Biofilm formation is a critical determinant in the pathopoiesis of Pseudomonas aeruginosa. It could significantly increase bacterial resistance to drugs and host defense.
KEYWORDS: β-propeller, Pseudomonas aeruginosa, biofilm formation, new drug target, virulence
ABSTRACT
Biofilm formation is a critical determinant in the pathopoiesis of Pseudomonas aeruginosa. It could significantly increase bacterial resistance to drugs and host defense. Thus, inhibition of biofilm matrix production could be regarded as a promising attempt to prevent colonization of P. aeruginosa and the subsequent infection. PpgL, a periplasmic gluconolactonase, has been reported to be involved in P. aeruginosa quorum-sensing (QS) system regulation. However, the detailed function and catalysis mechanism remain elusive. Here, the crystal structure of PpgL is described in the current study, along with biochemical analysis, revealing that PpgL is a typical β-propeller enzyme with unique metal-independent lactone hydrolysis activity. Consequently, comparative analysis of seven-bladed propeller lactone-catalyzing enzymes and mutagenesis studies identify the critical sites which contribute to the diverse catalytic and substrate recognition functions. In addition, the reduced biofilm formation and attenuated invasion phenotype resulting from deletion of ppgL confirm the importance of PpgL in P. aeruginosa pathogenesis. These results suggest that PpgL is a potential target for developing new agents against the diseases caused by P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa is a well-known nosocomial pathogen that can cause various acute and chronic infections in humans, especially among patients with compromised immunity (1). Owing to its ability to produce biofilm on various surfaces, P. aeruginosa infects a broad range of host organisms and develops extreme resistance to antibiotic treatment and immune responses (2–4). In this sense, targeting biofilm biogenesis may provide an alternative to traditional antibiotic therapy against P. aeruginosa infection, indicating the need for exploring the function and mechanism of factors that affect biofilm formation (5, 6).
Many pathogenic bacteria utilize quorum sensing (QS) to coordinate virulence factor production and biofilm biosynthesis (7). P. aeruginosa relies on multiple sets of connected QS systems, including las, iqs, pqs, and rhl, to respond to environmental stress and to control virulence gene expression (8–11). Previous study has demonstrated that the PA4204 gene in P. aeruginosa is crucial for normal growth and fitness, and deletion of PA4204 dramatically reduced the production of QS signal molecules, including N-acyl homoserine lactones and pseudomonas quinolone signal (PQS) (12). Similar phenotypes and increased swarming motility and susceptibility to hydrogen peroxide were observed in Pseudomonas syringae by deleting its PA4204 homolog, Psyr_1712 (13). These reports suggest that PA4204 might play an important role in P. aeruginosa biofilm formation.
The protein encoded by PA4204 is located at the periplasm and has gluconolactonase activity and thus was named PpgL (12). PpgL was predicted to belong to the β-propeller fold family, which is characterized by a regular compact structure and diverse cellular functions (14, 15). Importantly, many β-propeller domain-containing proteins are potent drug targets because of their suitable size, shape, multiple binding sites, and high affinity to small molecules (16). Therefore, investigating the structural and functional properties of PpgL would provide significant guidance for understanding its physiological roles and potential applications.
In this study, we present the crystal structure of PpgL and reveal its catalysis mechanism on a structural basis. The biochemical analysis suggests that PpgL adopts a unique active site in comparison to other metal-dependent β-propeller type lactonases (17–20). Moreover, functional studies verify the regulatory roles of PpgL in bacterial biofilm formation and surface attachment, suggesting that it may be a suitable target for developing new anti-infective agents against P. aeruginosa.
RESULTS
Pseudomonas aeruginosa PpgL adopts a typical seven-bladed β-propeller fold.
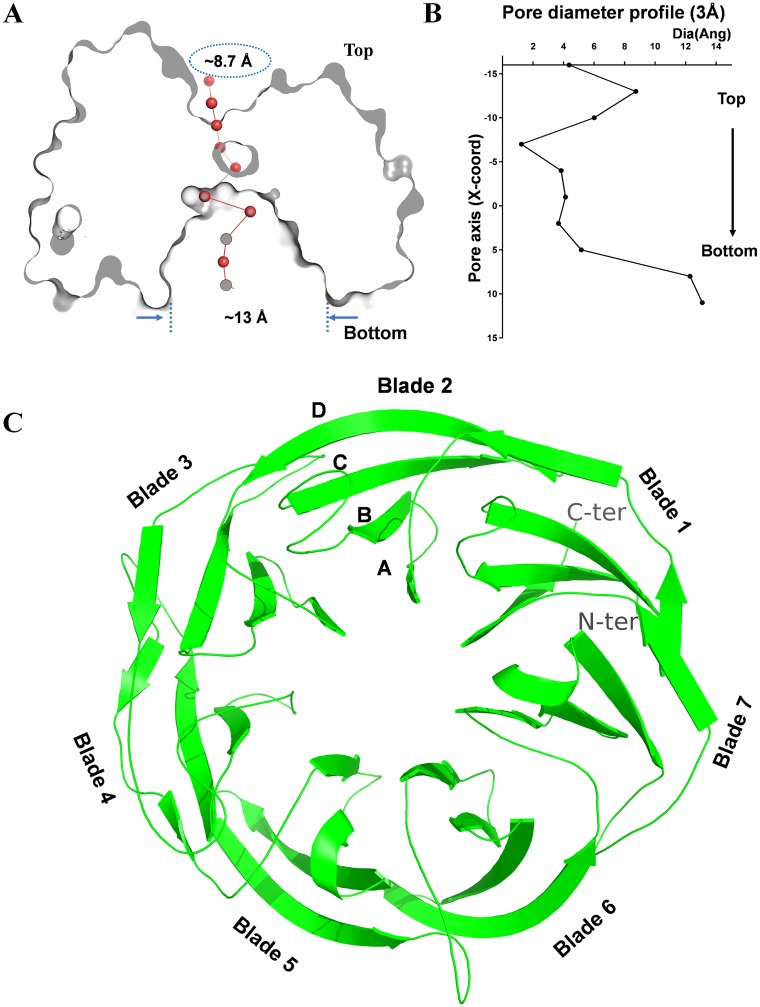
Recombinant PpgL was purified with high purity (see Fig. S1A and B in the supplemental material) and crystallized in space group P 41 21 2. The structure was solved by molecular replacement and refined to 1.65-Å resolution; the detailed crystallographic and refinement statistics are shown in Table S1. There are two molecules in one asymmetric unit, and each molecule presents a “doughnut” shape with a diameter of 57.2 Å and a height of 41.7 Å (Fig. 1A and B). PpgL resembles a typical β-propeller fold consisting of seven antiparallel, highly twisted, four-stranded β-meanders. These blade-shape β-meanders are arranged in a circular fashion around a central pseudorotation axis and eventually close into a ring via a 3 + 1 arrangement of β strands from the first and last blades (Fig. 1C). Many β-propeller proteins display a central channel; the narrow top is the entrance to the active site, while the wider bottom is referred to as the exit port (21). The central pore of PpgL has a funnel shape with a smaller charged pocket that is 8.7 Å wide and 7.1 Å deep on the “top face” and a larger bottom cavity that is 13.1 Å wide and 12.1 Å deep (Fig. 1A and B). The minimum diameter of the channel is only 1.2 Å and filled with ordered waters, making it impossible for large molecules to pass through.
FIG 1.
Overall structure of PpgL from P. aeruginosa. (A) View rotated 90° about the horizontal axis. The structure is sliced through the center to highlight the depression and the tunnel located on the top and bottom face of PpgL, respectively. (B) Pore diameter profile at 3-Å steps. (C) Ribbon diagram of PpgL viewed down the central axis. As observed in the 1.65-Å structure, the blades are numbered 1 to 7 with the 3 + 1 arrangement of β strands from the first and last blades: the C-terminal strand and the N-terminal part of the 7th blade are indicated. Strands in each blade are labeled A to D.
Despite the overall rigidity of the core structure, the loops on the top and bottom surfaces display a high degree of variation among β-propeller proteins, especially for those with six and seven blades, providing a structural basis for protein interaction or ligand trapping (15, 21). In PpgL, the loops on the top side demonstrate a larger accessible surface area (8,276.3 Å2) than that from the bottom side (7,410.3 Å2), indicating that the entry point of the substrate is from the top side and that the smaller pocket serves as the reaction center.
PpgL exhibits gluconolactonase activity in a metal-independent manner.
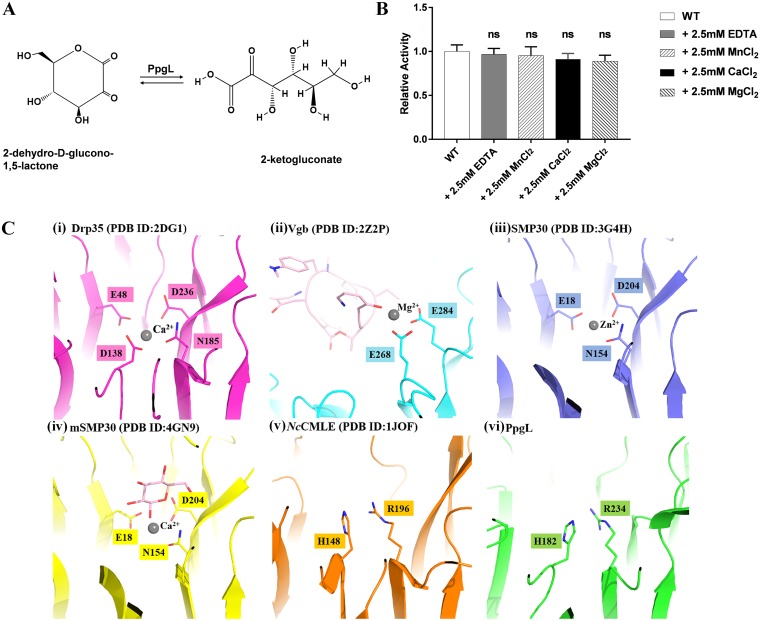
PpgL has been identified as a gluconolacotonase that may convert 2-dehydro-d-glucono-1,5-lactone into 2-ketogluconate and was implicated to be important for glucose metabolism in P. aeruginosa (Fig. 2A) (12). In most of the known β-propeller enzymes, including hydrolase, isomerase, and lyase, divalent metal cofactors are necessary for the activity by coordinating the substrate oxygen atoms and acting as a general acid to facilitate the hydrolysis (18–20, 22–28). In order to verify the metal dependence of PpgL, we examined the effects of EDTA and different divalent metal ions (2.5 mM MgCl2, CaCl2, and MnCl2) on its cleavage activity toward d-glucono-δ-lactone (PpgL was pretreated with 2.5 mM EDTA). As shown in Fig. 2B, the relative activities of PpgL of the control and other groups are not significantly different, revealing that PpgL has no metal dependence, in contrast to other β-propeller hydrolases.
FIG 2.
PpgL is a metal-independent gluconolactonase. (A) Hypothetical reaction that PpgL catalyzes in P. aeruginosa. (B) Relative activity of PpgL in the presence of 2.5 mM concentrations of different metal ions or EDTA. Activity was measured using d-glucono-δ-lactone as the substrate. Each bar represents the mean of three independent measurements (±SEM). ns, not significant. (C) The metal binding sites of different metal-dependent/-independent lactone-forming/-hydrolyzing enzymes, including Drp35 (PDB 2DG1) from Staphylococcus aureus (i), Vgb (PDB 2Z2P) from Staphylococcus aureus (ii), SMP30 (PDB 3G4H) from Homo sapiens (iii), mSMP30 (PDB 4GN9) from Mus musculus (iv), NcCMLE (PDB 1JOF) from Neurospora crassa (v), and PpgL (PDB 6IGB) (vi).
In strict metal-dependent lactone-forming/-hydrolyzing enzymes, the active site is always next to the metal binding site and composed of residues with carboxylic acid or carboxamide groups (Fig. 2C). In this mechanism, metal ions are directly involved in the hydrolytic reaction by stabilizing the protonated substrate intermediates. However, there is no equivalent metal-binding motif in P. aeruginosa PpgL; alternatively, two basic residues, His182 and Arg234, were found at the corresponding cation-binding region (Fig. 2C). Thus, the charged amino groups of the two residues could act as proton acceptors and functionally substitute for the metal ion. This explanation is also supported by the metal-tolerant Staphylococcus cohnii Vgb, a virginiamycin B lyase which catalyzes the linearization of the cyclic antibiotic (29). Importantly, a metal-free catalysis mechanism has been reported in Neurospora crassa 3-carboxy-cis,cis-muconate lactonizing enzyme (NcCMLE) (Fig. 2C) (17), providing evidence that the metal-independent catalysis is a unique mechanism in PpgL and suggesting that distinct active-site profiles account for the unforeseen diverse functions in propeller enzymes.
Comparisons with other seven-bladed propeller lactone-catalyzing enzymes.
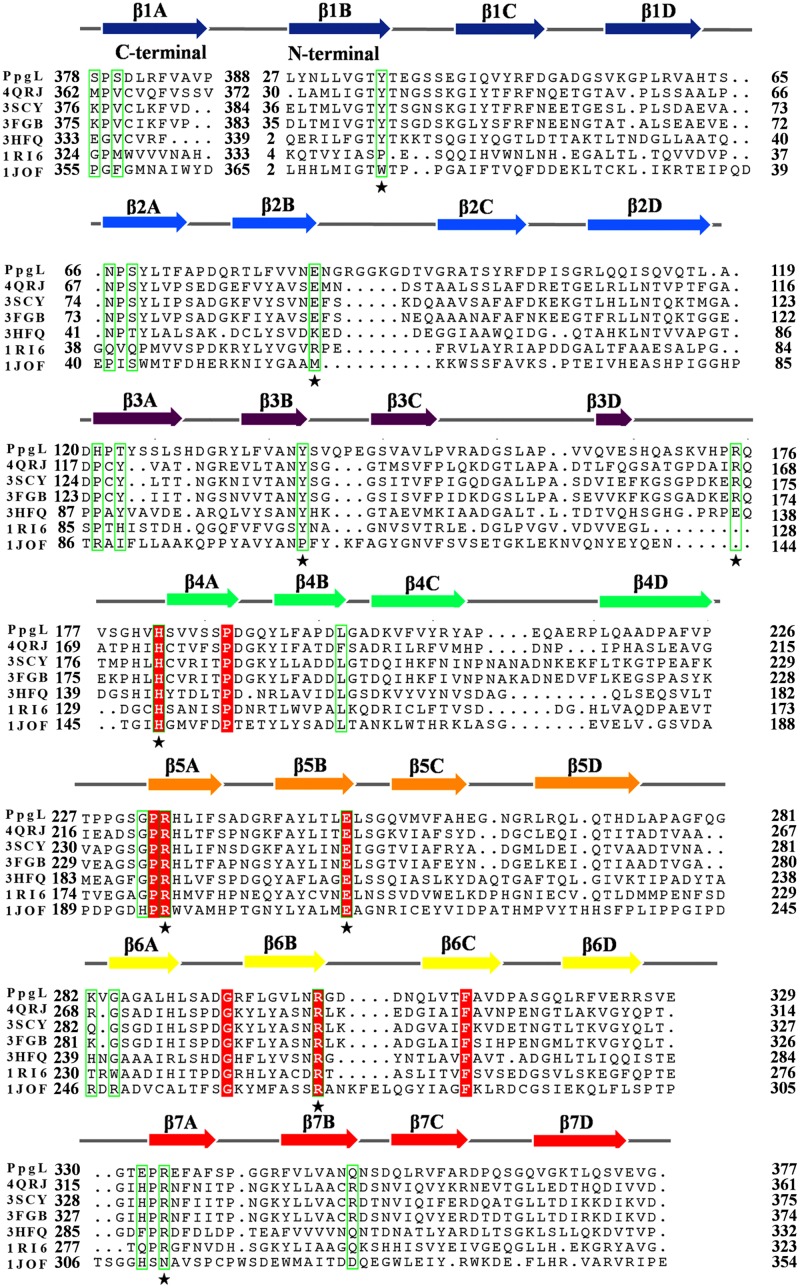
Both NcCMLE and PpgL belong to the seven-bladed β-propeller fold family and are rare examples of metal-independent lactone-catalyzing enzymes. To further elucidate their general features, especially the structural basis that determines the reaction preference, we chose five putative β-propeller isomerases or lactonases from the DALI server results (30) and aligned them with NcCMLE and PpgL. As shown in Fig. 3, despite the diverse sequence identities (15.24% to 36.94%), the chosen seven-bladed β-propeller proteins share an overall structure (root mean square deviation [RMSD], 1.389 to 2.065 Å). It is worth noting that residues corresponding to PpgL His182 and Arg234 are absolutely conserved among all of the sequences, implying that the metal-independent catalysis might be a common property in these β-propeller enzymes. Using the CAVER program (31), we explored the routes that connect the central tunnel to the extracellular top surface in each structure. The major accessible pockets are always positioned next to the two conserved histidine and arginine residues (Fig. S2). Thus, the structural and mechanistic similarities indicate a general substrate-binding pattern and similar active-site locations. Besides His182 and Arg234, there are two more charged residues, Glu250 and Arg302 (residue numbering is as in PpgL), that are fully conserved among the aligned sequences (Fig. 3). The equivalent residues in NcCMLE have been predicted to be critical for catalysis (17). Therefore, it is reasonable to believe that these four residues are essential for the functions of the aligned β-propeller proteins.
FIG 3.
Comparisons with other seven-bladed propellers. Sequence alignment of the following seven-bladed propeller lactone-catalyzing enzymes from bacterial species: BACUNI_04672 (PDB 4QRJ) from Bacteroides uniformis, BF1038 (PDB 3SCY) from Bacteroides fragilis, lp_2219 (PDB 3HFQ) from Lactobacillus plantarum, YbhE (PDB 1RI6) from Escherichia. coli, Q89ZH8 (PDB 3FGB) from Bacteroides thetaiotaomicron, and NcCMLE (PDB 1JOF) from Neurospora crassa. Completely conserved sites (red highlighting plus star) and 21 sites (green box) are indicated.
Conformational rearrangement of the loops and residues may have dramatic effects on the size and geometry of the orthostatic binding pockets in different β-propeller enzymes (21). Despite this, according to the structural comparison, we could find that residues involved in substrate binding and/or catalysis almost locate at the same sites among “blades”: two sites at the beginning of the A strand and one site at the end of the B strand (Fig. 3). Because of the local conformational and steric constraints imposed by the main chain configuration, the side chains in those positions point toward the reaction center. The high internal sequence symmetry of the seven-β-propeller architecture generates 21 sites for the diverse enzymatic activities (Fig. 3; Fig. S3). In PpgL, 14 out of the 21 sites have large side chains, providing candidate residues for the next analysis.
Substrate binding modeling and activity assay.
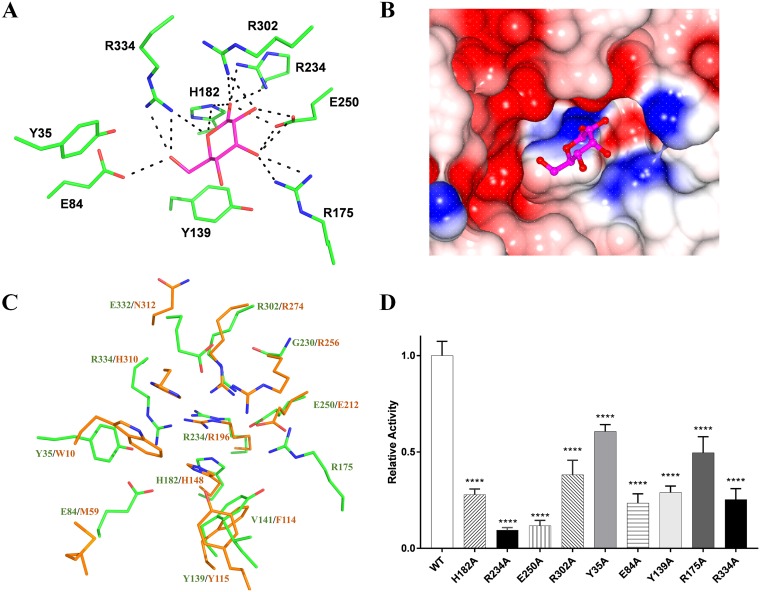
To further identify the critical residues in PpgL catalysis, we attempted to crystallize the complex of PpgL with substrates d-glucono-δ-lactone or (S)-5-oxo-2-tetrahydrofurancarboxylic acid, but all attempts failed. Thus, we docked the d-glucono-δ-lactone on the top shallow depression of PpgL by using the Autodock 4.0 tool (32). The generated poses were clustered according to their relative RMSD of CA atoms (RMSD, 2.0 Å tolerance); the lowest energy cluster with the most poses (39 out of 50) was selected to find the relatively reliable pose. The best-docked complex is presented graphically in Fig. 4A and B. As predicted above, His182 and Arg234 could coordinate the lactone’s carbonyl oxygen, while another two conserved residues, Glu250 and Arg302, also interact with the carbonyl oxygen atoms and determine the binding preference. In addition, the docking result elucidates more details regarding the catalytic mechanism: Arg175 stabilizes the substrate binding by forming a hydrogen bond with the hydroxyl oxygen; Tyr35, His182, Glu84, and Arg334 may function as the catalytic base and acid or the electrophile for reaction; and Tyr139 plays a critical role as a hydrophobic platform to stack with the lactone ring.
FIG 4.
The modeled d-glucono-δ-lactone-bound PpgL complex and the residues identified to be important to gluconolactonase activity. (A) The d-glucono-δ-lactone docked into the PpgL substrate binding pocket using the AutoDock suite represents NcCMLE, and the putative hydrogen bonds are shown with dashed lines. (B) The surface potential presentation of the active-site cleft with the substrate docked is shown. (C) Superposition of the PpgL and NcCMLE active sites. The PpgL (green) and CMLE (dark gold) active-site residues around the docked substrates are shown, indicating the different active pockets consisting of four conserved active sites and nonconserved residues. (D) Relative activities of wild-type and mutant PpgL. Activity was measured using the d-glucono-δ-lactone as mentioned before. Each bar represents the mean of three independent measurements (±SEM). ****, P < 0.0001.
PpgL catalyzes a reaction opposite to that of NcCMLE, providing a good example to study the relationship between the active-site architecture and the lactone bond-forming/-breaking properties. The four highly conserved residues (Arg302, Glu250, Arg234, and His182) of PpgL superpose perfectly with their equivalents in NcCMLE structures (Fig. 4C), supporting the idea that the two enzymes evolve from a common ancestor. Moreover, Trp10′, Phe114′, and Tyr115′ from NcCMLE can find similar hydrophobic counterparts in PpgL (Tyr35, Tyr139, and Val141), leaving the rest of the nonconserved residues (Glu84/Met59′, Arg334/His310′, Glu332/Asn312′, and Gly230/Arg256′) and the Arg175 from the extended loop of PpgL as the possible structural basis for explaining the different catalysis properties.
Thus, in the PpgL active site, nine resides (Tyr35, Glu84, Tyr139, Arg175, His182, Arg234, Glu250, Arg302, and Arg334) were identified and predicted to be crucial for its catalysis. In order to verify their functions, we introduced an Ala substitution on those sites. As Fig. 4D shows, Arg234Ala and Glu250Ala nearly eliminated the lactonase activity against d-glucono-δ-lactone, whereas mutations at two other highly conserved sites, His182 and Arg302, retained about 30% to 40% activity, suggesting that Arg234 and Glu250 have more effect on the reaction. As expected, significant activity reduction was also observed in Tyr139Ala, highlighting the importance of the C–H···π interaction between lactone and Tyr139. Consistent with the structural analysis, mutations in Glu84 and Arg334 obviously decreased PpgL activity to a degree similar to that observed with His182Ala, while the Tyr35Ala and Arg175Ala mutations resulted in relatively less reduction.
PpgL is associated with P. aeruginosa biofilm formation and invasion.
It is well known that the QS systems regulate the expression of multiple virulence factors, such as pyocyanin and biofilm formation. Previous studies have demonstrated that ppgL deletion in P. aeruginosa resulted in reduced QS signal molecule production and growth defect, guiding us to explore the relationship of ppgL and biofilm production (12, 13). However, in our study, the in-frame ppgL deletion mutant in strain PA14 had no impact on the growth rate (Fig. S4). To further assess how mutations in PpgL affect the bacterial function, we investigated biofilm production in different mutants. We constructed a ppgL knockout strain and complementation plasmid pME6032-ppgL. As for ppgL mutant strains, different point mutation pME6032-ppgL plasmids were constructed and then transformed into P. aeruginosa PA14-△ppgL by conjugation (the overexpression of the ppgL gene can be induced by IPTG [isopropyl-β-d-thiogalactopyranoside]) (Fig. S5).
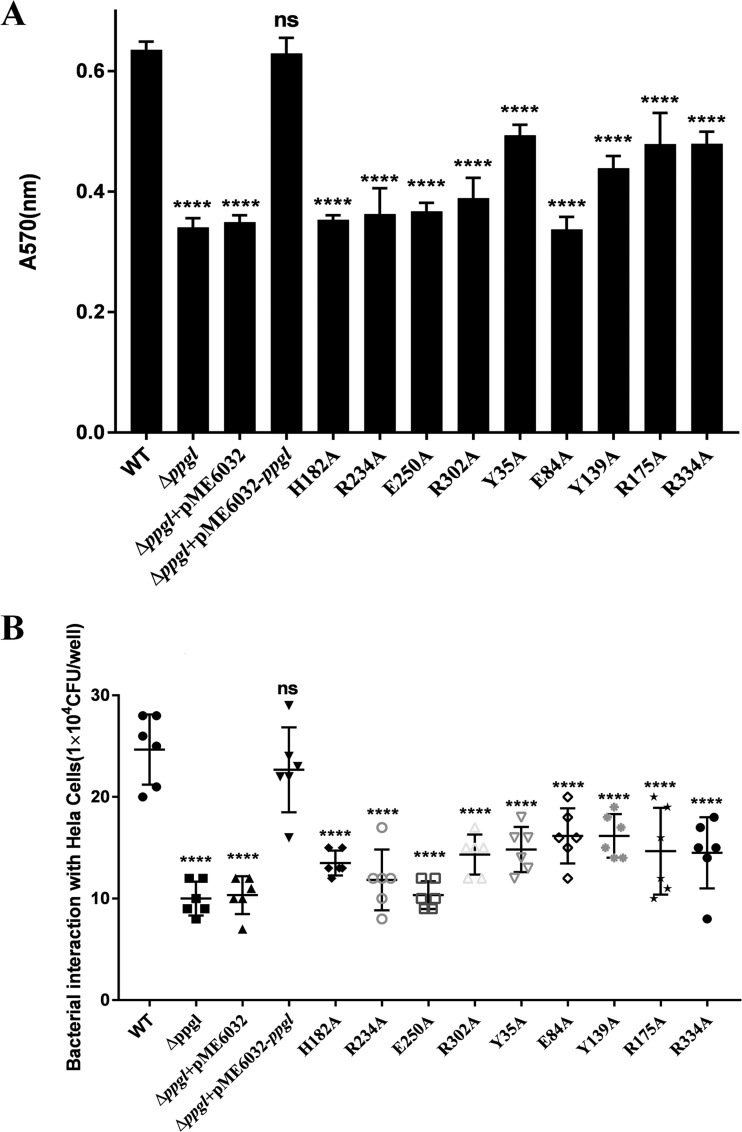
As Fig. 5A shows, PA14-ΔppgL bacterial biofilm production decreased approximately 50% compared with that of the wild-type (WT) strain, and the defect could be rescued by introducing a complementary plasmid, pME6032-ppgL. Consistent with the structural and biochemical assays, the H182A, R234A, E250A, R302A, and E84A mutants notably decreased biofilm production to the same level as the PA14-ΔppgL strain (P < 0.0001), while the remaining mutants, the Y35A, Y139A, R175A, and R334A mutants, showed variable reductions in biofilm synthesis. These studies further reveal that ppgL modulates P. aeruginosa biofilm formation.
FIG 5.
Effect of ppgL knockout and mutants on biofilm formation and virulence. (A) Biofilm formation of the ΔppgL knockout and a ΔppgL(pME6032-ppgL) complementation strain compared with that of the wild type, PA14 (WT). Quantification of biofilm biomass was done via crystal violet staining, and the A570 was measured using a microplate reader. Data are shown as the changes relative to the values for PA14 and are representative of three independent experiments. (B) Bacterial invasion (gentamicin-surviving assay) of HeLa cells upon 1 h of infection at a multiplicity of infection of 10 with P. aeruginosa PA14 as well as mutant strains. Data are shown as the changes relative to the values for PA14 and are representative of three independent experiments (±SEM). ****, P < 0.0001; ns, not significant.
Besides roles in promoting antibiotic resistance, biofilm formation is also required for bacterial attachment to biotic or abiotic surfaces (33). Thus, we conducted a gentamicin survival assay using human HeLa cells to elucidate the effects of ppgL mutations on the invasion ability of P. aeruginosa PA14 (Fig. 5B). The results showed that the PA14-ΔppgL strain displayed an attenuated invasion ability (P < 0.0001), while this ability was restored by introducing a complementary plasmid, pME6032-ppgL. H182A and E250A mutants exhibited invasion abilities similar to that of PA14-ΔppgL. Remarkably, other mutants with reduced PpgL activity also exhibited significantly reduced invasion abilities (P < 0.0001) (Fig. 5B), revealing that the specificity of protein-substrate recognition is indispensable to the invasion. All of the above-described data reveal that the lactonase activity of ppgL is vital for P. aeruginosa virulence.
DISCUSSION
PpgL represents a novel class of β-propeller gluconolactonase.
Gluconolactonase (EC 3.1.1.17) catalyzes the hydrolysis of 1,5-gluconolactone to gluconic acid. Until now, most of the solved gluconolactonase structures adopt a six-bladed β-propeller fold and require divalent metal for catalytic activity (19, 24, 34). In contrast, propeller lactonases with a seven-bladed fold have larger average channel diameters, resulting in structural variations in their top pocket (21). In such instances, two basic residues (His182 and Arg234 in PpgL) replaced the role of the metal ion, to interact with the lactone ring and decrease its ester pKa value. Corresponding to the importance of the metal cofactor of six-bladed β-propeller gluconolactonase, the two basic residues are critical for its seven-bladed counterparts. Importantly, the high degree of sequential and structural conservation of the His and Arg could be a structural signature in this new type of metal-independent β-propeller enzyme (Fig. 3).
The increased blade number also shows a correlation with greater distance and space from the channel center, providing a structural basis for accommodating more potential substrates, while the expanded surface area and volume of the loops on the top allow diverse strategies to evolve multiple enzyme properties. According to the structural comparison between NcCMLE and PpgL, the nonconserved positions around active sites are suspected to contribute to the catalytic differences. Furthermore, among those nonconserved residues, His310′/Arg256′ of NcCMLE and Arg334/Arg175 of PpgL, have been proven to be vital for the activity; thus, further investigations focusing on those sites would be helpful to explain the distinct reaction mechanisms.
The regularly distributed sites on the edges of the top pocket are responsible for diverse functions but similar ligand-binding patterns.
The core structural rigidity but functional versatility is characteristic of the β-propeller protein (15, 21, 35). Prospective studies have concluded that propellers are replicated from single blades (35); thus, we may expect that the functional motifs were amplified along with the increment of the blade units. Based on our analysis, all of the catalytic resides in PpgL locate at the three conserved positions of each blade and the 21 sites in seven-bladed propeller proteins may serve as potent and specific “hot spots” for the emergence of diverse enzymatic activities within a structurally similar framework (Fig. S3). As Fig. 3 shows, in addition to the invariable four residues, other regularly distributed sites may be expected to be starting points for elucidating additional specific functions or engineering novel biocatalysts.
Because of the prevalence and impact on diverse cellular systems, many β-propeller domain-containing proteins have been successfully targeted by chemical probes and predicted to be potential drug targets (16). In seven-bladed propeller enzymes, the general reaction profile ensures virtual identification of the central pocket for the screening of small molecules that modulate activity (Fig. S3). Consequently, targeting the predictable catalytic sites and adjacent cavity of seven-bladed propeller enzymes provides feasible strategies to maximize their application.
Implications of the role of PpgL in P. aeruginosa virulence.
PpgL and its homolog PspL (encoded by Psyr_1712 of Pseudomonas syringae) are responsible for the sugar metabolism and QS signal molecule production (12, 13). This distinctive metal-independent catalytic enzyme is probably associated with the alternative systems that complement the typical quorum quenching enzymes such as N-acyl homoserine lactone lactonase (36). The existence of an exquisite pathway regulating QS signaling in Pseudomonas strains may result from the unique metabolism of lactone-type compounds. The current study highlights the physiological importance of PpgL in P. aeruginosa biofilm formation and invasion, encouraging further investigations into its relationships to PQS.
MATERIALS AND METHODS
Protein expression and purification.
The PA4204 gene (without signal peptide) was amplified from P. aeruginosa PAO1 genomic DNA using 2×I5 MIX (TsingKe, Beijing, China) and subcloned into pET-22b plasmid. The protein and its mutants were produced in E. coli strain BL21(DE3) (TransGen Biotech). Cells were grown on Luria-Bertani (LB) medium and grown at 37°C in an incubator shaker (ZQWY-200T0; Zhichu Instrument Co., Ltd.) to an optical density at 600 nm of 0.8 to 1.0 and induced with 0.4 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 16 h at 16°C. The cells were lysed with a high-pressure cracker (Union-Biotech, Shanghai, China) in 25 mM Tris-HCl (pH 8.0) and 150 mM NaCl. The sample was then centrifuged at 18,000 rpm for 30 min, and the supernatant was loaded onto a nickel-nitrilotriacetic acid column (Qiagen) preequilibrated with buffer A (20 mM Tris-HCl, pH 8, 150 mM NaCl) containing 10 mM imidazole, washed with buffer B containing 30 mM imidazole, and eluted with buffer C containing 300 mM imidazole. The protein was further purified by gel filtration on an ENrich SEC 650 column which had been previously equilibrated with 25 mM Tris-HCl (pH 8.0) and 150 mM NaCl.
Crystallization and data collection.
Protein at a concentration of 10 to 20 mg/ml in 25 mM Tris-HCl buffer (pH 8.0) was used for crystallization experiments. Initial crystallization experiments were carried out by a sitting-drop vapor diffusion method and using four commercial crystallization screens from Hampton Research and Rigaku (Index HT, Crystal Screen HT, WIZARD HT, XTAL QUEST HT). PpgL crystals were obtained in several conditions. After optimization, the final crystallization condition was 0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate (pH 4.6), 30% (wt/vol) polyethylene glycol (PEG) monomethyl ether (MME) 2000. Crystals grew in approximately 2 to 3 days and were transferred to a cryoprotectant solution (reservoir solution with 25% PEG MME 350) prior to flash-cooling in liquid nitrogen. X-ray data were collected with a charge-coupled device (CCD) camera on BL-19U stations of the National Center for Protein Sciences Shanghai (NCPSS) beamline.
Structure determination and refinement.
Data processing and scaling were carried out using the HKL2000 software package (37). The data were processed to a resolution of 1.65 Å in space group P 41 21 2, with unit cell parameters of a = 91.763 Å, b = 91.763 Å, and c = 170.708 Å. The phase problem was solved by molecular replacement using the PHENIX suite with a putative 6-phosphogluconolactonase structure from Bacteroides uniformis ATCC 8492 (PDB code 4QRJ) as a template (38). The process of structure building and refinement was monitored using COOT (39). Water molecules were automatically added by PHENIX. The figures were prepared using PyMOL (https://pymol.org/2/).
Enzyme activity assays.
Gluconolactonase activity was determined using the colorimetric assay described by Hucho and Wallenfels (40) and d-glucono-δ-lactone as the substrate (Sigma), with slight variations (40). Briefly, the substrate solution was prepared with cold buffer (4°C) immediately before the assay, and a 98.5-μl volume of Bicine buffer containing 2.5 mM Bicine (pH 8.3), 0.2 M NaCl, and 0.25 mM cresol purple indicator was mixed with PpgL (0.5 μl, 0.5 μg). After the addition of 1 μl d-glucono-δ-lactone (5 mM final concentration in the reaction system) to 98.5 μl of the mixture in the cuvette, the absorption at 570 nm was measured at 25°C. The assays were performed using a spectrophotometer (BioTek).
Modeling of ligand-bound PpgL.
Autodock 4.0 was used to perform molecular docking of PpgL in complex with the d-glucono-δ-lactone by performing the flexible side chain method (41). The receptor structure was obtained from the PpgL structure by selecting a single protein subunit and removing all waters. The ligand was obtained by ChemDraw (http://www.chemdraw.com.cn/) and modified by PHENIX. Docking simulations were performed using the Lamarckian genetic algorithm, and 50 poses were generated. Results were clustered with AutoDock, and the low-energy docking score conformation was chosen.
Construction of P. aeruginosa ppgL deletion mutants.
The sacB-based strategy was employed to construct P. aeruginosa ppgL mutant strains (42, 43). PCRs were performed to amplify the target fragment sequences upstream (450 bp) and downstream (450 bp) from the P. aeruginosa PA14 chromosomal DNA, while the suicide plasmid pEX18Gm was linearized with gene-specific primers. The two PCR products were recombined with a ClonExpress II one-step cloning kit (Vazyme Biotech Ltd., Nangjing, China), and the resulting plasmid, pEX18Gm-ppgL, was then used to perform the deletion. All of these primers are listed in Table S2 in the supplemental material. The vector was then transformed into E. coli S17-1 and mobilized into P. aeruginosa PA14 by conjugation to transfer the suicide plasmids from an E. coli S17-1 donor to the P. aeruginosa recipient, PA14. Colonies were screened by nutritional selection, sucrose (10%) sensitivity, and gentamicin resistance, which typically indicates a double-crossover event and, thus, the occurrence of gene replacement. The ppgL gene mutant strains were further confirmed by DNA sequencing.
Construction of complementation plasmid pME6032-ppgL and mutants.
In order to construct the complementation plasmid pME6032-ppgL, PCR-amplified ppgL (full length) was cloned into plasmid pME6032 using the ClonExpress II one-step cloning kit (Vazyme Biotech Ltd., Nanjing, China). The recombinant plasmid and pME6032 were transformed into E. coli S17-1 and then mobilized into P. aeruginosa PA14-△ppgL, respectively. The PA14-△ppgL strains carrying pME6032-ppgL and plasmid pME6032 were screened using LB agar with 60 μg/ml tetracycline. For expression of ppgL in the P. aeruginosa strains, the PA14-△ppgL strain carrying plasmid pME6032-ppgL or plasmid pME6032 was induced by the addition of 0.1 mM IPTG, and then biofilm formation and cell invasion assays were conducted. As for the different point mutants, we constructed mutational pME6032-ppgL plasmids and transformed them into P. aeruginosa PA14-△ppgL by the same method as that used for pME6032-ppgL. All of these primers are listed in Table S2.
Quantitative PCR and gene expression analysis.
To rule out the possibility that the abundance of the mutant PpgL proteins was reduced compared to that of the wild type, we carried out reverse transcription-quantitative PCR (qRT-PCR) analysis for the expression of ppgL. The PA14-△ppgL strains carrying pME6032-ppgL, plasmid pME6032, and different mutations were induced by the addition of 0.1 mM IPTG and grown on Luria-Bertani (LB) medium in a 37°C incubator shaker to a density at 600 nm of 0.8 to 1.0, with the PA14 WT as a control. The different bacterial RNAs were extracted by using TRIzol (Invitrogen, USA) as previously reported (44). First-strand cDNA was synthesized by using the PrimeScript RT reagent kit (TaKaRa, Beijing, China) and then amplified by using TB Green Premix Taq II (TaKaRa, Beijing, China) with primers targeting ppgL. The oprL gene (PA0973) was used as a normalizer in this test, and the results are shown in Fig. S5. The primer sequences are listed in Table S2.
Biofilm formation assay.
Biofilm formation was determined as previously described (43). Briefly, overnight bacterial cultures were diluted 100-fold in fresh LB medium. The cell suspension (1 ml) was transferred into each well of a 24-well polyvinylchloride (PVC) plate (Sigma) and incubated at 37°C. After incubation for 48 h, the medium was removed and the wells were washed twice with sterilized phosphate-buffered saline (PBS). The cells adhering to the wells were stained with 0.1% crystal violet for 30 min and then washed twice with PBS. The cell-bound dye was eluted in 2 ml of 95% ethanol, and the absorbance of the eluted solution was measured using a microplate reader at 570 nm.
Bacterial survival in human HeLa cells.
Human HeLa cells (obtained from the ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco, Auckland, New Zealand) for survival experiments (45, 46). The HeLa cells were seeded at 2 × 105 cells per well in a 24-well tissue culture plate and cultured at 37°C under 5% CO2 overnight. Then, the HeLa cells were infected with 1 × 106 cells of Pseudomonas aeruginosa strain PA14 and its derived mutants for 1 h in 5% CO2 at 37°C. The cells were washed three times with PBS and incubated for an additional 1 h in DMEM containing 150 μg/ml of gentamicin in order to kill extracellular bacteria. After this step, the cells were washed with PBS three times and lysed in 0.025% SDS and then diluted with PBS for CFU counting on LB agar plates to determine the number of viable intracellular bacteria.
Statistical analyses.
Statistical analyses were performed with GraphPad Prism version 6, and P values for the differences between means were determined by the one-way analysis of variance (ANOVA) statistical test with equal variances. Results were considered significant if the P value was <0.05 (*), <0.01 (**), <0.001 (***), or <0.0001 (****).
Accession number(s).
The atomic coordinates of the refined structures have been deposited in the Protein Data Bank under the PDB code 6IGB.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the National Natural Science Foundation of China (grant no. 81670008 and 81871615).
We thank the National Center for Protein Sciences Shanghai (NCPSS) beamline BL19U for beamtime allowance. We also thank the staff members of the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL-17U and BL-18U, Shanghai, People’s Republic of China, for assistance during data collection.
Y.-J.S., R.B., K.-L.W., Y.-L.S. and B.S. designed the study. Y.-J.S., K.-L.W., and Y.-L.S. performed the experiments. Y.-J.S., K.-L.W., T.L., J.G., H.L., and Y.-B.Z. analyzed the data. Y.J-.S., C.-C.L., L.-H.H., N.-L.Z., C.Z., J.Y., D.-F.D., C.L., and J.G. wrote the manuscript. R.B., J.-H.H., Q.-X.Z., Q.H., and X.-Y.M. provided supervision.
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00847-18.
REFERENCES
- 1.Winstanley C, O’Brien S, Brockhurst MA. 2016. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol 24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 4.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 5.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. PNAS 110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. 2017. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 15:740. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Zhang L. 2015. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul W, Miguel C. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12:182–191. [DOI] [PubMed] [Google Scholar]
- 10.Schuster M, Greenberg E. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarighi S, Wei Q, Cámara M, Williams P, Fletcher MP, Kajander T, Cornelis P. 2008. The PA4204 gene encodes a periplasmic gluconolactonase (PpgL) which is important for fitness of Pseudomonas aeruginosa. Microbiology 154:2979–2990. doi: 10.1099/mic.0.2008/018465-0. [DOI] [PubMed] [Google Scholar]
- 13.Tarighi S, Taheri P. 2011. The role of a periplasmic gluconolactonase (PpgL)-like protein in Pseudomonas syringae pv. syringae B728a. World J Microbiol Biotechnol 27:1303–1311. doi: 10.1007/s11274-010-0577-2. [DOI] [PubMed] [Google Scholar]
- 14.Xu C, Min J. 2011. Structure and function of WD40 domain proteins. Protein Cell 2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulop V, Jones DT. 1999. Beta propellers: structural rigidity and functional diversity. Curr Opin Struct Biol 9:715–721. doi: 10.1016/S0959-440X(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 16.Schapira M, Tyers M, Torrent M, Arrowsmith CH. 2017. WD40 repeat domain proteins: a novel target class? Nature Rev Drug Discovery 16:773–786. doi: 10.1038/nrd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajander T, Merckel MC, Thompson A, Deacon AM, Mazur P, Kozarich JW, Goldman A. 2002. The structure of Neurospora crassa 3-carboxy-cis,cis-muconate lactonizing enzyme, a beta propeller cycloisomerase. Structure 10:483. doi: 10.1016/S0969-2126(02)00744-X. [DOI] [PubMed] [Google Scholar]
- 18.Korczynska M, Mukhtar TA, Wright GD, Berghuis AM. 2007. Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase. Proc Natl Acad Sci U S A 104:10388. doi: 10.1073/pnas.0701809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CN, Chin KH, Wang HJ, Chou SH. 2008. The first crystal structure of gluconolactonase important in the glucose secondary metabolic pathways. J Mol Biol 384:604–614. doi: 10.1016/j.jmb.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka Y, Morikawa K, Ohki Y, Yao M, Tsumoto K, Watanabe N, Ohta T, Tanaka I. 2007. Structural and mutational analyses of Drp35 from Staphylococcus aureus. J Biol Chem 282:5770. doi: 10.1074/jbc.M607340200. [DOI] [PubMed] [Google Scholar]
- 21.Chen CKM, Chan NL, Wang AHJ. 2011. The many blades of the β-propeller proteins: conserved but versatile. Trends Biochem Sci 36:553–561. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Kumar V, Yadav AN, Verma P, Sangwan P, Saxena A, Kumar K, Singh B. 2017. beta-Propeller phytases: diversity, catalytic attributes, current developments and potential biotechnological applications. Int J Biol Macromol 98:595–609. doi: 10.1016/j.ijbiomac.2017.01.134. [DOI] [PubMed] [Google Scholar]
- 23.Lipka M, Filipek R, Bochtler M. 2008. Crystal structure and mechanism of the Staphylococcus cohnii virginiamycin B Lyase (Vgb). Biochemistry 47:4257–4265. doi: 10.1021/bi7015266. [DOI] [PubMed] [Google Scholar]
- 24.Chakraborti S, Bahnson BJ. 2010. Crystal structure of human senescence marker protein 30; insights linking structural, enzymatic and physiological functions. Biochemistry 49:3436–3444. doi: 10.1021/bi9022297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aizawa S, Senda M, Harada A, Maruyama N, Ishida T, Aigaki T, Ishigami A, Senda T. 2013. Structural basis of the γ-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase. PLoS One 8:e53706. doi: 10.1371/journal.pone.0053706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moshe BD, Sussman JL, Maxwell CI, Klaudia S, Kamerlin SCL, Tawfik DS. 2015. Catalytic stimulation by restrained active-site floppiness–the case of high density lipoprotein-bound serum paraoxonase-1. J Mol Biol 427:1359–1374. doi: 10.1016/j.jmb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Chufán EE, De M, Eipper BA, Mains RE, Amzel LM. 2009. Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure 17:965–973. doi: 10.1016/j.str.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochiai A, Itoh T, Mikami B, Hashimoto W, Murata K. 2009. Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases. J Biol Chem 284:10181. doi: 10.1074/jbc.M807799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhtar TA, Koteva KP, Hughes DW, Wright GD. 2001. Vgb from Staphylococcus aureus inactivates streptogramin B antibiotics by an elimination mechanism not hydrolysis. Biochemistry 40:8877–8886. doi: 10.1021/bi0106787. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Laakso LM. 2016. Dali server update. Nucleic Acids Res 44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. 2012. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol 8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 34.Carper WR, Mehra AS, Campbell DP, Levisky JA. 1982. Gluconolactonase: a zinc containing metalloprotein. Experientia 38:1046–1047. doi: 10.1007/BF01955358. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhuri I, Soding J, Lupas AN. 2008. Evolution of the beta-propeller fold. Proteins 71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 36.Wang LH, Weng LX, Dong YH, Zhang LH. 2004. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J Biol Chem 279:13645–13651. doi: 10.1074/jbc.M311194200. [DOI] [PubMed] [Google Scholar]
- 37.Otkinowski Z, Minor W. 1997. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X.. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hucho F, Wallenfels K. 1972. Glucono-δ-lactonase from Escherichia coli. Biochim Biophys Acta 276:176–179. doi: 10.1016/0005-2744(72)90018-6. [DOI] [PubMed] [Google Scholar]
- 41.Bianco G, Forli S, Goodsell DS, Olson AJ. 2016. Covalent docking using Autodock: two-point attractor and flexible sidechain methods. Protein Sci 25:295–301. doi: 10.1002/pro.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahn CE, Charkowski AO, Willis DK. 2008. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods 75:318–324. doi: 10.1016/j.mimet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Chi E, Mehl T, Nunn D, Lory S. 1991. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect Immun 59:822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenshine I, Duronio V, Finlay BB. 1992. Tyrosine protein kinase inhibitors block invasin-promoted bacterial uptake by epithelial cells. Infect Immun 60:2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.