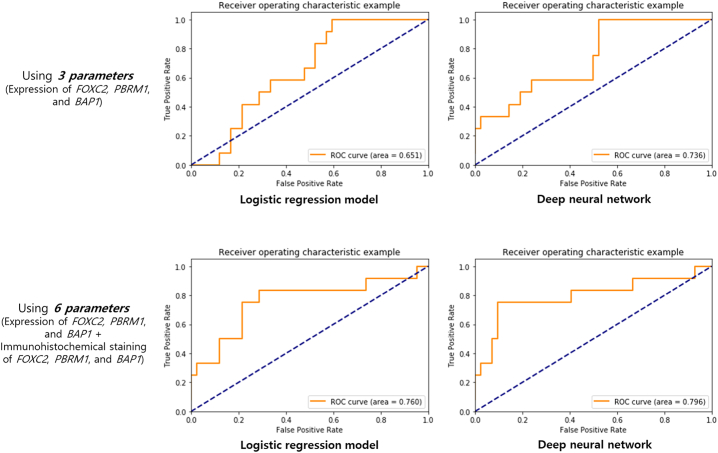
Table 3 summarizes the performance of the logistic regression model and DNN model derived from the expression levels of FOXC2, PBRM1, and BAP1. Using only these three genes, both the logistic regression and DNN models could effectively predict aggressive ccRCC with accuracy of 0.555 and 0.537, respectively. The AUC of the logistic regression and DNN models also showed good predictive power at 0.651 and 0.736, respectively (Fig. 1). Furthermore, performance of the logistic regression model and DNN model using immunohistochemical staining of FOXC2, PBRM1, and BAP1 in addition to expression levels of FOXC2, PBRM1, and BAP1 was also shown in Table 3. Using 6 parameters, the accuracy and AUC increased to 0.759 and 0.852, and 0.760 and 0.796 in logistic regression and DNN models, respectively.
Table 3.
Performance of prediction models of aggressive clear cell renal cell carcinoma.
| Logistic regression model | Deep neural network model | ||
|---|---|---|---|
| Using 3 parameters (Expression of FOXC2, PBRM1, and BAP1) | Accuracy | 0.555 | 0.537 |
| Area under the curve | 0.651 | 0.736 | |
| Using 6 parameters (Expression of FOXC2, PBRM1, and BAP1 + Immunohistochemical staining of FOXC2, PBRM1, and BAP1) | Accuracy | 0.759 | 0.852 |
| Area under the curve | 0.760 | 0.796 |
Fig. 1.
Receiver operating characteristic curve from the logistic regression model and deep neural network model for predicting aggressive clear cell carcinoma.
To test the combinatorial assessment of the expression of FOXC2, PBRM1, and BAP1, a multiple logistic regression model was constructed. We interpreted the effect of independent variables on the probability of detecting aggressive clinical T1 stage ccRCC by calculating the probability (Pr) for the aggressive group for different expression levels of the three genes. The regression equation was:
For the multiple logistic regression model that used 6 parameters, including expression of FOXC2, PBRM1, and BAP1 and immunohistochemical staining of FOXC2, PBRM1, and BAP1, the regression equation was:
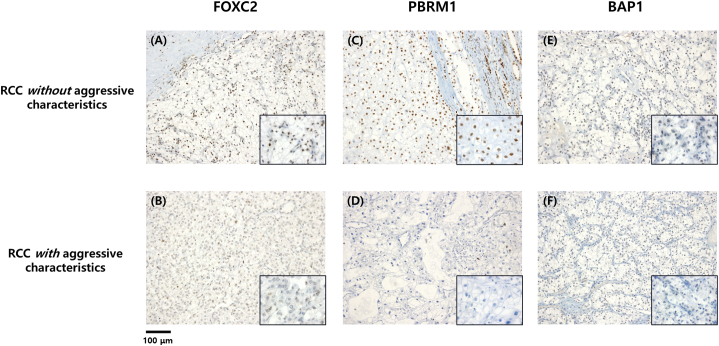
Consistently, representative immunohistochemistry stains of non-aggressive ccRCC demonstrated robust expression of FOXC2, PBRM1, and BAP1, while aggressive ccRCC samples were all negative for FOXC2, PBRM1, and BAP1 staining with significant difference both in univariate and multivariate analysis (Table 2 and Fig. 2).
Fig. 2.
Immunohistochemical findings of FOXC2, PBRM1, and BAP1 (A) Positive expression of FOXC2 (B) Negative expression of FOXC2 (C) Positive expression of PBRM1 (D) Negative expression of PBRM1 (E) Positive expression of BAP1 (F) Negative expression of BAP1, Scale bars indicate 100 μM (original magnification x100; inset x400).
4. Discussion
Through this study, we identified molecular biomarkers associated with aggressive clinical T1 stage ccRCC of ≤7-cm, which were used to propose an optimal, rapid, and inexpensive approach that could help stratify aggressive tumors among patients diagnosed with clinical T1 stage ccRCC. To our knowledge, this is the first prediction model developed for aggressive clinical T1 stage ccRCC using molecular biomarkers. This risk prediction tool can provide clinicians with accurate and evidence-based information to estimate the malignant potential of each case, which would help guide the management of clinical T1 stage ccRCC.
ccRCC is the most common subtype of RCC, and most patients who present with clinical T1 stage ccRCC will have excellent oncological outcomes following resection or active surveillance. However, rare cases are significantly aggressive and can even be lethal once the cancer progresses to a metastatic state or the disease recurs. Currently, management of clinical T1 stage ccRCC depends on the surgeon's discretion and preference of the patients, based only on information related to predicting the malignant potential of the cancer. This has been a complex and at times challenging task owing to the lack of definite information about the malignant potential of ccRCC.
To ease this burden, several nomograms have been developed to predict recurrence and cancer-specific survival; however, these are based solely on clinical and pathological parameters, and are limited by the time-dependency of certain factors and inter-observer variability [[11], [12], [13], [14], [15]]. Thus, identification of biomarkers specific to ccRCC will not only provide more objective parameters for identifying ccRCC with aggressive characters but will also provide new insight into the molecular pathogenesis, which could lead to the development of novel targeted cancer therapies.
Few studies have attempted to uncover the fundamental genetic events leading to tumor initiation and cancer-specific outcomes in RCC to date [8,15,16]. Some reports have shown that loss of the predominant tumor suppressor VHL alone is insufficient and additional genetic events are required [21,22]; the chromosome 3p21 locus has been proposed as a candidate region for finding such additional tumor suppressors [23]. Indeed, a recent large-scale high-throughput sequencing study identified several recurrently mutated genes in ccRCC, PBRM1, SETD2, and BAP1, which are located in the frequently lost 3p21 locus and function in the epigenetic regulation of gene expression [16].
PBRM1, encoding a SWI/SNF chromatin-remodeling complex component, was also reported to be associated with pancreatic cancer [24]. In ccRCC, mutations of PBRM1 were detected in 32.5% (198/609) of the MSKCC cohort [16] and in 33.0% (67/203) of the sample analyzed by Brandon et al. [8], which included the MSKCC cohort along with data from three publicly available cohorts: The Caner Genome Atlas, University of Tokyo, and The International Cancer Genome Consortium. Analysis of the MSKCC cohort further showed a significant association of PBRM mutations with higher T stages and earlier invasion in smaller tumors, but there was no association detected with inferior clinical outcomes [16]. Thus, the authors suggested that inactivation of PBRM1 is likely associated with an early, essential event in kidney tumorigenesis.
BAP1 has been reported to be associated with uveal melanoma and mesothelioma [25,26]. BAP1 mutation in uveal melanoma is associated with an aggressive subtype; however, in mesothelioma, BAP1 mutation is not associated with inferior clinical outcomes [27]. BAP1 was reported to be a critical gatekeeper for disease progression [28]. Peña-Llopis et al. [29] reported the mutually exclusive characteristics of PBRM1 and BAP1 mutations, in which BAP1-mutant tumors were associated with worse survival and a higher Fuhrman grade than PBRM1-mutant tumors [28]. This finding was supported by the MSKCC cohort study, which indicated an association of BAP1 mutations with poor prognostic factors such as a higher T stage, higher nuclear grade, large size, more necrosis, and the presence of metastatic disease at presentation [16].
Based on this background, PBRM1 mutations, which do not impact clinical outcome, appear to play a principal role in tumor initiation, while BAP1 mutations are more strongly associated with worse oncological outcomes that likely occur during disease progression. This can explain why PBRM1 and BAP1 emerged as biomarkers for predicting aggressive ccRCC. Since the tumor size in our study was small in all cases, PBRM1 showed a significant association. Moreover, BAP1 emerged as a significant factor in our model to predict ccRCC with aggressive characteristics, in line with its reported association with disease progression and a worse prognosis. FOXC2, which was also identified as a biomarker of synchronous ccRCC in our previous study [10], was found to be downregulated in aggressive ccRCC in the present study. FOXC2 upregulation has been associated with cancer metastasis and epithelial-mesenchymal transition [30]; however, Hader et al. [31] recently reported that FOXC2 upregulation acts as a checkpoint to inhibit epithelial cell dedifferentiation and/or to activate epithelial cell redifferentiation during kidney repair. This finding is consistent with a study by Bard et al. [32] that identified FOXC2 along with several other transcription factors as candidate regulators of the mesenchyme-epithelium transition. Other genes such as SETD2 and KDM5C that have previously been reported to be associated in the development of ccRCC were not associated with aggressive features in our study.
Although previous studies have focused on identification mutation profiles of ccRCC, our model is more practical and suitable for clinical settings since we used qRT-PCR for determining gene expression levels. Performing next-generation sequencing on multiple tumor samples for each patient in a clinical setting is unrealistic due to cost and time limitations. Moreover, the tumor-associated mutations identified with such high-throughput methods are not all necessarily associated with clinical outcome, and changes in DNA are not always reflected by RNA expression [15]. Therefore, using qRT-PCR would not only be cost-effective but also could more readily provide a significant association with clinical outcomes. Adding immunohistochemical staining results in addition to expression levels by qRT-PCR increased accuracy and AUC significantly, however, immunohistochemistry requires time and cost although we tried to simplify the evaluation process of immunohistochemical staining. Therefore, we believe that whether to use immunohistochemical staining included models which have higher accuracy and AUC or to use only RNA expression models which are more cost-effective would depend on the choice of clinicians.
In this study, we examined the potential association of molecular markers with aggressive tumors in clinical T1 stage ccRCC from post-operative FFPE tissues. Thus, this risk assessment tool would allow for stratifying patients with clinical T1 stage ccRCC who underwent surgery for their follow-up schedule; that is, patients with more aggressive features should be more closely monitored in postoperative follow-up. Ultimately, preoperative risk stratification would be beneficial given the strong association of these genes with adverse oncological outcomes, which would permit determining the expression status in clinical T1 stage ccRCC and then could guide treatment planning accordingly, such as whether to perform partial or radical nephrectomy or ablation, or close observation only.
Despite these advantages, there are several limitations of this study that should be mentioned. First, there the intratumoral heterogeneity in ccRCC tumors can be substantial [33], even in such relatively small localized tumors [8]; however, this should be considered as a tradeoff for model development and discovery, and is a worthy challenge that should be addressed in optimizing this tool in future studies. Second, owing to the limitation of the retrospective study and significantly small number of cases of aggressive ccRCC, the sample size was small and we used cutoff of size 7 cm. It would be more interesting to focus on small renal mass of size <4 cm and intermediate grade tumors (Fuhrman grade 2 and 3) and prospective clinical trial would focus on these cases. Third, as mentioned above, this study was based only on postoperative FFPE samples. To better guide treatment planning such as whether to perform surgery or ablation, or close observation only, a preoperative molecular risk prediction model should be developed based on frozen tissues from renal mass biopsy samples. Since ae prospective clinical trial is now underway (ClinicalTrials.gov Identifier: NCT03694912), the validation of our models will be reported in the near future, including mutation profiles of the selected genes, FOXC2, PBRM1, and BAP1, for aggressive ccRCC.
5. Conclusion
Patients diagnosed with clinical T1 stage ccRCC with aggressive tumors, defined as those who exhibit synchronous metastasis, recurrence, or cancer-specific death, showed significantly lower expression levels of FOXC2, PBRM1, and BAP1. Using these molecular biomarkers, we developed risk prediction models that could assist in stratifying patients with clinical T1 stage ccRCC. Moreover, identification of these biomarkers will provide further guidance for basic and clinical studies designed to better understand the pathogenesis of aggressive clinical T1 stage ccRCC.
Conflict of Interest
None.
Acknowledgements
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI17C1095).
References
- 1.Lipworth L., Tarone R.E., McLaughlin J.K. The epidemiology of renal cell carcinoma. J Urol. 2006;176:2353–2358. doi: 10.1016/j.juro.2006.07.130. [DOI] [PubMed] [Google Scholar]
- 2.Znaor A., Lortet-Tieulent J., Laversanne M. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67:519–530. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Pantuck A.J., Zisman A., Belldgrun A.S. The changing natural history of renal cell carcinoma. J Urol. 2001;166:1611–1623. [PubMed] [Google Scholar]
- 4.Kane C.J., Mallin K., Ritchey J. Renal cell cancer stage migration: analysis of the National Cancer Data Base. Cancer. 2008;113:78–83. doi: 10.1002/cncr.23518. [DOI] [PubMed] [Google Scholar]
- 5.Smaldone M.C., Corcoran A.T., Uzzo R.G. Active surveillance of small renal masses. Nat Rev Urol. 2013;10:266–274. doi: 10.1038/nrurol.2013.62. [DOI] [PubMed] [Google Scholar]
- 6.Campbell S.C., Novick A.C., Belldegrun A. Guideline for management of the clinical T1 renal mass. J Urol. 2009;182:1271–1279. doi: 10.1016/j.juro.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Motzer R.J., Jonasch E., Agarwal N. Kidney Cancer, version 2.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15:804–834. doi: 10.6004/jnccn.2017.0100. [DOI] [PubMed] [Google Scholar]
- 8.Manley B.J., Reznik E., Ghanaat M. Characterizing recurrent and lethal small renal masses in clear cell renal cell carcinoma using recurrent somatic mutations. Urol Oncol. 2017;17:30549–30555. doi: 10.1016/j.urolonc.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rini B.I., Campbell S.C., Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 10.Ahn J., Han K.S., Heo J.H. FOXC2 and CLIP4: a potential biomarker for synchronous metastasis of ≤7-cm clear cell renal cell carcinomas. Oncotarget. 2016;7:51423–51434. doi: 10.18632/oncotarget.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank I., Blute M.L., Cheville J.C. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 12.Leibovich B.C., Blute M.L., Cheville J.C. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;97:1663–1671. doi: 10.1002/cncr.11234. [DOI] [PubMed] [Google Scholar]
- 13.Zisman A., Pantuck A.J., Dorey F. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19:1649–1657. doi: 10.1200/JCO.2001.19.6.1649. [DOI] [PubMed] [Google Scholar]
- 14.Sorbellini M., Kattan M.W., Snyder M.E. A postoperative prognostic nomogram predicting recurrence for patients with conventional clear cell renal cell carcinoma. J Urol. 2005;173:48–51. doi: 10.1097/01.ju.0000148261.19532.2c. [DOI] [PubMed] [Google Scholar]
- 15.Rini B., Goddard A., Knezevic D. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: development and validation studies. Lancet Oncol. 2015;16:676–685. doi: 10.1016/S1470-2045(15)70167-1. [DOI] [PubMed] [Google Scholar]
- 16.Hakimi A.A., Ostrovnaya I., Reva B. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–3267. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eble J.N., Sauter G., Epstein J. IARC Press; Lyon: 2004. World Health Organization classification of tumors. Pathology and genetics of tumours of the urinary system and male genital organs. [Google Scholar]
- 18.Edge S.B., Compton C.C. The American joint committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrman S.A., Lasky L.C., Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Ingimarsson JP1, Sigurdsson M.I., Hardarson S. The impact of tumour size on the probability of synchronous metastasis and survival in renal cell carcinoma patients: a population-based study. BMC Urol. 2014;14:72. doi: 10.1186/1471-2490-14-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandriota S.J., Turner K.J., Davies D.R. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 22.Rankin E.B., Tomaszewski J.E., Haase V.H. Renal cyst development in mice with conditional inactivation of the von Hippel–Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clifford S.C., Prowse A.H., Affara N.A. Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes Chromosomes Cancer. 1998;22:200–209. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Shain A.H., Giacomini C.P., Matsukuma K. Convergent structural alterations define SWItch/sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109:E252–E259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bott M., Brevet M., Taylor B.S. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harbour J.W., Onken M.D., Roberson E.D. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zauderer M.G., Bott M., McMillan R. Clinical characteristics of patients with malignant pleural mesothelioma harboring somatic BAP1 mutations. J Thorac Oncol. 2013;8:1430–1433. doi: 10.1097/JTO.0b013e31829e7ef9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapur P., Peña-Llopis S., Christie A. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peña-Llopis S., Vega-Rubín-de-Celis S., Liao A. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mani S.A., Yang J., Brooks M. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hader C., Marlier A., Cantley L. Mesenchymal-epithelial response to injury: the role of Foxc2. Oncogene. 2010;29:1031–1040. doi: 10.1038/onc.2009.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bard J.B., Lam M.S., Aitken S. A bioinformatics approach for identifying candidate transcriptional regulators of mesenchyme-to-epithelium transitions in mouse embryos. Dev Dyn. 2008;237:2748–2754. doi: 10.1002/dvdy.21652. [DOI] [PubMed] [Google Scholar]
- 33.Gerlinger M., Rowan A.J., Horswell S. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]